Abstract
THAP1 is a DNA binding protein that has been recently associated with DYT6 dystonia, a hereditary movement disorder involving sustained, involuntary muscle contractions. A large number of dystonia-related mutations have been identified in THAP1 in diverse patient populations worldwide. Previous reports have suggested that THAP1 oligomerizes with itself via a C-terminal coiled-coil domain, raising the possibility that DYT6 mutations in this region might affect this interaction. In this study we examined the ability of wild-type THAP1 to bind itself and the effects on this interaction of the following disease mutations: C54Y, F81L, ΔF132, T142A, I149T, Q154fs180X, and A166T. The results confirmed that wild-type THAP1 associated with itself and most of the DYT6 mutants tested, except for the Q154fs180X variant, which loses most of the coiled-coil domain due to a frameshift at position 154. However, deletion of C-terminal residues after position 166 produced a truncated variant of THAP1 that was able to bind the wild-type protein. The interaction of THAP1 with itself therefore required residues within a 13-amino acid region (aa 154–166) of the coiled-coil domain. Further inspection of this sequence revealed elements highly consistent with previous descriptions of leucine zippers, which serve as dimerization domains in other transcription factor families. Based on this similarity, a structural model was generated to predict how hydrophobic residues in this region may mediate dimerization. These observations offer additional insight into the role of the coiled-coil domain in THAP1, which may facilitate future analyses of DYT6 mutations in this region.
Keywords: THAP1, DYT6, dystonia, dysphonia, leucine zipper, dimerization, transcription factor
Introduction
DYT6 dystonia is a hereditary movement disorder involving sustained, involuntary muscle contractions for which few treatment options exist. Affected individuals typically exhibit a characteristic clinical phenotype of generalized or segmental dystonia primarily of the upper limbs and cranial muscles (for review, see Ozelius and Bressman 2011). Laryngeal dystonia is a common feature, which can result in severe speech defects, such as dysarrthria or dysphonia. The causative gene was recently identified as THAP1 (thanatos-associated [THAP] domain-containing apoptosis-associated protein 1), which encodes a DNA binding protein of largely unknown function (Fuchs et al. 2009). Since the initial discovery of two distinct, DYT6-related mutations in THAP1 in Amish-Mennonite families, further screening has now identified over 45 different mutations in THAP1 in genetically diverse populations (Fuchs et al. 2009; Bressman et al. 2009; Djarmati et al. 2009; Bonetti et al. 2009; Paisan-Ruiz et al. 2009; Xiao et al. 2010; Groen et al. 2010; Houlden et al. 2010; Söhn et al. 2010; de Carvalho Aguiar et al. 2010; Cheng et al. 2011; Zittel et al. 2010; Clot et al. 2011; Schneider et al. 2011; Jech et al. 2011). As a result, THAP1 has rapidly emerged as a significant cause of familial dystonia worldwide. The vast majority of the mutations identified to date are present in the heterozygous state, producing an autosomal dominant disease with an estimated lifetime penetrance rate of approximately 60% (Ozelius and Bressman 2011).
THAP1 is an atypical zinc finger protein with a highly conserved, N-terminal C2CH module (CX2–4CX35–53CX2H), termed THAP, which defines a family of over 100 proteins in diverse species, including 12 human THAPs (Roussigne et al. 2003a and b; Clouaire et al. 2005). This metal-coordinating signature encodes a DNA-binding domain that recognizes an 11-nucleotide sequence within the promoters of target genes, the best characterized of which are cell cycle-related genes such as RRM1 (Clouaire et al. 2005, Cayrol et al. 2007, Bessiere et al. 2008; Campagne et al. 2010). Although DNA binding by a recombinant, isolated THAP domain has been demonstrated in cell-free systems (Clouaire et al. 2005; Bessiere et al. 2008), it is hypothesized that, in vivo, full-length THAP1 may form oligomeric complexes, either with itself or other proteins, to achieve full transcriptional activity (Campagne et al. 2010).
Most of the known DYT6 mutations identified to date cluster within the N-terminal THAP domain and are believed to negatively affect DNA binding. That observation has raised the hypothesis that disease pathogenesis, at least for these particular mutations, may involve transcriptional dysregulation due to insufficient DNA binding by the various mutant forms (Tamiya et al. 2009; Bragg et al. 2011). Yet an increasing number of THAP1 mutations have been detected in its C-terminus, downstream of the DNA binding module (Bressman et al. 2009; Djarmati et al. 2009; Bonetti et al. 2009; Xiao et al. 2010; Groen et al. 2010; Houlden et al. 2010; Söhn et al. 2010; Clot et al. 2011; Cheng et al. 2011). The key structural feature of this region is a predicted coiled-coil domain, a common motif which typically mediates protein: protein interactions (Fuchs et al. 2009; Bonetti et al. 2009; Burkhard et al. 2001; Parry et al. 2008). Yeast 2-hybrid screens have indicated that THAP1 oligomerizes with itself via this domain (Rual et al. 2005; Lanati et al. 2010). However, oligomerization of THAP1 in mammalian cells has not been established, and it remains unclear whether any dystonia-related mutations in THAP1 affect its ability to bind itself.
In the present study we addressed this question by examining the ability of THAP1 variants bearing different epitope tags to associate with each other when expressed in cultured cells. A number of different DYT6-related mutants were evaluated, including five which target residues within the C-terminal coiled-coil domain and two which affect the N-terminal DNA binding module. This analysis identified a DYT6 mutant which loses the ability to bind wild-type THAP1, while also revealing a 13-amino acid (aa) sequence which appears necessary for THAP1 dimerization. This region overlaps with a predicted nuclear localization signal (NLS) and, as a result, proved equally critical for nuclear translocation by THAP1. A potential structural model of this domain was therefore generated to predict the spatial segregation between residues likely to mediate dimerization versus ones believed to interact with the cellular nuclear import machinery. Collectively these data provide further insight into the role of the C-terminal domain in THAP1, which may assist in future efforts to predict functional consequences of different DYT6-related mutations in this region.
