Abstract
We report on the effectiveness of CID, HCD, and ETD for LC-FT MS/MS analysis of peptides using a tandem linear ion trap-Orbitrap mass spectrometer. A range of software tools and analysis parameters were employed to explore the use of CID, HCD, and ETD to identify peptides isolated from human blood plasma without the use of specific “enzyme rules”. In the evaluation of an FDR-controlled SEQUEST scoring method, the use of accurate masses for fragments increased the numbers of identified peptides (by ~50%) compared to the use of conventional low accuracy fragment mass information, and CID provided the largest contribution to the identified peptide datasets compared to HCD and ETD. The FDR-controlled Mascot scoring method provided significantly fewer peptide identifications than with SEQUEST (by 1.3–2.3 fold) at the same confidence levels, and CID, HCD, and ETD provided similar contributions to identified peptides. Evaluation of de novo sequencing and the UStags method for more intense fragment ions revealed that HCD afforded more sequence consecutive residues (e.g., ≥7 amino acids) than either CID or ETD. Both the FDR-controlled SEQUEST and Mascot scoring methods provided peptide datasets that were affected by the decoy database and mass tolerances applied (e.g., the identical peptides between the datasets could be limited to ~70%), while the UStags method provided the most consistent peptide datasets (>90% overlap) with extremely low (near zero) numbers of false positive identifications. The m/z ranges in which CID, HCD, and ETD contributed the largest number of peptide identifications were substantially overlapping. This work suggests that the three peptide ion fragmentation methods are complementary, and that maximizing the number of peptide identifications benefits significantly from a careful match with the informatics tools and methods applied. These results also suggest that the decoy strategy may inaccurately estimate identification FDRs.
Keywords: CID, HCD, ETD, FT MS/MS, FDR, protein UStags, de novo sequencing, peptides, peptidomic analysis, blood plasma
INTRODUCTION
Dissociation or fragmentation of protein and polypeptide ions is central to tandem mass spectrometry (MS/MS) analysis of intact proteins and their proteolytic peptide products. Advances in mass spectrometry instrumentation have enabled the integration of multiple fragmentation methods such as CID, HCD, and ETD (1–3) with high-precision mass measurements of the resultant fragment ions using Fourier transform mass analyzers (4,5) for improved characterization. For example, low energy CID fragmentation in ion traps, which is widely utilized for peptide analysis, has been complemented by the use of higher energy collision conditions (HCD).
A number of studies have investigated HCD, ETD, and CID for protein characterization (6–19). For example, HCD facilitated iTRAQ-based peptide quantification because of its ability for better detection of small reporter ions (6–8). ETD also has proved beneficial to iTRAQ-based quantification in some cases (9), as well as for the analysis of post-translational modifications, e.g., phosphorylation (10,11), glycosylation (12), ubiquitination (13), disulfides (14,15), and protein isoforms (e.g., histone) (16). However, for broad analysis of peptides, ETD tends to provide fewer peptide identifications than CID (17,18); alternating use of ETD and CID can improve sequence coverage, but affords significantly smaller improvements in the number of peptides identified (17). Similarly, HCD has been reported to be a minor contributor to increasing the number of peptide identifications in an alternating HCD-CID approach (7). The combined use of ETD and CID also has been reported, but is less effective for increasing the number of peptide identifications from complex proteomic samples compared to duplicate analyses performed using CID only (18). Although both ETD and CID have been applied to dissociate intact protein sequences (19,20), it remains unclear which is better in terms of number of identifications provided.
Both the effectiveness of the fragmentation methods and the data analysis tools are important for peptide/protein identification. A data-dependent decision tree derived for efficient acquisition of MS/MS spectra revealed that ETD is less effective for low charge state (CS) (e.g., +2) peptides, while CID is less effective for high CS (e.g., > +5) peptides when OMSSA was utilized (18). However, reports indicate that ETD can contribute significant identifications for low CS peptides compared to CID (17), while CID has also been effectively used for dissociation of CS > +10 (MW >30 kDa) protein ions (19). With regard to search engines, Mascot and SEQUEST are considered less effective for ETD spectra, which led to the development of an alternative low-resolution-based MS-GF algorithm (21). Compared to Mascot, the MS-GF algorithm better utilized the complementary information from CID and ETD and improved peptide identifications for analyses that made use of sequence specific enzymes (e.g., trypsin and Lys-N) (21). Yet in spite of efforts to date, the effectiveness remains uncertain of different approaches for identification from e.g., the use of non-specific enzymes, as well as the trade-offs associated with the use of CID, HCD, and ETD and combinations of these methods for improving peptide identification.
In this work, we investigated CID, ETD, and HCD performance as part of our efforts to develop approaches for more effectively studying aberrant protein degradomic activity associated with diseases, e.g., breast cancer (22–24). Considering potential preferences/biases of peptide identification methods, we investigated these fragmentation methods using a large range of peptide lengths and termini, and applied different database search and peptide validation methods with multiple parameters. Importantly, the collective results from this joint comparison of fragmentation performance and peptide identification methods are useful for determining which combinations can improve identification rates for peptidomic-degradomic analyses and proteomics applications in general.
METHODS
Samples
Approval for the study was obtained in accordance with federal regulations. A human blood plasma sample was purchased from Equitech-Bio Inc (Kerrville, TX). The peptides were isolated using affinity chromatography (IgY12 LC10 AC column, Agilent, Palo Alto, CA) and size exclusion chromatography (Superdex 200 10/300 GL SEC column, GE Healthcare, Piscataway, NJ) as described previously (22). Isolated plasma peptide samples were stored at −80°C prior to the LC-CID/ETD/HCD FT MS/MS measurements.
A bovine serum albumin (BSA) Glu-C digest used for optimization of CID, HCD, and HCD fragmentations of various CS peptides was prepared as follows: 100 μg BSA (Sigma, St Louis, MI) was dissolved in 150 μL 25 mM ammonium bicarbonate (pH 8), then reduced in 5 mM dithiothreitol at 60°C for 30 min followed by alkylation with 20 mM iodoacetamide at 37°C in dark for 30 min. The resultant mixture was diluted 10-fold with 25 mM ammonium bicarbonate, after which sequencing grade endoproteinase Glu-C (Roche Applied Science, Indianapolis, IN) was added at an enzyme/protein ratio of 1:100. The resulting mixture was incubated at 25°C for 3 h. Prior to MS analysis, a solvent mixture used for LC mobile phase A was added to the digest at a ratio of 1:9 (v/v), and the resultant solution was vortexed.
High-resolution LC separations
Isolated peptides were separated using high-resolution LC as described previously (22). Briefly, the column used with a 20 Kpsi LC system consisted of a 100 cm × 100 μm i.d. capillary column containing C4-bonded silica particles (Sepax Technologies, Inc. Newark, DE) (25). The sample (50 μg) was loaded onto the LC column and separated with a mobile phase gradient from mobile phase A (acetonitrile/H2O/acetic acid, 10:90:0.2, v/v/v) to B (acetonitrile/isopropyl alcohol/H2O/acetic acid/trifluoroacetic acid, 60:30:10:0.2:0.1, v/v/v/v/v) performed over 600 min.
CID, HCD, and ETD FT MS/MS measurements
LTQ-Orbitrap Velos mass spectrometers (Thermo Fisher Scientific, San Jose, CA) with CID, HCD, and ETD capability were employed for FT MS/MS analyses. The heated capillary temperature and spray voltage were held at 290°C and 2.0 kV, respectively. FT MS and FT MS/MS were obtained with AGC targets of 1×106 and 3×105, respectively, at 60K resolution and with 2 micro scans. A 400≤m/z≤ 2000 survey scan was followed by FT MS/MS of the three most intense ions from the survey scan (monoisotopic precursor selection not enabled). Each most intense ion (precursor) was fragmented sequentially with CID, ETD, and HCD prior to the next precursor selected. The fragmentation methods were optimized using a direct infusion of the BSA Glu-C digest at ~500 nL/min. For HCD and CID, a normalized collision energy from 30 to 35% was optimized with an activation time of 0.25 ms for CID and 0.10 ms for HCD; while for ETD, the reaction time from 50 to 500 ms was optimized for the instrument default CS 2 and with the supplementary activation enabled. The instrument default and the optimized CID, HCD, and ETD conditions (see Results below) were applied for fragmentation of the most intense ions from the survey scan with an isolation window of 6 m/z units and a minimal signal of 2000. Dynamic exclusion was enabled with no repeat counts, using a 3 m/z tolerance and a duration cycle of 5 min. Mass calibration was performed according to the method provided by the instrument manufacturer.
FT MS/MS data analysis
Two combined protein databases were constructed for identification of peptides in this work. Database 1 was generated by combining the IPI human protein database (ftp://ftp.ebi.ac.uk/pub/databases/IPI/, ipi.HUMAN.v3.39) with its reverse decoy database that was achieved by reversing each of the IPI database protein sequences, Database 2 was generated by combining the IPI database with its scrambled decoy database that was obtained by scrambling each of the IPI database protein sequences such that it maintained the same protein length and proportion of amino acid residue with a randomized sequence. Each combined protein database contained 139,462 entries (i.e., 69,731 entries from the IPI database plus the same number of entries from either the reversed or scrambled decoy databases).
