Abstract
Infection of metazoan cells with some viruses alters the balance of cellular mRNA export to favor viral RNA export and to retain cellular transcripts in the nucleus. Here, evidence is presented to show that the herpes simplex virus 1 (HSV-1) essential regulatory protein ICP27, which inhibits host cell-splicing, resulting in the accumulation of unspliced transcripts in the nucleus, mediates RNA export of viral intronless mRNAs. ICP27 was shown to shuttle between the nucleus and cytoplasm through a leucine-rich nuclear export signal, which alone was able to direct the export of the heterologous green fluorescent protein. In vivo UV irradiation studies demonstrated that ICP27 could be crosslinked to poly(A)+ RNA in the nucleus and the cytoplasm, supporting a role in export. Furthermore, the amount of hnRNP A1, which has been implicated in the export of cellular spliced mRNAs, that was bound to poly(A)+ RNA in HSV-1-infected cells was reduced compared with uninfected cells. In addition, it was demonstrated that ICP27 bound seven intronless HSV-1 transcripts in both the nucleus and the cytoplasm, and export of these transcripts was diminished substantially during infection with an ICP27 null mutant virus. In contrast, ICP27 did not bind to two HSV-1 mRNAs that undergo splicing. Finally, binding of ICP27 to RNA in vivo required an arginine-glycine region that resembles an RGG box. These results indicate that ICP27 is an important viral export factor that promotes the transport of HSV-1 intronless RNAs.
Keywords: RNA export, intronless RNA, RNA binding, NES, RGG box, herpes simplex virus gene expression
Eukaryotic RNAs are synthesized in the nucleus by RNA polymerase I, II, or III (Pol I, II, III) after which they are processed and transported to their sites of action, predominantly in the cytoplasm. The exchange of macromolecules between the nucleus and the cytoplasm occurs through nuclear pore complexes (NPCs). Different classes of RNAs appear to leave the nucleus by distinct pathways (Jarmolowski et al. 1994; Fischer et al. 1995; for review, see Gorlich and Mattaj 1996; Corbett and Silver 1997) through an active, energy-dependent, saturable mechanism (for review, see Nakielny et al. 1997). Recently, a number of studies have shown that RNA transport through the NPC is mediated by proteins associated with the RNAs, and these proteins contain specific signals to use receptor-mediated export through the NPC to transport their cargo (Fischer et al. 1995; Michael et al. 1995; Bogerd et al. 1996; Fridell et al. 1996b; Kim et al. 1996; Lee et al. 1996; Palmer and Malim 1996; Dobbelstein et al. 1997). A feature of these proteins is that they shuttle continuously between the nucleus and the cytoplasm. The best candidates to mediate export of metazoan mRNAs are the abundant hnRNPs, and especially hnRNP A1. It has been demonstrated that hnRNP A1 shuttles between the nucleus and the cytoplasm through a sequence, designated M9, which serves as both the export and import signal (Fritz et al. 1995; Izaurralde et al. 1997). Furthermore, hnRNP A1 was found associated with poly(A)+ RNA in both the nucleus and the cytoplasm (Piñol-Roma and Dreyfuss 1992). Most compelling, a protein in Chironomus tentans termed hrp36, which is similar to mammalian hnRNP A1, was found to associate with Balbiani ring RNA concomitant with transcription, and to remain associated as the Balbiani ring RNA particles were translocated through the nuclear pore (Visa et al. 1996).
Perhaps the best characterized RNA export protein is HIV-1 Rev. Rev shuttles between the nucleus and the cytoplasm (Kalland et al. 1994; Meyer and Malim 1994; Richard et al. 1994), promoting the export of unspliced and partly spliced HIV env RNA (Fischer et al. 1994) by binding to a stem–loop structure within the env intron, termed the Rev response element (RRE) (Malim et al. 1989; Kjems et al. 1991). The nuclear export signal (NES) of Rev has been identified (Fischer et al. 1995; Szilvay et al. 1995), and consists of a short stretch of amino acids that is rich in leucine residues. Similar sequences that function as NESs have been identified in PKI, the heat-stable inhibitor of cAMP-dependent protein kinase (Wen et al. 1995), in the Rex protein of HTLV-1 (Bogerd et al. 1996; Kim et al. 1996; Palmer and Malim 1996), in the Rev proteins of visna virus and equine infectious anemia virus (Meyer et al. 1996), in adenovirus E4–34-kD (Dobbelstein et al. 1997), in transcription factor IIIA (Fridell et al. 1996b), and in the yeast mRNA transport protein Gle 1 (Murphy and Wente 1996). Using the yeast two-hybrid system, a human protein called Rip (Fritz et al. 1995) or Rab (Bogerd et al. 1995) that interacted specifically with Rev was identified. This protein possesses FG repeats that are characteristic of a subclass of nucleoporins, suggesting an involvement of Rip/Rab in Rev export (Fridell et al. 1996a), however, direct interaction with the NES has not been demonstrated. Recently, three groups have provided evidence that CRM1 (chromosome maintenance region 1), a protein that shares sequence similarity with the karopherin β family of import proteins, forms a complex with a leucine-rich NES and functions as an essential export receptor (Fornerod et al. 1997; Ossareh-Nazari et al. 1997; Stade et al. 1997). Furthermore, it has been shown that CRM1 bridges the interaction between Rev and the nuclear pore complex (Neville et al. 1997). Accordingly, CRM1 has been named exportin 1 (for review, see Ullman et al. 1997).
In metazoans, most pre-mRNAs require an intron for efficient processing and export (Liu and Mertz 1995). Pre-mRNAs are retained largely in the nucleus, and it has been hypothesized that certain splicing factors may interact with nuclear structures, or the spliceosomes may prevent the interaction of RNA with export factors, therefore holding RNA in the nucleus until spliced (for review, see Nakielny et al. 1997). Retroviruses require viral structural products that are encoded by unspliced transcripts, and therefore, have evolved both cis-acting signals (Kiss-Laszlo and Hohn 1996) and trans-acting export factors (Cullen 1992) to facilitate the export of intron-containing mRNAs. In contrast, the export of many cellular RNAs appears to be directly dependent on splicing (Liu and Mertz 1995, and references therein). In this context, herpes simplex virus type 1 (HSV-1) presents an interesting situation for probing mammalian RNA export pathways. HSV-1 expresses over 80 transcripts during viral lytic infection, and only four of these undergo splicing, the remainder being intronless. In addition, HSV-1 infection inhibits host-cell splicing (Hardy and Sandri-Goldin 1994), resulting in the accumulation of pre-mRNA in the nucleus (Hardy and Sandri-Goldin 1994) and decreased accumulation of spliced cellular mRNAs in the cytoplasm (Hardwicke and Sandri-Goldin 1994). The HSV-1 protein responsible for these effects is the essential immediate-early protein termed ICP27. Recent studies have shown that ICP27 can shuttle between the nucleus and the cytoplasm (Soliman et al. 1997) and that ICP27 affects the nuclear export of two HSV-1 intron-containing transcripts (Phelan et al. 1996). The goal of this study was to determine whether ICP27 functions as a viral export protein involved in the transport of HSV-1 intronless RNAs. Here, it is demonstrated that ICP27 shuttles via a leucine-rich NES, and that it binds poly(A)+ RNA in both the nucleus and the cytoplasm. Most interestingly, ICP27 was shown to bind to intronless HSV-1 RNAs, but no binding was seen with two HSV-1 transcripts that undergo splicing. Export of the HSV-1 intronless RNAs was reduced greatly during infections where ICP27 was not expressed. Furthermore, binding of hnRNP A1 to poly(A)+ RNA was reduced significantly during wild-type HSV-1 infection. Finally, in vivo RNA binding by ICP27 required an arginine-rich sequence that resembles an RGG box. These results suggest that ICP27 is an important viral transport factor that mediates export of intronless viral RNAs.
