Abstract
The growth of amyloid fibrils is studied by replica exchange molecular dynamics in an implicit solvent. Our data indicate that extremely long simulation times (at least a few hundred ns) are necessary to study the thermodynamics of fibril elongation in detail. However some aspects of the aggregation process are already accessible on the time scales available in the present study. A peak in the specific heat indicates a docking temperature of Tdock ≈ 320 K. Irreversible locking requires lower temperatures with the locking temperature estimated as Tlock ≈ 280 K. In our simulation the fibril grows from both sides with the C-terminal of the incoming monomer attaching to the C-terminal of the peptides in the fibril forming a β-sheet on the fibril edge. Our simulation indicates that the C-terminal is crucial for aggregation.
INTRODUCTION
A hallmark of Alzheimer's disease are amyloid fibrils in brain tissue and blood vessels. The fibrils are built out of 40 to 42 residue long Aβ peptides, which in turn are the product of sequential cleavage of the amyloid precursor protein, a transmembrane glycoprotein of undetermined function.1 The propensity of forming fibrils depends on the length of the peptides and is largest for Aβ42. In recent work we have studied in silico the early stages in oligomerization of Aβ peptides2, 3 and the dependence of the observed mechanisms on sequence (i.e., mutations) and environment.4 Small oligomers of Aβ peptides are also the focus of many other studies, both computational and experimental ones.5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 This is because they are likely candidates for the neurotoxic agents in the pathology of Alzheimer's disease while as part of the unsolvable fibrils Aβ peptides appear to be less harmful. This suggests that treatment strategies should aim at decreasing the concentration of harmful oligomers, by either dissolving such oligomers or enhancing fibril growth.
Despite its potential medical importance, the mechanism of fibril formation is only poorly understood. Structures of amyloid beta fibrils have been resolved17, 18 but experimental investigations of the growth process are challenging. Measurements of kinetic rate constants indicate a dock-lock mechanism. In a first step, the Aβ peptide docks reversibly to the amyloid template before becoming locked in a second step.20 Sometimes a third and intermediate step during the docking phase is assumed.21 The growth process has also been probed by various computational studies that study how monomers of Aβ-fragments add to preformed oligomers.22, 23, 24, 25, 26, 27 For instance, Buchete and Hummer28 proposed from all-atom molecular dynamics simulations of Aβ40 a three-step process, where strong hydrophobic interactions align the C-terminal segments of the incoming peptide with such in the fibril encouraging β-strand formation in the C-terminal segment of the incoming monomer. The N-terminal strand forms afterwards and is less stable. Finally, in a third step, the loop from 27ASN to 30ALA, connecting C-terminal and N-terminal strand, with its hydrophilic residues assumes the form observed in fibrils. The role of C-terminal β-sheet formation as driving force for fibril elongation is also supported by work of Masman et al.29 who studied the same molecule but with different force field. On the other hand, Takeda and Klimov19 found that the N-terminal strand segment of incoming monomer forms parallel hydrogen bond with the corresponding segments of peptides in the fibril.
In the present work, we try to add to the previous computational investigations by studying the interaction between a full-length Aβ42 peptide, the more fibrillogenic variant, and the fibril through all-atom replica exchange molecular dynamics simulations with an implicit solvent. As experiments have indicated20, 21 that the Aβ peptide deposits to the fibril predominantly as a monomer, we simulate the interaction between a monomer and a pre-structured fibril, and compare our observed monomer structures with the solid-state NMR-structures of Aβ peptides in a fibril. Unlike Buchete et al.28 and Masman et al.29 we study directly the fibril elongation process, not the time-reversed process of aggregate dissociation. By simulating the full-length Aβ42 peptide we also go beyond previous work that studied similar questions but relied on short fragments such as Aβ(16–22),22, 23 Aβ(35–40),24, 26 or Aβ(37–42).27
Our data indicate that fibril elongation is an extremely slow process that starts with attachment of the C-terminus of the incoming monomer to the C-terminus of a peptide in the fibril. In a zipper-like way the C-terminal end of the monomer docks to the fibril in a random-coil configuration. This docking transition is observed at T ≈ 320 K. In a next step, the residues in this segment form a β-sheet with the C-terminal residue of the edge peptide in the fibril, tightly locked in now to the fibril. The associate locking temperature is at T ≈ 280 K. Only afterwards, the peptide forms the characteristic turn around residues 21ASN-30ALA with the N-terminal strand.