Materials and Methods
Cell culture and DNA transfection
Human embryonic kidney (HEK)-293T and osteosarcoma (U2OS) cells (both from ATCC; Rockville, MD, USA) were grown in Dulbecco’s modified Eagle’s Medium (DMEM; Gibco/Invitrogen; Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Mediatech; Manassas, VA, USA) and penicillin/streptomycin (Sigma; St. Louis, MO, USA) at final concentrations of 100 U and 0.1 mg per ml, respectively. Cultures were maintained in a humidified incubator at 37ºC and 5% CO2. For immunoprecipitation/western blotting, cells were plated at an approximate density of 5 × 105 cells/well in 6-well culture dishes. For immunolocalization, cells were seeded onto glass coverslips at an approximate density of 5 × 104 cells/well in 24-well culture dishes. After 24 hrs, cells were transfected with cDNAs using TransIT-2020® reagent (MirusBio; Madison, WI, USA) according to manufacturer’s instructions.
Plasmids, antibodies, and related reagents
The construct encoding full-length human wild-type THAP1 (wtTHAP1) with an N-terminal V5 epitope tag in pcDNA3.1 (Invitrogen) has been previously described (Fuchs et al. 2009). A second construct encoding full-length human wtTHAP1 with C-terminal myc/FLAG epitope tags in pCMV6-Entry was obtained from Origene (Rockville, MD, USA). Expression constructs for DYT6 mutants C54Y, F81L, and Q154fs180X have also been previously described (Fuchs et al. 2009; Gavarini et al. 2010). All were generated via QuikChange™ mutagenesis (Stratagene; La Jolla, CA, USA) using V5-wtTHAP1 as template to produce N-terminal V5-tagged mutant constructs in pcDNA3.1. The same protocol was used to generate N-terminal V5-tagged expression constructs for four additional DYT6 mutants: ΔF132, T142A, I149T, and A166T. A truncated variant of wtTHAP1, engineered to remove amino acids 167–213 from the encoded protein (THAP1Δ167–213), with C-terminal myc/FLAG epitope tags in pCMV6-Entry was obtained from Origene. All constructs were confirmed by sequencing.
Primary antibodies used in the present study consisted of (1) mouse monoclonal antibodies against V5 (Sigma), Hsp70 (Santa Cruz Biotechnology; Santa Cruz, CA, USA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Millipore, Billerica, MA, USA), and histone deacetylase-1 (HDAC1; Cell Signaling Technology; Danvers, MA, USA); (2) rabbit polyclonal antibodies against V5 (Sigma), FLAG (Cell Signaling), and calnexin (Enzo Life Science; Plymouth Meeting, PA, USA), lamin A/C (Epitomics), and lamin B receptor (Epitomics); and (3) a rabbit monoclonal antibody against histone H3 (Cell Signaling). Immunoprecipitation studies were performed with agarose beads directly coupled to the anti-V5 or anti-FLAG monoclonal antibodies (Sigma). For immunoblotting, horseradish peroxidase (HRP)-linked secondary antibodies against mouse- and rabbit-IgG were obtained from GE Healthcare (Piscataway, NJ, USA). Additional reagents used for immunostaining included Cy3-conjugated anti-mouse IgG, Alexa Fluor® 488-coupled anti-rabbit IgG, wheatgerm agglutin-Alexa Fluor® 488, and TO-PRO®-3-iodide (all from Invitrogen).
THAP1 dimerization assays
The ability of THAP1 to associate with itself was assayed in vitro by coimmunoprecipitation. HEK-293T cells were cotransfected with two expression constructs: wtTHAP1-FLAG and a V5-tagged THAP1 variant (wt or DYT6 mutant). Control cells were transfected with wtTHAP1-FLAG alone. Cells were collected 16–18 hrs after transfection, washed 3X in ice-cold phosphate buffered saline (PBS), and lysed for 30 min with intermittent vortexing in ice-cold RIPA buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS) with 1X protease inhibitor (Complete Mini™; Roche; Indianapolis, IN, USA). After brief sonication, lysates were centrifuged at 16,000 × g to collect residual debris. Aliquots of each supernatant were retained to monitor transgene expression in total extracts, while the remaining volume was transferred to anti-V5-coupled agarose beads and tumbled at 4ºC. After 90 min, beads were collected by centrifugation, washed 3 × 10 min in RIPA buffer, and heated at 95ºC for 10 min in an equal volume of 2X Laemmli buffer (150 mM Tris-HCl pH 6.8, 5% SDS, 5% β-mercaptoethanol, 20% glycerol) to elute precipitated proteins.
SDS-PAGE and western blotting were performed as previously described (Bragg et al. 2004a). Briefly, equal amounts of total protein were resolved by electrophoresis on 4–12% bis-acrylamide gels (Invitrogen) and transferred to nitrocellulose (BioRad; Hercules, CA, USA). Membranes were blocked in 10% nonfat milk powder in TBS-T (150 mm NaCl, 50 mm TRIS, pH 7.9, 0.5% TWEEN), then probed with polyclonal antibodies against V5 (bait), FLAG (prey) or a monoclonal antibody against Hsp70 (loading control). All primary antibodies were diluted 1:1000 in TBS-T + 2% milk. Blots were washed 3X in TBS-T, incubated in HRP-conjugated secondary antibodies (1:10,000 in TBS-T + 2% milk), and visualized by chemiluminescence with SuperSignal West Pico Substrate™ (Pierce/Thermo Scientific; Rockford, IL, USA).
