Abstract
This article proposes a joint model for longitudinal measurements and competing risks survival data. The model consists of a linear mixed effects sub-model with t-distributed measurement errors for the longitudinal outcome, a proportional cause-specific hazards frailty sub-model for the survival outcome, and a regression sub-model for the variance-covariance matrix of the multivariate latent random effects based on a modified Cholesky decomposition. A Bayesian MCMC procedure is developed for parameter estimation and inference. Our method is insensitive to outlying longitudinal measurements in the presence of non-ignorable missing data due to dropout. Moreover, by modeling the variance-covariance matrix of the latent random effects, our model provides a useful framework for handling high-dimensional heterogeneous random effects and testing the homogeneous random effects assumption which is otherwise untestable in commonly used joint models. Finally, our model enables analysis of a survival outcome with intermittently measured time-dependent covariates and possibly correlated competing risks and dependent censoring, as well as joint analysis of the longitudinal and survival outcomes. Illustrations are given using a real data set from a lung study and simulation.
Keywords and phrases: Joint model, Competing risks, Bayesian analysis, Cholesky decomposition, Mixed effects model, MCMC, Modeling random effects covariance matrix, Outlier
1. INTRODUCTION
In the past decades, much work has been done in the field of joint modeling of longitudinal and survival data. Joint models have been proposed to adjust inferences on longitudinal measurements in the presence of non-ignorable missing values due to dropout (Schluchter, 1992; DeGruttola and Tu, 1994; Little, 1995; Hogan and Laird, 1997; Henderson et al., 2000; Elashoff et al., 2007, 2008); to solve difficulties in Cox proportional hazards model arising from time-dependent covariates which are possibly missing at some event times or subject to substantial measurement error (Faucett and Thomas, 1996; Wulfsohn and Tsiatis, 1997; Faucett et al., 1998; Wang and Taylor, 2001; Xu and Zeger, 2001; Song et al., 2002; Brown and Ibrahim, 2003; Tseng et al., 2005; Ye et al., 2008); and to assess covariates effects on both endpoints simultaneously (Henderson et al., 2000; Zeng and Cai, 2005; Elashoff et al., 2007, 2008; Liu et al., 2008).
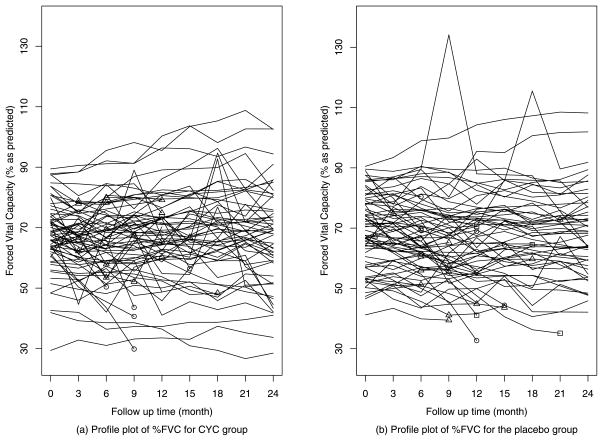
Most joint models in the literature assume a normal random error for the longitudinal sub-model. The normal model, however, can be sensitive to outliers or heavy-tailed data. For example, the profile plot of repeated measured FVC (Forced Vital Capacity) in Figure 1 reveals several suspiciously large values. We demonstrate in Section 3 that the normal model is sensitive to the extreme values. We also observe in a simulation study (see Tables 2, 3) that the regression and random effects parameters in a normal model can be seriously biased in the presence of outliers.
Figure 1.
(a)–(b) Profile plots of observed %FVC for CYC group vs. placebo group including potential outlying measurements: ○ for treatment failure or death; △ for informatively censored events; □ for noninformatively censored events.
Table 2.
Comparison of the joint model with a normal distribution versus that with a t-distribution for εij (sample size = 250)
| m | Parameter | True | Normal distribution | t-distribution | |||||
|---|---|---|---|---|---|---|---|---|---|
| Bias | SE | CP | Bias | SE | CP | ||||
| 250 | Longitudinal | ||||||||
| Fixed effects | |||||||||
| β0 | 10 | 0.295 | 0.352 | 0.655 | 0.020 | 0.137 | 0.903 | ||
| β1 | −1 | 0.059 | 0.735 | 0.927 | −0.009 | 0.221 | 0.952 | ||
| β2 | 1.5 | 0.272 | 0.340 | 0.807 | −0.015 | 0.155 | 0.949 | ||
| Survival | |||||||||
| Fixed effects | |||||||||
| γ11 | 0.8 | −0.004 | 0.121 | 0.953 | −0.011 | 0.130 | 0.923 | ||
| γ12 | −1 | 0.042 | 0.286 | 0.927 | 0.002 | 0.248 | 0.954 | ||
| γ21 | 0.5 | −0.025 | 0.128 | 0.927 | −0.012 | 0.119 | 0.929 | ||
| γ22 | −1 | 0.026 | 0.260 | 0.960 | −0.034 | 0.255 | 0.959 | ||
| Random effects | |||||||||
| ν2 | 0.5 | 0.147 | 0.688 | 0.895 | −0.004 | 0.308 | 0.939 | ||
|
|
2.5 | −0.117 | 1.640 | 0.876 | 0.109 | 0.561 | 0.944 | ||
| σuv1 | 1.5 | −0.018 | 0.890 | 0.931 | 0.036 | 0.514 | 0.949 | ||
|
|
1 | 0.253 | 0.713 | 0.949 | 0.063 | 0.585 | 0.959 | ||
|
|
0.5 | 0.719 | 0.967 | 0.887 | 0.039 | 0.259 | 0.938 | ||
| σuv0 | −0.4 | −0.492 | 2.905 | 0.840 | 0.002 | 0.297 | 0.908 | ||
|
|
0.5 | 0.054 | 0.530 | 0.895 | −0.018 | 0.416 | 0.929 | ||
Note: The bold numbers represent relatively large biases.
