Abstract
Human neural progenitor cells differentiated from human embryonic stem cells offer a potential cell source for studying neurodegenerative diseases and for drug screening assays. Previously, we demonstrated that human neural progenitors could be maintained in a proliferative state with the addition of leukemia inhibitory factor and basic fibroblast growth factor. Here we demonstrate that 96 hours after removal of basic fibroblast growth factor the neural progenitor cell culture was significantly altered and cell replication halted. 14 days after the removal of basic fibroblast growth factor, most cells expressed MAP2 and TUJ1, markers characterizing a post-mitotic neuronal phenotype as well as neural developmental markers Cdh2 and Gbx2. Real-time PCR was performed to determine the ionotrophic receptor subunit expression profile. Differentiated neural progenitors express subunits of glutamatergic, GABAergic, nicotinic, purinergic and transient receptor potential receptors. In addition, sodium and calcium channel subunits were also expressed. Functionally, virtually all the NP cells tested under whole-cell voltage clamp exhibited delayed rectifier potassium channel currents and some differentiated cells exhibited tetrodotoxin-sensitive, voltage-dependent sodium channel current. Action potentials could also be elicited by current injection under whole-cell current clamp in a minority of cells. These results indicate that removing basic fibroblast growth factor from the neural progenitor cell cultures leads to a post-mitotic state, and has the capability to produce excitable cells that can generate action potentials, a landmark characteristic of a neuronal phenotype. This is the first report of an efficient and simple means of generating human neuronal cells for ionotrophic receptor assays and ultimately for electrically active human neural cell assays for drug discovery.
Keywords: human embryonic stem cells, human neural progenitors, glutamate receptors, sodium channels
1
Human embryonic stem cells (hESCs) can differentiate into human neural progenitor (hNP) cells in the absence of bone morphogengic protein signaling and in the presence of basic fibroblast growth factor (bFGF). Once derived, bFGF is required for hNP cell replication over extended culture (Carpenter et al., 2003, Shin et al., 2006, Elkabetz et al., 2008). These hNP cells present an ideal neural cell source for high throughput screening of a large variety of pharmacological compounds (Kiryushko et al., 2004) and the differentiated progeny of these cells were recently used to screen for compounds that act as a potentiator of α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) activity (McNeish et al., 2010). Achieving efficient and cost effective human in vitro neuronal differentiation through the minimal use of growth factors and other alterations in the differentiation procedures could facilitate neural drug discovery and developmental toxicology studies using hESC neural derivatives and enabling higher throughput in these assays.
Of particular interest to the field of drug development are the ion channels, which are required for normal functional neural activity. In vivo, mature functional neurons develop from progenitors by migrating into place, developing neurotransmitter specificity, developing electrical excitability and forming axons, dendrites and functional synaptic connections. Ion channels also have been shown to play an essential role in many aspects of neural development (Yen et al., 1993).
Developmental characterization of many ion channel subunits has been done in rodents; however, characterization in human tissue is lacking (Jeng et al., 1991, Numakawa et al., 2002). Fully functional neurons have been derived from mouse embryonic stem cells (mESCs) [reviewed by (Wu et al., 2007)], but other studies have reported that cells differentiated from mESCs had the morphology, synaptic contacts and other biochemical markers characteristic of differentiated neurons, but lacked the voltage-dependent sodium channels required for functional synaptic transmission (Benn et al., 2001, De Filippis et al., 2007). hNP cell differentiation in vitro may provide a good experimental system for elucidating the factors necessary for regulation of ion channels throughout neural development.
Ionotropic glutamate receptors are critical for neural migration (Behar et al., 1999), synaptogensis (Gallo et al., 1995) and neural survival (Platel et al., 2008). Glutamate receptors are also important in mediating excitotoxicity and apoptosis (Buckingham et al., 2008, Milanese et al., 2010). Glutamate receptor mediated excitotoxicity occurs in a wide range of maladies including but not limited to stroke, traumatic brain injury and seizure, and could be associated with neurological diseases such as Huntington’s disease, amyotrophic lateral sclerosis or Parkinson’s disease (Erceg et al., 2008, Ladewig et al., 2008, Blandini, 2010, Tallaksen-Greene et al., 2010).
Previously, we derived a karotypically normal, stable, adherent monolayer of hNP cells from WA09 hESCs (Shin et al., 2006). These cells have been characterized for their ability to maintain multipotency and for their expression of neural stem cell marker NESTIN and lack of expression of stem cell marker POU5F1 (Dhara et al., 2008). The hNP cells have also been previously differentiated into all three subtypes of the neural lineage: neurons, oligodentrocytes and astrocytes (Shin et al., 2006). Upon further differentiation in basal conditions (without bFGF) but with leukemia inhibitory factor (LIF), greater than 90% of the cells were TUJ1 and microtubule-associated protein 2 (MAP2) positive (Shin et al., 2006, Harrill et al., 2010). To define the potential of hNP cells and derivatives in functional neural drug discovery, glutamate receptor activity as well as function of other ion channel subtypes requires electrophysiological studies.
Here, we demonstrate that post-mitotic differentiated hNP cells express developmental regionalization genes, as well as markers of functional neural receptors. These differentiated hNP cells can evoke action potentials that can be blocked with tetrodotoxin (TTX) as well as increase the intracellular calcium response when exposed to AMPA receptor potentiator, cyclothiazide. GABAergic and glutamatergic ionotrophic receptor expression was found to be up regulated as early as after two weeks of hNP cell differentiation. These results suggest that these differentiated hNP cells are capable of differentiation into a functional neuronal phenotype.
2 Experimental Procedures
2.1 hESC Cultures
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. WA09 (WiCell Research Institute, Madison, WI). hESC line WA09 (H9) was obtained from WiCell Research Insitute in Madison, WI. These hESCs are an NIH approved line derived from embryos donated from in vitro fertilization clinics by James Thompson (Thomson et al., 1998). hESCs were cultured on mouse embryonic fibroblast (MEF; Harlan, Indianapolis, IN) feeders inactivated by mitomycin C in medium consisting of Dulbecco’s modified Eagle medium (DMEM)/F12 medium (Invitrogen, Carlsbad, CA) supplemented with 20% knockout serum (Invitrogen), 2 mM L-glutamine (Invitrogen), 0.1 mM non-essential amino acids (Invitrogen), 50 units/ml penicillin (Invitrogen), 50 μ/ml streptomycin (Invitrogen), 0.1mM (β-mercaptoethanol and 4 ng/ml bFGF (R&D, Minneapolis, MN). They were maintained in 5% CO2 and at 37°C. Cells were passaged every 3 days by mechanical dissociation and re-plated on fresh feeder layer cultures to prevent undirected differentiation with daily medium changes as previously described (Kalia et al., 2004).
2.2 hNP Cell Cultures
Human neural progenitor (hNP) cells were derived from hESC line WA09 by our laboratory as previously described (Shin et al., 2006). Briefly, after one week of culture on MEF feeder layers, WA09 hESCs were cultured with derivation medium containing DMEM/F12 medium supplemented with 2 mM L-glutamine, 2 U/mL penicillin, 2 μg/mL streptomycin, N2 (Invitrogen), and 4 ng/ml bFGF (R&D) for 7 days. Rosettes were selected utilizing hook passaging from culture dishes and re-plated on polyornithine (20 μg/ml) and laminin (5 μg/ml) coated dishes. These rosettes were propagated for 3 days on polyornithine and laminin coated dishes in growth medium consisting of Neurobasal medium (Invitrogen) supplemented with 2 mM L-glutamine, 2 U/mL penicillin, 2 μg/mL streptomycin, B27 (Invitrogen), 20 ng/mL bFGF, and 10 ng/mL LIF (Millipore, Billerica, MA). Medium was changed every other day and cells were passaged every fourth day or as needed. Cells used for this experiment were passages 22-39. To initiate hNP cell differentiation 24 hours after the last passage, growth medium was switched to growth medium lacking bFGF (differentiation media). Differentiation medium was changed every three days. Cells were collected at 14 days, 35 days and 125 days for analysis.