Results
HSV-1 ICP27 shuttles between the nucleus and cytoplasm through a leucine-rich NES
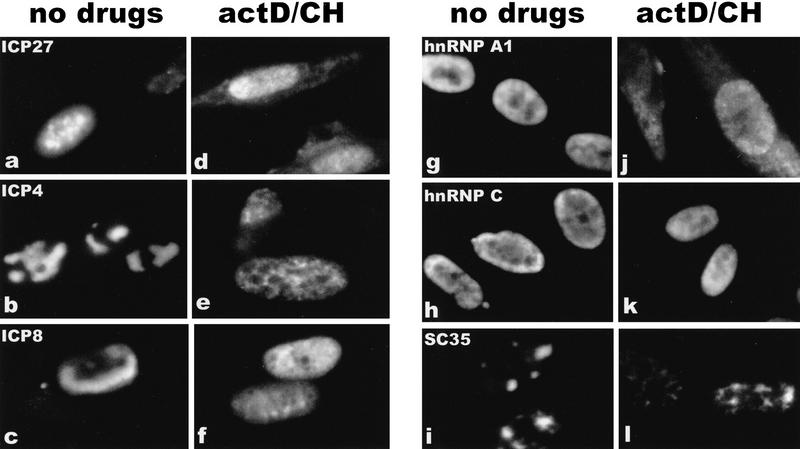
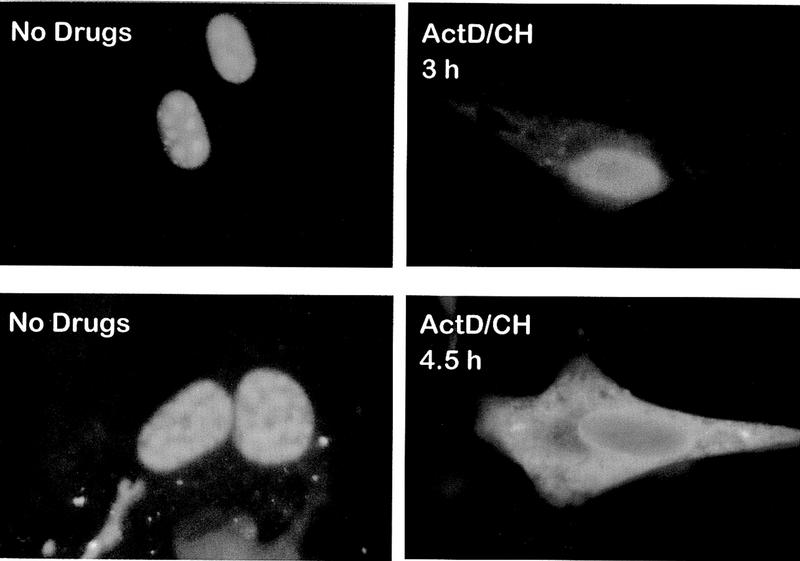
To confirm that ICP27 shuttles and to map the region of ICP27 that is required for export, the intracellular distribution of ICP27 during viral infection was monitored. Cells were infected with wild-type HSV-1 strain KOS either in the absence of inhibitors or in the presence of the RNA Pol II inhibitor actinomycin D and the protein synthesis inhibitor cycloheximide. Actinomycin D was added because it has been demonstrated that the inhibition of transcription interferes with the nuclear import of some proteins, whereas export continues (Piñol-Roma and Dreyfuss 1992; Meyer and Malim 1994); therefore, it is possible to visualize the cytoplasmic accumulation of rapidly shuttling proteins. Cycloheximide was added to insure that cytoplasmic staining resulted from nuclear protein export, not newly synthesized protein. The predominantly nuclear localization of ICP27 in the absence of inhibitors (Figs. 1a and 2, left hand panels) was seen to change to a pronounced cytoplasmic fluorescence by 3 hr after the addition of actinomycin D (Figs. 1d and 2, top right), with most of the protein found in the cytoplasm by 4.5 hr after treatment (Fig. 2, bottom right). A coalescent staining pattern over a more diffuse nuclear staining was seen for ICP27 in untreated cells (Figs. 1 and 2). This is because ICP27 redistributes snRNPs and some of the protein co-localizes with these condensed structures (Phelan et al. 1993; Sandri-Goldin et al. 1995). Staining after actinomycin D treatment was diffuse throughout the nucleus and cytoplasm, indicating that ICP27 leaves these structures. Under the conditions of drug treatment, two other HSV-1 proteins remained nuclear. ICP4, the major transcriptional transactivator of HSV-1, and ICP8, the major DNA-binding protein required for DNA replication, were found associated with viral transcription-replication complexes in untreated cells (Fig. 1b,c) but had a less structured, more diffuse nuclear staining after actinomycin D treatment (Fig. 1e,f) when these complexes disassemble. In addition, hnRNP A1 moved to the cytoplasm (Fig. 1g,j) as originally shown by Piñol-Roma and Dreyfuss (1992). In contrast, hnRNP C and splicing factor SC35, which do not shuttle, remained in the nucleus (Fig. 1h-l).
Figure 1.
The HSV-1 regulatory protein ICP27 accumulates in the cytoplasm of infected cells treated with actinomycin D. Cells were infected with HSV-1 strain KOS. Cells shown in a–c and g–i were not treated with inhibitors and were fixed 7 hr after infection. Actinomycin D (10 μg/ml) and cycloheximide (100 μg/ml) were added to the cells shown in d–f and j–l beginning 4.5 hr after infection for a period of 3 hr. Immunofluorescent staining was performed with monoclonal antibodies to the HSV-1 proteins ICP27, ICP4 and ICP8, and to the cellular proteins, hnRNP A1, hnRNP C, and SC35, as indicated.
Figure 2.
The majority of ICP27 in HSV-1- infected cells moves to the cytoplasm after a longer treatment with actinomycin D. Cells infected with HSV-1 KOS were fixed 7 hr after infection in the absence of inhibitors (left-hand panels) or 3 or 4.5 hr after the addition of actinomycin D (right-hand panels). Cells were stained with ICP27 monoclonal antibody H1113.
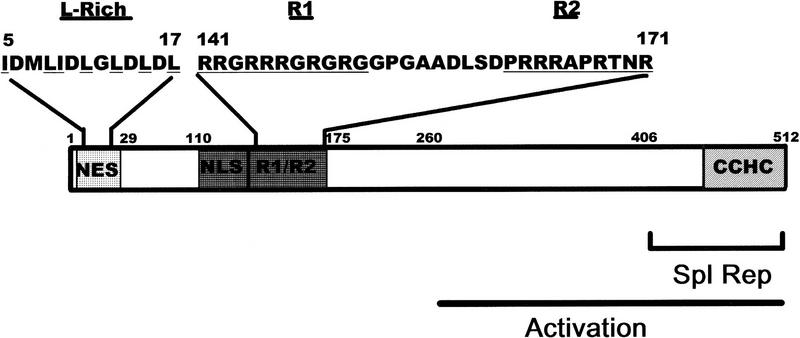
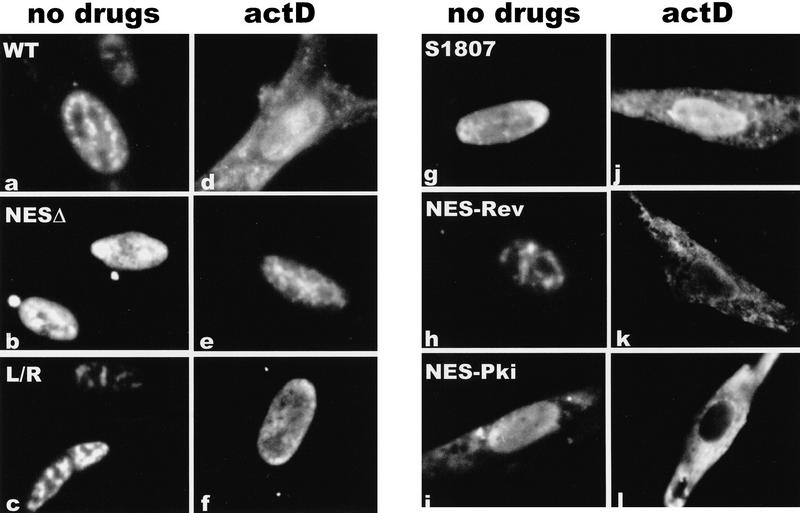
The predicted amino acid sequence of ICP27 contains a leucine-rich region in the amino terminus from residues 5 to 17 (Fig. 3). This sequence resembles the leucine-rich NESs first identified in Rev and PKI (Fischer et al. 1995; Szilvay et al. 1995; Wen et al. 1995). To determine whether this region functioned as an NES, we constructed ICP27 mutants that deleted amino acids 4 to 27, substituted leucine residues in the NES with arginine residues, or replaced amino acids 8 to 17 with the NES of Rev or PKI. Cells were transfected with each of these mutants and were either untreated or actinomycin D was added 24 hr after transfection. Both the wild-type protein and a mutant that has an insertion in the carboxy-terminal splicing repressor region of ICP27 (Sandri-Goldin and Hibbard 1996) moved to the cytoplasm following the addition of actinomycin D (Fig. 4a,d,g,j). A similar movement to the cytoplasm was seen with several other mutants in the splicing repressor and activator regions (data not shown). In contrast, deletion of the putative NES (Fig. 4b,e) or substitution of the leucine residues with arginine residues (Fig. 4c,f) resulted in a protein that remained nuclear. Exclusively nuclear staining was also seen in a mutant in which proline residues were substituted for the leucine residues (data not shown). Cytoplasmic fluorescence was seen again when either the Rev or PKI NES replaced the ICP27 NES. In fact, cytoplasmic staining could be seen with the PKI NES in the absence of actinomycin D. This would suggest that the NES of PKI is a stronger signal than the NLS of ICP27. These data demonstrate that the amino-terminal leucine-rich region is necessary for the export of ICP27.
Figure 3.