METHODS
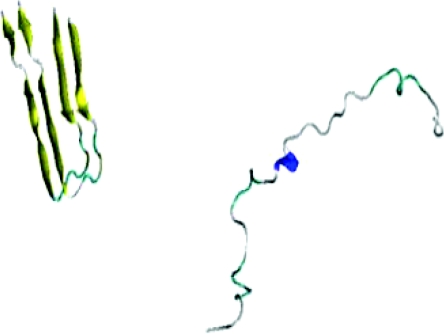
In our all-atom simulations, we probe the interaction between an incoming Aβ monomer and a pre-formed fibril. For the fibril we took the structure (PDB-ID 2BEG.pdb) of the pentameric aggregate as obtained by NMR.18 In order to reduce the computational effort we deleted the edging chains and therefore approximate the fibril by two Aβ peptide fragments consisting of residues 17LEU-42ALA kept in a fixed structure as shown in Fig. 1. The peptides in the fibril are in a bent β- sheet hairpinlike structure with two antiparallel beta sheets and the polar region at the turn. The N-terminal strand includes the central hydrophobic core. The polar region lies in the middle of the 16LYS-35MET sequence. A salt-bridge (23ASP-28LYS) helps stabilize the fibril structure. The backbones of fibril peptides are constrained with the a force constant of 2 kcal∕(mol Å2). On the other hand, the incoming monomer is a full sized Aβ42 peptide. We choose as start configuration a random coil generated by the LEAP subroutine in the AMBER software package30 followed by a short MD run at 600 K, and put at a distance of 40 Å to the fibril fragment. The interacting system of Aβ42 monomer and fibril fragment is kept together by a restraining force that is zero if the distance r between their center of mass is smaller than 40 Å, and otherwise takes the form k(R − r)2 with R = 40 Å and a force constant k = 3 kcal∕(mol Å2). The atomic charges and atom types are assigned by the antechamber tool in the AMBER software package.30 We select the AMBER99SB force field31 with the generalized Born solvent model.32 Chirality restraints are applied on the monomer backbone for temperatures higher than 400 K.
Figure 1.
Fibril fragment and initial monomer conformation used in our simulations.
Simulations of folding and aggregation of proteins are hindered by their rough energy landscape, leading to prohibitively long simulation times when all-atom simulations are used. One way to alleviate this problem are generalized ensemble simulations. In our case, we use replica exchange molecular dynamics (REMD) (Ref. 33) with velocity rescaling34 by the Sander module in AMBER11. Before starting the REMD simulations, we run each replica for 500 ps to equilibrate them to their individual temperatures. At start, 24 replicas were distributed in a temperature range from 270 K to 600 K. After 80 ns we have analyzed the specific heat curve and decided from its form at high temperature that we could lower the upper temperature limit to Tmax ≈ 400 K. Consequently, we have reduced the number of replicas to twelve, distributed between 270 K and 395.6 K. The observed peak in specific heat around 320 K supports in hindsight this choice. The temperature distribution is listed in Table 1. After 256 ns, we analyzed the replica exchange rates and flow of replicas through temperature space, and optimized the temperature distribution accordingly, and continued the simulation until 415 ns. This final distribution of temperatures is also listed in Table 1. Replica exchanges between all pairs of neighboring replicas were attempted every 20 ps and subject to the Metropolis criterion. The average acceptance rate was 25%. The cumulative simulation time is 5.94 μs. A cutoff of 16 Å was used for the van der Waals interactions. The simulations were performed in a N-V-T ensemble, with the temperature coupled by a Langevin thermostat with a collision frequency of 2.0 ps−1. The bonds of molecules were constrained by using the SHAKE algorithm. The integration time step was 2 fs, and coordinates and velocities were saved every 4 ps for subsequent analysis.
Table 1.