Table 3.
Comparison of the joint model with a normal distribution versus that with a t-distribution for εij (sample size = 500)
| m | Parameter | True | Normal distribution | t-distribution | |||||
|---|---|---|---|---|---|---|---|---|---|
| Bias | SE | CP | Bias | SE | CP | ||||
| 500 | Longitudinal | ||||||||
| Fixed effects | |||||||||
| β0 | 10 | 0.249 | 0.239 | 0.594 | 0.004 | 0.091 | 0.935 | ||
| β1 | −14 | −0.064 | 0.413 | 0.934 | −0.026 | 0.157 | 0.936 | ||
| β2 | 1.5 | 0.231 | 0.239 | 0.791 | −0.008 | 0.114 | 0.907 | ||
| Survival | |||||||||
| Fixed effects | |||||||||
| γ11 | 0.8 | 0.007 | 0.085 | 0.955 | −0.004 | 0.081 | 0.947 | ||
| γ12 | −1 | −0.002 | 0.191 | 0.938 | 0.007 | 0.173 | 0.947 | ||
| γ21 | 0.5 | −0.005 | 0.084 | 0.934 | −0.010 | 0.087 | 0.931 | ||
| γ22 | −1 | 0.011 | 0.182 | 0.934 | −0.021 | 0.170 | 0.955 | ||
| Random effects | |||||||||
| ν2 | 0.5 | −0.027 | 0.299 | 0.910 | −0.006 | 0.192 | 0.931 | ||
|
|
2.5 | −0.375 | 0.540 | 0.796 | 0.072 | 0.388 | 0.951 | ||
| σuv1 | 1.5 | −0.013 | 0.457 | 0.959 | −0.009 | 0.385 | 0.943 | ||
|
|
1 | 0.153 | 0.457 | 0.951 | −0.015 | 0.396 | 0.927 | ||
|
|
0.5 | 1.266 | 1.129 | 0.575 | 0.050 | 0.154 | 0.939 | ||
| σuv0 | −0.4 | −0.204 | 0.699 | 0.868 | −0.022 | 0.170 | 0.923 | ||
|
|
0.5 | −0.051 | 0.292 | 0.877 | −0.012 | 0.232 | 0.907 | ||
This paper proposes a new joint model of longitudinal and competing risks survival data. Our model consists of three sub-models: a linear mixed effects sub-model with a t-distributed measurement error, a cause-specific hazards sub-model for the survival outcome, and a variance-covariance sub-model for the multivariate latent random effects based on a modified Cholesky decomposition. A Bayesian MCMC method is developed for estimation and inference. Our model has several distinct features from existing models. The inference resulting from the linear mixed effects sub-model with a t-distributed measurement error is more robust to outlying longitudinal measurements or heavy-tailed data than its normal-error counterpart. Secondly, the cause-specific hazards sub-model for the survival outcome allows one to account for possibly correlated competing risks and dependent censoring. The random effects of the two sub-models induce the correlation between the longitudinal and survival outcomes. Finally, by extending the idea of Pourahmadi (1999), the sub-model for the variance-covariance matrix of the multivariate latent random effects based on a modified Cholesky decomposition allows for high-dimensional heterogeneous covariance matrices of the multivariate random effects and the resulting estimated covariance matrices are guaranteed to be positive definite. Our method also provides a framework for testing the homogeneous random effects assumption of a homogeneous joint model.
It is worth noting that compared to other robust methods such as Huber’s robust regression (Huber, 1973), the t-distribution based method is mathematically more tractable and computationally simple. The use of the t-distribution in applications with heavy-tailed or outlying data is well-developed in the literature. Lange et al. (1989) and Liu (1996) used it in linear and nonlinear regressions. Rosa et al. (2003) applied a class of normal/independent distributions (Lange and Sinsheimer, 1993), which include the t-distribution as a special case, in linear mixed effects models with ignorable missing data. Li et al. (2009) extended the idea to a robust joint model for longitudinal and survival data with possible non-ignorable missing data due to some terminating events. Our approach is distinct from Li et al. (2009) in several aspects. Our Bayesian MCMC method can easily handle high-dimensional random effects that otherwise complicates the likelihood-based method of Li et al. (2009). Secondly, Li et al. (2009) assume a homogeneous variance-covariance matrix for the random effects, while we allow heterogeneous random effects. Finally, the homogeneous variance-covariance assumption is untestable under the Li et al. (2009) model. Our model provides a useful framework to test this assumption.
This paper is organized as follows: in Section 2 we define the robust joint model and derive a Bayesian estimation procedure. In Section 3, we illustrate our method using a data set from scleroderma lung study (Tashkin et al., 2006). In Section 4, the performance of our method is examined by simulation. A discussion of the method is provided in Section 5. Technical details of the MCMC algorithm are deferred to the Appendix.