Multiple mass tolerances were employed to optimize the search results from the experiments, with specific attention paid to particular scan types. The spectral parsing program Extract_MSn (version 5.0, Thermo Fisher Scientific) was used to create input files for the SEQUEST (version 27, revision 12, Thermo Fisher Scientific) tandem mass spectral database search. Extract_MSn utilizes the scan header information to apply the appropriate charge state and parent mass values for each MS/MS spectrum searched. As the species being studied may include polypeptides up to the small protein level, an upper mass tolerance of 25 KDa was allowed for SEQUEST input file creation. After input files were created, they were filtered by scan mode type (e.g., CID vs. HCD vs. ETD) using MSMS Spectra Preprocessor (http://omics.pnl.gov/software/MSMSSpectraPreprocessor.php) for the subsequent SEQUEST database search. The SEQUEST database search was completed using search modes of monoisotopic precursor tolerances from 5 Da to 50 ppm coupled with monoisotopic fragment ion tolerances from 1 to 0.05 Da. CID and HCD spectra employed b-type and y-type ions, while the ETD spectra employed c-type and z-type ions. No amino acid modifications were considered and no enzyme was specified. The top hit output for each spectrum was used for the next peptide validation. When peptide validation was performed using SEQUEST scores, the top hits were filtered with a relative correlation score ΔCn > 0.1 and the filtered hits were accepted as peptide identifications when their correlation scores (Xcorr) were higher than the thresholds that allowed generating a desired FDR value (26) for each charge state (i.e., the FDR-controlled SEQUEST scoring method).
Mascot analysis was completed on a local Mascot server (version 2.3.01, Matrix Science Inc., Boston, MA) configured with the combined databases described above. No enzyme rule was applied and neither static nor dynamic modifications were assumed. Peptide mass tolerances of 5 Da and 50 ppm and a fragment mass tolerance 0.05 Da were used for monoisotopic masses with a 13C option set to ‘2’ (allowing for correction of de-isotoping errors of 1 and 2 Da). CID and HCD spectra were searched using Instrument option ‘ESI-FTICR’, and ETD spectra were searched using Instrument option ‘ETD-Trap’. The peptide charge states were specified in the MGF input file which was created from the corresponding DTA files using an in-house built function DtaTextToMGFConverter (http://omics.pnl.gov/software/DtaToMGFConverter.php). The peptide-centric report was extracted using the built-in Export function without any cutoff applied (i.e., the counts inferred for Mascot represented its upper-limit). The database search top hit for each spectrum was used for peptide validation (achieved when the peptide score was higher than the threshold needed to obtain the desired FDR for each charge state; i.e., the FDR-controlled Mascot scoring method).
When unique sequence tags (UStags) (27) were used to validate peptides, the top ten hits output from the SEQUEST database search were considered putative peptide candidates. Peaks in CID and HCD MS/MS spectra were assigned as b or y fragments, and peaks in ETD MS/MS spectra as c or z fragments for each of peptide candidates, using ICR2LS (http://ncrr.pnl.gov/software/) with a mass tolerance of 10 ppm. Assigned fragment peaks were used to construct amino acid sequences, and the resultant sequences were searched against the combined databases. When the sequences were unique within the protein database, the candidates were considered validated as the identified peptides.
A de novo sequencing method (28) was used to investigate capabilities for generating consecutive fragments using CID, HCD, and ETD. Briefly, fragment ions in FT MS/MS spectra were first de-isotoped (29) and then only those fragments having ≥3 isotopic peaks (i.e., intense ions) were transformed to provide their neutral masses. Sequencing was completed using a mass tolerance of 0.005 Da, and the sequences were searched against the combined databases. The database hits were accepted as identified peptides when their molecular masses agreed with the precursors agreed to within 10 ppm.
The peptide datasets obtained are provided in Supporting Tables.
RESULTS
Optimization of CID, HCD, and HCD conditions for peptide fragmentation
The performance of an LTQ Orbitrap Velos mass spectrometer equipped with CID, HCD, and ETD was optimized using a BSA tryptic digest prior to LC-MS/MS analysis of plasma peptides. For CID and HCD, increasing the normalized collision energy from 30% to 35% at activation times of 0.25 ms and 0.10 ms (the instrument default values), respectively, had little influence on the resultant spectra for CS 2–4 BSA peptides. For ETD, the reaction time for the default CS 2 was >250 ms to achieve the effective fragmentation of CS 2–4 BSA peptides (Supporting Figure 1), significantly longer than the instrument default of 100 ms. We also evaluated the ETD reaction time using another LTQ-Orbitrap Velos mass spectrometer whose filament was cleaned prior to the experiment, and again observed that >250 ms reaction time was required for effective dissociation of CS 2–4 BSA peptides. Furthermore, when a 100 ms reaction time was used to analyze plasma peptides, most ETD spectra displayed poor fragmentation of precursors. Therefore, the spectral dataset acquired with the normalized collision energy of 35% for CID and HCD and 300 ms reaction time for ETD was used for the study of CID, HCD, and ETD performance below.
CID, HCD, and ETD FT MS/MS spectral dataset
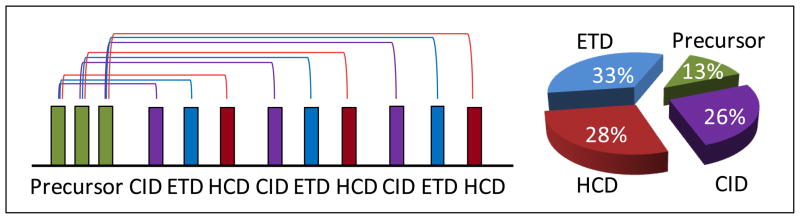
Figure 1 shows the data acquisition mode used to evaluate CID-, HCD-, and ETD-FT MS/MS performance for analyzing plasma peptides. In a 600-min LC-FT MS/MS experiment, 28,321 total spectra were acquired: 2836 FT MS spectra were acquired from precursor survey scans and 8495 FT MS/MS spectra for each CID, HCD, and ETD fragmentation methods. ETD required slightly longer acquisition time than HCD, and HCD a slightly longer time than CID to acquire the same number of spectra, as illustrated by Figure 1. All acquired spectra are in the public repository PRIDE (http://www.ebi.ac.uk/pride/).
Figure 1.
Utilization of CID, HCD, and ETD fragmentation methods for FT MS/MS analysis of peptides. Each precursor from the survey scan was successively fragmented by CID, HCD, and ETD prior to analysis of the next precursor. Acquisition time distributions are given for a 600-min high-resolution reverse-phased LC separation with CID-, HCD-, and ETD-FT MS/MS analysis. An Orbitrap Velos mass spectrometer was used for this analysis under the conditions described in the text.
CID, HCD, and ETD peptide datasets obtained with different peptide identification methods
Table 1 gives the numbers of peptides identified in all datasets for the different peptide identification methods and parameters. Two commercial database search engines (i.e., SEQUEST and Mascot) and a de novo sequencing method developed in house (28) were employed to search peptide ion fragment spectra. Peptides were validated using FDR-controlled scoring methods (score systems built into SEQUEST and Mascot), UStags, and sequence length. These methods are detailed in the Methods section and information for each of identified peptides is given in Supporting Tables.
Table 1.
The number of peptides identified from CID, HCD, and ETD spectral datasets with various fragmentation and peptide identification methods.*
| Methods | Number of peptides |
|---|---|
| SEQUEST | |
| [5Da, 1Da]_r | |
| CID | 494 |
| HCD | 438 |
| ETD | 186 |
| CID+HCD | 631 |
| CID+ETD | 560 |
| HCD+ETD | 516 |
| CID+HCD+ETD | 678 |
| [3Da, 1Da] _r | |
| CID | 473 |
| HCD | 386 |
| ETD | 194 |
| CID+HCD | 579 |
| CID+ETD | 561 |
| HCD+ETD | 494 |
| CID+HCD+ETD | 656 |
| [1Da, 1Da] _r | |
| CID | 551 |
| HCD | 479 |
| ETD | 214 |
| CID+HCD | 673 |
| CID+ETD | 631 |
| HCD+ETD | 564 |
| CID+HCD+ETD | 733 |
| [5Da, 0.05Da] _r | |
| CID | 749 |
| HCD | 665 |
| ETD | 439 |
| CID+HCD | 916 |
| CID+ETD | 895 |
| HCD+ETD | 809 |
| CID+HCD+ETD | 1012 |
| [3Da, 0.05Da] _r | |
| CID | 715 |
| HCD | 631 |
| ETD | 447 |
| CID+HCD | 869 |
| CID+ETD | 861 |
| HCD+ETD | 779 |
| CID+HCD+ETD | 961 |
| [50ppm, 0.05Da]_r | |
| CID | 639 |
| HCD | 529 |
| ETD | 416 |
| CID+HCD | 725 |
| CID+ETD | 763 |
| HCD+ETD | 676 |
| CID+HCD+ETD | 813 |
| [5Da, 05Da]_s | |
| CID | 725 |
| HCD | 618 |
| ETD | 428 |
| CID+HCD | 858 |
| CID+ETD | 849 |
| HCD+ETD | 755 |
| CID+HCD+ETD | 948 |
| [3Da, 05Da]_s | |
| CID | 703 |
| HCD | 621 |
| ETD | 422 |
| CID+HCD | 843 |
| CID+ETD | 825 |
| HCD+ETD | 753 |
| CID+HCD+ETD | 929 |
| [50ppm, 05Da]_s | |
| CID | 797 |
| HCD | 617 |
| ETD | 562 |
| CID+HCD | 896 |
| CID+ETD | 947 |
| HCD+ETD | 814 |
| CID+HCD+ETD | 1014 |
| Mascot | |
| [5Da, 0.05Da]_r | |
| CID | 324 |
| HCD | 329 |
| ETD | 276 |
| CID+HCD | 450 |
| CID+ETD | 449 |
| HCD+ETD | 441 |
| CID+HCD+ETD | 536 |
| [50ppm, 0.05Da]_r | |
| CID | 355 |
| HCD | 445 |
| ETD | 362 |
| CID+HCD | 538 |
| CID+ETD | 514 |
| HCD+ETD | 569 |
| CID+HCD+ETD | 635 |
| [5Da, 0.05Da]_s | |
| CID | 351 |
| HCD | 330 |
| ETD | 277 |
| CID+HCD | 451 |
| CID+ETD | 478 |
| HCD+ETD | 452 |
| CID+HCD+ETD | 550 |
| [50ppm, 0.05Da]_s | |
| CID | 404 |
| HCD | 413 |
| ETD | 319 |
| CID+HCD | 536 |
| CID+ETD | 519 |
| HCD+ETD | 524 |
| CID+HCD+ETD | 611 |
| UStags | |
| [5Da, 0.05Da]_r | |
| CID | 531 |
| HCD | 605 |
| ETD | 180 |
| CID+HCD | 765 |
| CID+ETD | 602 |
| HCD+ETD | 651 |
| CID+HCD+ETD | 797 |
| [50ppm, 0.05Da]_r | |
| CID | 455 |
| HCD | 555 |
| ETD | 176 |
| CID+HCD | 659 |
| CID+ETD | 518 |
| HCD+ETD | 596 |
| CID+HCD+ETD | 685 |
| [5Da, 0.05Da]_s | |
| CID | 518 |
| HCD | 600 |
| ETD | 187 |
| CID+HCD | 751 |
| CID+ETD | 585 |
| HCD+ETD | 647 |
| CID+HCD+ETD | 782 |
| [50ppm, 0.05Da]_s | |
| CID | 510 |
| HCD | 590 |
| ETD | 180 |
| CID+HCD | 733 |
| CID+ETD | 575 |
| HCD+ETD | 635 |
| CID+HCD+ETD | 762 |
| De novo sequencing | |
| CID | 206 |
| HCD | 314 |
| ETD | 39 |
| CID+HCD | 395 |
| CID+ETD | 233 |
| HCD+ETD | 332 |
| CID+HCD+ETD | 410 |
All SEQUEST and Mascot peptides were identified at 2 % FDR; UStags and de novo sequencing-identified peptides had 0 false positives estimated from decoy strategy; [xxx, yyy]_r (or s) represent that the precursor mass tolerance xxx and fragment mass tolerance yyy were used for searching against the reverse (r) or the scrambled (s) decoy database-combined protein database for peptide identification.