2.3 Cell Proliferation Analysis by Carboxyfluorescein succinimidyl ester (CFSE)
Cell proliferation was analyzed using the CellTrace™ CFSE Cell Proliferation Kit (Invitrogen) following manufacturer’s instructions. Briefly, cells were incubated for 10 minutes in 10 μM of CFSE solution at 37°C. CFSE staining was quenched with ice-cold media. Cells were washed, re-suspended in fresh hNP cell media, re-plated at a density of 1×106 cells per 35mm dish and incubated for 0, 24 and 48 hours. At each time point, cells were harvested, washed twice with PBS+/+ (phosphate buffered saline containing calcium and magnesium; ThermoScientific, Whatham, MA) and analyzed on Dako CyAn flow cytometer (Beckman Coulter, Brea, CA) equipped with a 488nm laser. Non-stained cells were used as a control. CFSE data were analyzed using the FlowJo software (TreeStar, Ashland, OR) proliferation model. The half-life of mean fluorescence was determined by fitting the mean fluorescence with the following equation:
where I is the mean fluorescence intensity, I0 is the fluorescence intensity at steady state, τ is the characteristic time of half intensity, A is the amplitude and t is time.
2.4 Immunocytochemistry
Cells were fixed with 2% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in PBS+/+ for 20 minutes and processed for immunocytochemistry. Cells were washed 3 times with PBS+/+ followed by 3 washes for 5 minutes each with permeabilization buffer consisting of 25 μL (.05%) Tween 20 (EMD Chemicals, Gibbstown, NJ) in a 50 mL high salt buffer solution consisting of 1M Tris base and 0.25M NaCl. Cells were then blocked in 6% goat serum (Jackson Immuno Research Laboratories, West Grove, PA) for 45 minutes. The following primary antibodies were used: mouse anti NESTIN (1:200, Neuromics, Edina, MN), mouse anti TUJ1 (1:200, Neuromics) and mouse anti MAP2 (1:500, Millipore). AlexaFluor 488 and 594 goat anti-mouse (1:1000, Invitrogen) were used as secondary antibodies to visualize primary antibody staining. Cell nuclei were stained using 4′,6-diamidino-2-phenylindole DAPI (Invitrogen). Fluorescence was visualized using spinning disk confocal microscope (Olympus, Center Valley, PA).
2.5 Real Time Polymerase Chain Reaction (RT-PCR)
RNA was extracted from cell cultures using the Qiashredder and RNeasy kits (Qiagen, Valencia, CA) according to manufacturer’s instructions. RNA quality and quantity were measured using a RNA 600 Nano Assay (Agilent Technologies, Santa Clara, CA) and the Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA (5 μg) was reverse-transcribed using the cDNA Archive Kit (Applied Biosystems Inc., Foster City, CA) according to manufacturer’s instructions. Reverse transcriptase reactions were initially incubated at 25°C for 10 minutes and subsequently at 37°C for 120 minutes. Quantitative real-time PCR was carried out on an ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Inc) utilizing the Taqman real time PCR kit (Applied Biosystems). For calculation of relative fold change values, initial normalization was achieved against endogenous 18S ribosomal RNA using the ΔΔCT method of quantification (Applied Biosystems Inc.) (Livak and Schmittgen, 2001). Average fold change from four independent runs was calculated as 2ΔΔCT. Significance of average fold change was determined by 2-way ANOVA with a Tukey’s Pair-Wise post-hoc test (Statistical Analysis Software, SAS, Cary, NC). Treatments where a p-value was <0.05 were considered to be significantly different.
Primers for PCR were selected using Primer Blast (National Center for Biotechnology Information, Bethesda, MD) and were as follows: Gapdh sense GAGTCAACGGATTTGGTCGT antisense TTGATTTTGGAGGGATCTCG (GeneID: 12597), Mapk sense TTCCAAGGGCTACACCAAGT antisense CAGTCCTCTGAGCCCTTGTC (GeneID: 5594), Akt sense AACACCATGGACAGGGAGAG, antisense CAAACTCGTTCATGGTCACG (GeneID: 207), Ncam sense CAGGTCATTGTGAATGTGCC antisense TGCCCATCCAGAGTCTTTTC (GeneID: 4684), Src sense AGCACAACCTGACCATCCTC antisense CCACCAGTCTCCCTCTGTGT (GeneID: 6714), Cdh2 sense CTCCGCGGCCCGCTATTTGT antisense CCAGAAGCCTCTACAGACGCCTGA (GeneID: 1000), Neurod sense CTAACGCCCGGGAGCGGAAC antisense TGCGGCGGAGGCTTAACGTG (GeneID: 4760), Gbx2 sense CGAGCGCGTCTATGAGCGCA antisense GACAGCCCCGACGAGCGAAG (GeneID: 2637), Foxg1 sense ACGAGAAGCCGCCGTTCAGC antisense TTGAAGGCCAGCTTGGCCCG (GeneID: 2290). PCR reactions were run for 35 cycles where melting was done at 95°C for 180s, annealing done at 57°C for 30s and elongation done at 72°C for 30s. Initial melting and final elongation were done for 180s at 95°C and 10 minutes at 72°C, respectively.
2.6 Fluorometric Imaging Plate Reader (FLIPR) Assay
FLIPR assays were performed using Molecular Devices Calcium 4 assay kit (Molecular Devices, Sunnyvale, CA) following manufacturer’s directions. Briefly, 2X Calcium 4 dye was prepared in a buffer containing 145 mM NaCl, 10 mM glucose, 5 mM KCl, 1 mM MgSO4, 10 mM HEPES and 2 mM CaCl2 and diluted with an equal volume of Locke’s buffer to make the dye incubation medium. At the start of the assay, culture media from each well was replaced with 200 μl of dye incubation medium and incubated for 1 hour at 37°C and 5% CO2. Plates were then brought to room temperature over the course of 30 minutes. A final concentration of 100 μM AMPA, 100 μM kainic acid or 100 μM NMDA was added to the cell plates and a fluorescence intensity baseline was recorded using the Flexstation 3 plate reader (Molecular Devices). Then, half-log dilutions of cyclothiazide or veratridine were added and fluorescence intensity recorded. The highest concentration of cyclothiazide or veratridine used was 50 μM. For dose response analysis, total area under the curve for responses was normalized to the response of 50 μM cyclothiazide or veratridine. Dose response curves were fit to a three parameter model with a Hill coefficient of 1 using Prism software (GraphPad, La Jolla, CA). Significance was determined by 2-way ANOVA with a Tukey’s post-hoc test for multiple comparisons (SAS). Treatments where a p-value was <0.05 were considered to be significantly different.
2.7 Whole Cell Patch Clamp
hNP cells were grown in the absence of bFGF on a substrate of poly-ornithine/laminin for up to one month. hNP cells with significant neurite growth 23 days after removal of bFGF were subjected to whole cell voltage clamp using an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA) and pClamp 9.2 data acquisition software for electrophysiology (Molecular Devices, Sunnyvale, CA). The extracellular solution consisted of 139 mM NaCl, 3 mM KCl, 16 mM glucose, 1.8 mM CaCl2, 0.5 mM MgSO4, 0.5 mM NaH2PO4, 1 mM NaHCO3, 2 mM HEPES, 1 mM NaPyruvate (Invitrogen), 0.1 mM choline chloride, and phenol red. The pH was titrated at 7.25 with NaOH and the osmolarity was 300 mOsm. The intracellular solution consisted of: 135 mM Kgluconate, 0.1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 1 mM EGTA, 2 mM MgATP, 0.4 mM NaGTP, titrated to pH 7.29 and had an osmolarity of 281 mOsm. In some cases, an equimolar amount of Csgluconate was substituted for Kgluconate in order to block currents through potassium channels. Experiments were carried out at 30°C in a humidified atmosphere of 5% CO2/ 95% O2. In voltage clamp experiments, the holding potential was kept at -60 mV, but was hyperpolarized to -100 mV 50 ms prior to the depolarization step to allow for recovery from fast sodium channel inactivation. Current clamp experiments were done with minimal current injection to maintain a membrane potential of approximately -70 mV prior to injection of the depolarizing current pulse. Drugs were locally perfused around the cell using a glass pipette with a 500 μm diameter opening positioned 100 μm from the cell.