2. MODEL AND ESTIMATION
2.1 Robust joint model
Suppose there are m subjects in the study. For the ith subject at time t, the longitudinal outcome Yi(t) follows a linear mixed effects model:
| (1) |
where and Zi(t) are vectors of covariates associated with the fixed effects β (p × 1) and the random effects Ui (q×1) respectively. Assume that the measurement error εi(t) is t(0, σ2, k) distributed with k degrees of freedom. Assume further that εi(t) ⊥ Ui and εi(t1) ⊥ εi(t2) for any t1 ≠ t2.
During follow-up, each subject may experience one of g distinct competing causes of failure or may be right censored. Let Ci = (Ti, Di) be the competing risks survival data on subject i, where Ti is the failure or censoring time, and Di assumes a value from 0, 1, … , g, with Di = 0 indicating a noninformative censored event and Di = k indicating the kth failure type, k = 1, … , g. Dependent (or informative) censoring is treated as one of the g types of failures. The cause-specific hazards sub-model for the competing risks survival data is specified as follows:
| (2) |
The function is the instantaneous failure rate from cause k at time t given the vector of covariates and the latent unknown factor vi, in the presence of all other failure types. The regression coefficient νk represents the effect of the latent variable vi with ν1 set to 1 to ensure identifiability. The parameter γk represents the effects of the observed covariates on cause k. We further assume that the kth baseline hazard is a step function, , for , where is a partition of (0, ∞) and Sk indicates the number of steps for the kth baseline hazard. The competing risks model is one of the useful methods to handle dependent censoring. The identification of dependent competing risks models in which each risk has a mixed proportional hazard specification with regressors, and the risks are dependent by way of the unobserved heterogeneity, or frailty components, has been proved by Abbring and Van den Berg (2003).
Assume that
| (3) |
Similar to Pourahmadi (1999), we model the covariance matrices Σi through a modified Cholesky decomposition , where Hi is a diagonal matrix with positive entries and Mi is the lower triangular matrix with −φi,jl as its (j, l)th entry. This decomposition has a clear statistical interpretation: the below-diagonal entries of Mi are the negatives of generalized autoregressive parameters (GARP), φi,jl, in the autoregressive model
| (4) |
The diagonal entries of Hi are the innovation variances (IV) and we have cov(eij, ejk) = 0 if j ≠ k (1 ≤ j, k ≤ q + 1 and i = 1, … , m). The GARPs and the logarithms of the IVs are modeled with linear and log link functions:
| (5) |
where ai,jl and bij are covariates, and η1 and η2 are low-dimensional parameter vectors. For example, ai,jl and bij may contain group indicators, implying that the random effects covariances are heterogeneous. The homogeneous random effects assumption in existing joint models becomes a testable assumption within our model framework. Furthermore, the resulting estimated covariance matrix is guaranteed to be positive definite. The latent association between the longitudinal measurements and survival outcomes can be assessed by testing the hypothesis Σ Uvi = 0.
2.2 Likelihood
The standard maximum likelihood method involves integrating out latent variables from the log-likelihood function which is difficult when dealing with high-dimensional variables. We develop a Bayesian estimation procedure and a Markov chain Monte Carlo (MCMC) method for estimation and inference. To make the sampling of σ2 and β easier, we use the fact that the t-distributed error can be represented as , where τij |k ~ Gamma(k/2, k/2) is independent of Ui and εij ~ N (0, σ2). We define the parameter set in the joint model as: Ω = {β, σ2, γ, ν, λ0, η1, η2}, where γ = (γ1,γ2, … , γg ), ν = (ν2, … , νg ) and . We assume that for each subject i the longitudinal data are independent of the survival data, given all the parameters in O, latent factors θ = (W, τ), and covariates (Xi, Zi). For simplicity, we assume k is prespecified. Thus, the full likelihood function for Ω, conditional on the data (Yi, Ci) for i = 1, … , m and covariates, is:
| (6) |
It is convenient to work directly with the joint distribution of the observed data (Y, C) and the unobservable random vector θ, conditional on Ω, which facilitates the MCMC implementation. The conditional joint density of (Y, C) and θ is:
| (7) |
where and
| (8) |
2.3 Estimation and inference
Our Bayesian method involves a combination of direct sampling from the full conditional distribution, Metropolis-Hastings (MH) sampling (Hastings, 1970; Chib and Greenberg, 1995) and adaptive rejection sampling (ARS) (Gilks and Wild, 1992). We estimate the parameters by their posterior medians. Approximate 95% probability intervals are based on the 2.5th and 97.5th percentiles. Standard errors are obtained from the standard deviations of the posterior samples. The convergence of the Gibbs sampler is monitored by examining time series plots of the parameters over iteration and the Gelman and Rubin (1992) approach of using multiple chains.
We assume independent priors for O. We specify Normal priors for the parameters β, γ, ν, η1 and η2, leading to conjugate posteriors for β and some components of η1. We use an inverse Gamma prior for the measurement error variance σ2 and a gamma prior for each step of the kth baseline hazard function λ0k by which conjugate posterior distributions are easy to obtain. Because the full conditional distributions of the parameters β, σ2, and , (s = 1, … , Sg, k = 1, … , g) are standard distributions, drawing random variates from their full conditional distributions is straightforward. The full conditional distribution of the random variate τij given k is also known. For other parameters and the random effects (Ui, vi), we either use a Metropolis-Hastings step with the normal approximation to the full conditional distribution as the candidate distribution or apply the adaptive rejection sampling technique. The technical details on the sampling distributions are given in the Appendix.