To compare the relative amount of wtTHAP1-FLAG immunoprecipitated by each V5-THAP1 variant, intensity values for immunoreactive bands were determined by densitometry using ImageJ software and normalized as follows. In total cell lysate samples, values for each FLAG-reactive band (wtTHAP1-FLAG input) were normalized to values for the corresponding Hsp70-reactive band to correct for variations in gel loading. In immunoprecipitate eluate samples, values for each FLAG-reactive band (wtTHAP1-FLAG output) were normalized to values for the corresponding V5-reactive band to correct for potential differences in V5-THAP1 expression and agarose bead volume. The ratio of normalized wtTHAP1-FLAG output to normalized wtTHAP1-FLAG input was then calculated for each V5-THAP1 variant. Ratios for each DYT6 mutant were expressed as fold changes relative to the ratio for V5-wtTHAP1 and then averaged across four independent experiments. Data were analyzed by student’s t-test with Bonferroni correction.
As an additional measure of dimerization, the interaction was also probed in pull-down format using recombinant wtTHAP1-FLAG expressed and purified from HEK-293T cells (Origene) as bait. 2.5 μg aliquots of recombinant wtTHAP1-FLAG were first immobilized on anti-FLAG agarose beads (Sigma) by tumbling at 4ºC. After 2 hrs, beads were collected via centrifugation and washed 3X in RIPA buffer diluted 1:1 with PBS. Lysates from HEK-293T cells expressing each of the V5-THAP1 variants were added to agarose-wtTHAP1-FLAG resins. As a negative control, lysate from cells overexpressing V5-wtTHAP1 was incubated with anti-FLAG beads in the absence of recombinant wtTHAP1-FLAG. Samples were tumbled at 4ºC overnight. The next day, bound proteins were eluted by heating at 95ºC for 10 min in 2X Laemmli buffer. SDS-PAGE and western blotting was performed as described above.
Immunofluorescence
U2OS cells on coverslips were transfected for 16–18 hrs with V5-THAP1 expression constructs. To visualize cellular morphology, intact cells were labeled in culture with wheat germ agglutinin-Alexa® 488 (WGA-488; 1:500 in serum-free DMEM) for 15 min, washed gently in phosphate-buffered saline (PBS, pH 7.4) to remove excess dye, and then fixed in 4% paraformaldehyde in PBS for 15 min. After washing 3X in PBS, cells were permeabilized in 0.1% Nonidet P-40 in PBS for 20 min, rinsed again in PBS, and then incubated in blocking buffer (10% normal goat serum + 1% bovine serum albumin in PBS) for 1 hr to saturate nonspecific binding sites. Coverslips were probed with a monoclonal anti-V5 antibody (1:1000 in blocking buffer) overnight at 4ºC. The next day cells were washed thoroughly in PBS and reacted with goat anti-mouse IgG coupled to Cy3 (1:1000 in blocking buffer). Coverslips were rinsed 3X in PBS, counterstained with TO-PRO®-3-iodide (5 μM in PBS) for 20 min to visualize nuclei, rinsed again, and then inverted onto glass slides using gelvatol mounting medium with an anti-fade agent, 15 μg/ml 1,4-diazabicyclo (2.2.2) octaine (DABCO; Aldrich, Milwaukee, WI, USA). Images of labeled cells were captured on a Zeiss LSM 5 Pascal laser-scanning confocal microscope at a final magnification of 100X under oil immersion.
For double labeling experiments, the initial WGA-488 labeling step was omitted. Coverslips were incubated overnight with the monoclonal anti-V5 antibody combined with polyclonal antibody against either lamin A/C or lamin B receptor (both at 1:500 in blocking buffer). The next day cells were reacted with Cy3-coupled anti-mouse IgG and goat-anti-rabbit IgG-Alexa 488 (1:1000 in blocking buffer) and then processed as described above.
Cellular fractionation
To determine if the interaction of wtTHAP1 with itself was restricted to a particular subcellular domain, U2OS cells expressing wtTHAP1-FLAG in the absence or presence of wtTHAP1-V5 were fractionated into different cellular compartments (cytoplasm, membrane, nuclear soluble extract, and nuclear chromatin-bound extract) prior to coimmunoprecipitation. Fractionation was achieved via differential detergent extraction using Subcellular Protein Fractionation Kit™ (Pierce/Thermo Scientific) according to manufacturer’s instructions. Aliquots of each compartmental extract were retained for SDS-PAGE and western blot, with the remaining volume used for coimmunoprecipitation, as outlined above. Following SDS-PAGE, blots were probed for V5 (bait) and FLAG (prey) as described above as well as the following markers: GAPDH (cytoplasm), calnexin (membrane), HDAC1 (nuclear soluble), and histone H3 (nuclear chromatin-bound).
In silico analyses
The boundaries of the coiled-coil domain in wtTHAP1 were predicted using different algorithms described by Lupas et al. (1991) and McDonnell et al. (2006). The NLS was predicted via PredictNLS (Nair et al. 2003) and NLSMapper (Kosugi et al. 2009a and b). Predicted secondary structures of wild-type and mutant THAP1 variants were compared using the PSIPRED Protein Structure Prediction Server (Jones 1999; Bryson et al. 2005). Helical wheel plots were constructed via HeliQuest (Gautier et al. 2008; http://heliquest.ipmc.cnrs.fr/). Three-dimensional Richardson diagrams of a partial segment of the coiled-coil domain of wtTHAP1 were generated with Pymol™ software (Schrodinger, Inc.) using the parallel coiled-coil domain of residues 164–191 of Protein Data Bank (PDB) 3Q0X as a model.