3 Results
3.1 hESC-derived hNP cell differentiation
hNP cells were maintained on poly-ornithine/laminin coated plates in an adherent monolayer and were characterized by a stable expression of neural stem cell marker NESTIN in almost all cells (Figure1A). Upon removal of bFGF from the culture medium, hNP cell differentiation ensued. Immunocytochemical analysis indicated that at 14 days after removal of bFGF, immature neural cell marker TUJ1 was expressed in almost all cells (Figure 1B). MAP2 expression was observed 21 days after removal of bFGF (Figure 1C) and is an indicator of neuron formation. Terminal differentiation is marked by the transition from a proliferating cell type to a post-mitotic cell type. CFSE dye can be absorbed by the parent cell and has approximately half of the fluorescence expressed in each daughter cell with each cell division. Thus, total population doubling is represented as a decrease in relative fluorescent units (RFU) per cell by 50% of the parent cells. Proliferation was measured in hNP cells in the presence of bFGF every 8 hours for 48 hours after the addition of the CFSE dye (Figure 1D). After bFGF was removed from the culture medium for 14 days, proliferation was measured with the CFSE dye every 24 hours for 96 hours (Figure 1E). The half-life of mean fluorescence approximates the doubling time for cell proliferation. Fitting the decay of mean fluorescence data points, we observed a half-decay time for the hNP cell population at 6.8 hrs (Figure 1F). In contrast, the mean fluorescence for the hNP cell population cultured in the absence of bFGF did not decay at all, suggesting that little or no proliferation has taken place (Figure1F). In summary, when bFGF was removed from the culture medium the cell cycle of hNP cells was arrested in less than 96 hours followed by an increase in pan neuronal marker expression.
Figure 1. Human neural progenitor cells exhibit signs of neuronal maturation.
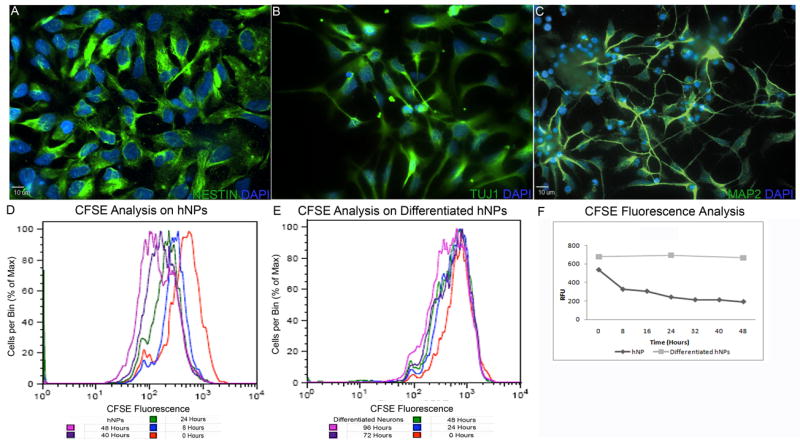
hNP cells maintained in proliferation media express neural stem cell marker NESTIN (green, A). When subjected to neural differentiation media for 14 days, differentiated hNP cells express immature neural marker TUJ1 (green, B) and after 21 days differentiated hNP cells express intermediate filament marker MAP2 (green, C). Cessation of proliferation is a characteristic of terminal differentiation. hNP cell mean fluorescence per cell decreases with a half time constant of 6.8 hrs (equation 1; D, F). Differentiated hNP cells did not show any decrease in mean fluorescence per cell indicating little or no proliferation over a 96 hour period (E,F). hNP – human neural progenitor
Another sign of differentiation into a neuronal lineage would be machinery, which regulates neurotransmitter production. Quantitative RT-PCR was used to compare the abundance of transcripts from hNP cells cultured in the presence and absence of bFGF. Results were normalized to GAPDH expression. Expression of Dat, Sert and Gad1 peaked at 14 days but declined thereafter (Figure 2A); whereas, VACht expression plateaued 35 days after removal of bFGF (Figure 2A). Expression of markers of synapse formation also indicate neuronal lineage. Synaptic marker syntaxin 1A was stably expressed at its highest level at the 14 day time point but declined between the 35 and 125 day time points (Figure 2A). Synaptophysin peaked at 35 days after the removal of bFGF, but also declined thereafter (Figure 2A). This followed a general trend where by day 125 in culture gene transcript levels were lower than at previous time points. This may reflect a general decrease in cellular function and health given that LIF was the only growth factor supporting these cultures and these cells lacked glia as evidenced by the high proportion of cells expressing TUJ1 and MAP2. Transcripts for endosomal markers Rab5a and CD146 were not significantly altered during differentiation (Figure 2A). Potentially the most widely known and important potassium genes for drug safety studies analysis of a candidate compound is Kv11.1 (hERG) potassium channel. This gene along with the potassium chloride transporter Kcc2, initially increased by day 14, but expression decreased significantly by 125 days after bFGF removal (Figure 2A) again suggesting that day 125 cultured cells may be declining.
Figure 2. Differentiated neural progenitor cells express markers of developmental progression.
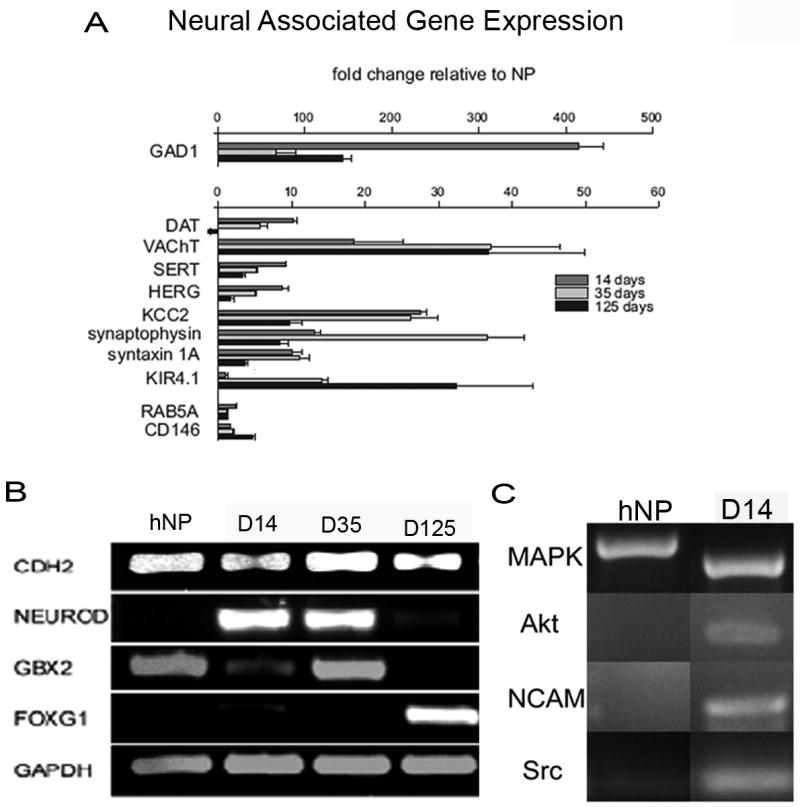
After differentiation of hNP cells for 14, 35 or 125 days, there is an increase in presynaptic receptors and proteins necessary for functional neurotransmitter release, Dat, Sert, Herg, Kcc2, synaptophysin, syntaxin1A and Kir4.1 (A). Embryonic neural development markers Cdh2 and Gbx2 are observed in hNP cells and differentiated hNP cells at 14, 35 and 125 days (Cdh2) or 35 days (Gbx2, B). Differentiation marker NeuroD is expressed in hNP cells at 14 and 35 days of exposure to differentiation media but not in hNP cells exposed to bFGF., Telencephalic neurogenesis marker Foxg1 is seen in 125 day differentiated hNP cells (B). Pathways involved in neural development include Mapk and Pi3k. Markers of these pathways, Mapk, Akt, Ncam and Src are expressed in 14 day differentiated hNP cells and Mapk is expressed in hNP cells (C). hNPs – human neural progenitor
The developmental marker Gbx2 was expressed in hNP cells cultured in bFGF and LIF and in hNP cells after bFGF was removed for 14 and 35 days but was not expressed in hNP cells 125 days after bFGF removal (Figure 2B). Transcription factor Foxg1 is present at the initiation of telecephalon development from the single cell neuroepithelium and may work co-operatively with FGF signaling during development (Hebert and Fishell, 2008). Yet, significant expression of this transcription factor was not found until the 125 day time point (Figure 2B). Cdh2, calcium dependent cell to cell adhesion marker, was observed at all time points in the absence and presence of bFGF (Figure 2B). Neural differentiation maker Neurod was expressed at 14 and 21 days post differentiation (Figure 2B).
Regionalized differentiation and signaling pathway genes were expressed in the undifferentiated hNP cells and after bFGF was removed. Mapk is a signaling factor important for neural synaptic plasticity and hESC self-renewal and cell cycle maintenance (Thomas and Huganir, 2004, Binetruy et al., 2007) and transcripts were present in both hNP cells and differentiated hNP cells (Figure 2C). Akt, a signaling factor involved in axon elongation and neuron polarity (Yoshimura et al., 2006, Read and Gorman, 2009), Ncam, a cell adhesion molecule involved in directed growth of axons in neural development and presynaptic function (Skaper, 2005) and in triggering neurite outgrowth through intracellular signaling cascades (Kiryushko et al., 2004) and Src, important in fully developed neurons for up regulating ion channel expression and in gating synaptic plasticity and potentiation (Kalia et al., 2004) were only expressed in day 14 differentiated hNP cells (Figure 2C).