The initial values of the parameters for sampling are obtained by modeling the longitudinal data and survival data separately by a linear mixed model and a cause-specific proportional hazards model. The initial value for (s = 1, … , Sk, k = 1, … , g) can be obtained by drawing a random variate from the gamma full conditional distribution described in the Appendix.
3. AN EXAMPLE
We apply the developed method to analyze a data set from a scleroderma lung study (SLS) (Tashkin et al., 2006) to evaluate the effectiveness of oral cyclophosphamide (CYC) for scleroderma lung disease. The SLS enrolled 158 patients, which were randomized to receive either CYC (79 patients) or placebo (79 patients) for 12 months. An additional year of follow-up was performed to determine if CYC effects persisted after treatment. The primary outcome is forced vital capacity (FVC, % predicted) which was measured at 3-month intervals from the baseline. Treatment failure was defined as a ≥ 15% (absolute) decrement in %FVC from baseline occurring at least 3 months into treatment that was sustained for at least 1 month.
Our analysis is based on the 6–24 months %FVC scores of 141 patients. The longitudinal profile of %FVC over time for the two groups (Figure 1) reveals some potential outliers. We are particularly concerned how these outlying data points would affect the joint model inference. We observe 14 treatment failures or deaths, 32 informative dropouts and 5 noninformative dropouts. A dropout is noninformative if there is no evidence showing that the dropout is related to the disease or the treatment, and informative otherwise. Since the informative dropout is potentially related to the patient’s disease condition, it causes not only non-ignorable missing data in %FVC, but also dependent censoring for treatment failure or death.
We consider two baseline factors in our joint model when assessing the CYC treatment effects: baseline %FVC (FVC0), and lung fibrosis (FIB0). It is suggested by clinicians that the beneficial effects of CYC on pulmonary function continue to increase after stopping treatment at 12 months and eventually begin to wane after 18 months. Therefore, we fit the following linear spline mixed effects model with a change point at month 18 for the longitudinal measurements %FVC:
| (9) |
where Ui = (Ui1, Ui2)T is the subject-specific random effects and the εij is the mutually independent measurement errors. Since we include baseline %FVC as a fixed effect covariate, we don’t consider random intercept to avoid possible confounding effects and Zij = (Timeij, (Timeij − 18)+).
A cause-specific competing risks sub-model was applied to model disease-related dropout (risk 1) and treatment failure or death (risk 2):
| (10) |
and
| (11) |
The latent variables from both sub-models are assumed to have a multivariate normal distribution with mean zero and variance-covariance matrices
We first test the homogeneous random effects covariance matrix assumption by considering subject-dependent covariates for aijl and bij. Specifically, we choose aijl = bij = (1, CYCi), which allows heterogeneous covariance matrices for different treatment groups, and test the null hypothesis by examining if the 95% credible interval of CYC effects contains zero for all the GARP and IV parameters. None of the 95% CIs for CYC of all the covariance parameters exclude zero (results not shown here), suggesting no obvious violation of the homogeneous random effects covariances assumption.
We use independent noninformative prior distributions for all the parameters with relatively large variances. A 3-step baseline hazard function, with the time points defining the steps being equallysplit percentiles of the observed event times, is utilized for the informatively censored events and the event of treatment failure or death. Sensitivity analyses with 4- and 5-step baseline hazard functions are conducted and show no significant difference. The corresponding priors for the parameters are β0 ~ N (70, 103) and βl ~ N (0, 103) for l = 1, … , 9; σ2 ~ IG(10−3, 10−3); γkr ~N (0, 103) for k = 1, 2 and r = 1, … , 5; for s = 1, … , Sk and S1 = S2 = 3; ν2 ~ N (0, 105); and each element of η1 and η2 ~ N (0, 105). We use 30,000 iterations of MCMC sampling chains following a 15,000-iteration “burn-in” period.
We identify 7 possible outlying data points by examining the residuals from our robust joint model. Table 1 compares results from a normal joint model with and without the outliers together with a model with a t-distributed error (k = 3) using all data points. The estimation procedure for the normal joint model is the same except that there is no need to estimate parameter τij (Huang, 2008). First, the outliers are observed to be influential to the parameter estimates for the longitudinal endpoint. The t-model with all data and the normal model without outliers yield consistent conclusions. For example, both methods reveal a significant time trend in the placebo group before 18 months (β1), which indicates a more steep decrease before 18 months in %FVC scores than that estimated from the normal joint model with all the data points. They also both identify a significant interaction (β8) between the time trend (before 18 months) and the treatment group. In contrast, these effects are not significant using the normal method with all data points. Secondly, comparable estimates at the survival endpoint are obtained for all three models. Thirdly, all models identify a negative significant covariance ΣU1v between the random slope before 18 months in the longitudinal model and the latent variable of the survival model, and a positive significant covariance ΣU2v between the random slope after 18 months in the longitudinal model and the latent variable from the survival model, which indicates a dependence between the longitudinal measurement %FVC and the survival endpoint. We also observe a significant positive coefficient ν2 which suggests a latent positive association between the two competing risks. Finally, the negative sign of ΣU1v and positive sign of ΣU2v together with the positive ν2 indicate that before month 18, there is a lower risk of treatment failure or death and informative dropout for patients with higher than average increasing rate of %FVC over time; after 18 months, the trend is reversed due to the negative association between the two slopes. In summary, the longitudinal sub-model with a normal error is not robust against potential outliers in the longitudinal data, although the outliers may not have much impact on the estimation of the survival model parameters.