Schematic representation of ICP27 coding region showing the putative NES. The 512-amino-acid coding region of ICP27 is represented. The putative NES from residues 5–17 is shown as are the two arginine-rich regions, R1 (residues 141–151) and R2 (162–171) that follow the NLS (residues 110–137) (Hibbard and Sandri-Goldin 1995; Mears et al. 1995). The activation region extends from residues 260–512 (Hardwicke et al. 1989) and the splicing repressor region, which includes a cysteine–histidine–zinc-finger-like domain (CCHC) encompasses residues 406–512 (Sandri-Goldin and Hibbard 1996).
Figure 4.
The leucine-rich region is required for the export of ICP27 to the cytoplasm. Cells were transfected with plasmids expressing wild-type ICP27 (WT), a mutant in the splicing repressor region (S1807), or mutants in the putative NES. NESΔ has a deletion of amino acids 4–27, which encompasses the NES. In L/R, leucine residues at positions 8, 11, 13, and 15 were replaced by arginine residues. NES–Rev has a substitution of the Rev NES, and NES–Pki has a substitution of the PKI NES. Cells in a–c and g–i were not treated with inhibitors, whereas cells in d–f and j–l were treated with actinomycin D for 3 hr.
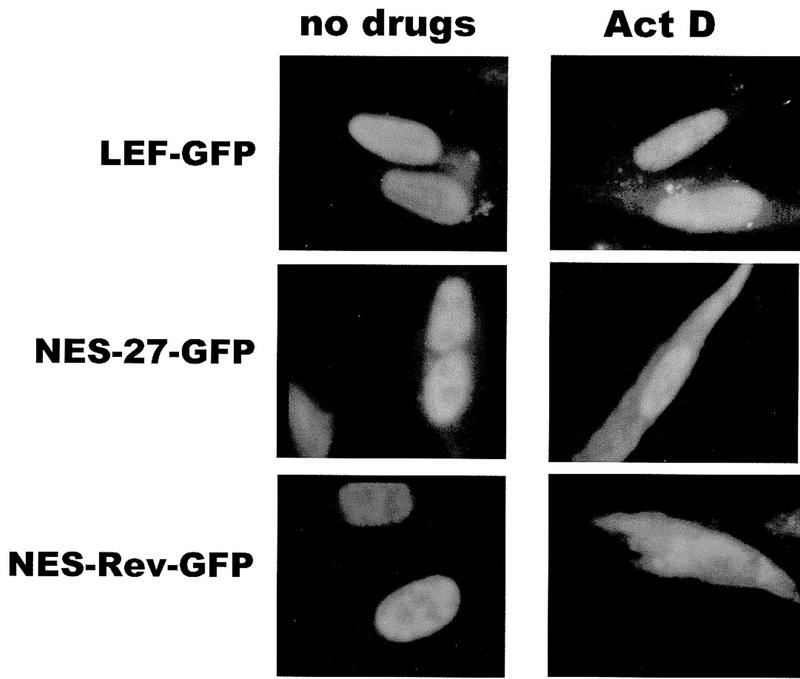
To determine whether the NES of ICP27 could export a heterologous protein as the Rev and PKI signals were shown to enable the export of ICP27, a green fluorescent protein (GFP) import–export substrate was constructed. A plasmid that contains the NLS from the transcription factor LEF (lymphoid enhancer factor) inserted into the carboxyl terminus of GFP was used (Prieve et al. 1996). LEF–GFP was found to be localized to the nucleus in the presence and absence of actinomycin D (Fig. 5, top panels). When the NES of ICP27 from amino acids 5–17 was inserted in the amino terminus of LEF–GFP, a defined cytoplasmic fluorescence was seen after the addition of actinomycin D (Fig. 5, middle panels). A similar result was seen when the Rev NES was fused to LEF–GFP. Therefore, the NES of ICP27 is sufficient to export a heterologous protein with a heterologous NLS, confirming its role as an export signal.
Figure 5.
The ICP27 NES can serve as an export signal on a heterologous protein with a heterologous import signal. The ICP27 NES from amino acids 5–17 was fused to the amino terminus of GFP, which contains an NLS from transcription factor LEF (Prieve et al. 1996). The Rev NES was also added to GFP–LEF. Actinomycin D was added for 3 hr to cells shown in the right-hand panels.
ICP27 binds poly(A)+ RNA in vivo in both the nucleus and the cytoplasm, but the amount of hnRNP A1 bound is reduced during infection
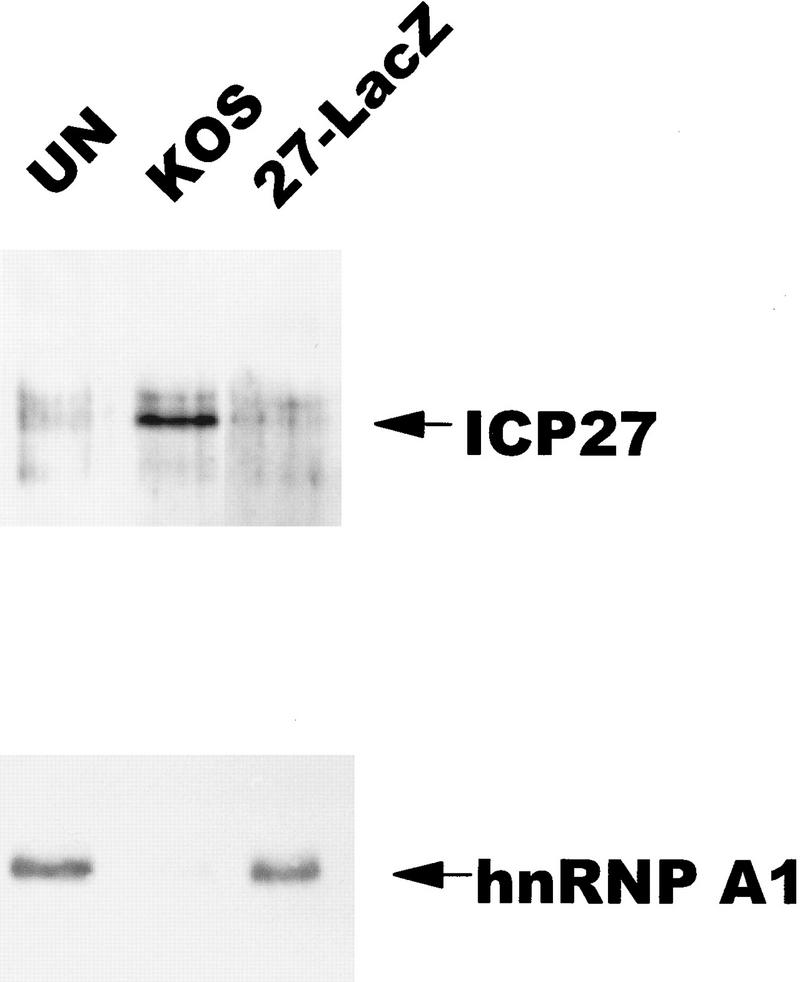
To determine if the purpose of ICP27 shuttling was to export RNA, in vivo UV-cross-linking studies were performed, similar to those described initially for hnRNP A1 (Piñol-Roma and Dreyfuss 1992). Uninfected cell monolayers, or cells infected with HSV-1 KOS or the ICP27 null mutant 27–LacZ were exposed to UV light to induce covalent protein–RNA cross-links in vivo as described (Piñol-Roma and Dreyfuss 1992). Total RNA was isolated, and poly(A)+ RNA was selected by oligo(dT)–cellulose chromatography under protein denaturing conditions so that only proteins that were bound to the RNA during irradiation would copurify (Piñol-Roma and Dreyfuss 1992). Following digestion with RNase, bound proteins were fractionated and immunoblot analysis was performed with a monoclonal antibody to ICP27. As seen in Figure 6, (top), ICP27 was recovered with the poly(A)+ RNA from KOS-infected cells, indicating that ICP27 bound poly(A)+ RNA in vivo. Surprisingly, probing the same blot with antibody 4B10 showed that although hnRNP A1 was recovered with the poly(A)+ RNA in equivalent amounts from uninfected cells and 27–LacZ-infected cells, it was barely detectable in the poly(A)+ RNA fraction from wild-type infected cells. This suggests that ICP27 expression during HSV-1 infection results in reduced binding of hnRNP A1 to RNA.
Figure 6.
ICP27 can be bound covalently to poly(A)+ RNA by in vivo UV irradiation. 293 cell monolayers that were uninfected (UN), infected with wild-type HSV-1 KOS, or 27–LacZ were irradiated with UV light and RNA was isolated and fractionated by oligo(dT)–cellulose chromatography under protein-denaturing conditions. Associated proteins were released by RNase digestion. Immunoblot analysis was performed with anti-ICP27 monoclonal antibodies. Subsequently, the membrane was probed with anti-hnRNP A1 antibody 4B10.