3.2 Glutamate receptor expression in hNP cells and differentiated hNP cells
The expression of ionotropic glutamate receptors might also be an indicator of neuronal maturation. These receptors are composed of three distinct families: NMDA, kainate and AMPA receptors. The hNP cells and differentiated hNP cells cultured in the absence of bFGF for 2 weeks were analyzed for mRNA expression of subunits of each glutamate receptor subtype relative to hESCs. Significant increases (p<0.05) in Grin2b were seen in hNP cells (20 fold) and differentiated hNP cells (25 fold) relative to hESCs (Figure 3A). Additionally, Grin1 and Grin2d were significantly increased (p<0.05) only in differentiated hNP cells relative to hESCs, but not in undifferentiated hNP cells (Figure 3A). Of the kainate receptors, Grik4 and Grik5 were significantly (p<0.05) increased only in undifferentiated hNP cells relative to hESCs (Figure 3B); whereas, Grik2 was significantly (p<0.05) increased only in hNP cells where bFGF had been removed (Figure 3B). AMPA receptor subunits were also examined. Gria1 and Gria4 were up regulated in hNP cells relative to hESCs (Figure 3C). Two week differentiated hNP cells showed significant (p<0.05) up regulation of Gria2 and Gira4 relative to hESCs (Figure 3C).
Figure 3. Expression of ionotropic glutamatergic receptor subunits, sodium and calcium channels in hNP cells and differentiated hNP cells.
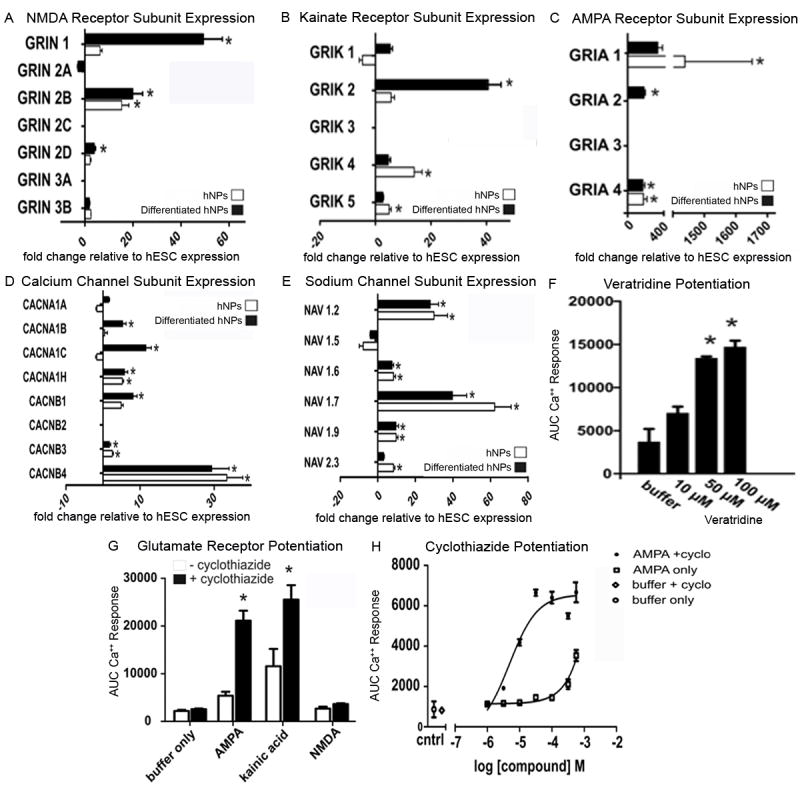
Differentiated hNP cells show up-regulation of Grin1, Grin2b and Grin2d NMDA subunits relative to hESCs while hNP cells only show up-regulation of Grin2d (A) relative to hESCs. hNP cells have increased expression of kainate subunits Grik4 and Grik5 relative to hESCs, while differentiated hNP cells Have high expression of Grik2 relative to hESCs (B). The AMPA receptor subunit transcripts Gria1 and Gria4 are increased relative to hESCs in proliferating hNP cells, while Gria2 and Gria4 are up-regulated relative to hESCs in differentiated hNP cells (C). Expression of various calcium channel subunits (D) and sodium channel subunits (E) is similar to expected in vivo expression. Sodium channel response can also be attenuated with the addition of veratridine (F). Addition of cyclothiazide to neural cultures yields a dose-dependent increase of the calcium response in the FLIPR assay in with AMPA and kainic acid but not with NMDA (G,H). This increase responds in a dose related manner (H). AMPA - α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate, NMDA - N-methyl-D-aspartic acid, hESCs – human embryonic stem cells; hNPs – human neural progenitor cells, * significantly different (p>0.05) relative to hESCs
To determine if functional glutamate channels exist in differentiated hNP cells, calcium influx in response to AMPA, kainic acid or NMDA application was measured on hNP cells, 14 days after the removal of bFGF. Figure 3G indicates that NMDA could not depolarize differentiated or undifferentiated hNP cells enough to cause significant calcium influx above background. In contrast, AMPA and kainic acid can cause calcium influx which can be potentiated by AMPA receptor specific modulator, cyclothiazide (50 μM, Figure 3G). Calcium influx was detected in the presence of cyclothiazide in calcium activity as measured (Figure 3H).
3.3 Voltage-Gated Channel Expression in hNP cells
Voltage dependent calcium and sodium channels play a critical role in action potential generation and synaptic transmission. Calcium and sodium channel subunit expression in hNP cells cultured in the presence and absence of bFGF was evaluated using real-time PCR. The following subunit transcripts were significantly (p<0.05) increased in hNP cells relative to hESCs: Cacna1h, Cacnb3 Cacnb4 (Figure 3D). Subunits significantly (p<0.05) up regulated in day 14 differentiated in hNP cells relative to hESCs were Cacna1b, Cacn1c, Cacna1h, Cacnb3 and Cacnb4 (Figure 3D). Sodium subunits Nav1.2, Nav1.4, Nav1.7, Nav1.9 were significantly (p<0.05) up regulated in both hNP cells and day 14 differentiated hNP cells relative to hESCs (Figure 3E). Additionally, Nav2.3 was significantly (p<0.05) up regulated in hNP cells relative to hESCs (Figure 3E). In support of this, increasing concentrations of a sodium channel activator veratridine in a FLIPR assay on differentiated hNP cells show an increasing calcium response (Figure 3F).
Sodium channel activity in differentiated hNP cells was measured using whole cell voltage clamp. 81 total hNP cells cultured in the absence of bFGF from 4 to 27 days were analyzed. Of these, 34 exhibited no fast inward currents in response to a step depolarization indicating the absence of functional voltage gated sodium channels (Figure 4G). The remaining cells yielded between 0.04 - 1.5 nA of inward current in response to the step depolarization (Figures 4B and 4G). These currents inactivated rapidly in all cases (Figures 4B and 4C) and could be abolished with the addition of 1 μM TTX (n = 3 cells; Figure 4C). Additionally, voltage-dependent steady state inactivation (n = 11 cells; Figure 4D) and recovery from fast inactivation (n = 5 cells; Figure 4E) were also measured in all positive cells and observed in several of the 47 positive cells. A subset of these cells was subjected to current clamp and action potentials were elicited by current injection (n = 8 cells, Figure 4F). In support of this, increasing concentrations of a sodium channel activator veratridine in a FLIPR assay on differentiated hNP cells show an increasing calcium response (Figure 4H). This probably resulted from voltage-gated sodium channel depolarization of cells that subsequently allowed calcium influx through calcium channels. These data indicate that differentiation of hNP cells by removal of bFGF can lead to a neuronal phenotype that can depolarize the cell and generate action potentials.
Figure 4. Differentiated hNP cells exhibit voltage-dependent, TTX-sensitive inward currents that generate action potentials.