Distribution of temperatures for the replica exchange molecular dynamics simulation. Note that we used in the first 80 ns additional 12 replicas extending the temperature distribution toward higher temperatures: 409.5, 424.0, 439.0, 454.0, 454.5, 470.5, 487.2, 504.4, 522.2, 540.6, 559.7, 597.5, and 600.0. All temperatures are in Kelvin.
| 1–256 ns | 256–415 ns |
|---|---|
| 270.0 | 270.0 |
| 279.5 | 279.5 |
| 289.4 | 289.4 |
| 299.6 | 299.6 |
| 310.2 | 310.2 |
| 321.2 | 315.5 |
| 332.5 | 321.0 |
| 344.3 | 333.0 |
| 356.4 | 348.0 |
| 369.0 | 362.0 |
| 382.1 | 377.0 |
| 395.6 | 395.0 |
RESULTS AND DISCUSSION
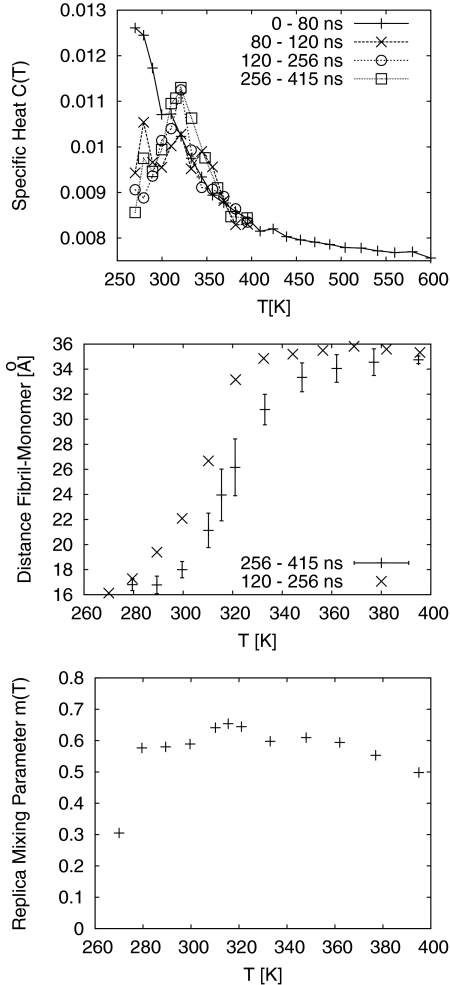
Computational studies of protein folding and aggregation are limited by sampling difficulties that only partially can be overcome by replica-exchange techniques and similar approaches. Hence, any computational investigation of fibril growth needs to monitor carefully the convergence toward the equilibrium distribution. One suitable quantity for this purpose is the specific heat C(T), shown in Fig. 2a and defined by
| (1) |
and kB the Boltzmann constant. This is because C(T) measures the fluctuations in energy, a quantity that is especially sensitive to deviations from the equilibrium. The first 80 ns in our system was simulated with 24 replicas distributed over a range of temperatures from 270 K to 600 K, see Table 1. The corresponding curve clearly indicates that our system is far from equilibrium. However, the slow decrease of C(T) for temperatures larger than ≈400 K suggests that an upper temperature limit of 400 K is sufficient to allow the system crossing of all relevant energies, and we reduced temperature range and number of replicas accordingly. This choice seems to be justified by the peak in specific heat at T ≈ 320 K that subsequently appears with increasing simulation time. Such a peak is usually a signal for a transition between a high-temperature phase characterized by disordered states of high energy, and a low-temperature phase dominated by ordered low-energy configurations. Note that it takes at least 120 ns before such peak appears. The two specific heat curves, calculated from the trajectories between 120 ns–256 ns and 256 ns–415 ns, are very similar, which suggests that the equilibrium distribution has been reached for times larger than 120 ns. Unfortunately, this is not the case. Analyzing the exchange rate between neighboring temperatures and the flow of replicas through temperature space we optimized our temperature distribution (keeping the number of replicas (12) unchanged) after 256 ns (see Table 1). Figure 2b displays the average distance d between incoming monomer and the fibril fragment. The curve evaluated over the time interval between 256 ns and 415 ns differs substantially from the one calculated from trajectories between 120 ns and 256 ns. Hence, we decided to discard the first 256 ns of our simulations. However, even with these long times we cannot be sure that our simulations has indeed approached equilibrium. Figure 1c displays the mixing of replicas as each temperature, defined by
| (2) |