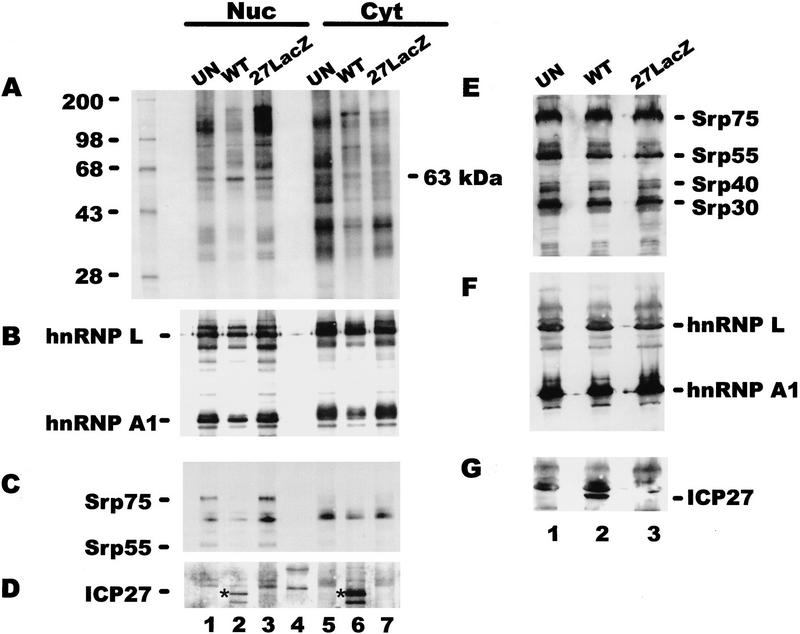
To investigate whether ICP27 bound poly(A)+ RNA in both the nucleus and the cytoplasm therefore, suggesting a role in RNA export, and to determine whether binding of other cellular RNA-binding proteins was affected by ICP27 expression, in vivo UV cross-linking was performed followed by nuclear and cytoplasmic fractionation. In this experiment cells were labeled with 32Pi for 4 hr before irradiation. In this way, phosphoproteins and other proteins bound covalently to RNA were labeled so that the repertoire of proteins bound to poly(A)+ RNA could be visualized. A complex array was seen in both the nuclear and cytoplasmic fractions from uninfected cells, and a similar pattern with some differences in band intensity and some additional bands could be seen in fractions from 27–LacZ-infected cells (Fig. 7A). The pattern appeared to be less complex in both the nuclear and cytoplasmic KOS-infected cell fractions, and a 63-kD band was visible in both the nuclear and cytoplasmic lanes. ICP27 is a 63-kD phosphoprotein, but there was a cellular band near this position in uninfected and 27–LacZ samples, making identification of ICP27 difficult. Immunoblot analysis with anti-ICP27 antibody, however, demonstrated that ICP27 was recovered with both the nuclear and cytoplasmic poly(A)+ RNA from wild-type-infected cells (Fig. 7D). In fact, at this relatively late time after infection, most of the protein recovered was bound to cytoplasmic RNA. This result suggests that ICP27 is involved in RNA export during HSV infection. The blot was also probed with monoclonal antibodies to hnRNP A1 and L. HnRNP L has been shown to bind a sequence present in the HSV-1 intronless thymidine kinase (TK) transcript (Liu and Mertz 1995). Nearly equivalent amounts of hnRNP L were found in the nuclear and cytoplasmic poly(A)+ RNA fractions from uninfected, KOS-infected and 27–LacZ-infected cells (Fig. 7B). In contrast, the amount of hnRNP A1 seen in samples from KOS-infected cells was reduced about fivefold. To determine if the total amount of hnRNP A1 was reduced during wild-type infection, perhaps by degradation, Western blot analysis was performed on total protein lysates taken from a portion of each sample before UV irradiation. The amounts of hnRNP A1 were equivalent in all three samples, as were the amounts of hnRNP L. These results suggest that ICP27 expression during HSV-1 infection results in decreased binding of hnRNP A1 to poly(A)+ RNA but it does not affect overall levels. This would suggest that HSV-1 RNA export does not involve hnRNP A1.
Figure 7.
ICP27 binds poly(A)+ RNA in vivo in both the nucleus and the cytoplasm. 293 cells, which were mock-infected or infected with HSV-1 KOS or 27–LacZ were labeled with 32Pi for 5 hr beginning 1 hr after infection. Following UV irradiation, RNA–protein complexes were purified from nuclear and cytoplasmic fractions under protein denaturing conditions, and poly(A)+ RNA was selected using oligo(dT)–cellulose. In addition, a portion of each cell suspension was lysed directly in electrophoresis sample buffer before irradiation. Proteins were electrophoresed and transferred to nitrocellulose membranes. Immunoblots of proteins that copurified with poly(A)+ RNA are shown in A–D. Immunoblots of total protein lysates are shown in E–G. (A) The membrane with proteins that copurified with poly(A)+ RNA was exposed to x-ray film overnight to visualize the labeled proteins. The left-hand lane contains 14C-labeled standards. (B) After autoradiography, the blot was treated with anti-hnRNP L and anti-hnRNP A1 antibodies. Bands were visualized using ECL, and the exposure was 30 sec. (C) The blot was washed, then treated with anti-SR protein antibody mAb104. The ECL exposure was 15 min. (D)The blot was washed and treated with anti-ICP27 antibody H1119. Because this was the third successive probing of the membrane, several background bands were seen. Lane 4 contains prestained molecular mass standards that were used to orient the membrane. The position of ICP27, detected after a 30-sec exposure, is marked by an asterisk. The lower band seen in both the nuclear and cytoplasmic fractions (lanes 2,6) is probably a proteolytic product of ICP27. (E) The membrane with total protein lysates was treated with mAb104. (F) The blot was subsequently probed with anti-hnRNP L antibody and anti-hnRNP A1 antibody. (G) The blot was probed with anti-ICP27 antibody. Some residual background, probably from hnRNP L was seen; however, the 63-kD ICP27 band was clearly distinguishable in the wild-type (WT) lane.
A possible explanation for the decreased binding of hnRNP A1 to poly(A)+ RNA during wild-type infection is that hnRNP A1 may not come into contact with HSV-1 mRNA because the majority of HSV-1 transcripts are intronless. Furthermore, ICP27 impairs host-cell splicing (Hardy and Sandri-Goldin 1994) and causes the redistribution of splicing factors into large globular structures that move to the periphery of the nucleus (Phelan et al. 1993; Sandri-Goldin et al. 1995). Therefore, factors that associate with RNA during splicing would be expected to be less abundant in poly(A)+ RNA samples from HSV-1-infected cells when ICP27 is expressed. To determine if this was the case, the blot was treated with mAb104, which is specific for a family of SR-domain proteins involved in splicing (Roth et al. 1991). At least four members of this family were present in equal abundance in uninfected and HSV-1-infected cells as seen in the total protein lysates (Fig. 7E). In contrast, only Srp75 and Srp55 were clearly detectable among the proteins that copurified with poly(A)+ RNA and only in nuclear samples from uninfected and 27–LacZ-infected cells (Fig. 7C). The nuclear location of these proteins was expected because SR proteins would be bound to RNA undergoing processing. In addition, because ICP27 impairs splicing, reduced binding in KOS-infected cells would be expected (Fig. 7C).