Results
THAP1 functional domains and DYT6 mutations
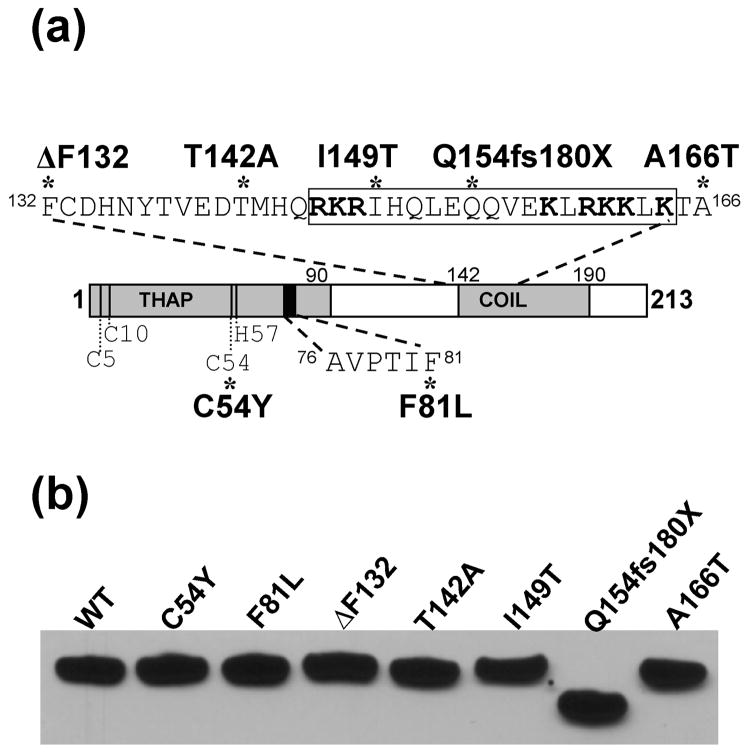
Fig. 1A depicts the DYT6 mutations examined here, relative to known functional domains in THAP1. The collection included five missense mutations (C54Y, F81L, T142, I149T, and A166T), an in-frame codon deletion (ΔF132), and a single nucleotide deletion that produces a frameshift and premature stop codon (Q154fs180X). Four of these mutations (C54Y, F81L, ΔF132, Q154fs180X) have been associated with early onset, generalized dystonia, while two (T142A and A166T) have been linked to late onset, focal laryngeal dystonia. The I149T mutation produced an intermediate phenotype, consisting of mild, multi-focal dystonia with late onset. All mutations were present in the heterozygous state, except for the ΔF132 mutation which was detected on both alleles. Four of these mutations (F81L, I149T, Q154fs180X, and A166T) have been previously documented (Fuchs et al. 2009; Bressman et al. 2009; Xiao et al. 2010; Van Gerpen et al. 2010). Identification of the three novel mutations (C54Y, ΔF132, and T142A) is being described in detail elsewhere (Fuchs, T. in preparation).
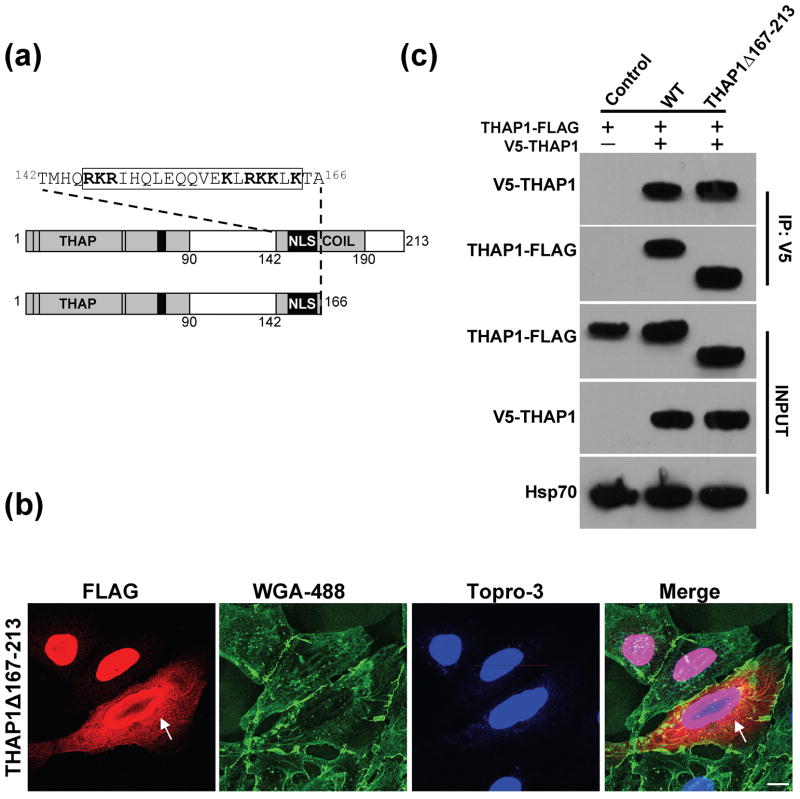
Fig. 1. DYT6 mutations in THAP1.
(A) Schematic depicting THAP1 functional domains and DYT6 mutations examined in this study. The N-terminal THAP domain (THAP) encodes the DNA-binding module including four zinc ligands (C5, C10, C54, H57) and a motif, AVPTIF, which is conserved among THAP proteins and critical for DNA binding. The C-terminus contains a predicted coiled-coil domain (COIL) within which is a predicted nuclear localization signal (NLS; boxed residues). This sequence resembles a bipartite NLS consisting of two clusters of basic arginines and lysine residues (bold) separated by a 9-aa linker. DYT6 mutants tested here included 2 variants in the N-terminal DNA binding domain (C54Y and F81L) and 5 within the C-terminus (ΔF132, T142A, I149T, Q154fs180X, A166T). (B) Expression of V5 fusions of wild-type or mutant THAP1 in HEK-293T cells as detected via western blot with anti-V5 antibody. All variants were expressed with similar efficiency and migrated at the predicted molecular weight.
The conserved N-terminal THAP domain comprises a zinc-dependent DNA binding module spanning the first 90 amino acids (Roussigne et al. 2003a). Specific features of this domain include four zinc coordinating ligands at positions 5, 10, 54, and 57 as well as a motif, 76AVPTIF81, which is highly conserved among THAP proteins and critical for DNA binding (Roussigne et al. 2003a and b; Clouaire et al. 2005, Bessiere et al. 2008). For this study we included two mutations targeting critical residues in the THAP domain: C54Y, which introduces a substitution at one of the zinc ligands (Gavarini et al. 2010); and F81L, which targets the last residue in the 76AVPTIF81 motif (Fuchs et al. 2009). Four mutations (T142A, I149T, Q154fs180X, and A166T) fall within the predicted coiled-coil domain in the C-terminus of THAP1, while the remaining variant, ΔF132, is located in close proximity to the N-terminal boundary of this motif. Within the coiled-coil domain is a predicted NLS (Osmanovic et al. 2011), which resembles a bipartite sequence consisting of two clusters of predominantly basic amino acids, 146RKR148 and 158KLRKKLK164, separated by a 9-aa linker. In a classical bipartite NLS, similar clusters mediate binding to carrier proteins which trigger active transport into the nucleus (Görlich et al. 1994; Lange et al. 2007). The I149T mutation involves a substitution within the NLS linker region, while the Q154fs180X variant disrupts the longer stretch of basic residues due to the frameshift.