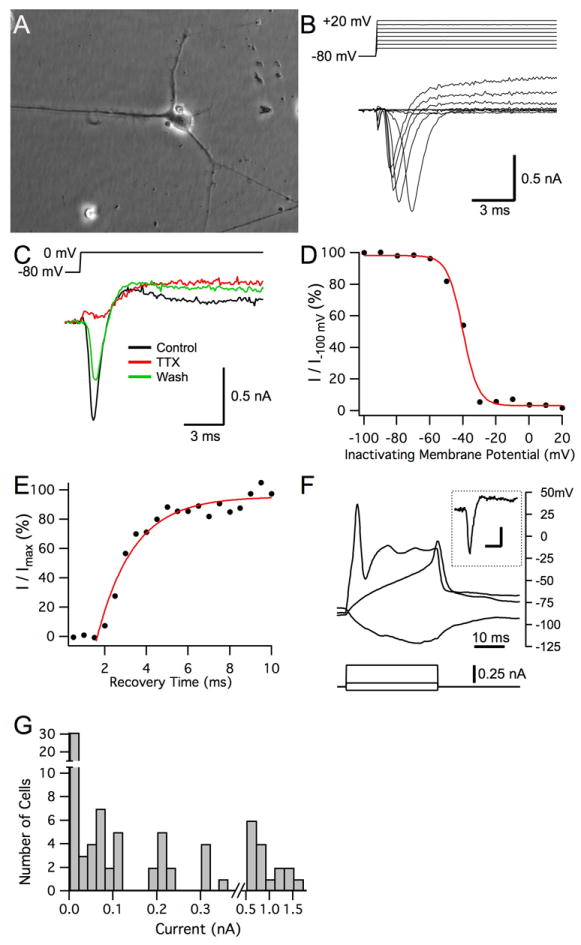
Differentiated hNP cells grown on a substrate of laminin for up to one month and differentiated hNP cells with significant neurite growth were isolated after 23 days of differentiation (A). This cell was subjected to whole cell voltage clamp utilizing a potassium gluconate based intracellular solution and elicited both voltage gated inward and outward currents in response to depolarizing voltage steps (B, C). Inward currents from another cell (potassium gluconate intracellular) were abolished by local application of 1 μM TTX (red trace) while outward currents remained. Inward current recovered as TTX washed out of the region (green trace; D). A different cell exhibited classic sodium channel steady state inactivation by showing voltage activated inward currents that inactivated in response to a 50 ms pre-pulse at different membrane potentials. The experiment was done 27 days after the removal of bFGF. A cesium gluconate based intracellular solution was used for this experiment to block outward potassium currents. The membrane potential for half maximal inactivation by standard Boltzman fitting (red line) was -40.1 mV with a slope of 4.7 (E). Recovery from fast inactivation utilizing a paired pulse protocol in the same cell as C. The single exponential time constant for recovery of inactivation was 1.7 ms (red line; F). A different cell elicited an overshooting action potential upon current injection under whole cell current clamp utilizing a potassium gluconate based intracellular solution. Inset: Response of the same cell under voltage clamp to a change in membrane potential from -80 mV to -10 mV elicited a peak current of 457 pA. Scale bars for inset: 5 ms, 0.2 nA (G). Histogram of maximum peak current amplitudes elicited on depolarization from all successfully patched differentiated hNP cells under voltage-clamp. Left-most bar indicates cells exhibiting no inward current in response to the depolarization.. TTX – tetrodotoxin; hNP – human neural progenitor, bFGF- basic fibroblast growth factor
The 58% success rate for finding voltage-gated sodium channel function in cells tested by whole-cell voltage clamp (Figure 4G), does not reflect the true proportion of sodium channel positive cells in our differentiated hNP cells, but rather our ability to morphologically distinguish these cells from negative cells by eye. An example of the morphology of a sodium channel positive cell is shown in Figure 4A. All of the positive cells were phase bright with a few long processes; however some of the negative cells also expressed long processes potentially due to their timing in the differentiation process at the time of measurement.
All cells examined with a potassium gluconate intracellular solution exhibited voltage-dependent outwardly rectifying currents (Figure 4B). These currents were not observed for cells in which a cesium gluconate based intracellular solution was used indicating that both hNP cells cultured in the presence and absence of bFGF had functional potassium channels.
3.4 Expression of other ionotropic receptors in hNP cells
Although glutamate is the main excitatory transmitter in the CNS, characterization of other neurotransmitter receptors will help determine the neuronal phenotype. We used real-time PCR to measure the relative mRNA expression levels of nicotinic receptor subunits. Relative to hESCs, Chrna3, Chrna5, and Fam7a3 had significantly increased transcript levels in both differentiated and undifferentiated hNP cells relative to hESCs (p<0.05, Figure 5A). Chrna1, Chrna4, Chrnb4 were significantly increased only for differentiated hNP cells relative to hESCs (p<0.05). One subunit transcript, Chrna10, had a lower level of expression in both hNP cells and day 14 differentiated hNP cells relative to hESCs (Figure 5A).
Figure 5. Expression of nicotinic, purinergic, transient receptor potential and GABAergic subunits.
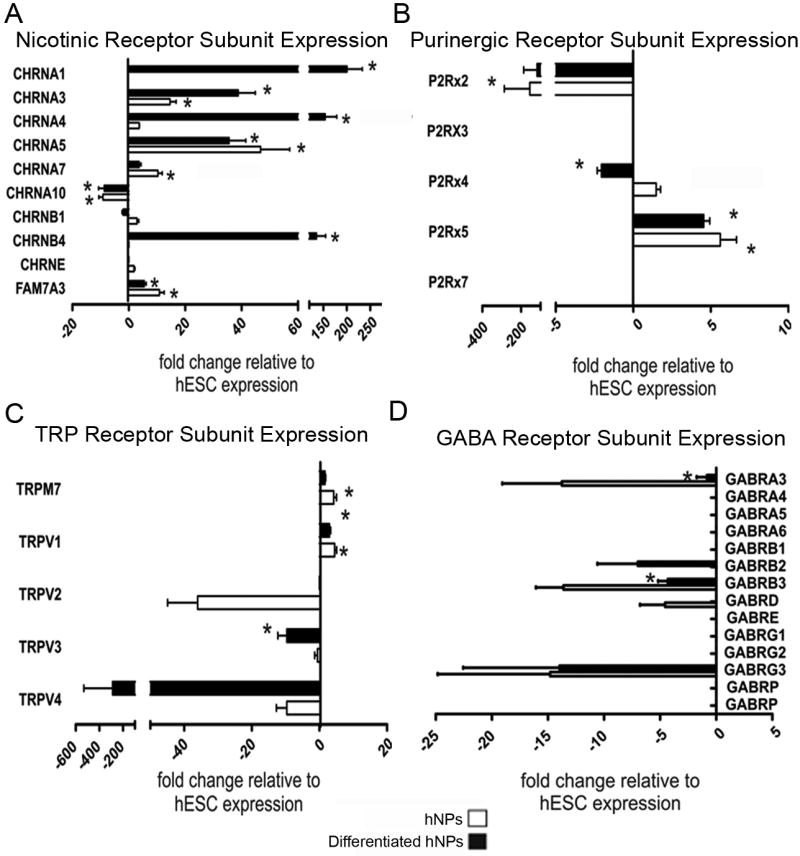
Ionotropic receptors are responsible for elicting response to several stimuli. Nicotinc receptors (A) are most commonly associated with nicotine addiction. Purinergic receptors (B) are newly discovered and are linked to ATP activation. TRP channels (C) are sensory channels which transmit hot/cold responses, mechanical response or mineral regulation. GABA is the primary inhibitory neurotransmitter in the brain and has a role in modulating signals transmitted in the brain (D). TRP – tranisent receptor potential, GABA - gamma-aminobutyric acid, ATP – adenosine triphosphate * significantly different (p>0.05) relative to hESCs
Purinergic receptors are ion channels which are activated by ATP and regulate cellular secretions and sensory transmission (Apolloni et al., 2009). P2rx5 expression was significantly (p<0.05) higher in both differentiated and undifferentiated hNP cells relative to hESCs (Figure 5B); however, lower levels of P2rx2 transcripts were measured from differentiated and undifferentiated hNP cells relative to hESCs (Figure 5B).
TRP channels are ion channels, which are relatively non-selective for monovalent and divalent cations and can be activated by polyunsaturated fatty acids (Talavera et al., 2008). Trpv1 subunit transcripts were significantly (p<0.05) higher in both differentiated and undifferentiated hNP cells relative to hESCs (Figure 5C), while Trpv1 transcripts were only higher in undifferentiated hNP cells is also significantly (p<0.05) up regulated in differentiated hNP cells relative to hESCs. RNA levels of the Trpv3 subunit in differentiated and undifferentiated hNP cells were significantly reduced in comparison to hESC cells (p<0.05, Figure 5C).