ICP27 binds to HSV-1 intronless RNAs but not to HSV-1 RNAs that undergo splicing
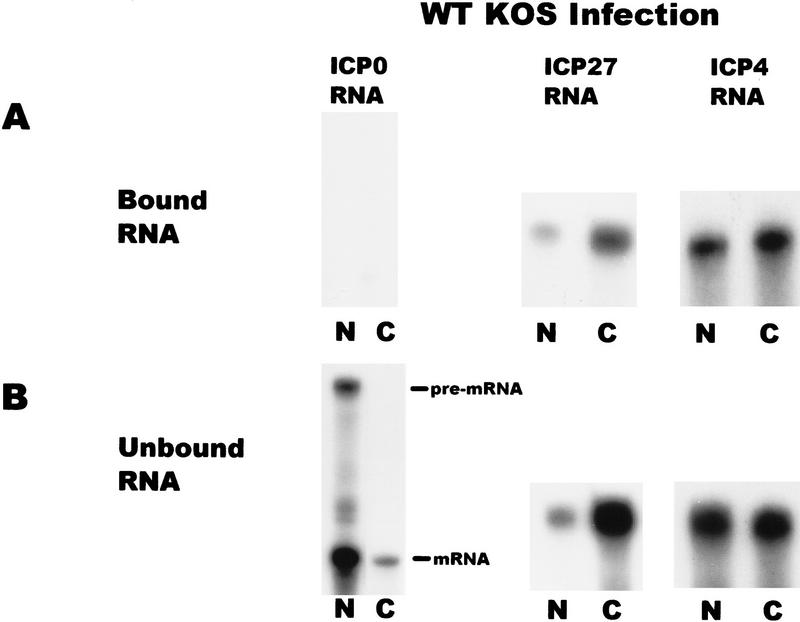
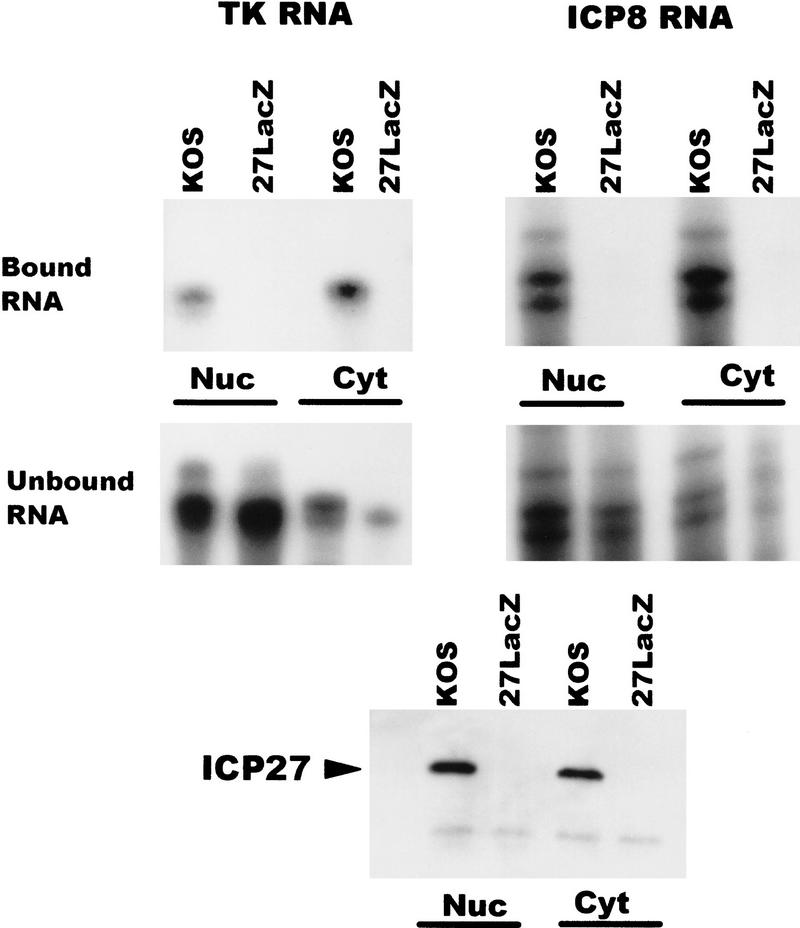
To determine to which RNAs ICP27 binds, in vivo UV cross-linking was performed on wild-type HSV-1-infected cells and nuclear and cytoplasmic fractions were prepared. ICP27 was immunoprecipitated using two anti-ICP27 monoclonal antibodies. The antigen–antibody complexes were bound to protein A–Sepharose beads and washed extensively. RNA that remained bound to the beads because of covalent association with ICP27 was termed the bound fraction, whereas, RNA in the supernatants was termed the unbound fraction. Both fractions were digested with proteinase K and RNase protection analysis was performed. A probe specific for ICP27 RNA showed that ICP27 bound its own intronless, immediate-early transcript in both the nuclear and cytoplasmic fractions (Fig. 8). ICP4 RNA, another intronless, immediate-early transcript was also recovered in the bound nuclear and cytoplasmic fractions. In contrast, ICP0 RNA was not detected in the bound RNA fractions. The ICP0 pre-mRNA contains two introns and ICP27 has been shown to reduce splicing of this transcript during infection so that pre-mRNA accumulates in the nucleus (Hardy and Sandri-Goldin 1994). This was seen in the unbound RNA fraction (Fig. 8). Therefore, ICP27 bound two intronless, immediate-early RNAs but not an immediate-early RNA that undergoes splicing.
Figure 8.
ICP27 binds two HSV-1 intronless transcripts in the nucleus and cytoplasm but does not bind an HSV-1 spliced mRNA. Cells infected with HSV-1 KOS for 5 hr were irradiated with UV light, and nuclear and cytoplasmic fractions were prepared. ICP27 and RNA bound by UV cross-linking was immunoprecipitated with antibodies H1119 and H1113. Antigen–antibody complexes were bound to protein A–Sepharose beads and washed extensively. RNA in the supernatant was termed the unbound fraction, and material that remained bound to the beads was termed the bound fraction. RNase protection analysis was performed using antisense probes specific for the HSV-1 transcripts encoding ICP0, ICP27, and ICP4. The autoradiographs of the protected bound and unbound RNA shown are at different exposures. The amount of ICP27 and ICP4 RNA recovered in the bound fraction represented 1%–3% of the unbound RNA in three different experiments.
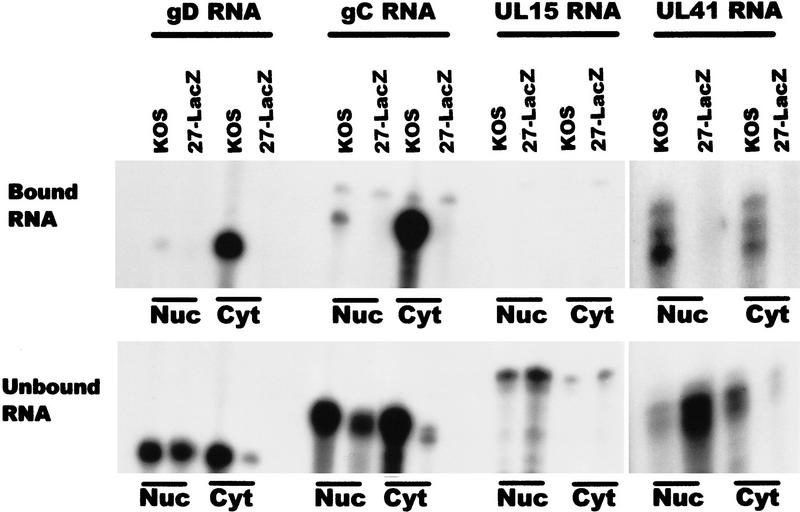
To identify additional transcripts to which ICP27 binds, immunoprecipitations were performed after UV cross-linking wild-type KOS- and 27–LacZ-infected cells. Interestingly, the amount of ICP27 protein that was precipitated in the nuclear and cytoplasmic fractions was equivalent (Fig. 9, bottom), even though immunofluorescent staining of cells that were not treated with actinomycin D showed a predominant nuclear fluorescence (Fig. 1). This could reflect the larger volume of the cytoplasm, which could appear to dilute the actual concentration of ICP27 normally present in the cytoplasm. Additionally, it is possible that some of the nuclear protein that is associated with splicing complex factors precipitated when the lysates were cleared, therefore reducing the amount of nuclear ICP27 that was immunoprecipitated. The early intronless mRNAs encoding TK and the major single-stranded DNA-binding protein (ICP8) were recovered with the bound RNA in the nuclear and cytoplasmic fractions in samples from KOS-infected cells (Fig. 9). As expected, RNA was not recovered bound to protein A beads in the absence of ICP27 during infection with 27–LacZ virus. Three intronless HSV-1 late mRNAs, those encoding glycoprotein D (gD), glycoprotein C (gC), and the virion host shutoff factor, vhs (UL41) were recovered in the bound nuclear and cytoplasmic fractions from KOS-infected cells (Fig. 10). The UL15 transcript, however, which is spliced, was not found in the bound RNA (Fig. 10), again suggesting that ICP27 mediates intronless RNA export, whereas spliced transcripts use the cellular mRNA export pathway.
Figure 9.
ICP27 binds two HSV-1 early intronless mRNAs and facilitates their transport to the cytoplasm. Cells infected with HSV-1 KOS or 27–LacZ for 6 hr were treated with UV light, divided into nuclear and cytoplasmic fractions, and immunoprecipitation was performed. Cells were labeled with [35S]methionine during infection to allow monitoring of immunoprecipitated ICP27 protein (bottom). RNase protection analysis of the bound (top) and unbound (middle) RNA fractions was performed with antisense probes specific for mRNAs encoding TK and ICP8, respectively.
Figure 10.
ICP27 is required for the efficient export of late intronless RNAs. HSV-1 KOS or 27–LacZ-infected cells were UV irradiated and immunoprecipitations were performed. The antisense probes used were specific for HSV-1 late intronless mRNAs encoding gD, gC, UL41, and UL15, a late RNA that contains an intron. RNA recovered in the bound fractions (top) represents ∼ 1%–2% of the RNA in the unbound fractions (bottom).
The bound and unbound RNA fractions in Figures 8–10 are shown at different exposures, and the actual amount of RNA bound for each transcript was calculated to be 1%–3% of the unbound RNA. This would suggest that ICP27 binds RNA, rapidly exports it to the cytoplasm where it is released, and the protein returns to the nucleus. In fact, the amount of bound cytoplasmic RNA recovered was greater than the nuclear recovery for several of the transcripts analyzed, especially TK, ICP8, gD, and gC. This could indicate that export of these transcripts is inefficient in the absence of ICP27. That this was the case can be seen in the unbound RNA fractions from 27–LacZ-infected cells (Figs. 9 and 10). Whereas abundant amounts of TK, ICP8, gD, gC, and UL41 RNA were found in the unbound nuclear RNA from 27–LacZ-infected cells, substantially lower amounts of these transcripts were recovered in the cytoplasmic fractions. ICP27 has been shown to be required for efficient HSV-1 early gene expression (Uprichard and Knipe 1996) and to be essential for late gene expression (Sacks et al. 1985; Rice and Knipe 1990). These data indicate that ICP27 regulates HSV-1 gene expression in part by facilitating the export of HSV-1 intronless RNAs. This premise is supported by the finding that NESΔ and NES-L/R were unable to complement 27–LacZ virus infection (data not shown), indicating that nuclear export is an essential function for ICP27 during viral infection.