Western blot analysis confirmed that V5-fusions of each THAP1 variant were expressed with similar efficiency in HEK-293T cells and migrated at the predicted molecular weights during SDS-PAGE (Fig. 1B). All variants were detected as single immunoreactive bands at approximately 32 kDa, except for the Q154fs180X truncated mutant which displayed slightly increased mobility as expected. wtTHAP1-FLAG was also efficiently expressed at similar levels to V5-wtTHAP1 (data not shown).
Dimerization of THAP1 and effects of DYT6 mutations
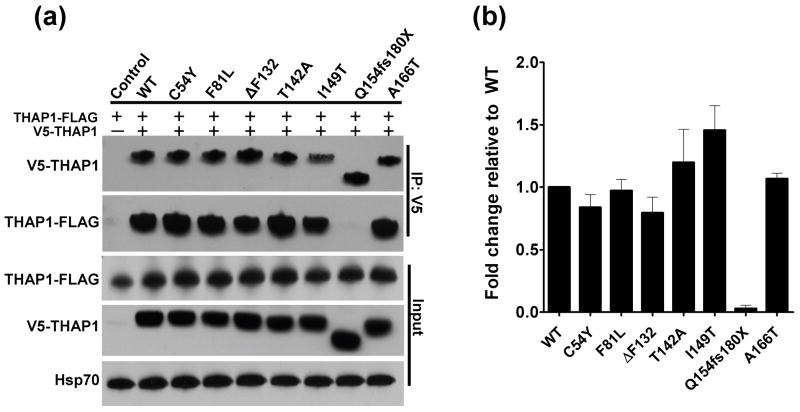
The ability of THAP1 to bind itself in cultured cells was assayed by co-expressing variants bearing different epitope tags (either V5 or FLAG) in HEK-293T cells followed by immunoprecipitation (Fig. 2A). In lysates from cells co-expressing wtTHAP1-FLAG and V5-wtTHAP, anti-V5 agarose beads effectively coimmunoprecipitated wtTHAP1-FLAG, indicating an association between the tagged fusion proteins. In control lysates, wtTHAP1-FLAG expressed alone was not immunoprecipitated by anti-V5 beads, confirming the specificity of the interaction. Coimmunoprecipitation was also performed in the opposite direction, using anti-FLAG agarose beads, with similar results (data not shown). Most of the V5-tagged mutant proteins also coimmunoprecipitated wtTHAP1-FLAG, with the sole exception being the Q154fs180X mutant. This variant was robustly expressed and immunoprecipitated itself, as indicated by detection of V5-reactive bands in total cell lysate and eluate samples, respectively, but it did not coimmunoprecipitate any detectable wtTHAP1-FLAG. Compared to V5-wtTHAP1, there was a tendency across experiments for the T142A and I149T mutants to immunoprecipitate higher amounts of wtTHAP1-FLAG, whereas the ΔF132 mutant immunoprecipitated slightly less (Fig. 2B). Yet aside from the Q154fs180X defect, none of these differences achieved statistical significance.
Fig. 2. Wild-type THAP1 binds itself and most DYT6 mutants, but not Q154fs180X.
(A) Representative immunoblot demonstrating coimmunoprecipitation of wild-type THAP1-FLAG (wtTHAP-FLAG) by different V5-tagged THAP1 variants in lysates from transfected HEK-293T cells. No wtTHAP1-FLAG was immunoprecipitated by anti-V5 from control lysates expressing wtTHAP1-FLAG alone. All of the V5-tagged THAP1 variants coimmunoprecipitated wtTHAP1-FLAG except for the Q154fs180X mutant, which lost the interaction. (B) Relative differences in levels of wtTHAP1-FLAG immunoprecipitated by each V5-tagged THAP1 variant. Data for each DYT6 mutant are expressed as mean fold changes relative to V5-wtTHAP1, +/− standard errors from four independent experiments. Normalization of densitometry values is described in Materials and Methods.
As an additional confirmation of THAP1 dimerization, the analysis was repeated in a pull-down format. V5-THAP1 variants were expressed separately in HEK-293T cells, and lysates were reacted with anti-FLAG beads to which purified recombinant wtTHAP1-FLAG was immobilized. Consistent with the coimmunoprecipitation results, all V5-THAP1 variants except for the Q154fs180X mutant were successfully pulled down by the wtTHAP1-FLAG bait protein (Fig. S1).
Intracellular localization of THAP1 variants
Given that the two of the mutations tested disrupt residues within the predicted NLS, we hypothesized that nuclear translocation might be disrupted in these variants. In the case of the Q154fs180X mutant, a defect in nuclear entry could potentially account for its inability to bind wtTHAP1, as some transcription factors have been shown to dimerize only in the nucleus (Metallo and Schepartz, 1997, Virbasius et al., 1999, Kohler and Schepartz, 2001). For that reason we characterized the nuclear vs. cytoplasmic distributions of each variant in U2OS cells via immunofluorescence.