CID, HCD, and ETD contributions for peptide identification evaluated from the FDR-controlled SEQUEST scoring method
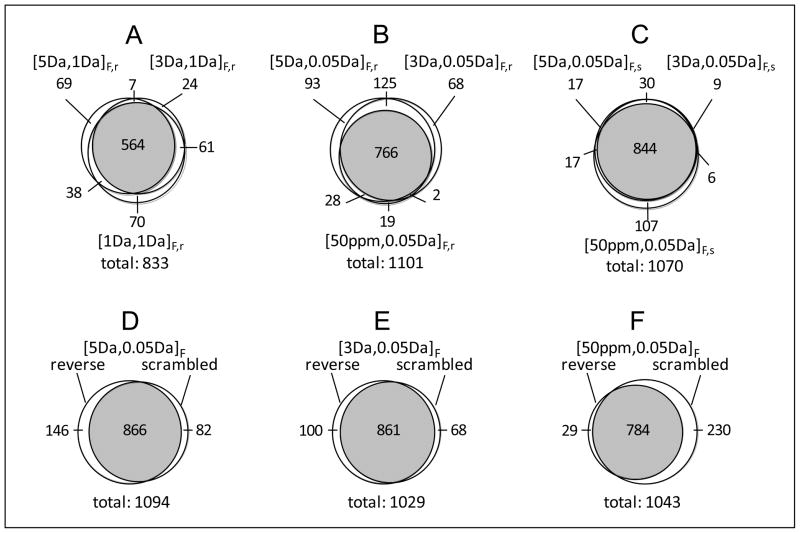
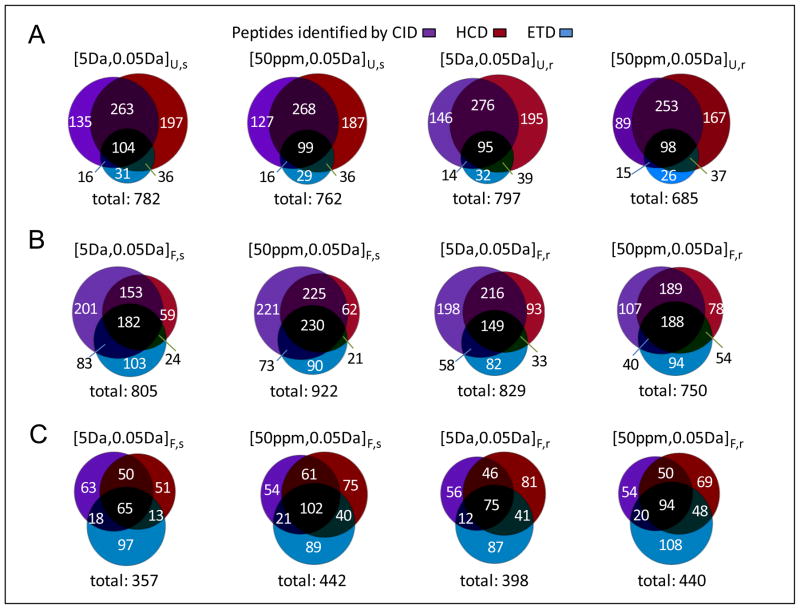
The contributions of CID, HCD, and ETD to peptide identifications were dependent on the mass tolerance (or mass accuracy), as well as on the decoy database used for the SEQUEST database search (Table 1). Based on the same FDR level, reducing the fragment mass tolerance from 1 Da (i.e., a mass tolerance typically used for analysis of conventional ion trap MS/MS spectra) to 0.05 Da (a mass tolerance that can be provided by FT MS/MS) resulted in a 49% increase in the total number of peptides identified from CID, HCD, and ETD spectra when Database 1 (i.e., forward plus reverse decoy database) was used for peptide identification. This increase shows that use of accurate MS/MS can significantly improve the peptide analysis coverage for a given number of spectra acquired. Depending on the combined database applied for peptide identification, reducing the precursor mass tolerance did not necessarily lead to an increase in the number of identified peptides. For example, reducing the precursor mass tolerance from 5 Da to 50 ppm resulted in a 20% increase in the number of peptides identified with Database 1, in contrast to a 7% decrease with Database 2 (i.e., forward plus scrambled decoy database). This example highlights that use of the same mass tolerances to search CID, HCD, and ETD spectra with different combined databases can result in significantly different numbers of total peptide identifications. The overlaps among peptide datasets identified with different mass tolerances and decoy databases were limited to 68–83% (Figure 2), although all peptide datasets were obtained at the same estimated FDR. Similar overlaps were observed for individual peptide subsets identified with the different decoy databases (Supporting Figure 2).
Figure 2.
The overlaps of peptide datasets identified from the SEQUEST method. (A)–(C) respectively present overlaps of peptide datasets obtained with use of 1 Da fragment mass tolerance for searching against combined database 1 (forward plus reverse database), with use of 0.05 Da fragment mass tolerance for searching against combined database 1, and with use of 0.05 Da fragment mass tolerance for searching against combined database 2 (forward plus scrambled database); (D)–(E) present overlaps between peptide datasets obtained with searching against combined database 1 (‘reverse’ labeled) and combined database 2 (‘scrambled’ labeled) using specific mass tolerances; subscripts F,r and F,s of mass tolerance respectively represent the peptide datasets identified from the FDR-controlled scoring method with searching against combined databases 1 and 2; all peptide identifications were achieved on a 2% FDR level. The significances of other symbols are the same as for Table 1.
Several factors affected CID, HCD, and ETD peptide datasets identified using the FDR-controlled SEQUEST scoring method. Table 2 lists some examples that depict the influence of database search mass tolerance on peptide identification. Peptides identified in one dataset using a specified mass tolerance and decoy database but not in another dataset using a different mass tolerance (e.g., 3 Da versus 50 ppm) and decoy database (e.g., forward sequences combined with reversed versus scrambled sequences), can be due to changes in ΔCn values (highlighted in yellow in the table), changes in the peptide CS, changes in peptide candidates with the same or different CS (highlighted in green and brown, respectively), and changes in the Xcorr threshold required by a specific FDR (highlighted in blue). Changes of ΔCn values for both the IPI protein (or ‘correct’) database hits and the decoy (or ‘incorrect’) database hits with precursor mass tolerances were predictable as these values were assigned from comparison of candidates added for the mass tolerance allowed. ΔCn>0.1 was specified for peptide identification in this work, and any changes to this criteria led to different peptide identifications. Additionally, erroneous peptide molecular mass values derived from the spectral parsing software package Extract_MSn due to incorrect precursor CS or monoisotope identification would be expected to result in variations among peptide candidates assigned to specific spectra. As a result detected peptides that had been identified with high scores (e.g., Xcorr>8) may be missed. The influence of this factor may potentially be reduced by de-isotoping the high-resolution FT MS/MS spectra prior to database searching (e.g., with tools such as Decon_MSn available at http://omics.pnl.gov/software/). Note that changes to the Xcorr threshold were required to identify peptides with a specific FDR. (All Xcorr thresholds required for 2% FDR are provided in Supporting Tables). This factor was inherent in the FDR-controlled scoring method and by selecting the same Xcorr threshold to filter database search hits output using different mass tolerances, which resulted in peptide datasets having different FDR values. For example, use of Xcorr>2.3 (ΔCn>0.1) to filter CID CS 2 peptides resulted in FDR values of 0% and 1.3% for searches against databases 1 and 2, respectively, with mass tolerances of 5 Da for precursors and 0.05 Da for fragments. The influence of the decoy database on peptide identification stemmed from comparing ‘correct’ and ‘incorrect’ database hits (e.g., the order of the hits and ΔCn scores) that were the basis for the FDR-controlled SEQUEST scoring method to validate peptides.