Table 1.
Analysis of 6–24 months scleroderma lung study data
| Normal distribution | t-distribution (κ = 3) | |||
|---|---|---|---|---|
| With outliers | Without outliers | With outliers | ||
| Estimate (95%CI) | Estimate (95%CI) | Estimate (95%CI) | ||
| Longitudinal outcome %FVC | ||||
| Time (β1) | −0.13 (−0.30, 0.05) | −0.18 (−0.33, −0.02) | −0.20 (−0.36, −0.03) | |
| Time18 (β2) | 0.23 (−0.25, 0.74) | 0.15 (−0.25, 0.53) | 0.22 (−0.13, 0.56) | |
| FVC0 (β3) | 0.92 (0.83, 1.03) | 0.88 (0.79, 0.98) | 0.93 (0.85, 1.01) | |
| FIB0 (β4) | −2.05 (−3.22, −0.94) | −1.83 (−2.84, −0.81) | −1.88 (−2.75, −0.99) | |
| CYC (β5) | −1.38 (−3.66, 1.00) | −1.00 (−2.89, 0.88) | −1.12 (−2.83, 0.59) | |
| FVC0 × CYC (β6) | 0.11 (−0.04, 0.25) | 0.15 (−0.02, 0.27) | 0.09 (−0.01, 0.20) | |
| FIB0 × CYC (β7) | 2.03 (0.43, 3.69) | 1.74 (0.32, 3.19) | 2.25 (0.92, 3.52) | |
| Time × CYC (β8) | 0.26 (−0.01, 0.51) | 0.23 (0.01, 0.45) | 0.24 (0.01, 0.47) | |
| Time18 × CYC (β9) | −0.64 (−1.33, 0.07) | −0.43 (−0.96, 0.13) | −0.41 (−0.92, 0.09) | |
| p-value for H0:Overall CYC Effects=0 | 0.040 | 0.022 | 0.003 | |
| Cause-specific hazards (informatively censored events) | ||||
| FVC0 (γ11) | −0.06 (−0.13, −0.01) | −0.06 (−0.12, −0.01) | −0.06 (−0.12, −0.01) | |
| FIB0 (γ12) | 0.21 (−0.28, 0.77) | 0.21 (−0.29, 0.76) | 0.21 (−0.27, 0.79) | |
| CYC (γ13) | 0.24 (−0.61, 1.20) | 0.29 (−0.55, 1.21) | 0.25 (−0.57, 1.18) | |
| FVC0 × CYC (γ14) | 0.11 (0.03, 0.18) | 0.10 (0.03, 0.18) | 0.11 (0.04, 0.19) | |
| FIB0 × CYC (γ15) | 0.13 (−0.58, 0.83) | 0.12 (−0.61, 0.81) | 0.08 (−0.63, 0.78) | |
| Cause-specific hazards (treatment failure of death) | ||||
| FVC0 (γ21) | 0.01 (−0.07, 0.11) | 0.02 (−0.06, 0.10) | 0.01 (−0.07, 0.09) | |
| FIB0 (γ22) | 0.25 (−0.68, 1.22) | 0.22 (−0.62, 1.13) | 0.25 (−0.67, 1.20) | |
| CYC (γ23) | −1.24 (−3.20, 0.35) | −1.19 (−3.29, 0.22) | −1.20 (−3.50, 0.34) | |
| FVC0 × CYC (γ24) | −0.06 (−0.20, 0.08) | −0.07 (−0.20, 0.07) | −0.05 (−0.19, 0.09) | |
| FIB0 × CYC (γ25) | −0.55 (−2.26, 1.07) | −0.52 (−2.12, 0.94) | −0.51 (−2.22, 1.02) | |
| Random effects | ||||
| ν2 | 3.34 (1.34, 8.35) | 3.28 (1.42, 8.81) | 3.25 (1.18, 7.84) | |
| ΣU11 | 0.27 (0.20, 0.36) | 0.28 (0.21, 0.37) | 0.25 (0.19, 0.33) | |
| ΣU12 | −0.36 (−0.61, −0.18) | −0.32 (−0.53, −0.17) | −0.27 (−0.45, −0.15) | |
| ΣU22 | 1.60 (0.89, 2.59) | 0.93 (0.51, 1.61) | 0.73 (0.39, 0.1.25) | |
|
|
0.40 (0.07, 1.59) | 0.31 (0.05, 1.16) | 0.35 (0.06, 1.31) | |
| Covariance of Ui and vi | ||||
| ΣUv1 | 0.25 (−0.53, −0.09) | −0.25 (−0.50, −0.09) | −0.23 (−0.47, −0.08) | |
| ΣUv2 | 0.69 (0.24, 1.50) | 0.42 (0.11, 1.02) | 0.39 (0.11, 0.93) | |
| Model fit | ||||
| DIC | 5693.08 | 5295.18 | 5089.35 | |
Note: The bold numbers indicate the significant results (p-value < 0.05).