In early development, the GABA neurotransmitter acts as an excitatory signal. As the CNS matures, GABA becomes the primary inhibitory neurotransmitter in the CNS (Laurie et al., 1992). Gabra3 and Gabrb3 were significantly (p<0.05) reduced in differentiated hNP cells relative to hESCs (Figure 5D).
4 Discussion
Here we developed a protocol for differentiation of hNP cells in minimal conditions that resulted in significant increases in gene transcript levels for neuronal markers, ion channels and ionotrophic receptors. VAChAT required only the removal of bFGF and the presence of LIF for sustained expression. Although these same conditions yielded an initial increase in DAT expression, the expression diminished completely by 125 days in vitro as was true for most transcription levels measured. The addition of GDNF and LIF to these cultures supported DAT expression (Young et al., 2010). Therefore, it is important to determine if in the absence of bFGF and non-inductive signaling, neuronal cells with functional ion channel, ionotrophic receptor and electrophysiological characteristics of interest to drug discovery can be generated under these basal conditions. Previously, spontaneous firing and repetitive action potentials has required either co-culture with murine hippocampal cells (Ladewig et al., 2008)or astrocytes (Wu et al., 2007) or the addition of morphogens (Erceg et al., 2008).
4.1 Comparing in vitro gene expression in hNP cells to neuron and brain development in vivo
Early regulation of the midbrain-hindbrain boundary and the development of the midbrain and the cerebellum are controlled by Gbx2 (Li and Joyner, 2001) which was expressed in hNP cells as well as in cultures after bFGF was removed for 14 and 35 days. The differentiation of the telencephalon is regulated by Foxg1’s modulation of brain morphogenic protein and fibroblast growth factor 8 signaling (Martynoga et al., 2005, Shen et al., 2006). In human cells bFGF promotes neural induction but not differentiation. In addition, it is not clear how the lack of bFGF signaling influences Foxg1 expression in differentiating human neural cells. The factors expressed in the differentiated hNP cells are representative first (35 days after bFGF removal) of midbrain/hindbrain development (Gbx2) then later (125 days after bFGF removal) of proper telencephalon development (Foxg1).
Following regional organization, Cdh2 is involved in regulation of cortical neuron differentiation in the subventricular zone (Yagita et al., 2009). Cdh2 was expressed in both hNP cells and differentiated hNP cells at day 14, 35 and 125 of differentiation. NeuroD expression is part of a regulatory pathway that controls early neural differentiation and glutamatergic neurogenesis (Katayama et al., 1997, Lee, 1997, Hevner et al., 2006). This should be absent from undifferentiated precursors, yet immediately precede glutamate receptor expression. hNP cell differentiation somewhat mimics this time course, as NeuroD is not expressed in undifferentiated hNP cells but is strongly expressed 14 and 35 days after the removal of bFGF (Figure 2B). Expression of these genes in the hNP cells differentiated in culture may be indicative of early segmentation of the midbrain and hindbrain regions.
NMDA receptor subunits are important for the pruning of developing synapses (Yen et al., 1993) and are also instrumental in the initiation of many forms of synaptic plasticity (Stoneham et al., 2010). Most functional NMDA receptor subunits contain both Grin1 and Grin2 subunits. Grin1 expression was increased in hNP cells, 14 days after initialization of differentiation by removal of bFGF. This coincides with strong NeuroD expression. The Grin2 subunits are expressed postnatally (Grin2a and Grin2c) or embryonically (Grin2b and Grin2d) in mice (Watanabe et al., 1993, Takai et al., 2003) and as neurons develop postnatally, Grin2b subunit is swapped for the Grin2a subunit (Liu et al., 2004). In the differentiated hNP cells used in these studies, we see an increase in expression of the Grin2b and Grin2d subunits but not the Grin2a or Grin2c subunits (Figure 3A).
NMDA receptor activity was found to be at background levels when monitored with a FLIPR assay (Figure 3G) despite the increase in expression of some NMDA receptor subunits at this 14 day time point. Thus, it is possible that differentiated hNP cells are incapable of forming functional NMDA receptors at this time point. However, it is also possible that stimulation of these cultures with 100 μM NMDA was not sufficient to depolarize the cells enough to overcome voltage-dependent blockage by 1 mM magnesium.
AMPA receptors are responsible for the initial rapid depolarization in the excitatory postsynaptic potential and are expressed at most excitatory synapses. AMPA receptors lacking Gria2 are calcium permeable while those containing Gria2 have low calcium permeability (Geiger et al., 1995). Modulation of excitatory synaptic strength at most synapses is mediated by the movement of AMPA subunits into and out of the postsynaptic density (Clem and Barth, 2006). Gria1-4 expression becomes segregated into distinct brain regions at E14 (Gallo et al., 1995). In hNP cells Gria1 and Gria4 are highly expressed in both differentiated and undifferentiated hNP cells but that Gria2 is only increased after differentiation is initiated by the removal of bFGF (Figure 3C). The fact that Gria2 transcript levels increase in older cultures suggests that these conditions do allow normal AMPA receptor development in hNP cells.
The physiological relevance of kainate receptors is still relatively un-elucidated. However, they are present both presynaptically and postsynaptically in various regions of the CNS. There are two classes of kainate receptors: Grik1 - 3 can form channels alone or in combination with one another, but Grik4 and Grik5 can only form functional channels in the presence of either Grik1, 2 or 3 (Gallyas et al., 2003, Ren et al., 2003). Grik1 peaks around birth and (Bahn et al., 1994). This expression profile is somewhat mimicked in hNP cells where expression of Grik4 and 5 are strongest in undifferentiated hNP cells but undergo a modest decline upon differentiation (Figure 3B).
The fact that the FLIPR assay can measure AMPA receptor calcium influx may suggest that hNP cells also have functional voltage-gated calcium channels. We observed that voltage-gated N-type calcium channels α subunits Cacna1b and Cacna1c as well as β subunit Cacb1 had increased expression after removal of bFGF. Most N-type calcium channel subunits are expressed beginning at E18 in vivo with the majority of expression occurring postnatally (Jones et al., 1997).
Voltage-gated sodium channels play a major role in increasing cell excitability. Generation of action potentials is initiated by sodium channel activation, and is a primary characteristic used to define functional neurons. Expression of Nav1.2 is high in both differentiated and undifferentiated hNP cells. This is consistent with the constitutive expression of Nav1.2 throughout CNS development. However, Nav1.6, Nav1.7 and Nav1.9 were expressed at similar levels in both differentiated and undifferentiated hNP cells; whereas, in the CNS Nav1.6, Nav1.7, and Nav1.9 are expressed predominately in differentiated neurons and increase postnatally (Benn et al., 2001). The subunits up regulated in the hNP cells and differentiated hNP cells used in this study (Nav1.6, Nav1.7, Nav1.9 and Nav2.3) all have roles in modulating neural differentiation, neural excitability and neural development (Benn et al., 2001, Mechaly et al., 2005). Here electrophysiological results indicate that differentiated hNP cells can generate inward currents that were blocked with TTX and can generate action potentials (Figure 4).
GABAA receptors are chloride channels that are activated by the neurotransmitter, GABA. Responses generated by GABA receptors are inhibitory in the mature brain; however, early in embryogenesis the concentration of intracellular chloride is higher in some immature neurons such that chloride flows out of the cell through GABA receptors creating an excitatory response, which can evoke glutamate release (Culiat et al., 1994, Gunther et al., 1995). Gabrb2, Gabrb3 and Gabrg2 subunits are expressed throughout development while Gabrg1 and Gabrg3 expression decreases as brain development ensues (Laurie et al., 1992, Paysan et al., 1994). The results here demonstrate a decrease in all GABAA receptor subunits in hNP cells and differentiated hNP cells relative to hESCs.
GABAA receptor activity can be altered by currents elicited through sodium channels as sodium and chloride ions are transported into GABAergic neurons via a sodium/chloride co-transporter. bFGF has been shown to increase the expression of glutamate receptor subunits; however, it has no effect on GABA receptor subunit expression (Numakawa et al., 2002). GABAA receptor expression has been shown to reduce the proliferation of neuroblasts and stem cells and to reduce migration (Platel et al., 2008).
The TRP channels transmit sensory information involved in hot/cold sensation, modulation of pain and protection of neurons from oxidative stress (Talavera et al., 2008). They are generally associated with the peripheral nervous system, but their expression in hNP cells was largely reduced in comparison to hESC cultures (Figure 5C). These data suggest that hNP cells may not easily differentiate into peripheral nervous system lineages using these minimal conditions of bFGF removal.