Table 2.
Examples showing influence of database search mass tolerance on peptide identification.*
| [1Da, 0.05Da]_r accepted peptide | [3Da, 0.05Da]_r rejected candidate | Xorr1 | Xcorr2 | ΔCn1 | ΔCn2 | CS1 | CS2 |
|---|---|---|---|---|---|---|---|
| A.VLPSPPTVPVIPVL.P | A.VLPSPPTVPVIPVL.P | 2.522 | 2.522 | 0.190 | 0.047 | 2 | 2 |
| I.WTKMADTNSVATVE.I | I.WTKMADTNSVATVE.I | 3.640 | 3.640 | 0.155 | 0.084 | 2 | 2 |
| E.TQISEDFVDIQTDI.E | E.TQISEDFVDIQTDI.E | 3.889 | 3.889 | 0.107 | 0.043 | 2 | 2 |
| G.FIHKIPGLVTLKKLPCVSFA.G | G.FIHKIPGLVTLKKLPCVSFA.G | 3.493 | 3.493 | 0.127 | 0.043 | 3 | 3 |
| L.SSRQLGLPGPPDVPDHAAYHPFRR.L | L.SSRQLGLPGPPDVPDHAAYHPFRR.L | 4.240 | 4.240 | 0.128 | 0.030 | 4 | 4 |
| R.HTFMGVVSL.G | R.HTFMGVVSL.G | 2.893 | 2.893 | 0.103 | 0.103 | 2 | 2 |
| K.ALGISPFHEHAE.V | K.ALGISPFHEHAE.V | 2.902 | 2.902 | 0.190 | 0.190 | 2 | 2 |
| D.TVTAPQKNLK.S | D.TVTAPQKNLK.S | 2.975 | 2.975 | 0.250 | 0.241 | 2 | 2 |
| H.IANVERVPFDAATLH.T | H.IANVERVPFDAATLH.T | 3.007 | 3.007 | 0.127 | 0.127 | 2 | 2 |
| A.DEREPTESTQQLNKPEVLEVTLNRPFLF.A | A.DEREPTESTQQLNKPEVLEVTLNRPFLF.A | 4.566 | 4.566 | 0.307 | 0.271 | 4 | 4 |
| [3Da, 0.05Da]_r accepted peptide | [50ppm, 0.05Da]_r rejected candidate | Xcorr1 | XCorr2 | ΔCn1 | ΔCn2 | CS1 | CS2 |
|---|---|---|---|---|---|---|---|
| S.DAFHKAFLEVNEEGSEAAASTAVVIAGR.S | I.GDKGECVITPSTDVKFDPGLKSKNKLN.Y | 8.589 | 2.651 | 0.568 | 0.012 | 3 | 3 |
| A.ALLSPYSYSTTAVVTNPKE.- | T.TIDASSIGIVQPELTLEQE.D | 7.412 | 3.076 | 0.421 | 0.034 | 3 | 3 |
| R.HTFMGVVSLGSPSGEVSHPR.K | R.LMYLEAMISGVAWVDIPSS.Y | 5.921 | 3.293 | 0.388 | 0.097 | 3 | 3 |
| R.EVQGFESATFLGYFK.S | T.QNITSFLFPNEQASK.I | 4.612 | 1.857 | 0.519 | 0.103 | 2 | 2 |
| H.GLTTEEEFVEGIYK.V | L.VGVSNDCLQYGLGYK.D | 5.414 | 2.161 | 0.494 | 0.034 | 2 | 2 |
| K.AFLEVNEEGSEAAASTAVVIAGR.S | T.ETEEEDDGMNDMN.H | 7.618 | 0.367 | 0.566 | 0.077 | 3 | 2 |
| T.FEYPSNAVEEVTQNNFRLL.F | H.BSVTFGSAACSLRLS.G | 7.394 | 0.205 | 0.560 | 0.905 | 3 | 2 |
| F.SPEKSKLPGIVAEGRDDLYVSDAFHK.A | K.DBBBIRALWELA.K | 6.955 | 0.289 | 0.434 | 0.000 | 4 | 2 |
| L.FGPDLKLVPPMEEDYPQFGSPK.- | F.ARAVPQTAAVCAVPHGN.G | 6.915 | 1.177 | 0.458 | 0.022 | 3 | 2 |
| R.TVVQPSVGAAAGPVVPPCPGRIRHFKV.- | S.LSVAKSLDPQQAN.S | 5.914 | 1.684 | 0.307 | 0.160 | 4 | 2 |
Peptide datasets [3Da, 0.05Da]_r, [1Da, 0.05Da]_r, and [50ppm, 0.05Da]_r showing in Table 1 were used for these examinations; terms “accepted peptide” and “rejected candidate” respectively represent the peptide identified and the candidate recommended from the database search, but rejected for identification for the indicated peptide dataset; Xcorr1 and Xcorr2, ΔCn1 and ΔCn2, and CS1 and CS2 respectively represent Xcorr, ΔCn, and CS for the accepted peptide and rejected candidate. Different color highlights indicate sources that affect peptide identification from the FDR-controlled SEQUEST scoring method (see descriptions in the text).
In general, CID provided the largest contribution to the FDR-controlled SEQUEST-identified peptide datasets, and ETD, the smallest (Table 1). However, mass tolerance and decoy database also influenced relative contributions of CID, HCD, and ETD to identified peptides. For example, HCD and ETD provided ~25% and ~13% additional (extra) identifications, respectively, compared to CID peptide subsets when 1 Da was used as the database search fragment mass tolerance, and ~20% and ~22% additional identifications, respectively, when 0.05 Da fragment mass tolerance was used for peptide identification. The estimated ETD relative contributions increased by >60% when the database search fragment mass tolerance of 1 Da, which typically has been applied for analysis of low-resolution ion trap MS/MS spectra, was reduced to 0.05 Da. The improvement in ETD performance was again observed when combined database 2 was used for peptide identification.
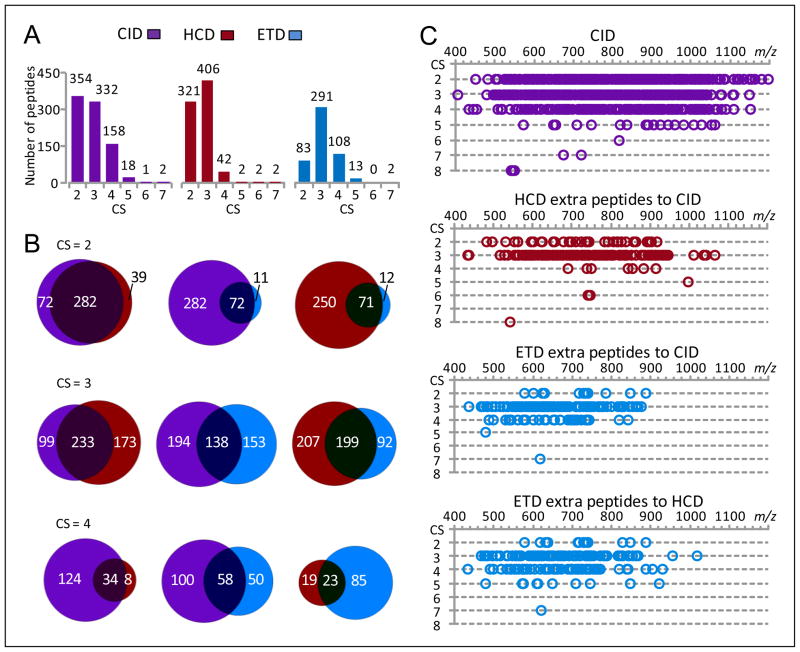
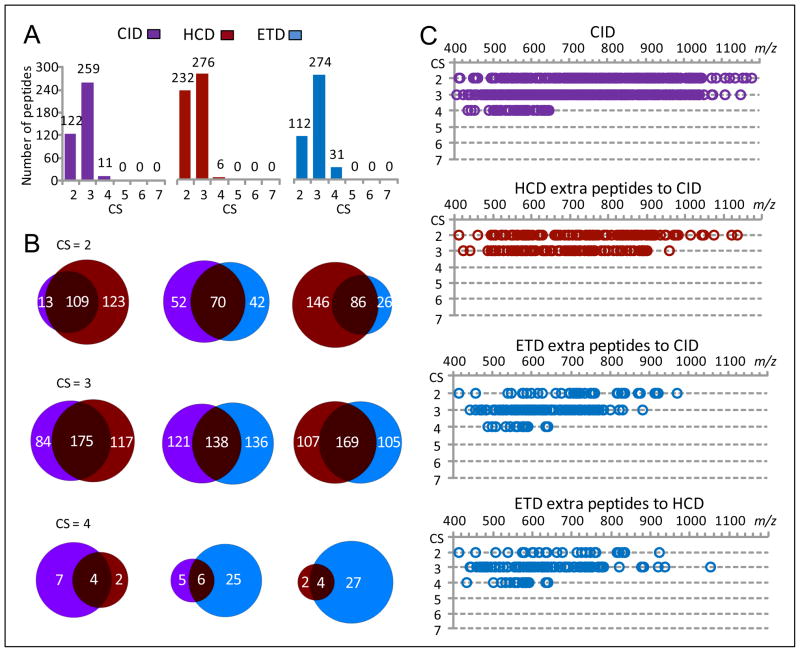
The CID, HCD and ETD contributions to the identification of various CS peptides using the FDR-controlled SEQUEST scoring method are shown in Figure 3. CID contributed more CS 2 and CS 4–5 peptides than HCD, and more CS 2–5 peptides than ETD (Figure 3A). The complementary role of HCD to CID was primarily for CS 3 peptides where additional identifications resulted, while ETD mainly provided additional CS 3 and 4 peptide identifications to the CID peptide subsets (Figure 3B). CID peptides were identified in an m/z range of 400–1200 (Figure 3C), which included the small m/z ranges where HCD and ETD contributions were most significant (e.g., 400–950 and 400–880). The m/z range for ETD contributions narrowed with increased peptide CS (e.g., 400–880 for CS 2 peptides and 450–750 for CS 3 peptides).