The models are assessed using the Deviance Information Criterion (DIC) (Spiegelhalter et al., 2002). The robust joint model gives the lowest DIC of 5089.35, which indicates the best fit. We note that there are several versions of DIC for missing data models (Celeux et al., 2006; Chen, 2006). Here we use the DIC constructed from the conditional distribution while treating both Ω and W as parameters because it is easy to compute. We conduct a small simulation to evaluate the DIC which selects 147 times out of 200 datasets and the effective dimension is always positive.
4. SIMULATION STUDY
We carry out a simulation study to assess the performance of our robust joint model and compare it to the joint model with a normal measurement error. The longitudinal measurements are simulated from the following random slope model:
| (12) |
where τij = 0, 0.15, 0.3, … , 3, represents the scheduled visit time and X2i ~ Bernoulli(0.5) is a group indicator. We generate 1% outliers in the placebo group from normal distributions N (60, 100) and N (−60, 100) with rates 30% and 70%, respectively. The rest of the data have measurement error εij ~ N (0, 5). We simulate two competing risks
failure times with the following cause-specific hazards:
| (13) |
| (14) |
where X1 ~ N (2, 1.0), and X2 is shared with the longitudinal model. We use constant baseline hazards of 0.12 and 0.25 for risk 1 and risk 2, respectively, to generate the survival data. The random effects are generated from the multivariate normal distribution with covariance matrices Σi which are decomposed into the GARPs and IVs modeled with covariates aijl = bij = (1, X2i). In other words, the covariance matrices are different in the two groups: strong positive correlation in one group and strong negative correlation in the other. The parameter values are given in Table 2. With this setup, the rate of risk 1 is approximately 0.44, the rate of risk 2 is 0.36 and the censoring rate is 0.20. Longitudinal responses are missing after the observed or censored event times. The average number of total longitudinal observations is 7.8 per subject. We use a vague prior of N (0, 105) for each component of β, γ, ν, η1 and η2, IG(10−3, 10−3) for σ2, and Γ(10−3, 10−3) for λ0. The simulation is based on 200 Monte Carlo samples with a sample size of 250. The MCMC sampling is run using 5, 000 iterations, and the estimation results are based on the last 2, 500 iterations.
The bias, standard deviations of the posterior medians and coverage rate of 95% credible intervals are given in Table 2. In the presence of outliers, the joint model with a normal measurement error shows large bias for the longitudinal fixed effects β, the joint random effects covariance matrix parameters and ν2. In particular, the group effect β2 is overestimated since the outliers only exist in the placebo group and have a higher probability of being negative. Furthermore, these biases do not disappear for a large sample size of 500 (Table 3). In the contrast, our robust joint model yields much smaller biases for all the parameters. Both methods produce comparable estimates for the fixed effects for the competing risks survival endpoint.
5. DISCUSSION
We propose a robust joint model for longitudinal measurements and competing risks survival data with heterogeneous random effects. The robustness against potential outliers in the longitudinal measurements is achieved by specifying a t-distribution for the measurement error in the linear mixed effects sub-model. In addition, the proposed approach allows for high-dimensional random effects and heterogenous covariance matrices of the multivariate random effects, and the resulting estimated covariance matrices are guaranteed to be positive definite.
The t-distribution model is a robust model in two ways. First, with t-distributed random errors, τij reweights the observations according to their residuals, and thus we are able to obtain robust estimation by downweighting the outliers. Secondly, we carried out simulation to compare the two models (t-model and normal model) when there were no outliers, that is, the underlying measurement error followed a zero-mean normal distribution. There are almost unbiased estimates for all the parameters in both methods and the simulated coverage probabilities are close to 0.95, although the joint model with the t-distribution produces a little bit larger standard errors for the parameters at the longitudinal endpoint.
We did some sensitivity analyses of the estimates to mis-specification of the variance-covariance structure and found out that we may obtain biased parameter estimates for the survival endpoint when combining the information of the longitudinal outcome if the correlation of the two endpoints is incorrectly modeled. Therefore, ignoring the heterogeneity can result in biased estimates and invalid inference.
The t-distributed error with k = 3 has demonstrated nice properties for the purpose of guarding against outliers in the longitudinal data in our numerical study, even though it can also be estimated as a parameter. General principles of parsimony suggest that k be fixed for small data sets and estimated for large ones (Lange et al., 1989). Lange et al. (1989) also suggest that estimated values of degree of freedom below 1 should be regarded with suspicion. In addition to the t-distribution, other normal/independent distributions (Lange and Sinsheimer, 1993), such as the slash distribution or the contaminated normal distribution, can be adapted for the measurement error in our robust joint model. Rosa et al. (2003) pointed out that the t process is the most commonly used thick-tailed distribution for robust inference which is often a good alternative to a Gaussian distribution; the contaminated normal distribution is more flexible but at the expense of an additional parameter, while the slash distribution is rarely encountered in the literature despite its relatively easier implementation in hierarchical modeling.
Our model can be extended to clustered data. Clustered data arise frequently from multi-site clinical trials, in which each site can be viewed as a cluster, or from studies across families, in which each family may be treated as a cluster. The cluster effect can easily be incorporated as a random effect or as a design vector for the GARP/IV parameters in order to take into account the heterogeneity across the cluster.