where the sum goes over all 12 replicas, and ti is the total time that replica i has resided at temperature T. The mixing parameter m(T) approaches zero if a single replica sits most time at temperature T, and becomes maximal if all replicas stay equally long at that temperature. For a well-equilibrized replica-exchange simulation one would expect that m(T) ≈ const. However, Fig. 2c indicates a much smaller mixing of replicas at T = 270 K than at higher temperature. In fact, out of the 159 ns that we consider for our analysis, a single replica sat for 102 ns at this temperature. Almost all of the remaining time, this specific replica spent at the next higher temperature, T = 279.5 K. As we could not extend our simulation to obtain a better mixing of replicas, we decided to discard data for the two lowest temperatures (T = 270 K and T = 279.5 K), and to focus in our analysis on the higher temperatures where we are more confident in their reliability. Nevertheless, it is interesting to observe the long simulation times needed even in our much simplified model of fibril growth before equilibrium is approached. With an implicit solvent and AMBER force field these times appear to be of order a few hundred ns for each replica. This may be important to consider for simulations of this system. For instance, in Ref. 19 the cumulative simulation time is with 48 μs a factor of eight higher than our cumulative simulation time. However, this time is distribute over 10 independent runs with 24 replicas each, i.e., each replica is simulated for only 200 ns.
Figure 2.
The approach to equilibrium in our simulation is monitored through observing (a) the specific heat C(T), (b) the distance between incoming monomer, and fibril fragment; and (c) the mixing of replicas.
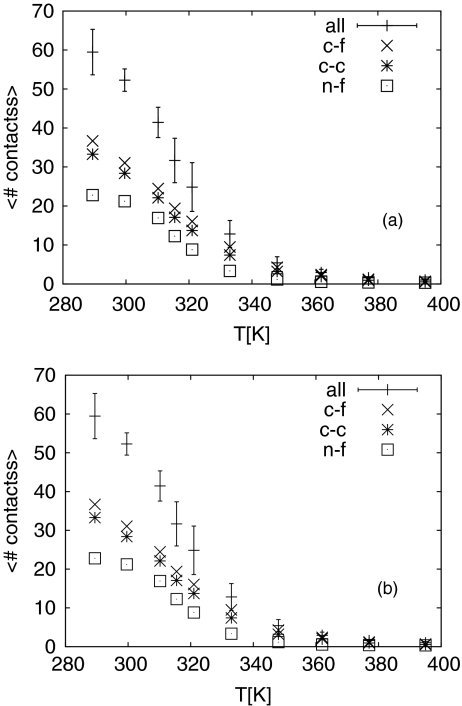
Figure 2b shows that the average distance between the incoming monomer and the fibril decreases with lowering temperatures, indicating that it is energetically favorable when the monomer is in close proximity to the fibril fragment. This gain in potential energy results from the van der Waal's interaction and hydrogen bond formation between monomer and the fibril fragment. This can be seen from Fig. 3a where we plot the contacts between incoming monomer and fibril. Here we define a contact as a Cα-atom of the incoming monomer being closer than 5 Å to a Cα-atom of the fibril fragment. Below T = 350 K the average number of contacts grows rapidly with decreasing temperature until approaching a plateau at around 300 K. A more careful analysis shows that the majority of contacts between monomer and fibril come from the C-terminal residues 31ALA to 42ALA. Average values for such contacts are also displayed in Fig. 3a. Residues in this segment are mainly hydrophobic, and the curve therefore describes the hydrophobic packing of the monomer to the fibril fragment. A similar behavior in temperature is seen for the average number of hydrogen bonds between monomer and fibril fragment, displayed in Fig. 3b. This curve describes the bonding between fibril-fragment and monomer by hydrogen bonds, and therefore a second mechanism for docking. The midpoint of the rapid increase in either contacts or hydrogen bonds with decreasing temperature is at T ≈ 320 K, and therefore corresponds to the peak in specific heat. Hence, this peak marks a docking transition. Note that this value is considerably lower than the one reported by the Klimov laboratory: Tdock = 360 K.19 It is also differs considerably from the values calculated from the first 256 ns of our simulations that we have discarded. Hence, the differences between the temperatures measured by us and by Klimov may reflect the difficulties in thermalizing the simulation. Besides differences in energy function other possible reasons for this large discrepancy are differences in the model (i.e., our choice of studying the docking of a full-size Aβ42 peptide to a truncated fibril fragment of rather small size), especially as we can expect that the geometry of the fibril edge will influence the growth process.19
Figure 3.