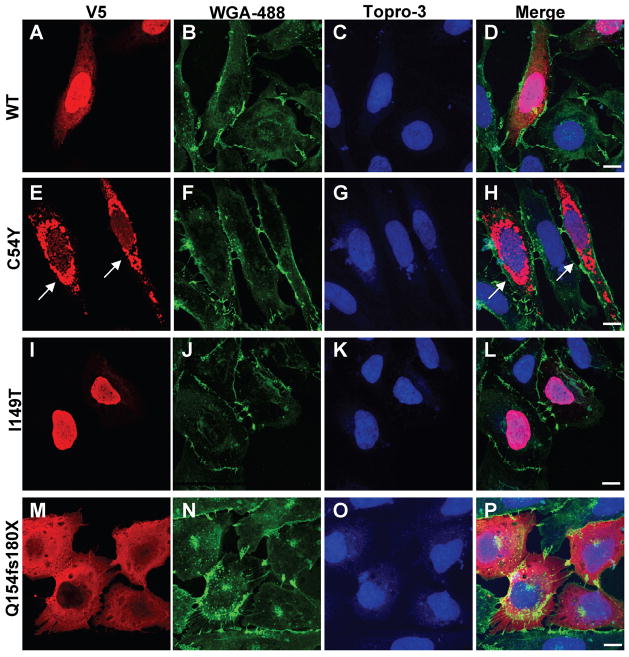
In cells expressing V5-wtTHAP1 (Fig. 3A–D), immunostaining with a monoclonal antibody against the epitope tag produced particularly strong labeling within the nucleus and, in many cases, diffuse immunoreactivity throughout the cytoplasm as well. The same pattern was observed in cells expressing wtTHAP1-FLAG and stained with an anti-FLAG monoclonal antibody (data not shown), suggesting that the cellular distribution of the THAP1 transgene product was most likely not altered by the epitope tag. Antibodies against either epitope tag produced no staining in untransfected cells, nor was any labeling observed in the absence of primary antibody (data not shown), confirming specificity of primary and secondary antibodies. The F81L, ΔF132, T142A and A166T mutants all produced similar staining patterns which did not differ significantly from that of V5-wtTHAP1 (Fig. S2). Although the I149T mutant introduces a substitution within the NLS linker sequence, it did not interfere with nuclear entry as indicated by strong nuclear labeling in cells expressing this variant (Fig. 3G–J). In contrast, cells expressing the Q154fsX180 variant displayed robust cytoplasmic staining with little to no immunoreactivity within the nucleus, suggesting a defect in nuclear translocation (Fig. 3L–N).
Fig. 3. Nuclear localization is altered in some DYT6 mutants.
V5-tagged THAP1 variants in U2OS cells localized via immunofluorescence using anti-V5 monoclonal antibody (red), wheatgerm agglutinin-Alexa488 (membranes; green), TO-PRO®-3-iodide (nuclei; blue). (A–D) V5-wtTHAP1 was expressed in both nucleus and cytosol of transfected cells. (E–H) The C54Y mutant was frequently observed in robustly labeled, perinuclear inclusions (arrows) which were never observed in cells expressing wtTHAP1 or any of the other DYT6 mutants. (I–L) Although the I149T mutation falls within the predicted NLS, it did not disrupt nuclear import as shown by strong nuclear labeling in cells expressing this mutant. (M–P) The Q154fs180X mutant was localized exclusively within the cytoplasm, indicating impaired nuclear entry due to disruption of the NLS. The remaining mutants (F81L, ΔF132, T142A, A166T) produced staining patterns similar to that of wtTHAP1 (Fig. S1). Images shown were captured by laser confocal microscopy at 100X final magnification under oil immersion. Scale bars = 10 μM.
The C54Y mutant produced an unexpected phenotype, consisting of perinuclear, aggregate-like structures which were robustly immunostained (Fig. 3E–F). In cells expressing the other V5-THAP1 variants, labeled structures in the perinuclear region were observed only rarely and tended to be smaller and more punctate than the C54Y+ aggregates. To determine if these structures involved the nuclear envelope (NE), U2OS cells expressing either V5-wtTHAP1 or V5-C54Y were double labeled for the epitope tag and one of two NE markers, lamin A/C (Fig. S3) or the laminB receptor (Fig. S4). Antibodies for both NE markers produced the expected staining patterns, consisting of prominent perinuclear immunoreactivity and diffuse labeling throughout the nucleoplasm. This intranuclear staining overlapped with that of the THAP1 variants, but neither NE marker co-localized significantly with the C54Y+ inclusions. Furthermore, staining for lamin A/C and lamin B receptor did not differ in cells expressing wtTHAP1 vs. C54Y, or in transfected vs. untransfected cells.