Finally, it is possible to extend our method to handle recurrent event data when each subject may repeatedly experience a certain event. Typical medical examples are multiple infection episodes and tumor recurrences. Zhang et al. (2008) considered a joint mixed-effects regression model for time series measures and recurrent events to analyze the air quality and respiratory symptom data. Their work may represent the first attempt to include a latent process in both the hazard and recovery rates of a recurrent event process. Extension of our joint model to incorporate recurrent event data would offer another promising approach. A possible choice of the sub-model for the recurrent event data is the semiparametric transformation models with random effects described by Zeng and Lin (2007).
APPENDIX A: FULL CONDITIONAL DENSITIES
This section provides details for the full conditional distributions of the parameters used in the Gibbs sampling algorithm. We use p(·) and p(·|·) to denote marginal and conditional densities, respectively. We denote the prior distribution by p0(·). Based on the modified Cholesky decomposition, the random effects vi can be written as where . Instead of sampling vi directly, we sample ei,q+1, leading to a faster convergence rate.
- Sample τij from
-
Sample σ2 from
where p0(σ2) = Γ(α1, α2).
-
Sample β from
where Si = Diag (τij) and p0(β) = N (β0, Σβ)
-
Sample the random effects Ui from
where , and is a q × q matrix consisting of the first q columns and rows of Mi, is a q × q matrix consisting of the first q columns and rows of Hi. We use the one-step Metropolis-Hastings algorithm to obtain the update in the sampling sequence with the normal density from the longitudinal data as the proposal density. The random effects Ui is obtained by first sampling a random variable form the conditional density based on the longitudinal data and then using the conditional likelihood contribution from the survival data to determine the acceptance of the new draw.
-
Sample η1 from
where Qi is a q × q1 matrix with first row Qi1 = 0 and jth row for j = 2, …, q. We sample η1 in two steps: sample the entries only involving Ui from the normal conditional density, sample the entries involving Ui and vi with adaptive rejection sampling. It is worth noting that for the homogeneous case where the variance-covariance matrix Σ is diagonal, we have Σ = H and M = I. In this case, there is no need to sample η1 and it should be set to zero in the sampling procedure.
-
Sample η2 from
We use a Metropolis-Hastings step with a normal approximation to the full conditional as the candidate distribution. For details, see Daniels and Pourahmadi (2002).
-
Sample γkr, k = 1, …, g, r = 1, …, R from
We use a Metropolis-Hastings step within the single component sampler to update the values of these parameters. For each of these parameters, we propose a normal density as the proposal density, which has the current value of the parameter as its mean and its standard deviation is set equal to four times the standard error of a maximum partial likelihood estimate from a standard Cox model (Wang and Taylor, 2001).
- Sample νk with ARS from
-
Sample ei,q+1 (i = 1, …, m) from
The sample is obtained by first sampling a candidate from the normal densities as its assumption and then using the conditional likelihood contribution from the survival data to determine the acceptance of the new draw.
-
Sample each piece of λ0k (k = 1, … g) from
where indicates the number of events occurring in the time interval , and , for s = 1, …, Sk
Contributor Information
Xin Huang, Email: xin@amgen.com, Amgen Inc.1120 Veterans Boulevard, Mailstop ASF3-3, South San Francisco, CA 94080, USA.
Gang Li, Email: vli@ucla.edu, Department of Biostatistics, School of Public Health, University of California at Los Angeles, Los Angeles, California, 90095, USA.
Robert M. Elashoff, Email: relashof@biomath.medsch.ucla.edu, Department of Biomathematics, University of California at Los Angeles, Los Angeles, California, 90095, USA.
References
- Abbring JH, Van den Berg GJ. The identifiability of the mixed proportional hazards competing risks model. Journal of the Royal Statistical Society B. 2003;65:701–710. [Google Scholar]
- Brown ER, Ibrahim JG. A bayesian semiparametric joint hierarchical model for longitudinal and survival data. Biometrics. 2003;59:221–228. doi: 10.1111/1541-0420.00028. [DOI] [PubMed] [Google Scholar]
- Celeux G, Forbes F, Robert CP, Titterington DM. Deviance information criteria for missing data models. Bayesian Analysis. 2006;4:651–674. [Google Scholar]
- Chen MH. Comments on article by celeux et al. Bayesian Analysis. 2006;4:677–680. [Google Scholar]
- Chib S, Greenberg E. Understanding the metropolis-hastings algorithm. The American Statistician. 1995;49:327–335. [Google Scholar]
- Daniels MJ, Pourahmadi M. Bayesian analysis of covariance matrices and dynamic models for longitudinal data. Biometrika. 2002;89:553–566. [Google Scholar]
- DeGruttola V, Tu XM. Modeling progression of cd4 lymphocyte count and its relationship to survival time. Biometrics. 1994;50:1003–1014. [PubMed] [Google Scholar]
- Elashoff R, Li G, Li N. An approach to joint analysis of longitudinal measurements and competing risks failure time data. Statistics in Medicine. 2007;26:2813–2835. doi: 10.1002/sim.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elashoff R, Li G, Li N. A joint model for longitudinal measurements and survival data in the presence of multiple failure types. Biometrics. 2008;64:762–771. doi: 10.1111/j.1541-0420.2007.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucett CL, Schenker N, Elashoff RM. Analysis of censored survival data with intermittently observed time-dependent binary covariates. Journal of the American Statistical Association. 1998;93:427–437. [Google Scholar]