ICP27 binds RNA in vivo through an RGG motif
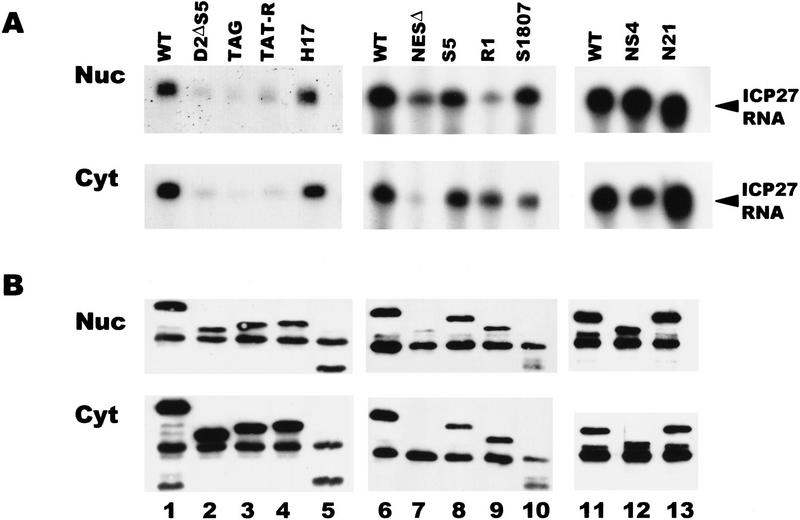
To determine which region of ICP27 is required for binding to RNA, in vivo UV cross-linking was performed on cells expressing ICP27 mutants with deletions or insertions in the major functional regions of the protein. Analysis of the predicted peptide sequence indicated that the most likely regions to be involved in RNA binding were one or both arginine-rich regions in the amino-terminal half of the protein from amino acids 141 to 171 or a zinc-finger-like region in the carboxyl terminus that is required for splicing inhibition (Fig. 3). The R-rich regions, termed R1 and R2, directly follow the NLS of ICP27 as mapped by Mears et al. (1995), which is an unusual NLS in that it is neither basic nor bipartite in structure. In fact, the R-rich regions were shown to increase the efficiency of import of ICP27 (Hibbard and Sandri-Goldin 1995). Cells were transfected with plasmids encoding various ICP27 mutants and were subsequently infected with 27–LacZ virus. This was done so that all other conditions of HSV-1 infection would occur, but transfected cells (∼10%–20% of the cells) would express mutant ICP27 protein during infection. The majority of the cells would be infected but not transfected, and these cells would express no ICP27. Binding of ICP27 to its own mRNA was monitored because this was the only HSV-1 transcript whose expression would be restricted to cells that were transfected and therefore would express ICP27 mutant protein.
ICP27 RNA was recovered in the nuclear and cytoplasmic bound fractions from cells expressing the mutants NS4 and S1807, which contain insertions in the zinc-finger region of the protein (Fig. 11A). The insertion in S1807 renders the mutant protein more susceptible to proteolytic cleavage in vivo, so that a smaller than full-length protein (45 kD) was recovered in the immunoprecipitations (Fig. 11B, lane 10), whereas full-length protein is synthesized in reticulocyte lysates (data not shown). NS4 contains a stop codon within the inserted sequences resulting in a protein that terminates at amino acid 460 at the start of the zinc-finger region (Fig. 3). Therefore, the zinc-finger region does not appear to be required for binding RNA. Similarly, N21, an insertion mutant and H17, a deletion mutant that map to the activation region of ICP27 (Hardwicke et al. 1989) bound RNA as efficiently as the wild-type protein (Fig. 11A). The NESΔ mutant bound RNA efficiently in the nucleus, but as this protein was undetectable in the cytoplasm, little bound RNA was recovered in the cytoplasmic fraction (Fig. 11A, lane 7). These results show that the NES, activation region and zinc-finger region are not required for binding. In contrast, three different mutants that lack the NLS and both R-rich regions, D2ΔS5, TAG, and TAT-R showed greatly reduced binding (Fig.11A, lanes 2–4). Because the D2ΔS5 mutant lacks the major NLS, the protein was localized to the nucleus inefficiently (Fig. 11B, lane 2). The TAG mutant contains the SV40 NLS (PKKKRKV) in the site of the deletion, and the TAT-R mutant contains the HIV Tat NLS and RNA-binding region (RKKKRRQRRA), which increase the nuclear localization of the mutant proteins (Hibbard and Sandri-Goldin 1995); however, RNA binding was reduced to the same background level seen with the D2ΔS5 mutant. This suggests that the NLS or one or both R-rich regions is required for efficient RNA binding. To distinguish among these possibilities, two other mutants were analyzed. The S5 mutant has a deletion of just the R2 region with both the NLS and R1 region intact, and the R1 mutant has a deletion of the NLS and the R2 region, but the R1 region is intact (Fig. 3). Both of these mutant proteins bound ICP27 RNA efficiently (Fig. 11A, lanes 8,9). In addition, mutant N6, which has a 5-amino-acid insertion (PGIPG) that disrupts the R2 sequence (Hardwicke et al. 1989), bound RNA efficiently (data not shown). These data suggest that the R1 region, which consists of arginine and glycine residues, and resembles RGG boxes found in other RNA-binding proteins (Birney et al. 1993; Liu and Dreyfuss 1995), is required for the binding of ICP27 to RNA in vivo. In accord with this finding, the RGG region was shown previously to be both required and sufficient for in vitro RNA-binding by ICP27 (Mears and Rice 1996).
Figure 11.
ICP27 binds RNA in vivo through an RGG motif. RSF cells transfected with different ICP27 mutants were infected with 27–LacZ and UV irradiated 6 hr later. RNase protections were performed with a probe specific for the ICP27 transcript. The region spanned by the probe is present in all of the ICP27 mutant RNAs. (A) RNA recovered in the nuclear and cytoplasmic bound fractions. (B) Immunoblot analysis of immunoprecipitated ICP27 protein. Wild-type ICP27 (lanes 1,6,11) migrates at 63 kD. The dark band in all lanes at 55 kD is heavy-chain IgG, which reacts with the secondary antibody and is present because immunoprecipitated proteins were transferred to the membrane.
Discussion
The experiments described here have provided strong evidence that an essential HSV-1 regulatory protein that acts predominantly at the post-transcriptional level (Sandri-Goldin and Mendoza 1992; Smith et al. 1992) mediates RNA export of viral intronless mRNAs. ICP27 was shown to possess a leucine-rich nuclear export signal in the amino terminus that enabled the protein to shuttle between the nucleus and the cytoplasm. In vivo UV irradiation studies demonstrated that ICP27 could be crosslinked to poly(A)+ RNA in both the nucleus and the cytoplasm, suggesting a role in RNA export. This was supported further by the finding that ICP27 bound seven intronless HSV-1 transcripts in vivo, and the export of these transcripts to the cytoplasm was substantially reduced during infection with 27–LacZ virus, where ICP27 is not expressed. These results, coupled with the finding that mutants in the NES cannot complement 27–LacZ viral infection, suggests that export of HSV-1 intronless mRNAs by ICP27 comprises at least part of its function as an essential regulator of viral gene expression.
The question arises as to why HSV-1 transcripts would require an export pathway that is distinct from the pathway for cellular mRNAs. Substantial evidence has been presented to show that hnRNP A1 has an important role in the export of cellular mRNAs. HnRNP A1 binds to 5′ splice site-like sequences (Burd and Dreyfuss 1994), which suggests that it exports mRNAs that undergo splicing. Interestingly, the amount of hnRNP A1 bound to poly(A)+ RNA was reduced significantly in wild-type HSV-1-infected cells, whereas the amount of hnRNP L that bound was similar in infected and uninfected cells. Liu and Mertz (1995) showed that hnRNP L binds to a cis-acting element in the intronless HSV TK mRNA, and these authors postulated that hnRNP L may have a role in the export of cellular and viral intronless mRNAs, whereas, hnRNP A1 mediates the export of mRNAs that are spliced. The majority of HSV-1 transcripts are intronless. In addition, HSV-1 has a relatively rapid and robust transcription pattern that is regulated by strong viral transcriptional activators. Following HSV-1 infection there is a rapid inhibition of cellular RNA PolII transcription followed by selective activation of the viral genome (Spencer et al. 1997). Therefore, viral intronless mRNAs become very abundant as infection progresses. If cellular mRNA export proteins recognize splice site sequences or associate with splicesomal complexes, these factors would not bind the abundant HSV-1 intronless RNAs. The finding that the amount of hnRNP A1 bound to poly(A)+ RNA was reduced in HSV-1-infected cells, whereas, the total amount of hnRNP A1 appeared unchanged would support that hypothesis. It is also possible that hnRNP A1 is modified following HSV-1 infection, perhaps by differential methylation of the RGG region. It has been proposed that selective methylation may have a role in the subcellular localization of pre-mRNA binding proteins (Henry and Silver 1996). Differences in the staining pattern of hnRNP A1 in infected versus uninfected cells were not seen (M.A. Hardwicke and R.M. Sandri-Goldin, unpubl.). hnRNP A1, however, is a very abundant protein, so that changes in the distribution of a percentage of the protein would be difficult to detect by immunofluorescent staining.