Figure 3.
The CID, HCD, and ETD contributions for identification of various charge state SEQUEST peptides. The peptide dataset [5Da, 0.05Da]F,r shown in Table 1 was used for this examination. (A) The numbers of various CS SEQUEST peptides identified from CID, HCD, and ETD spectra, (B) the overlaps of CS 2–4 peptides identified from CID, HCD, and ETD spectra, and (C) the m/z distributions of CS 2–8 peptides identified from CID spectra and extra peptides contributed from different fragmentation methods (peptides are plotted as a function of charge state and m/z).
CID, HCD, and ETD contributions for peptide identification evaluated from the FDR-controlled Mascot scoring method
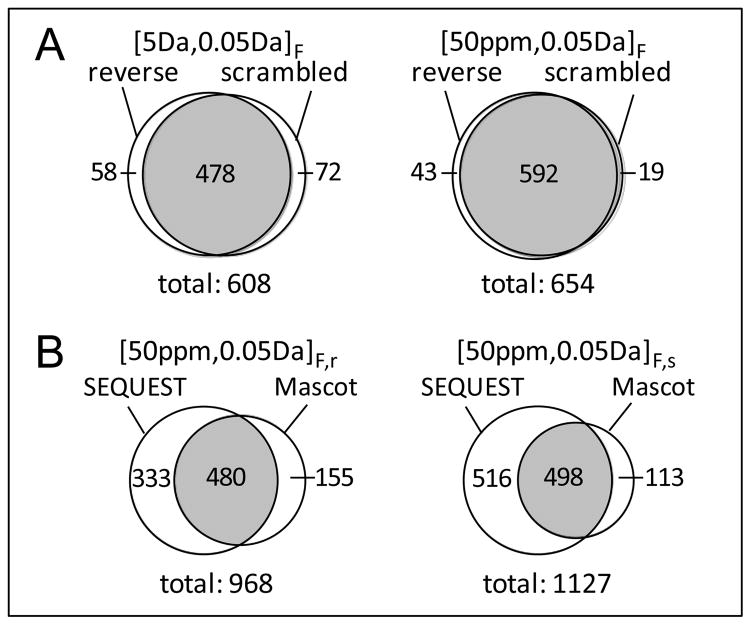
Mascot was examined for identification of peptides carrying multiple charges (see data shown in Figure 3) and having multiple terminal specificities (shown below) from CID, HCD, and ETD spectra with different mass tolerances and decoy databases (Table 1). The ratio of CID, HCD, and ETD peptide contributions obtained using the FDR-controlled Mascot scoring method changed from 1.0:1.3:1.0 to 1.0:0.9:0.8 when different mass tolerances and decoy databases were used for peptide identification. Note that any of the three fragmentation methods alone can contribute >10% identifications to the datasets. Reducing the database search precursor mass tolerance from 5 Da to 50 ppm led to an increase of 10–20% in peptide dataset size and the contents of the peptide datasets obtained were observed to vary with the use of different decoy databases (Figure 4A). Most significantly, the number of peptides identified with Mascot was only ~60% of that obtained with SEQUEST (Figure 4B) even though all of the peptide datasets were identified using the same low FDR (estimated from the decoy strategy).
Figure 4.
The overlaps of peptide datasets identified from the Mascot method. (A) The overlaps of Mascot peptide datasets identified with searching against different combined databases and (B) the overlaps of Mascot and SEQUEST peptide datasets identified with specific mass tolerances. The significances of symbols are the same as for Figure 2.
Contrary to SEQUEST, Mascot favored HCD for identification of CS 2 peptides and ETD for identification of CS 4 peptides (Figure 5). However, Mascot identified only a few CS ≥5 peptides from CID, HCD, and ETD spectra. For CS 3 peptides, CID, HCD, and ETD provided comparable numbers of identifications. Similar to the SEQUEST evaluation above, m/z ranges where CID, HCD, and ETD contributed CS 2–4 peptides were highly overlapped when using Mascot.
Figure 5.
The CID, HCD, and ETD contributions for identification of various charge state Mascot peptides. The peptide dataset [5Da, 0.05Da]F,r shown in Table 1 was used for this examination. (A) The numbers of various CS Mascot peptides identified from CID, HCD, and ETD spectra, (B) the overlaps of CS 2–4 peptides identified from CID, HCD, and ETD spectra, and (C) the m/z distributions of CS 2–7 peptides identified from CID spectra and extra peptides contributed from different fragmentation methods (peptides are plotted as a function of charge state and m/z).
CID, HCD, and ETD contributions for peptide identification evaluated from the UStags method
The UStags method identifies peptides based on analysis of long (typically ≥7 amino acids) peptide sequences (24). Table 3 gives the decoy database peptide (i.e., false) hits that match the most fragments and longest sequences observed during sequence analysis to search for UStags. The 6-residue sequences were the longest sequences that matched to the decoy databases, and these sequences were contributed by either CID or HCD, but not ETD. In total, only one 6-residue sequence from the reverse decoy database and two 6-residue sequences from the scrambled decoy database were matched. The 6-residue decoy sequence matched to the reverse database was the subsequence of a 7-residue decoy peptide A.LALDLFK.C. By using a 2.3 ppm mass measurement error for the precursor, all 7 residues in this decoy peptide could be determined. Because this decoy peptide has the same mass sequence as the IPI database peptide R.IAIDLFK.H (with the exchange of Ile for Leu), it was excluded based on its lack of uniqueness in the database, an important UStags requirement (27). The same situation was observed for the two decoy peptides from the scrambled database. The rest of the listed sequences contained ≤5 residues and were automatically excluded as UStags. Thus, the UStags approach rejected all false hits from the decoy databases, i.e., effectively validating peptides with 0% false positives. Table 3 also shows that use of a limited number (e.g., 3–5) of fragments or short sequences (e.g., ≤5 residues) in combination with molecular mass measurements for peptide identification, as suggested previously for the ‘peptide sequence tags’ concept (30,31), unavoidably incurs in matching either CID, or HCD, or ETD FT MS/MS spectra to decoy database peptides and results in false peptide identifications. This situation becomes more problematic with reduced fragment mass measurement accuracies.
Table 3.
Examples of decoy database peptides matched during spectra SEQUEST database search.*
| Reverse decoy database peptide | PME (ppm) | Fragment ion | Sequence | Fragment ion | Sequence |
|---|---|---|---|---|---|
| F.QLDLRMTSTSPSGPRVATQAGGP.L | −2.18 | y10 y11 y12 y13 y14 | SPSG | ||
| G.LSAELNHI.S | 3.06 | y2 y3 y4 y5 y6 y7 | SAELN | ||
| G.RLDVAVVHFR.S | 2.55 | y2 y4 y5 y6 y7 | VAV | ||
| K.IHLFFIHRTQEQLILHPEVFFK.V | 0.25 | b10 b11 b12 b13 | EQL | ||
| L.LAPLAVDTVSSPV.N | −1.52 | b4 b5 b10 b13 | y8 y9 y10 y11 y12 | APLA | |
| L.LFRSINFLTEPRHVSFPDE.T | 0.32 | b5 b6 b7 b8 b9 b10 | NFLTE | ||
| T.QTPEEMAELWGKPPRTQRLL.V | 2.86 | b6 b7 b8 b9 | MAEL | y11 y12 y13 y14 y15 | MAEL |
| A.LALDLFK.C | 2.33 | b2 b4 | y1 y2 y3 y4 y5 y6 | ALDLFK | |
| H.AKLPLPCKAVASARVKGGG.C | 1.58 | b2 b3 | y9 y10 y15 y16 y17 y18 y19 | AKLP | |
| I.VHGLVYVLSLYIITKF.K | 2.15 | b5 b8 b9 b10 b11 | SLY | ||
| K.LAYAQAALASEDVRMK.K | −6.02 | b2 b3 b4 b12 b13 b16 | YA | ||
| L.LAPLAVDTVSSPV.N | −1.52 | b2 b3 b4 | PL | y8 y9 y10 y11 y12 | APLA |
| R.FATSEELHEVIEGCSYIDQIY.K | 2.74 | y1 y2 y3 y17 y19 | QIY | ||
| R.GFARWDVVCISLP.P | 0.99 | b9 b10 b11 | IS | y2 y3 y4 | IS |
| D.DYGALLLRGSKSFAV.S | −2.64 | c6 c7 c8 c10 | LR | ||
| D.SWNVCVVSL.N | 4.24 | c4 c5 c6 c7 c8 | CVVS | ||
| Q.LVQSRLQRAISALRGL.R | −4.04 | c8 c9 c10 | AI | ||
| S.ATALILPSRGESDIK.N | 7.63 | c8 c13 c14 c15 | IK | ||
| T.REEDATQANSVARL.Q | 9.00 | c8 c9 c10 c11 c12 | NSVA |
| Scrambled decoy database peptide | PME (ppm) | Fragment ion | Sequence | Fragment ion | Sequence |
|---|---|---|---|---|---|
| E.ALDFIK.R | 2.64 | b2 b3 b4 b5 b6 | DFIK | y3 y4 y5 | LD |