Average number of (a) contacts and (b) hydrogen bonds between incoming monomer and fibril fragment as function of temperature. “All” marks that all contacts between the incoming monomer and fibril fragment are considered, “c-f” marks only contacts between the fibril and the C-terminal segment (31–42) of the monomer, “n-f” the contacts between fibril and residues (1–30) of the monomer, and “c-c” marks the case where only contacts between C-terminal residues in both monomer and fibril are considered.
Figure 3 shows also that the docking transition is driven by interactions between the C-terminal residues of the incoming monomer and the fibril fragment. The number of contacts or hydrogen bonds that involve these residues changes strongly with temperature while the other ones change less and approach quickly a plateau with decreasing temperature. The newly formed contacts or hydrogen bonds of C-terminal residues that appear below the docking transition temperature are mainly with C-terminal residues of the fibril fragment as can be seen from the corresponding curve in Fig. 3a. Contacts with other parts of the fibril fragment are less common and do not show the strong temperature dependence. Hence, we conclude that the docking transition involves a pairing of C-terminal strands of incoming monomer and fibril. While this observation is in agreement with N-methylation experiment to perturb the hydrogen bonds in N- and C-terminal β–sheets that show strong hydrophobic interactions between C-terminal sheets,35 it differs from what has been observed by the Klimov's group. They found that the N-terminal strand segment of incoming monomer forms parallel hydrogen bonds with the fibril.19 The difference may be due to the different models used. While we studied the more aggregation prone Aβ42, Klimov's group focused on the Aβ40 peptide. The same peptide was also used by us to study the dimerization process where we also found the N-terminal segment 17LEU-21ALA to be more important than the C-terminal segment in the association process. The additional C-terminal residues in Aβ42 may enhance binding with the fibril. Yan and Wang36 found that the Aβ42 C-terminus is more rigid than the one in the Aβ40 peptide. Hence, the C-terminus of Aβ42 may pay a smaller entropic price for forming a β-sheet with the fibril than the Aβ40 monomer. The promotion of β-sheet stability was also found in Ref. 37. Furthermore, the additional two hydrophobic residues 41ILE and 42ALA may increase the chance to form seed contacts that start the docking process. We speculate that once such contacts are formed they act as anchors increasing the chance for residues further away from the C-terminus to form contacts with the fibril, leading to a zipperlike docking of the incoming monomer with the fibril that starts with the C-terminus. However, our data do not provide us with enough evidence to prove conclusively such a zipperlike process.