Dimerization of THAP1 occurs in both the cytosol and nucleus
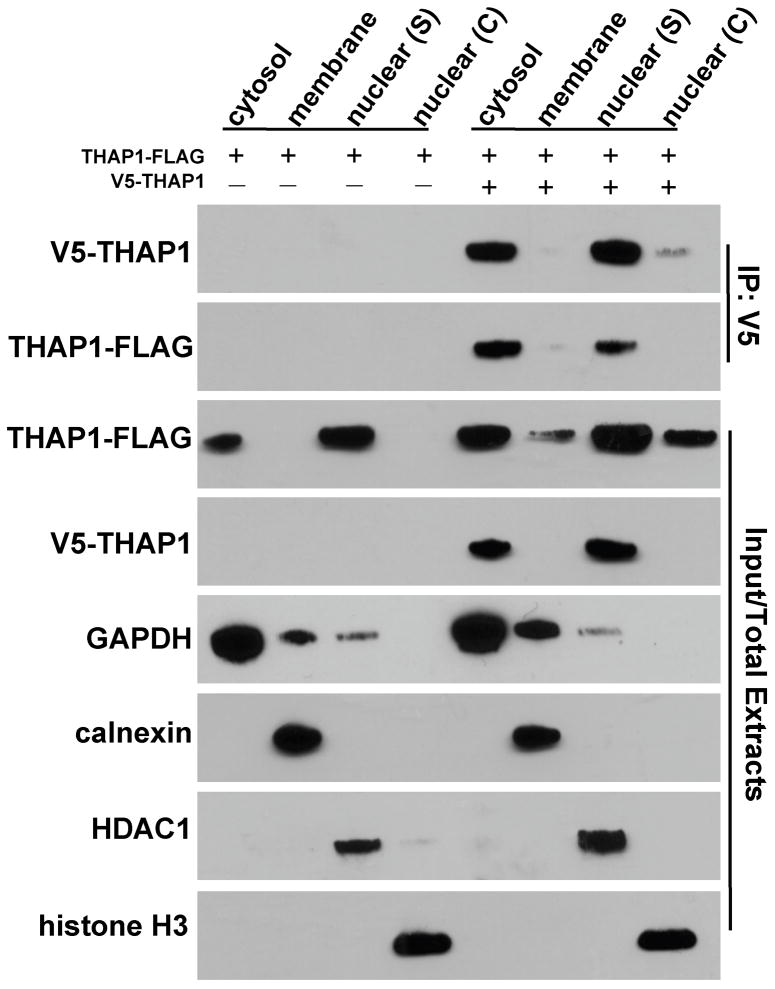
Based on the above results, we hypothesized that the inability of the Q154fs180X mutant to bind wtTHAP1 could indicate that (1) residues downstream of Q154 participate directly in the interaction between monomers; or (2) dimerization occurs only after nuclear translocation, which is abolished in the Q154fs180X mutant. To exclude the latter possibility, U2OS cells expressing wtTHAP1-FLAG in the absence or presence of V5-wtTHAP1 were first fractionated via differential detergent solubilization, followed by immunoprecipitation with anti-V5 beads from each of four cellular compartments: cytoplasm, membrane, nuclear soluble extract, and nuclear chromatin-bound extract. Immunoblotting for compartmental markers (GAPDH, calnexin, HDAC1, and histone H3) detected single bands of appropriate size for each and confirmed segregation of the different cellular domains across fractions (Fig. 4). The FLAG- and V5-tagged wild-type proteins were detected in both cytosolic and soluble nuclear extracts, consistent with the pattern observed via immunostaining (Fig. 3A–D). Furthermore, the coimmunoprecipitation between the two fusion proteins was observed in both cytosolic and soluble nuclear fractions. WTTHAP1-FLAG was therefore present within the cytosol and associated with its V5-tagged counterpart in this compartment. Thus the failure of the Q154fs180X mutant to dimerize could not be attributed to its aberrant cytoplasmic localization.
Fig. 4. Dimerization of THAP1 does not require nuclear localization.
Fractionation of U2OS cells expressing wtTHAP1-FLAG in the absence or presence of V5-wtTHAP1. Sequential detergent extraction generated four fractions: cytosol, membrane, nuclear soluble (S), and nuclear chromatin (C). Markers for each compartment (GAPDH, calnexin, HDAC1, histone H3) behaved as predicted. The V5- and FLAG-tagged fusions of wtTHAP1 were primarily detected in the cytosol and nuclear soluble fractions, although wtTHAP1-FLAG was also detected in membrane and nuclear chromatin fractions possibly due to higher expression level. Coimmunoprecipitation of wtTHAP1-FLAG by V5-wtTHAP1 was achieved from both cytosolic and nuclear soluble fractions, indicating that the interaction occurred in both cellular compartments and did not depend on nuclear import.
THAP1 deletion mutant (Δ167–213) binds the wild-type protein
Based on the fractionation analysis, we reasoned that the inability of the Q154fs180X mutant to bind wtTHAP1 most likely reflected a requirement for residues downstream of the mutation. To better define the role of the C-terminus in THAP1 dimerization, a FLAG-tagged deletion mutant was generated lacking the C-terminal region immediately downstream of the predicted NLS (THAP1Δ167–213-FLAG; Fig. 5A). Immunofluorescence of cells expressing this truncated mutant demonstrated its localization in both the nucleus and cytoplasm (Fig. 5B). The distribution of this variant mostly resembled that of wtTHAP1, although some cells were observed with prominent perinuclear immunoreactivity which in some respects appeared reminiscent of the C54Y mutant (Fig. 5B). In lysates from doubly transfected HEK-293T cells, V5-wtTHAP1 coimmunoprecipitated the truncated mutant, THAP1Δ167–213-FLAG, with similar efficiency as it did wtTHAP1-FLAG (Fig. 5C). Thus, the C-terminal region beyond position 166 was not necessary for THAP1 to bind itself.
Fig. 5. Wild-type THAP1 interacts with the deletion mutant, THAP1Δ167-213.
A) Schematic depicting a truncated THAP1 mutant which was generated to define residues required for THAP1 to bind itself. (B) THAP1Δ167–213-FLAG in U2OS cells was localized in both nucleus and cytoplasm, similar to wtTHAP1, although some cells were observed in which the transgene appeared to accumulate in the perinuclear region (arrows). Images captured at 100X magnification; scale bar = 10 μM. (C) V5-wtTHAP1 immunoprecipitated the FLAG-tagged deletion mutant with similar efficiency as it did wtTHAP1-FLAG, indicating that residues beyond position 166 are not necessary for THAP1 to bind itself.
Predicted structural consequences of Q154fs180X mutation
The observations that wtTHAP1 can bind the Δ167–213 deletion mutant but not the Q154fs180X variant suggested that its ability to dimerize was sensitive to substitutions within aa 154–166. This region also proved necessary for nuclear translocation, most likely due to the series of predominantly basic residues, 158KLRKKLK164, which in a classical bipartite NLS interact with the nuclear import machinery (Lange et al. 2007). We therefore questioned how this 13-aa region could participate in binding of THAP1 to itself while maintaining the NLS motif accessible for interactions involved in nuclear targeting. To address that question, in silico analyses were performed to compare the predicted structures of wtTHAP1 and the Q154fs180X mutant and generate a hypothetical model of THAP1 dimerization.