| E.ASRPMIDELFLISPL.D | 1.26 | b10 b11 b12 b13 b14 b15 | LISPL | ||
| F.IAHEEVKNMNKTSVGSP.M | 1.53 | y11 y12 y13 y14 y15 y16 | AHEEV | ||
| I.ALDFLK.Q | 2.64 | b2 b3 b4 b5 b6 | DFLK | y3 y4 y5 | LD |
| K.VSSVLSLLVEMCIIPRSE.K | 0.19 | b5 b6 b7 b8 | SLL | y9 y10 y11 y12 y13 | SLLV |
| L.NAIPYTQQLMLNPHLTEHTCGDFLPK.D | 1.74 | b10 b12 b24 | y14 y15 y16 y17 y18 | LMLN | |
| Q.ALDIFK.S | 2.64 | b2 b3 b4 b5 b6 | DIFK | y2 y3 y4 y5 | LDI |
| Y.LADLFK.H | 2.64 | b2 b3 b4 b5 b6 | DLFK | y2 y3 y4 | DL |
| D.EKNDHLLSSPNSGANSR.K | 0.30 | y7 y8 y9 y10 y11 y12 | LLSSP | ||
| K.PVGMNFAATWEAAASLIGKREA.I | −1.53 | y3 y5 y7 y8 y9 y10 y11 y12 | EAAAS | ||
| K.VSSVLSLLVEMCIIPRSE.K | 0.19 | b2 b4 b5 b6 b13 | LS | y3 y9 y10 y11 y12 y13 y15 | SLLV |
| R.IALLDFK.C | 2.33 | y1 y2 y3 y4 y5 y6 | ALLDFK | ||
| R.PYSYRRFHSHGIH.L | 1.95 | b2 b3 b4 b13 | SY | y9 y10 y11 y12 y13 | PYSY |
| R.QAGLNLARLCTSEELV.D | 2.09 | b1 b3 b4 b5 b6 b7 b16 | LNLA | y9 y11 y12 y13 | LN |
| Y.QFDKIASNVVNT.P | 3.30 | b1 b2 b3 b4 b5 b6 b11 | QFDKIA | ||
| A.ALRSAMRGEQLQK.Q | 4.01 | c3 c4 c5 c9 c13 | SA | z8 z9 z10 z12 z13 | SA |
| A.RQLGYATPCVAEVVFVAD.W | −0.76 | c10 c11 c12 c13 c14 c15 | AEVVF | ||
| G.ASAQEIGALLGLQAL.Q | −0.05 | c14 c15 | z5 z11 z12 z13 z14 z15 | ASAQ | |
| K.FRLEFKDEK.L | 3.89 | c2 c3 c4 c8 | LE | z5 z6 z7 z8 z9 | FRLE |
| P.GEVRLSRSKQVLLNFPEATR.V | −3.27 | z9 z10 z11 z12 z13 z16 z20 | SKQV | ||
| P.MALSQLKSQLELIK.R | 8.65 | c8 c9 c10 c11 c12 c13 c14 | QLELIK | z3 z4 z5 z6 z11 z12 z14 | QLE |
| R.GGEAASGPYPAPAGTSAAR.E | −3.01 | c13 c14 c15 c16 c17 | GTSA |
Both precursor and fragment masses were measured within a mass error of 10 ppm; PME: precursor mass error; yellow, blue, and pink respectively highlights the decoy peptides matched from CID, HCD, and ETD spectra.
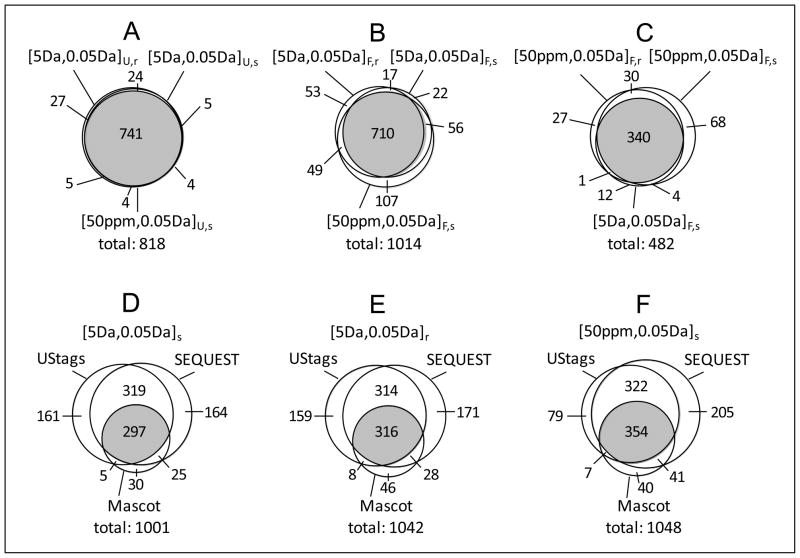
Given that results shown in Table 1 reveal SEQUEST could provide more potentially correct peptides than Mascot, peptide candidates output from the SEQUEST database search were used to search for UStags. Figure 6 compares the three methods with regard to identification of CID, HCD, and ETD peptides. The two scoring methods were controlled to produce 0% FDR so that the comparison was performed at the same peptide identification confidence levels. For the UStags method, precursor and fragment mass tolerances of [5Da, 0.05Da] consistently provided slightly (e.g., 2–5%) larger peptide datasets than [50ppm, 0.05Da], regardless of the decoy database applied for peptide identification (Figure 6A). This finding differs from the peptide datasets identified using the SEQUEST and Mascot scoring methods whereby dataset size varied based on changes to the database search precursor mass tolerance and the type of decoy database (Figures 6B and 6C).
Figure 6.
The CID, HCD, and ETD peptide datasets identified from the UStags method and comparisons with peptide datasets obtained from the SEQUEST and Mascot methods. (A)–(C) respectively represent the UStags peptide datasets, the SEQUEST peptide datasets (with 0% FDR), and the Mascot peptide datasets (with 0% FDR). Subscripts U,r and U,s of mass tolerances respectively represent the peptide datasets identified from the UStags method with searching against combined databases 1 and 2; the significances of other symbols are the same as for Figure 2.
Overall, the UStags-identified peptide datasets were 3–4% smaller than the SEQUEST identified peptide datasets, but >55% larger than the Mascot identified peptide datasets. The ratio of CID, HCD, and ETD contributions to the UStags-derived peptide datasets was approximately 1.0:1.2:0.4, regardless of the mass tolerance and the decoy database used. HCD was the largest single contributor to peptide identifications, although both CID and HCD contributed significant numbers of additional peptides to each other’s dataset (e.g., by 25–45%). This observation suggests that the combination of these two fragmentation methods should prove beneficial in maximizing the number of UStag peptide identifications. On the other hand, ETD had only a minor role in improving the number of peptides identified with the UStags method, with contributions of ~10% additional peptides to either CID or HCD peptide subsets. In contrast to the consistency of CID, HCD, and ETD contributions using the UStags method, their contributions varied for both SEQUEST and Mascot evaluations. For example, CID was the major contributor for SEQUEST-derived peptide datasets, and HCD and ETD could contribute additional 10–40% peptides to the CID subsets, whereas CID and HCD were the two major contributors to the Mascot peptide datasets, with ETD providing additional 10–20% peptides to either CID or HCD subsets. These results show significantly different CID, HCD, and ETD effectiveness resulting when different peptide identification approaches are used.
An analysis of peptide overlaps (Figure 7) reveals that the UStags method can provide practically identical peptide datasets (e.g., with 91% peptide overlaps) regardless of decoy database and database search mass tolerance applied during the search for initial putative peptide candidates (Figure 7A). The SEQUEST and Mascot scoring methods provided more variable or less stable peptide datasets (e.g., with ~70% peptide overlaps) that were sensitive to changes of the decoy database and database search mass tolerance (Figures 7B and 7C). In addition, the UStags and SEQUEST methods identified ~80% identical peptides and provided peptide datasets covering ~85% of the peptides obtained from the Mascot method compared to the Mascot method, which covered only ~40% of the peptides identified with either the UStags or SEQUEST methods (Figures 7D–7F).
Figure 7.
The overlaps of peptide datasets identified with the UStags, SEQUEST, and Mascot methods. (A)–(C) respectively represent overlaps of UStags peptide datasets, SEQUEST peptide datasets, and Mascot peptide datasets; (D)–(E) represent overlaps among UStags, SEQUEST, and Mascot peptide datasets identified with specific mass tolerances. The peptide datasets shown in Figure 6 are used for this examination.
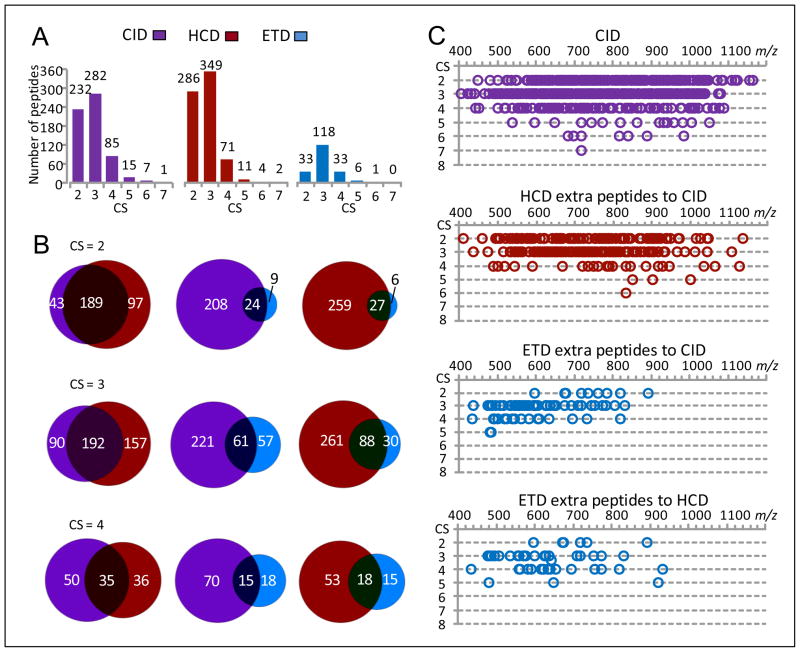
CID, HCD and ETD contributions towards identification of various CS peptides with the UStags method are shown in Figure 8. The advantage of HCD over CID is apparent for identification of CS 2 and 3 peptides. CID and HCD provided 25–50% additional identifications to each other’s CS ≥3 peptide subsets, revealing the significantly complementary role of these two fragmentation methods for identification of these peptides. ETD provided only a limited number of extra peptides. Similar to observations in Figures 3 and 5, m/z ranges where HCD and ETD made additional contributions towards peptide identifications were covered by ranges where CID performed well for the UStags peptide identification.