It has been well-documented that some viral infections alter the balance of cellular mRNA export pathways to favor viral export and to retain cellular transcripts in the nucleus. The E1B 55-kD and E4 34-kD adenoviral oncoproteins form a complex that promotes the export of viral mRNA late in infection, and inhibits the export of most cellular mRNAs (Dobbelstein et al. 1997, and references therein). Within this complex, the E4 34-kD protein directs both nuclear import and nuclear export and this protein contains an NES similar to the leucine-rich signal of Rev (Dobbelstein et al. 1997). When E4 34-kD is expressed alone, a predicted arginine-rich amphipathic α-helical region mediates nuclear retention of the protein. Association with E1B 55-kDa, however, abolishes this retention. It is hypothesized that the interplay between the two polypeptides contributes to the efficiency of the adenovirus export system (Dobbelstein et al. 1997). Another example of a viral protein that inhibits cellular mRNA export is the influenza virus NS1 protein (Fortes et al. 1994; Qui and Krug 1994), which binds the poly(A) tail of mRNAs and therefore, a generalized block of mRNA export from the nucleus ensues. In HSV-1 infection, ICP27 appears to promote both effects, the nuclear retention of unspliced cellular RNA and the export of viral intronless RNAs. In addition, the pre-mRNA for two spliced viral transcripts, ICP0 and UL15, was seen to accumulate in nuclear clumps (Phelan et al. 1996). In accord with these findings, neither ICP0 nor UL15 RNA was recovered in the fractions bound to ICP27 (Figs. 8 and 10), suggesting that these RNAs are retained in spliceosomal complexes. In fact, the amount of spliced ICP0 and UL15 mRNA in the unbound cytoplasmic fractions was at least 10 times lower than the amount present in the nuclear fraction in wild-type infections. This result suggests that export of spliced RNAs also may be inefficient during infection, perhaps as a consequence of competition with ICP27-mediated export of intronless RNA. Therefore, during HSV-1 infection, ICP27 may be sufficient to induce a shift in nucleocytoplasmic transport, such that viral intronless transcripts are favored.
It is likely that cellular proteins are also involved in the export of HSV-1 RNAs. For example, hnRNP L may have a role. Studies are underway to identify cellular proteins that interact with ICP27 as part of its export function and to determine the sequence or structural specificity of ICP27 RNA binding. In any event, the studies described here have defined an important role for an essential viral regulatory protein as an exporter of intronless viral mRNA.
Materials and methods
Cells, viruses, and recombinant plasmids
Rabbit skin fibroblasts (RSF) and 293 cells (American Type Culture Collection, Bethesda, MD) were grown as described (Sandri-Goldin and Mendoza 1992). Cells were infected with wild-type HSV-1 strain KOS or the ICP27 null mutant, 27–LacZ as described previously (Smith et al. 1992). Mutants H17 and N21 were described previously (Hardwicke et al. 1989), as were D2ΔS5, TAG, TAT-R, S5 and R1 (Hibbard and Sandri-Goldin 1995). Mutant S1807 was constructed by inserting three copies of a ClaI linker into a SmaI site at the position of amino acid 465 in the ICP27 coding sequence. NS4 was constructed by inserting an oligonucleotide encoding a stop codon into an NsiI site at amino acid 459. NESΔ was constructed by digesting the wild-type ICP27 plasmid with DrdI and NaeI, which results in the deletion of a fragment beginning five nucleotides upstream of the translation initiation site to the codon for amino acid 28. An oligonucleotide encoding the 5 upstream nucleotides and the first 3 amino acids was inserted into the site of the deletion, resulting in a deletion of amino acids 4–27. NESL/R was constructed by inserting an oligonucleotide encoding the upstream nucleotides and the first 27 amino acids of ICP27, but arginine codons were substituted for leucine codons at positions 8, 11, 13, and 15 (Fig. 3). NES–Rev and NES–PKI were made by inserting oligonucleotides encoding the amino terminus of ICP27 into NESΔ with the HIV Rev NES, LPPLERLTL, or the PKI NES, LALKLAGLDI, substituted for ICP27 amino acids 8–17. NES–27–GFP and NES–Rev–GFP have insertions of oligonucleotides encoding ICP27 amino acids 5–17 or the Rev NES into a BamHI site in the amino terminus of LEF–GFP (Prieve et al. 1996).
Virus infection, transfection, and radiolabeling
Cells were infected with HSV-1 KOS or 27–LacZ for the times indicated in each experiment. If 32Pi or [35S]methionine labeling was done, the procedure as described by Sandri-Goldin and Hibbard (1996) was performed. Cells were labeled for 5 hr beginning 1 hr after infection. When cells were transfected, lipofectamine (Life Technologies) was used at 50 μl per 100-mm-diameter tissue culture dish, and the plasmid DNA concentration was 10 μg per dish. Cells were infected with 27–LacZ at a multiplicity of infection of 10 beginning 24 ht after transfection.
Immunofluorescence staining
RSF cells were grown on glass coverslips for 16 hr and then infected with HSV-1 KOS. Transfections were performed on cells that were seeded onto coverslips 12 hr earlier. Actinomycin D (10 μg/ml) and cycloheximide (100 μg/ml) were added 4.5 hr after infection or 24 hr after transfection for a period of 3 or 4.5 hr as indicated. Cells were fixed in 3.7% formaldehyde in PBS for 10 min. Before staining, cells were permeabilized in PBS containing 0.5% NP-40. Indirect immunofluorescent staining was performed as described (Sandri-Goldin et al. 1995). Primary antibodies included the ICP27 monoclonal antibodies H1113 and H1119 (Goodwin Institute, Plantation, FL), used at a dilution of 1:500; anti-ICP4 monoclonal antibody H1114 at a dilution of 1:500; anti-ICP8 monoclonal antibody, H1115 at 1:500; anti-hnRNP A1 4B10 hybridoma supernatant (Piñol-Roma and Dreyfuss 1992) at 1:100; anti-hnRNP C monoclonal antibody, 4F4 (Piñol-Roma and Dreyfuss 1992) at 1:1000, and anti-SC35 hybridoma supernatant (Fu and Maniatis 1990) at a dilution of 1:50.
UV irradiation, cell fractionation, and RNA purification
The culture medium was removed from cell monolayers, which were washed twice with ice-cold PBS. Irradiation was performed for 4 min as described (Piñol-Roma and Dreyfuss 1992). Following UV irradiation, total RNA and bound protein were extracted by the guanidium thiocyanate method as described (Sandri-Goldin and Mendoza 1992). Nuclear and cytoplasmic RNA–protein fractions were prepared and isolated with guanidium thiocyanate (Sandri-Goldin and Mendoza 1992). After precipitation of the cross-linked RNA–protein complexes with LiCl, the pellets were resuspended in 10 mm Tris (pH 7.4), 1 mm EDTA, 0.5% SDS, and 1% β-mercaptoethanol. Samples were heated at 65°C for 5 min and adjusted to 0.5 m LiCl. Oligo(dT)–cellulose chromatography was performed under protein denaturing conditions (Piñol-Roma and Dreyfuss 1992), therefore, only proteins covalently bound to poly(A)+ RNA copurified. RNA samples were centrifuged and resuspended in PBS containing 2.5 mm Pefabloc and 2.5 mm leupeptin. Proteins cross-linked to RNA were released by digestion with 2.5 units of RNase A and 200 units of RNase T1 (Ambion) at 37°C for 60 min.
Immunoblotting procedures
Polyacrylamide gel electrophoresis and transfer of proteins to nitrocellulose were performed as described (Sandri-Goldin and Hibbard 1996). Enhanced chemiluminescence (ECL: Amersham Life Sciences) was used to visualize bands. Primary antibodies H1113 and H1119 were used at dilutions of 1:5000, anti-hnRNP A1 hybridoma supernatant 4B10, at a dilution of 1:4, anti-hnRNP L monoclonal antibody 4D10 (Liu and Mertz 1995), at a dilution of 1:5000, and anti-SR protein mAB104 hybridoma supernatant (Roth et al. 1991), at a dilution of 1:2. When membranes were reprobed with different primary antibodies, the membranes were washed extensively with PBS containing 0.1% Tween 20 between each probing, but the membranes were not stripped to avoid removing any of the proteins bound to the membranes.