- Faucett CL, Thomas DC. Simultaneously modeling of censored survival data and repeated measured covariates: A gibbs sampling approach. Statistics in Medicine. 1996;16:1663–1685. doi: 10.1002/(SICI)1097-0258(19960815)15:15<1663::AID-SIM294>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences(with discussion) Statistical Science. 1992;7:457–511. [Google Scholar]
- Gilks WR, Wild P. Adaptive rejection sampling for gibbs sampling. Applied Statistics. 1992;41:337–348. [Google Scholar]
- Hastings WK. Monte carlo sampling methods using markov chains and their applications. Biometrika. 1970;57:97–109. [Google Scholar]
- Henderson R, Diggle P, Dobson A. Joint modeling of longitudinal measurements and event time data. Biostatistics. 2000;4:465–480. doi: 10.1093/biostatistics/1.4.465. [DOI] [PubMed] [Google Scholar]
- Hogan JW, Laird NM. Model-based approaches to analysing incomplete longitudinal and failure time data. Statistics in Medicine. 1997;16:259–272. doi: 10.1002/(sici)1097-0258(19970215)16:3<259::aid-sim484>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Huang X. PhD Dissertation. University of California; Los Angeles: 2008. A general joint model for longitudinal measurements and competing risks survival data with heterogeneous random effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber PJ. Robust regression: asymptotics, conjectures and monte carlo. Annals of Statistics. 1973;1:799–821. [Google Scholar]
- Lange KL, Little RJA, Taylor JMG. Robust statistical modelling using the t distribution. Journal of the American Statistical Association. 1989;84:881–896. [Google Scholar]
- Lange KL, Sinsheimer JS. Normal/independent distributions and their applications in robust regression. Journal of the American Statistical Association. 1993;91:1461–1473. [Google Scholar]
- Li N, Elashoff RM, Li G. Robust joint modeling of longitudinal measurements and competing risks failure time data. Biometrical Journal. 2009;1:19–30. doi: 10.1002/bimj.200810491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RJA. Modeling the drop out mechanism in repeated measures studies. Journal of the American Statistical Association. 1995;90:1112–1121. [Google Scholar]
- Liu C. Bayesian robust multivariate linear regression with incomplete data. Journal of the American Statistical Association. 1996;91:1219–1227. [Google Scholar]
- Liu L, Ma JZ, O’Quigley J. Joint analysis of multilevel repeated measures data and survival: an application to the end stage renal disease (esrd) data. Statistics in Medicine. 2008;27:5679–5691. doi: 10.1002/sim.3392. [DOI] [PubMed] [Google Scholar]
- Pourahmadi M. Joint mean-covariance models with applications to longitudinal data: unconstrained parameterization. Biometrika. 1999;86:677–690. [Google Scholar]
- Rosa GJM, Padovani CR, Gianola D. Robust linear mixed models with normal/independent distributions and bayesian mcmc implementation. Biometrical Journal. 2003;45:573–590. [Google Scholar]
- Schluchter MD. Methods for the analysis of informatively censored longitudinal data. Statistics in Medicine. 1992;11:1861–1870. doi: 10.1002/sim.4780111408. [DOI] [PubMed] [Google Scholar]
- Song X, Davidian M, Tsiatis AA. A semiparametric likelihood approach to joint modeling of longitudinal and time-to-event data. Biometrics. 2002;58:742–753. doi: 10.1111/j.0006-341x.2002.00742.x. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit(with discussion) Journal of the Royal Statistical Society, Ser B. 2002;64:583–639. [Google Scholar]
- Tashkin DP, Elashoff RM, et al. Cyclophosphamide versus placebo in scleroderma lung disease. The New England Journal of Medicine. 2006;354:2655–2666. doi: 10.1056/NEJMoa055120. [DOI] [PubMed] [Google Scholar]
- Tseng YK, Hsieh F, Wang JL. Joint modelling of accelerated failure time and longitudinal data. Biometrika. 2005;92:587–603. [Google Scholar]
- Wang Y, Taylor JMG. Joint modeling longitudinal and event time data with application to acquired immunodeficiency syndrome. Journal of the American Statistical Association. 2001;96:895–905. [Google Scholar]
- Wulfsohn MS, Tsiatis AA. A joint model for survival and longitudinal data measured with error. Biometrics. 1997;53:330–339. [PubMed] [Google Scholar]
- Xu J, Zeger SL. Joint analysis of longitudinal data comprising repeated measures and times to events. Applied Statistics. 2001;50:375–387. [Google Scholar]
- Ye W, Lin XH, Taylor JMG. Semiparametric modeling of longitudinal measurements and time-to-event data – a two-stage regression calibration approach. Biometrics. 2008;64:1238–1246. doi: 10.1111/j.1541-0420.2007.00983.x. [DOI] [PubMed] [Google Scholar]
- Zeng D, Cai J. Simultaneous modelling of survival and longitudinal data with an application to repeated quality of life measures. Lifetime Data Analysis. 2005;11:151–174. doi: 10.1007/s10985-004-0381-0. [DOI] [PubMed] [Google Scholar]
- Zeng D, Lin DY. Semiparametric transformation models with random effects for recurrent events. Journal of the American Statistical Association. 2007;102:167–180. doi: 10.1080/01621459.2013.842172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ye Y, Diggle P, Shi J. Joint modeling of time series measures and recurrent events and analysis of the effects of air quality on respiratory symptoms. Journal of the American Statistical Association. 2008;103:48–60. [Google Scholar]