Purinergic receptors when bound with ATP induce fast synaptic potentials (Jarvis and Khakh, 2009). P2x4 and P2x7 subunits are expressed in microglia and can prevent excitotoxicity in motorneurons (Apolloni et al., 2009). In this in vitro model, P2x4 subunits are up regulated in hNP cells but down regulated in differentiated hNP cells while P2x7 is not expressed in hNP cells or differentiated hNP cells relative to hESCs. However, under the minimal culture conditions of this study, the P2x5 subunit is expressed in both hNP cells and differentiated hNP cells.
Nicotinic receptors mediate fast synaptic excitation at the neuromuscular junction and are found in neurons in the brain. The standard neural subunits are α2-10 and β2-4 and these subunits typically form heteromeric channels, although α7 and α9 can combine to form homomeric channels (Nashmi and Lester, 2006). All neuronal nicotinic subunits begin expression around E11 in the rat and the expression level remains relatively constant in adult tissue (Nashmi and Lester, 2006). In hNP cells, we observed that α5 was expressed at high levels in undifferentiated hNP cells and expression remained unchanged after bFGF removal. In contrast, α1, α3 and β4 transcripts were all induced after bFGF removal (Figure 5A).
4.2 Population Heterogeneity and Lineage
In vitro differentiation is still in its infancy. Although we know that some factors such as Foxg1 and Gbx2 are important for brain tissue development and cell migration in a three dimensional environment, we do not yet understand how the consequences of expression of these transcription factors in a two dimensional environment. In our study, we have found that a minority of hNP cells can elicit action potentials after induction of differentiation by removal of bFGF. This indicates that although all cells are in a uniform environment, they do not all differentiate into functioning neurons; however, neither do all cells in cultured in medium containing a full range of growth factor cocktails. Still, the fact that a minority of hNP cells are capable of generating action potentials in our minimal culture conditions provides a source of cells that can be maintained with the ease of a cell line and without the upkeep of primary neuronal culture. This may provide a cost effective way for screening large libraries of compounds in neural drug discovery provided the right pharmacological agents are used to isolate the screening to a specific response indicative of the expressed genes characterized here.
Acknowledgments
We thank Julie Nelson and the Center for Tropical and Emerging Global Diseases Flow Cytometry Facility for her technical expertise. Funding was provided by NIH R41N5053272-01 and DoD # N000140810989 (SLS) and National Institute of Neurological Disease and Stroke S11NS055883 (MB).
Abbreviations
- AMPA
α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate
- bFGF
basic fibroblast growth factor
- CFSE
Carboxyfluorescein succinimidyl ester
- CNS
central nervous system
- DAPI
4′,6-diamidino-2-phenylindole
- DMEM
Dulbecco’s modified Eagle medium
- GABA
gamma-aminobutyric acid
- hERG
human Ether-à-go-go related gene potassium channel
- hESCs
human embryonic stem cells
- hNPs
Human neural progenitors
- LIF
leukemia inhibitory factor
- MAP2
microtubule-associated protein 2
- MEF
mouse embryonic fibroblast
- NMDA
N-methyl-D-aspartic acid
- PBS+/+
phosphate buffered saline with calcium and magnesium
- RFU
relative fluorescence unit
- SAS
statistical analysis software
- TRP
transient receptor potential
- TTX
tetrodotoxin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apolloni S, Montilli C, Finocchi P, Amadio S. Membrane compartments and purinergic signalling: P2X receptors in neurodegenerative and neuroinflammatory events. FEBS J. 2009;276:354–364. doi: 10.1111/j.1742-4658.2008.06796.x. [DOI] [PubMed] [Google Scholar]
- Bahn S, Volk B, Wisden W. Kainate receptor gene expression in the developing rat brain. J Neurosci. 1994;14:5525–5547. doi: 10.1523/JNEUROSCI.14-09-05525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar TN, Scott CA, Greene CL, Wen X, Smith SV, Maric D, Liu QY, Colton CA, Barker JL. Glutamate acting at NMDA receptors stimulates embryonic cortical neuronal migration. J Neurosci. 1999;19:4449–4461. doi: 10.1523/JNEUROSCI.19-11-04449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn SC, Costigan M, Tate S, Fitzgerald M, Woolf CJ. Developmental expression of the TTX-resistant voltage-gated sodium channels Nav1.8 (SNS) and Nav1.9 (SNS2) in primary sensory neurons. J Neurosci. 2001;21:6077–6085. doi: 10.1523/JNEUROSCI.21-16-06077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binetruy B, Heasley L, Bost F, Caron L, Aouadi M. Concise review: regulation of embryonic stem cell lineage commitment by mitogen-activated protein kinases. Stem Cells. 2007;25:1090–1095. doi: 10.1634/stemcells.2006-0612. [DOI] [PubMed] [Google Scholar]
- Blandini F. An update on the potential role of excitotoxicity in the pathogenesis of Parkinson’s disease. Funct Neurol. 2010;25:65–71. [PubMed] [Google Scholar]
- Buckingham SD, Kwak S, Jones AK, Blackshaw SE, Sattelle DB. Edited GluR2, a gatekeeper for motor neurone survival? Bioessays. 2008;30:1185–1192. doi: 10.1002/bies.20836. [DOI] [PubMed] [Google Scholar]
- Carpenter MK, Rosler E, Rao MS. Characterization and differentiation of human embryonic stem cells. Cloning Stem Cells. 2003;5:79–88. doi: 10.1089/153623003321512193. [DOI] [PubMed] [Google Scholar]
- Clem RL, Barth A. Pathway-specific trafficking of native AMPARs by in vivo experience. Neuron. 2006;49:663–670. doi: 10.1016/j.neuron.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Culiat CT, Stubbs LJ, Montgomery CS, Russell LB, Rinchik EM. Phenotypic consequences of deletion of the gamma 3, alpha 5, or beta 3 subunit of the type A gamma-aminobutyric acid receptor in mice. Proc Natl Acad Sci U S A. 1994;91:2815–2818. doi: 10.1073/pnas.91.7.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis L, Lamorte G, Snyder EY, Malgaroli A, Vescovi AL. A novel, immortal, and multipotent human neural stem cell line generating functional neurons and oligodendrocytes. Stem Cells. 2007;25:2312–2321. doi: 10.1634/stemcells.2007-0040. [DOI] [PubMed] [Google Scholar]
- Dhara SK, Hasneen K, Machacek DW, Boyd NL, Rao RR, Stice SL. Human neural progenitor cells derived from embryonic stem cells in feeder-free cultures. Differentiation. 2008;76:454–464. doi: 10.1111/j.1432-0436.2007.00256.x. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erceg S, Lainez S, Ronaghi M, Stojkovic P, Perez-Arago MA, Moreno-Manzano V, Moreno-Palanques R, Planells-Cases R, Stojkovic M. Differentiation of human embryonic stem cells to regional specific neural precursors in chemically defined medium conditions. PLoS One. 2008;3:e2122. doi: 10.1371/journal.pone.0002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo V, Pende M, Scherer S, Molne M, Wright P. Expression and regulation of kainate and AMPA receptors in uncommitted and committed neural progenitors. Neurochem Res. 1995;20:549–560. doi: 10.1007/BF01694536. [DOI] [PubMed] [Google Scholar]
- Gallyas F, Jr, Ball SM, Molnar E. Assembly and cell surface expression of KA-2 subunit-containing kainate receptors. J Neurochem. 2003;86:1414–1427. doi: 10.1046/j.1471-4159.2003.01945.x. [DOI] [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Gunther U, Benson J, Benke D, Fritschy JM, Reyes G, Knoflach F, Crestani F, Aguzzi A, Arigoni M, Lang Y, et al. Benzodiazepine-insensitive mice generated by targeted disruption of the gamma 2 subunit gene of gamma-aminobutyric acid type A receptors. Proc Natl Acad Sci U S A. 1995;92:7749–7753. doi: 10.1073/pnas.92.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill JA, Freudenrich TM, Machacek DW, Stice SL, Mundy WR. Quantitative assessment of neurite outgrowth in human embryonic stem cell-derived hN2 cells using automated high-content image analysis. Neurotoxicology. 2010;31:277–290. doi: 10.1016/j.neuro.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Hebert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Jarvis MF, Khakh BS. ATP-gated P2X cation-channels. Neuropharmacology. 2009;56:208–215. doi: 10.1016/j.neuropharm.2008.06.067. [DOI] [PubMed] [Google Scholar]