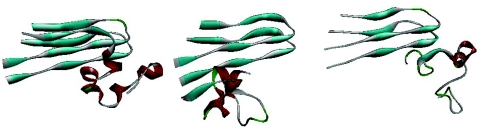
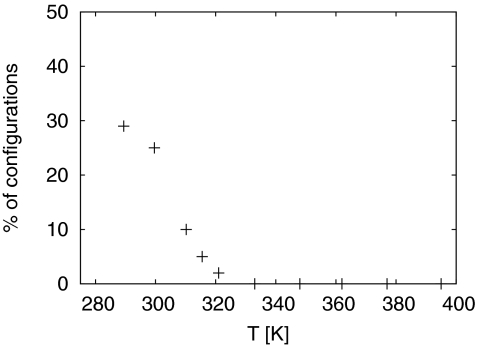
A clustering analysis at T = 289.5 K finds three cluster with fibril-like configurations. The centroids are shown in Fig. 4. Taking a cut-off distance of 3 Å these clusters together compromises 29% of conformations. The three clusters are made out of partially formed β–structures and differ by the position where the monomer attaches. In the remaining 71% of configurations the incoming monomer is a random coil without specific secondary structure. In these configurations, the incoming monomers can also form contacts with the fibril fragment. We conjecture that incoming monomers form contacts with the fibril fragments while being in a random-coil form, and therefore these random configurations represent typical monomer structures in the docked phase. On the other hand, the configurations of Fig. 4 with its stable hydrogen bonds between the two β-strands in monomer and fibril-fragment indicate the initial stages of the locking process. The percentage of such tightly bonded configurations decreases with temperatures as can be seen from Fig. 5. Here, we define a configuration as “fibril-like” if it is within 3 Å rmsd from the cluster centroids shown in Fig. 4. Our plot does not allow us to determine the locking temperature but indicates that it is well below the docking temperature of T ≈ 320 K.
Figure 4.
Representative fibril-like configurations found in our simulation at T=289.5 K.
Figure 5.
Percentage of fibril-like configurations as function of temperature T. Here, we define a configuration as “fibril-like” if it is within 3 Å rmsd from the cluster centroids shown in Fig. 4.
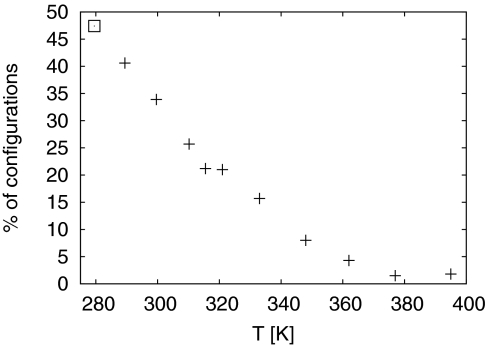
Note that we did not find configurations where the monomer is fully connected to the fibril fragment and in its correct form. This may be due to the long time scale of fibril growth but could also indicate that our fibril fragment is too small to provide a sufficient good template. For instance, the incoming monomer can form partial β-sheets with both sites of the fibril-fragment (see Fig. 4). A larger fibril fragment is likely required to ensure growth from only one side. However, even taking these limitations into account, our data suggest a nucleation process of aggregation that is in agreement with the mechanism proposed by Hummer.28 In this picture, the growth of fibril starts with the C-terminus of the incoming monomer forming contacts with the fibril. This leads to a docking of the monomer to the fibril. The monomer then locks to the fibril by forming a β-strand that is hydrogen-bonded to the fibril. A signal for the locking is therefore the percentage of configurations with “sheetness” in the C-terminal segment of the incoming monomer, a quantity that we show as function of temperature in Fig. 6. It displays the frequency of configurations with at least one C-terminal residue 31ALA-42ALA in a sheetlike form as determined by the program DSSP.38 Note, how the “sheetness” starts to increase around the docking temperature of Tdock ≈ 320 K. At T = 289 K its value is about 40%. An extrapolation suggests that the 50% point, marking the locking temperature, is at Tlock ≈ 280 K. We remark that while we excluded this temperature out of fear that our results at it will not be reliable, we measure at T = 279.5 K indeed a “sheetness” of 48%. The final step in the growth process is the rest of the incoming monomer folding into the turn and N-terminal strand form. Our previous work2 showed that even in the isolated monomer a U-like shape with a turn around residues 27ASN to 30ALA is energetically favorable. In the present work we observe only faint signals for this final step. At T = 289 K the “sheetness” for the N-terminal segment 18VAL-26SER is only ≈10%. Hence, this step seems to be the limiting process in our simulation.
Figure 6.
Percentage of configurations with “sheetness” in the C-terminal residues 31–42 as function of temperature T. The square marks the value at T = 279.5 K, temperature where our simulation may not have converged. The point is therefore shown only for illustration.