Figure 8.
The CID, HCD, and ETD contributions for identification of various charge state UStags peptides. The peptide dataset [5Da, 0.05Da]U,r shown in Figure 6A was used for this examination. (A) The numbers of various CS UStags peptides identified from CID, HCD, and ETD spectra, (B) the overlaps of CS 2–4 peptides identified from CID, HCD, and ETD spectra, and (C) the m/z distributions of CS 2–7 peptides identified from CID spectra and extra peptides contributed from different fragmentation methods (peptides are plotted as a function of charge state and m/z).
CID, HCD, and ETD contributions for de novo sequencing peptide identification
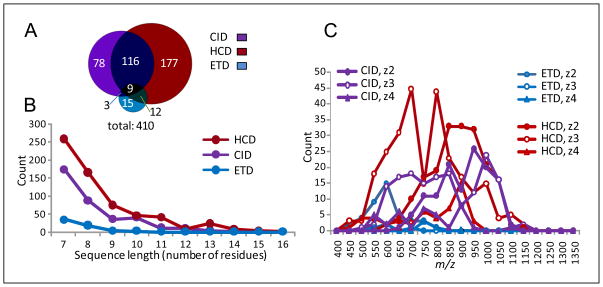
CID, HCD, and ETD spectral datasets were de novo sequenced using intense ions in the FT MS/MS spectra, and peptides were identified using sequences that had ≥7 consecutive residues sequenced and the molecular masses agreed with the precursors (i.e., <10 ppm mass tolerance; see Methods section). As shown in Table 3, use of ≥7-residue sequences for peptide identification greatly limits the number of false positives. Figure 9 shows CID, HCD, and ETD contributions to identified peptides. The ratio of CID, HCD, and ETD contributions was 1.0:1.5:0.2, indicating that HCD was better than CID, and that CID was significantly better than ETD for the de novo sequencing peptide identification (Figure 9A). HCD and CID contributed significant numbers of additional peptides to each other’s peptide subsets; however, ETD played only a minor role in contributing extra peptides to the CID and HCD subsets as it provided only a few ≥7-residue sequences. Most (91%) of de novo sequencing-identified peptides were covered by the UStags-identified peptide dataset, and a small fraction of the de novo sequencing-identified peptides were excluded from the UStags-identified dataset due to the lack of unique sequences required by the UStag method for unambiguous identification of peptides (27), even though the lengths of these sequences were ≥7 residues. The de novo sequencing method provided comparable numbers of peptide identifications to the Mascot method shown in Figure 6C, but the peptide overlap between the two methods was only ~60%.
Figure 9.
The CID, HCD, and ETD contributions for identification of various charge state peptides from the de novo sequencing method. (A) The CID, HCD, and ETD contributions to the de novo sequencing-identified peptide dataset, (B) the CID, HCD, and ETD capabilities to produce various lengths of sequences, and (C) the m/z distributions of CS 2–4 peptides identified from CID, HCD, and ETD spectra.
On average, HCD provided ~1 more residue than CID for sequences (Figure 9B). HCD performed best in an m/z range of 500–950, and HCD and CID became equivalently effective in the m/z range of 950–1000 (Figure 9C).
The physicochemical properties of peptides examined in this work
We further examined the GRAVY and pI values for the blood plasma peptidome peptides identified in this work using SEQUEST scoring, Mascot scoring, and UStags methods (Supporting Figure 3). These three different peptide identification methods provided no significant biases for peptide GRAVY and pI values, and the peptidome peptides identified herein had GRAVY and pI distributions similar to those observed for tryptic peptides in bottom-up proteomics analysis (32).
The MWs of identified peptidome peptides were distributed across 700–5500 Da (Supporting Figure 4A), which is broader than that for proteome tryptic peptides (32), and this distribution of peptidome peptides accounts for an overwhelming number of existing CS≥3 peptides (Figures 3, 5, and 8). Both SEQUEST and UStags methods were more effective than Mascot for identification of MW>2500 Da peptides. The identified peptides had multiple terminal cleavage specificities (Supporting Figure 4B), which excluded application of “enzyme rules” for spectral database searching and peptide validation. Nonetheless, a subset of the peptides identified in this study match typical tryptic cleavage patterns (e.g., Lys and/or Arg at peptide P1 position). Results (Supporting Figure 5) show an increased overlap between SEQUEST and Mascot identifications, while the ratios of CID, HCD, and ETD contributions to the peptide datasets remained similar to the overall observations described above.
DISCUSSION
Implementation of CID, HCD, and ETD fragmentation methods in combination with accurate FT MS/MS measurements in a single mass spectrometer provides opportunities for more confident and effective identification of peptides and will be increasingly applied for proteomics, peptidomics and other applications. In this work, we evaluated CID, HCD, and ETD FT MS/MS performance in conjunction with different identification approaches, including FDR-controlled SEQUEST and Mascot scoring methods, the UStags method, and a de novo sequencing method. Moreover, the peptides in this work stemmed from samples in which intracellular/intercellular proteases can generate peptides having multiple cleavage specificities. All of the methods examined are capable of identifying such peptides at low (e.g., 0–2%) FDR levels with no specific enzyme rules applied, however the size of resulting peptide datasets can be significantly different. For example, the number of peptides identified using Mascot was only 40–60% of that obtained using SEQUEST, an observation we attribute to a significant number of false negative identifications in the Mascot peptide datasets. Many peptides rejected by the Mascot method had molecular masses in agreement with the measured precursors (e.g., within 10 ppm mass errors) and had unique sequence tags, typically with ≥7 residues, similar to the criteria we apply with the USTags method. Mascot may be more effective for well defined small peptides, e.g., tryptic peptides that have specific cleavages and mostly carry two charges.
The overall effectiveness of CID, HCD, and ETD capabilities are by necessity intertwined with the methods and parameters used in their evaluation. Results obtained using the FDR-controlled SEQUEST scoring method indicated conventional CID fragmentation best as it provided 30–70 % more peptide identifications than HCD and ETD. However, results from the FDR-controlled Mascot scoring method indicated CID, ETD, and HCD were comparable, identifying similar numbers of peptides. Using the UStags method for peptide identification, HCD provided more peptide identifications than CID and ETD. Overall, these results suggest that CID generates more total fragment ions and provides higher SEQUEST scores (33), and thus more SEQUEST-identified peptides. In contrast, HCD was typically observed to produce more intense fragment ions that favor constructing fragment ladders that would provide more UStags-identified peptides, as well as better matches using our de novo sequencing method. Additionally, results show ETD as a relatively minor contributor to broad peptide identification when using FDR-controlled SEQUEST scoring, UStags, and de novo sequencing methods, while Mascot showed a slight improvement with ETD data. Thus, the use of only one specific peptide identification tool and parameter set provide biased conclusions regarding CID, HCD, and ETD fragmentation performance.
To some extent CID, HCD, and ETD each contributed unique peptides, varying with the peptide identification methods used. The use of all fragmentation methods undoubtedly resulted in improvements in coverage (Table 1); however, such improvements required ~3-fold increased analysis time (Figure 1). Alternating the selection of different fragmentation methods to acquire MS/MS data according to the precursor m/z values (e.g., the decision tree approach (18)), can reduce analysis time, but to some extent will miss peptides provided solely by a single fragmentation method, as the m/z ranges where CID, HCD and ETD made individual peptide contributions highly overlapped (Figures 3, 5, and 8). Thus to maximize peptide identifications, all three fragmentation methods should be applied if sufficient analysis time is available to also minimize “under-sampling” during MS/MS (e.g., 600 min used to identify ~1000 plasma peptidome peptides in this work). Selecting complementary fragmentation methods, e.g., CID and HCD for all precursors and then ETD for CS≥3, m/z < 650 precursors, can increase peptide identifications with only a modest decrease in analysis throughput. High-throughput FT MS/MS analysis can be achieved by selecting one or two alternating fragmentation methods that match well to the peptide identification method applied. We note that for the three fragmentation methods currently available, challenges still remain as we found a number of intense precursors which were not identified due to limited numbers of fragments observed under any fragmentation method. Additionally, we point out that the utility of CID, HCD, and ETD may vary for different applications; their effectiveness for modified peptides will be the subject of a future report.