Immunoprecipitation and RNase protection analysis
Following UV irradiation, cells were scraped into cold PBS, centrifuged and resuspended in 10 mm Tris (pH 7.4), 3 mm CaCl2, 2 mm MgCl2, 0.5% NP-40, 1.0 mm Pefabloc, 1.0 mm leupeptin, and 500 units of RNasin (Promega). The cell suspension was passed through a syringe with a 25-gauge needle five times to lyse the cells and the nuclei were harvested by centrifugation at 14,000g for 20 sec. The cytoplasmic supernatant was removed to a separate tube, and adjusted to 0.5 m NaCl. Nuclei were resuspended in high salt extraction buffer consisting of PBS containing 0.5 m NaCl, 0.5% NP-40, 1.0 mm Pefabloc, 1.0 mm leupeptin, and 500 units of RNasin. Proteins were extracted by placing the tubes on an end to end rotator for 40 min at 4°C. The extracts were cleared by centrifugation at 14,000g for 30 min at 4°C. Antibodies H1119 and H1113 were added to each supernatant (2 μl/ml) and tubes were placed on a rotator at 4°C for 60 min. Rabbit–anti-mouse IgG (Pierce, 2 μl/ml) was added to each tube to increase the efficiency of binding of the antigen-antibody complexes to protein A. After 60 min, the samples were divided into aliquots corresponding to each RNA probe to be used and a portion to be analyzed by SDS-PAGE. Protein A–Sepharose beads (50 μl of a 40% suspension) were added to all samples. The beads were prepared by extensive washing in high salt extraction buffer containing RNasin (1000 units). Complexes bound to the protein A–Sepharose beads were washed by five rounds of centrifugation and resuspension in high-salt extraction buffer, followed by two washes in PBS containing 0.5% NP-40 and one wash in PBS without NP-40. The supernatants from the first centrifugation and wash were combined as the unbound RNA fractions. Samples to be analyzed by SDS-PAGE were treated with RNase A and T1 as described above. Samples to be analyzed by RNase protection were digested with proteinase K (2.5 μg/μl) at 37°C for 60 min, then three volumes of inactivation/precipitation mixture (Ambion) was added. Samples were stored at −70°C, centrifuged at 14,000g for 30 min and resuspended in 20 μl of hybridization buffer (Ambion). Gel-purified RNA probes (5 × 105 cpm) were added. RNase protection assays were performed as described previously (Sandri-Goldin and Mendoza 1992). The hybridization temperature was 56°C for all HSV-1 RNA samples because of the high-GC content. The probes for ICP27, ICP0, and ICP8 were described previously (Hibbard and Sandri-Goldin 1995), as were the probes for ICP4, UL41 (Hardwicke and Sandri-Goldin 1994), and UL15 (Hardy and Sandri-Goldin 1994). A gD transcription plasmid (Hibbard and Sandri-Goldin 1995) was digested with BsaI to generate a 260-nucleotide antisense RNA on transcription with T7. The gC transcription vector (Hibbard and Sandri-Goldin 1995) was digested with NcoI to generate a 255-nucleotide probe. The TK probe was transcribed from a pGEM-2 plasmid containing a PstI to SstI fragment of the HSV-1 TK gene. The plasmid was digested with EcoRI before transcription with T7 polymerase, resulting in a probe that would protect a 300-nucleotide TK RNA fragment.
Acknowledgments
I thank Melissa Mazanet for excellent technical assistance; Gideon Dreyfuss (University of Pennsylvania) for providing antibodies to hnRNP A1, C, and L; and Marian Waterman (University of California, Irvine) for the plasmid encoding LEF–GFP. This work was supported by U.S. Public Health Service Grant AI21515 from the National Institutes of Allergy and Infectious Diseases.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
References
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd AM, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Madore S, Cullen BR. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved Structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–620. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M, Roth J, Kimberly WT, Levine AJ, Shenk T. Nuclear export of the E1B-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell RA, Bogerd AM, Cullen BR. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci. 1996a;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell RA, Fischer U, Luhrmann R, Meyer BE, Meinkoth JL, Malim MH, Cullen BR. Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1 Rev. Proc Natl Acad Sci. 1996b;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz CC, Zapp M, Green MR. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Hardwicke MA, Sandri-Goldin RM. The herpes simplex virus regulatory protein ICP27 can cause a decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwicke MA, Vaughan PJ, Sekulovich RE, O’Conner R, Sandri-Goldin RM. The regions important for the activator and repressor functions of the HSV-1 alpha protein ICP27 map to the C-terminal half of the molecule. J Virol. 1989;63:4590–4602. doi: 10.1128/jvi.63.11.4590-4602.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy WR, Sandri-Goldin RM. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MF, Silver PA. A novel methyltransferase (Hmt 1p) modifies poly(A)+-RNA-binding proteins. Mol Cell Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbard MK, Sandri-Goldin RM. Arginine-rich regions succeeding the nuclear localization region of the HSV-1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J Virol. 1995;69:4656–4667. doi: 10.1128/jvi.69.8.4656-4667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Jarmolowski A, Beisel C, Mattaj IW, Dreyfuss G. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by different factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalland K-H, Szilvay M, Brokstad KA, Saetrevik W, Haukenes G. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol Cell Biol. 1994;14:7436–7444. doi: 10.1128/mcb.14.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim FJ, Beeche AA, Hunter JJ, Chin DJ, Hope TJ. Characterization of the nuclear export signal of human T-cell lymphotrophic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol Cell Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Hohn T. Pararetro- and retrovirus RNA: Splicing and the control of nuclear export. Trends Microbiol. 1996;4:480–485. doi: 10.1016/s0966-842x(97)82909-5. [DOI] [PubMed] [Google Scholar]
- Kjems J, Brown M, Chang DD, Sharp PA. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc Natl Acad Sci. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes & Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Mertz JE. HnRNP L binds a cis-acting RNA sequence element that enables intron-independent gene expression. Genes & Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Mears WE, Rice SA. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J Virol. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears WE, Lam V, Rice SA. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J Virol. 1995;69:935–947. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BE, Malim MH. The HIV-1 Rev trans-activator shuttles between the nucleus and cytoplasm. Genes & Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- Meyer BE, Meinkoth JL, Malim MH. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious agent virus rev proteins: Identification of a family of nuclear export signals. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Murphy R, Wente SR. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Fischer U, Michael WM, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-βfamily member Crm 1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Palmer D, Malim MH. The human T-cell leukemia virus type 1 posttranscriptional trans-activator Rex contains a nuclear export signal. J Virol. 1996;70:6442–6445. doi: 10.1128/jvi.70.9.6442-6445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan A, Carmo-Fonseca M, McLauchlan J, Lamond AI, Clements JB. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan A, Dunlop J, Clements JB. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J Virol. 1996;70:5255–5265. doi: 10.1128/jvi.70.8.5255-5265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Prieve MG, Guttridge KL, Mungia JE, Waterman ML. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-nuclear localization sequence receptor proteins. J Biol Chem. 1996;271:7654–7658. doi: 10.1074/jbc.271.13.7654. [DOI] [PubMed] [Google Scholar]
- Qui Y, Krug RM. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice SA, Knipe DM. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J Virol. 1990;64:1704–1715. doi: 10.1128/jvi.64.4.1704-1715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard N, Iacampo S, Cochrane A. HIV-1 Rev is capable of shuttling between the nucleus and the cytoplasm. Virology. 1994;204:123–131. doi: 10.1006/viro.1994.1516. [DOI] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–591. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks WR, Greene CC, Ashman DP, Schaffer PA. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin RM, Mendoza GE. A herpes virus regulatory protein appears to act posttranscriptionally by affecting mRNA processing. Genes & Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- Sandri-Goldin RM, Hibbard MK. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum and the C-terminus appears to be required for this interaction. J Virol. 1996;70:108–118. doi: 10.1128/jvi.70.1.108-118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin RM, Hibbard MK, Hardwicke MA. The C-terminal repressor region of HSV-1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IL, Hardwicke MA, Sandri-Goldin RM. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- Soliman TM, Sandri-Goldin RM, Silverstein S. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CA, Dahmus ME, Rice SA. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. J Virol. 1997;71:2031–2040. doi: 10.1128/jvi.71.3.2031-2040.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (CRM1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Szilvay AM, Brokstad KA, Kopperrud R, Haukenes G, Kalland K-H. Nuclear export of the human immunodeficiency virus type 1 nucleocytoplasmic shuttle protein Rev is mediated by its activation domain and is blocked by transdominant negative mutants. J Virol. 1995;69:3315–3323. doi: 10.1128/jvi.69.6.3315-3323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman KS, Powers MA, Forbes DJ. Nuclear export receptors: From importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- Uprichard SL, Knipe DM. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J Virol. 1996;70:1969–1980. doi: 10.1128/jvi.70.3.1969-1980.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Bjorkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]