CONCLUSIONS
Using all-atom simulations with an implicit solvent we have studied the fibril-elongation process of β-amyloid peptides. Our data demonstrate the numerically difficulty of this process. Simulating our system more that 400 ns and with replica-exchange molecular dynamics, we observe still only the early steps of this process. Only after 250 ns the system starts even approaching equilibrium. This slow dynamics makes a numerical analysis of the growth process challenging. Our data indicates that at least a 1 μs of simulation time would be necessary to observe the elongation process completely. Nevertheless, even taking the limitations of the present study into account, we find some indications on the nature of the growth process that seems to support ideas by Hummer et al.28 Elongation of the fibril starts with attachment of the C-terminus of the incoming monomer to the C-terminus of a peptide in the fibril. This docking transition, during which the monomer is in a random coil configuration, is observed at T ≈ 320 K. In a next step, the residues in the docked C-terminal segment form a β-sheet with the C-terminal residue of the edge peptide in the fibril, and become tightly locked to the fibril. The associate locking temperature is likely at T ≈ 280 K. Only afterwards does the peptide fold into a U-shape and forms the N-terminal β-strand and the characteristic turn around residues 21ALA-30ALA. Only indirect evidence for the start of this process was found. Hence, this final step seems to be the limiting process and requires longer time scales than available in the present study.
ACKNOWLEDGMENTS
This work was supported, in part, by research Grant No. GM62838 of the National Institutes of Health (NIH) (USA).
References
- Selkov D. J., Nat. Cell Biol. 6, 1054 (2004). 10.1038/ncb1104-1054 [DOI] [PubMed] [Google Scholar]
- Anand P., Nandel F. S., and Hansmann U. H. E., J. Chem. Phys. 128, 165102 (2008). 10.1063/1.2907718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P., Nandel F. S., and Hansmann U. H. E., J. Chem. Phys. 129, 195102 (2008). 10.1063/1.3021062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P. and Hansmann U. H. E., Mol. Simul. 37, 440 (2011). 10.1080/08927022.2011.551879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkitadze M., Bitan G., and Teplow D. J., Neurosci. Res. (N Y) 69, 567 (2002). 10.1002/jnr.10328 [DOI] [PubMed] [Google Scholar]
- Baumketner A., Bernstein S. L., Wyttenbach T., Bitan G., Teplow D. B., Bowers M. T., and Shea J. E., Protein Sci. 3, 420 (2006). 10.1110/ps.051762406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H. and Chen J. Z., Proteins: Struct., Funct., Bioinf. 63, 555 (2006). 10.1002/prot.20846 [DOI] [PubMed] [Google Scholar]
- Wei G., Song W., Derreumaux P., and Mousseau N., Front. Biosci. 13, 5681 (2008). 10.2741/3109 [DOI] [PubMed] [Google Scholar]
- Melquiond A., Dong X., Mousseau N., and Derreumaux P., Curr. Alzheimer Res. 5, 244 (2008). 10.2174/156720508784533330 [DOI] [PubMed] [Google Scholar]
- Chebaro Y., Mousseau N., and Derreumaux P., Phys. Chem. B 113, 7668 (2009). 10.1021/jp900425e [DOI] [PubMed] [Google Scholar]
- Tarus B., Straub J. E., and Thirumalai D., J. Mol. Biol. 379, 815 (2008). 10.1016/j.jmb.2008.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarus B., Straub J. E., and Thirumalai D., J. Am. Chem. Soc. 128, 16159 (2006). 10.1021/ja064872y [DOI] [PubMed] [Google Scholar]
- Nikolaos S. G., Yan Y., Scott M. A., Wang C., and Angel E. G., J. Mol. Biol. 368, 1448 (2007). 10.1016/j.jmb.2007.02.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J. A. and Shen J. K., Biochemistry 49, 5290 (2010). 10.1021/bi100137y [DOI] [PubMed] [Google Scholar]