The use of different peptide identification methods (including different database search tools, mass tolerances, and decoy databases) to evaluate CID, HCD, and ETD performances revealed significant variation for peptide datasets identified by controlling FDR to attain a specific confidence level (see results Figures 2, 4 and 7). This finding highlights a concern for using an FDR decoy search strategy for peptide identification; i.e., different sets of peptides will be identified at the same low FDR level from the same set of spectra. Such inconsistency among peptide datasets raises questions concerning the accuracy of the FDR evaluation approach for degradomic-peptidomic analysis. Examination of the peptide datasets identified in this study using SEQUEST and 2% FDR provided ~9% incorrect peptides (i.e., molecular mass errors of ≫10 ppm; details will be reported elsewhere). This issue is of practical importance for both proteomic and peptidomic analyses, and especially for degradomic studies (24). The present work shows peptide identifications obtained from UStags through sequencing of individual residues results in more consistent peptide datasets when different mass tolerances or decoy databases are employed. This is as opposed to the iterative and cumbersome FDR-controlled SEQUEST and Mascot scoring methods, which result in larger variations in peptide dataset contents given similar search conditions. Here, FDR is an outcome of UStag peptide identification, rather than a variable used to control peptide identification. These characteristics of the UStags method make it much more tolerant to variables, including peptide identification cutoffs, database search comparison (relative) scores, and any decoy database used for peptide identification. The peptides advanced by UStags contained unique sequences that can unambiguously exclude other peptides (27), essential for degradomic-peptidomic analysis so as to confidently identify individual peptides.
We also note that the mass measurement accuracy achievable with Orbitrap mass analyzers (i.e., <10 ppm mass errors) was not fully utilized for this work. We are now exploring better utilization of such quality mass accuracy with de-convoluting of the high-resolution FT spectra, which is not feasible using the commercially available Extract_MSn program applied in this work. Initial results obtained using a software tool developed in house to deconvolute the high-resolution FT spectra for application of high-precision measurements (e.g., 2.5–10 ppm mass errors) and validate peptides have shown some improvements in peptide analysis coverage and dataset consistency (details will be reported elsewhere). In addition to mass measurement accuracy, spectral database search tools play an important role in improving both coverage and dataset consistency by providing better (more complete and correct) initial putative candidates for peptide identifications with either scoring or sequence analysis (e.g., UStags) methods. The peptide probability-based MSGF-DB (21) algorithm may have significant potential for this purpose but requires modifications to address datasets without application of enzyme rules.
Finally, we note the utility of applying multiple dissociation methods in conjunction with extended LC separations to establish optimal sets of highly confident peptide identifications without bias stemming from the use of a specific peptide fragmentation method. These confident peptide identifications are the key to obtaining high quality identifications in high throughput LC-MS-based measurement approaches, such as using AMT tags.
Supplementary Material
Acknowledgments
This research was supported by the NIH National Center for Research Resources (RR18522). Work was performed in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE/BER) national scientific user facility located on the campus of Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle for the DOE under contract DE-AC05-76RLO-1830.
Abbreviations
- CID
collision induced dissociation
- HCD
high energy collision dissociation
- ETD
electron transfer dissociation
- FT MS/MS
Fourier transform tandem mass spectrometry
- UStags
unique sequence tags
- FDR
false discovery rate
- CS
charge state(s)
Footnotes
Supporting Figures and Tables are included with this manuscript.
References
- 1.Hunt EF, Yates JR, III, Shabanowitz J, Winston S, Hauer CR. Protein sequencing by tandem mass spectrometry. Proc Natl Acad Sci USA. 1986;84:620–623. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. High-energy dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 3.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal Chem. 2009;81:7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köcher T, Pichler P, Schutzbier M, Stingl C, Kaul A, Teucher N, Hasenfuss G, Penninger JM, Mechtler K. High precision quantitative proteomics using iTRAQ an an LTQ Orbitrap: a new mass spectrometric method combining the benefits of all. J Proteome Res. 2009;8:4743–4752. doi: 10.1021/pr900451u. [DOI] [PubMed] [Google Scholar]
- 7.Dayon L, Pasquarello C, Hoogland C, Sanchez J-C, Scherl A. Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags. J Proteomics. 2010;73:769–777. doi: 10.1016/j.jprot.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 8.McAlister GC, Phanstiel D, Wenger CD, Lee MV, Coon JJ. Analysis of tandem mass spectra by FTMS for improved large-scale proteomics with superior protein quantification. Anal Chem. 2010;82:316–322. doi: 10.1021/ac902005s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phanstiel D, Zhang Y, Marto JA, Coon JJ. Peptide and protein quantification using iTRAQ with electron transfer dissociation. J Am Soc Mass Spectrom. 2008;19:1255–1262. doi: 10.1016/j.jasms.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu H, Zong C, Wang Y, Young GW, Deng N, Souda P, Li X, Whitelegge J, Drews O, Yang PY. Revealing the dynamic of the 20 S proteasome phosphoproteome. A combined CID and electron transfer dissociation approach. Mol Cell Proteomics. 2008;7:2073–2089. doi: 10.1074/mcp.M800064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W, Ross MM, Tessitore A, Ornstein D, VanMeter A, Liotta LA, Petricoin EF., III An initial characterization of the serum phosphoproteome. J Proteome Res. 2009;8:5523–5531. doi: 10.1021/pr900603n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snovida S, Bodnar ED, Viner R, Saba J, Perreault H. A simple cellulose column procedure for selective enrichment of glycopeptides and characterization by nano LC coupled with electron-transfer and high-energy collision-dissociation tandem mass spectrometry. Carbohydr Res. 2010;345:792–801. doi: 10.1016/j.carres.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Sobott F, Watt SJ, Smith J, Edelmann MJ, Kramer HB, Kessler BM. Comparison of CID versus ETD based MS/MS fragmentation for the analysis of protein ubiquitination. J Am Soc Mass Spectrom. 2009;20:1652–1659. doi: 10.1016/j.jasms.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Mentinova M, Han H, McLuckey SA. Dissociation of disulfide-intact somatostatin ions: the role of ions type and dissociation method. Rapid Commun Mass Spectrom. 2009;23:2647–2655. doi: 10.1002/rcm.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueberheide BM, Fenyö D, Alewood PF, Chait BT. Rapid sensitive analysis of cysteine rich peptide venom components. Proc Natl Acad Sci. 2009;106:6910–6915. doi: 10.1073/pnas.0900745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eliuk S, Maltby D, Panning B, Burlingame AL. High resolution electron transfer dissociation (ETD) studies of unfractionated intact histones from murine embryonic stem cells using online capillary LC separation: determination of abundant histone isoforms and post-translational modifications. Mol Cell Proteomics. 2010;9:824–837. doi: 10.1074/mcp.M900569-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molina H, Matthiesen R, Kandasamy K, Pandey A. Comprehensive comparison of collision induced dissociation and electron transfer dissociation. Anal Chem. 2008;80:4825–4835. doi: 10.1021/ac8007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5:959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunger MK, Cargile BJ, Ngunjiri A, Bundy JL, Stephenson JL., Jr Automated proteomics of E. Coli via top-down electron-transfer dissociation mass spectrometry. Anal Chem. 2008;80:1459–1467. doi: 10.1021/ac7018409. [DOI] [PubMed] [Google Scholar]
- 20.Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, Durazo A, Bowie JU, Hasan S, Baniulis D, Cramer WA, Faull KF, Whitelegge JP. Post-translational modifications of integral membrane proteins resolved by top-down Fourier-transform mass spectrometry with collisionally activated dissociation. Mol Cell Proteomics. 2010;9:791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Mischerikow N, Bandeira N, Navarro JD, Wich L, Mohammed S, Heck AJ, Pevzner PA. The generating function of CID, ETD and CID/ETD pairs of tandem mass spectra: Applications to database search. Mol Cell Proteomics. 2100;9:2840–2852. doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y, Liu T, Tolić N, Petritis BO, Zhao R, Moore RJ, Purvine SO, Camp DG, II, Smith RD. Strategy for degradomic-peptidomic analysis of the human blood plasma. J Proteome Res. 2010;9:2339–2345. doi: 10.1021/pr901083m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, Tolić N, Purvine SO, Smith RD. Identification of disulfide bonds in protein proteolytic degradation products using the de novo-protein unique sequence tags approach. J Proteome Res. 2010;9:4053–4060. doi: 10.1021/pr1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y, Tolić N, Liu T, Zhao R, Petritis BO, Gritsenko MA, Camp DG, II, Moore RJ, Purvine SO, Esteva FJ, Smith RD. Blood peptidome-degradome profile of breast cancer. PLoS ONE. 2010;5(10):e13133. doi: 10.1371/journal.pone.0013133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao R, Livesay EC, Udseth HR, Smith RD. Automated 20 kpsi RPLC-MS and MS/MS with chromatographic peak capacities of 1000–1500 and capabilities in proteomics and metabolomics. Anal Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 26.Peng J, Elias JE, Thoreen CC, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 27.Shen Y, Tolić N, Hixson KK, Purvine SO, Pasă-Tolić L, Qian WJ, Adkins JN, Moore RJ, Smith RD. Proteome-wide identification of proteomics and their modifications with decreased ambiguities and improved false discovery rates using unique sequence tags. Anal Chem. 2008;80:1871–1882. doi: 10.1021/ac702328x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen Y, Tolić N, Hixson KK, Purvine SO, Anderson GA, Smith RD. De novo sequencing of unique sequence tags for discovery of post-translational modifications of proteins. Anal Chem. 2008;80:7742–7754. doi: 10.1021/ac801123p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics. 2008;24:1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann M, Wilm M. Error-tolerance identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 31.Masselon C, Pasa-Tolić L, Anderson GA, Bogdanov B, Vilkov AN, Shen Y, Zhao R, Qian WJ, Lipton MS, Camp DG, II, Smith RD. Targeted comparative proteomics by liquid chromatography-tandem Fourier ion cyclotron resonance mass spectrometry. Anal Chem. 2005;77:400–406. doi: 10.1021/ac049043e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Y, Page JS, Smith RD. Advances in capillary liquid chromatography-mass spectrometry for proteomics. In: Grushka E, Grinberg N, editors. Chapter 2 in Advances in Chromatography. Vol. 47. CRC Press; 2009. pp. 31–58. [Google Scholar]
- 33.Eng JK, Mccormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequence in a protein database. J Am Soc Mass spectrum. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 34.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platform used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 35.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.