- Jeng SF, Wei FC, Chen PK. Successful replantation of an amputated nasal tip by microvascular anastomosis. Plast Reconstr Surg. 1991;87:1118–1120. doi: 10.1097/00006534-199106000-00017. [DOI] [PubMed] [Google Scholar]
- Jones OT, Bernstein GM, Jones EJ, Jugloff DG, Law M, Wong W, Mills LR. N-Type calcium channels in the developing rat hippocampus: subunit, complex, and regional expression. J Neurosci. 1997;17:6152–6164. doi: 10.1523/JNEUROSCI.17-16-06152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia LV, Gingrich JR, Salter MW. Src in synaptic transmission and plasticity. Oncogene. 2004;23:8007–8016. doi: 10.1038/sj.onc.1208158. [DOI] [PubMed] [Google Scholar]
- Katayama M, Mizuta I, Sakoyama Y, Kohyama-Koganeya A, Akagawa K, Uyemura K, Ishii K. Differential expression of neuroD in primary cultures of cerebral cortical neurons. Exp Cell Res. 1997;236:412–417. doi: 10.1006/excr.1997.3757. [DOI] [PubMed] [Google Scholar]
- Kiryushko D, Berezin V, Bock E. Regulators of neurite outgrowth: role of cell adhesion molecules. Ann N Y Acad Sci. 2004;1014:140–154. doi: 10.1196/annals.1294.015. [DOI] [PubMed] [Google Scholar]
- Ladewig J, Koch P, Endl E, Meiners B, Opitz T, Couillard-Despres S, Aigner L, Brustle O. Lineage selection of functional and cryopreservable human embryonic stem cell-derived neurons. Stem Cells. 2008;26:1705–1712. doi: 10.1634/stemcells.2008-0007. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Wisden W, Seeburg PH. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III. Embryonic and postnatal development. J Neurosci. 1992;12:4151–4172. doi: 10.1523/JNEUROSCI.12-11-04151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE. Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- Li JY, Joyner AL. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development. 2001;128:4979–4991. doi: 10.1242/dev.128.24.4979. [DOI] [PubMed] [Google Scholar]
- Liu XB, Murray KD, Jones EG. Switching of NMDA receptor 2A and 2B subunits at thalamic and cortical synapses during early postnatal development. J Neurosci. 2004;24:8885–8895. doi: 10.1523/JNEUROSCI.2476-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martynoga B, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- McNeish J, Roach M, Hambor J, Mather RJ, Weibley L, Lazzaro J, Gazard J, Schwarz J, Volkmann R, Machacek D, Stice S, Zawadzke L, O’Donnell C, Hurst R. High-throughput screening in embryonic stem cell-derived neurons identifies potentiators of alpha-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate-type glutamate receptors. J Biol Chem. 2010;285:17209–17217. doi: 10.1074/jbc.M109.098814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechaly I, Scamps F, Chabbert C, Sans A, Valmier J. Molecular diversity of voltage-gated sodium channel alpha subunits expressed in neuronal and non-neuronal excitable cells. Neuroscience. 2005;130:389–396. doi: 10.1016/j.neuroscience.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Milanese M, Zappettini S, Jacchetti E, Bonifacino T, Cervetto C, Usai C, Bonanno G. In vitro activation of GAT1 transporters expressed in spinal cord gliosomes stimulates glutamate release that is abnormally elevated in the SOD1/G93A(+) mouse model of amyotrophic lateral sclerosis. J Neurochem. 2010;113:489–501. doi: 10.1111/j.1471-4159.2010.06628.x. [DOI] [PubMed] [Google Scholar]
- Nashmi R, Lester HA. CNS localization of neuronal nicotinic receptors. J Mol Neurosci. 2006;30:181–184. doi: 10.1385/JMN:30:1:181. [DOI] [PubMed] [Google Scholar]
- Numakawa T, Yokomaku D, Kiyosue K, Adachi N, Matsumoto T, Numakawa Y, Taguchi T, Hatanaka H, Yamada M. Basic fibroblast growth factor evokes a rapid glutamate release through activation of the MAPK pathway in cultured cortical neurons. J Biol Chem. 2002;277:28861–28869. doi: 10.1074/jbc.M202927200. [DOI] [PubMed] [Google Scholar]
- Paysan J, Bolz J, Mohler H, Fritschy JM. GABAA receptor alpha 1 subunit, an early marker for area specification in developing rat cerebral cortex. J Comp Neurol. 1994;350:133–149. doi: 10.1002/cne.903500110. [DOI] [PubMed] [Google Scholar]
- Platel JC, Dave KA, Bordey A. Control of neuroblast production and migration by converging GABA and glutamate signals in the postnatal forebrain. J Physiol. 2008;586:3739–3743. doi: 10.1113/jphysiol.2008.155325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read DE, Gorman AM. Involvement of Akt in neurite outgrowth. Cell Mol Life Sci. 2009;66:2975–2984. doi: 10.1007/s00018-009-0057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Riley NJ, Needleman LA, Sanders JM, Swanson GT, Marshall J. Cell surface expression of GluR5 kainate receptors is regulated by an endoplasmic reticulum retention signal. J Biol Chem. 2003;278:52700–52709. doi: 10.1074/jbc.M309585200. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shin S, Mitalipova M, Noggle S, Tibbitts D, Venable A, Rao R, Stice SL. Long-term proliferation of human embryonic stem cell-derived neuroepithelial cells using defined adherent culture conditions. Stem Cells. 2006;24:125–138. doi: 10.1634/stemcells.2004-0150. [DOI] [PubMed] [Google Scholar]
- Skaper SD. Neuronal growth-promoting and inhibitory cues in neuroprotection and neuroregeneration. Ann N Y Acad Sci. 2005;1053:376–385. doi: 10.1196/annals.1344.032. [DOI] [PubMed] [Google Scholar]
- Stoneham ET, Sanders EM, Sanyal M, Dumas TC. Rules of engagement: factors that regulate activity-dependent synaptic plasticity during neural network development. Biol Bull. 2010;219:81–99. doi: 10.1086/BBLv219n2p81. [DOI] [PubMed] [Google Scholar]
- Takai H, Katayama K, Uetsuka K, Nakayama H, Doi K. Distribution of N-methyl-D-aspartate receptors (NMDARs) in the developing rat brain. Exp Mol Pathol. 2003;75:89–94. doi: 10.1016/s0014-4800(03)00030-3. [DOI] [PubMed] [Google Scholar]
- Talavera K, Nilius B, Voets T. Neuronal TRP channels: thermometers, pathfinders and life-savers. Trends Neurosci. 2008;31:287–295. doi: 10.1016/j.tins.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Tallaksen-Greene SJ, Janiszewska A, Benton K, Ruprecht L, Albin RL. Lack of efficacy of NMDA receptor-NR2B selective antagonists in the R6/2 model of Huntington disease. Exp Neurol. 2010;225:402–407. doi: 10.1016/j.expneurol.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M. Distinct spatio-temporal distributions of the NMDA receptor channel subunit mRNAs in the brain. Ann N Y Acad Sci. 1993;707:463–466. doi: 10.1111/j.1749-6632.1993.tb38099.x. [DOI] [PubMed] [Google Scholar]
- Wu H, Xu J, Pang ZP, Ge W, Kim KJ, Blanchi B, Chen C, Sudhof TC, Sun YE. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13821–13826. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita Y, Sakurai T, Tanaka H, Kitagawa K, Colman DR, Shan W. N-cadherin mediates interaction between precursor cells in the subventricular zone and regulates further differentiation. J Neurosci Res. 2009;87:3331–3342. doi: 10.1002/jnr.22044. [DOI] [PubMed] [Google Scholar]
- Yen LH, Sibley JT, Constantine-Paton M. Fine-structural alterations and clustering of developing synapses after chronic treatments with low levels of NMDA. J Neurosci. 1993;13:4949–4960. doi: 10.1523/JNEUROSCI.13-11-04949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Arimura N, Kaibuchi K. Signaling networks in neuronal polarization. J Neurosci. 2006;26:10626–10630. doi: 10.1523/JNEUROSCI.3824-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Assey KS, Sturkie CD, West FD, Machacek DW, Stice SL. Glial cell line-derived neurotrophic factor enhances in vitro differentiation of mid-/hindbrain neural progenitor cells to dopaminergic-like neurons. J Neurosci Res. 2010;88:3222–3232. doi: 10.1002/jnr.22499. [DOI] [PubMed] [Google Scholar]