- Urbanc B., Cruz L., Ding F., Sammond D., Khare S., Buldyrev S. V., Stanley H. E., and Dokholyan N. V., Biophys. J. 87, 2310 (2004). 10.1529/biophysj.104.040980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strodel B., Lee J. W. L., Whittleston C. S., and Wales D., J. Am. Chem. Soc. 132, 13300 (2010). 10.1021/ja103725c [DOI] [PubMed] [Google Scholar]
- Petkova A. T., Ishiil Y., Balbach J. J., Antzutkin O. N., Leapman R. D., Delaglio F., and Tycko R., Proc. Natl. Acad. Sci. U.S.A. 99, 16742 (2002). 10.1073/pnas.262663499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Döbeli H., Schubert D., and Riek R., Proc. Natl. Acad. Sci. U.S.A. 102, 17342 (2005). 10.1073/pnas.0506723102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T. and Klimov D. K., Biophys. J. 96, 442 (2009). 10.1016/j.bpj.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esler W. P., Stimson E. R., Jennings J. M., Vinters H. V., Ghilardi J. R., Lee J. P., Manthy P. W., and Maggio J. E., Biochemistry 39, 6288 (2000). 10.1021/bi992933h [DOI] [PubMed] [Google Scholar]
- Cannon M. J., Williams A. D., Wetzel R., and Myszka D. G., Anal. Biochem. 328, 67 (2004). 10.1016/j.ab.2004.01.014 [DOI] [PubMed] [Google Scholar]
- Nguyen P. H., Li M. S., Stock G., Straub J. E., and Thirumalai D., Proc. Natl. Acad. Sci. U.S.A. 104, 111 (2007). 10.1073/pnas.0607440104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone M. J., Hua L., Soto P., Zhou R., Berne B. J., and Shea J.-E., J. Am. Chem. Soc. 130, 11066 (2008). 10.1021/ja8017303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G., Straub J. E., and Thirumalai D., Proc. Natl. Acad. Sci. U.S.A. 106, 11948 (2009). 10.1073/pnas.0902473106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strodel B., Whittleston C. S., and Wales D. J., J. Am. Chem. Soc. 129, 16005 (2007). 10.1021/ja075346p [DOI] [PubMed] [Google Scholar]
- O'Brien E. P., Okamoto Y., Straub J. E., Brooks B. R., and Thirumalai D., J. Phys. Chem. B 113, 14421 (2009). 10.1021/jp9050098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Lei H., and Duan Y., J. Am. Chem. Soc. 127 13530 (2005). 10.1021/ja050767x [DOI] [PubMed] [Google Scholar]
- Buchete N. V. and Hummer G., Biophys. J. 92, 3032 (2007). 10.1529/biophysj.106.100404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masman M. F., Eisel U. L. M., Csizmadia I. G., Penke B., Enriz R. D., Marrink S. J., and Luiten P. G. M., J. Phys. Chem. B 113, 11710 (2009). 10.1021/jp901057w [DOI] [PubMed] [Google Scholar]
- Case D. A., Darden T. A., T. E.CheathamIII, Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., Walker R. C., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Tsui V., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Beroza P., Mathews D. H., Schafmeister C., Ross W. S., and Kollman P. A., AMBER 9, University of California, San Francisco, 2006.
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C., Proteins 65, 712 (2006). 10.1002/prot.21123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins G. D., Cramer C. J., and Truhlar D. G., J. Phys. Chem. 100, 19824 (1996). 10.1021/jp961710n [DOI] [Google Scholar]
- Hansmann U. H. E., Chem. Phys. Lett. 281, 140 (1997). 10.1016/S0009-2614(97)01198-6 [DOI] [Google Scholar]
- Sugita Y. and Okamoto Y., Chem. Phys. Lett. 314, 141 (1999). 10.1016/S0009-2614(99)01123-9 [DOI] [Google Scholar]
- Sciarretta K. L., Boire A., Gordon D. J., and Meredith S. C., Biochemistry 45(31), 9485 (2006). 10.1021/bi0605585 [DOI] [PubMed] [Google Scholar]
- Yan Y. and Wang C., J. Mol. Biol. 364, 853 (2006). 10.1016/j.jmb.2006.09.046 [DOI] [PubMed] [Google Scholar]
- Yang M. and Teplow D. B., J. Mol. Biol. 384, 450 (2008). 10.1016/j.jmb.2008.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. and Sander C., Biopolymers 22, 2577 (1983). 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]