Coiled-coil domains are supercoiled bundles of amphipathic α-helices in which amino acids are arranged in characteristic repeats of seven (heptad) or eleven (undecad) residues for left- or right-handed coils, respectively (Lupas 1997; Burkhard et al. 2001; Parry et al. 2008). Traditional nomenclature for a left-handed coiled-coil domain designates the position of each residue in a heptad as (abcdefg)n where n equals the number of heptads that generate the coiled-coil (McLachlan et al. 1975; Conway and Parry 1990; Lupas 1997). Figure 6 displays the sequences of wtTHAP1 and the Q154fs180X mutant which correspond to the first four heptads of the coiled-coil domain as predicted by two different methods (Lupas et al. 1991; McDonnel et al. 2006).
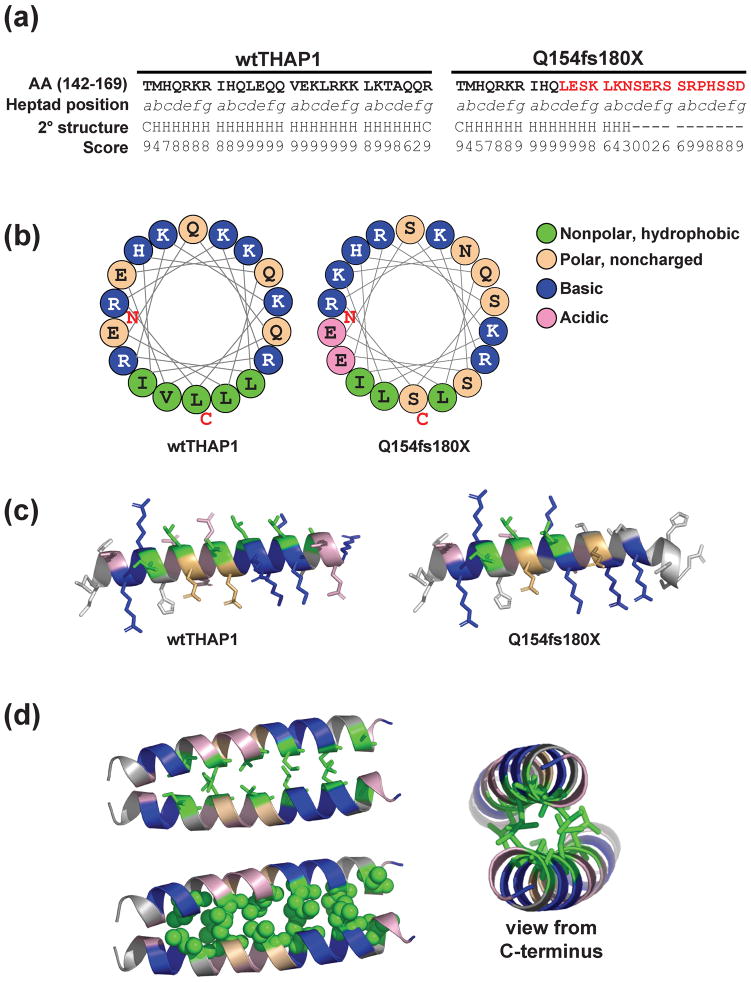
Fig. 6. Predicted structural model of THAP1 dimerization.
(A) Sequence of wtTHAP1 and the Q154fs180X corresponding to amino acids 142–169, representing the first four heptads of the predicted coiled-coil domain in wtTHAP1. Heptad position (abcdefg) is denoted below each residue, and the novel sequence encoded by the frameshift mutation is shown in red. Secondary structure analysis predicts a helical structure (H) for the wild-type sequence but not for most of the sequence encoded by the frameshift. Confidence scores for the secondary structure prediction determined by the PSIPRED Protein Structure Prediction Server are shown below each residue. (B) Helical wheel analysis of wtTHAP1 and Q154fs180X mutants. Examples shown represent residues 146–163. In wtTHAP1, clustering of hydrophobic residues on one face with basic/polar residues on the other suggests an amphipathic helix in this region. The Q154fs180X mutant loses this continuous stretch of hydrophobic residues due to polar serines which replace leucines at d3 and a4. (C) Richardson diagrams predicting the three-dimensional arrangement of hydrophobic (green), basic (blue), noncharged polar (tan), and acidic (pink) residues in this region. A potential hydrophobic interface is predicted in wtTHAP1 which is partially missing in Q154fs180X. (D) Predicted structural model of THAP1 dimerization at this potential hydrophobic interface. Model generated with Pymol software based on parallel coiled-coil domain of residues 164–191 of Protein Data Bank (PDB) 3Q0X. Side view of interacting monomers shown as stick (upper left) and space-filling (lower left) models. Alternate view from C-terminal (right) illustrates that basic arginine and lysine residues would be clustered on the outer, exposed faces of the predicted dimer structure.
Secondary structure analysis indicated that the sequence encoded by the frameshift mutation is not helical and may be disordered. In addition, the wtTHAP1 sequence contains five strongly hydrophobic residues (isoleucine, valine, or leucine) in an alternating 3-4-3 pattern at positions a and d in the second through fourth heptads (a2, d2, a3, d3, a4). Helical wheel projections of this region (Fig. 6B) demonstrated that these hydrophobic residues in wtTHAP1 are clustered on the same face of an amphipathic helix, segregated from the basic lysines and arginines. In the Q154fs180X mutant, this hydrophobic cluster is interrupted by polar serine residues which replace the leucines at d3 and a4. Richardson diagrams were generated to predict the three dimensional configuration of this domain in both the wild-type and mutant proteins (Fig. 6C). As suggested by the helical wheel plots, these models predicted a hydrophobic face in wtTHAP1 which is partially missing in the Q154fs180X mutant. In these projections, the basic lysine and arginine residues display exposed side chains which are spatially distinct from this hydrophobic region.
The arrangement of hydrophobic isoleucine, valine, and leucine residues in wtTHAP1 is consistent with previous descriptions of leucine zippers, which are common motifs formed by parallel coiled-coils that act as dimerization domains in various transcription factor families (Landschulz et al. 1988; Murre et al. 1989; Vinson et al. 2002; Elhiti and Stasolla 2009). We hypothesized that these residues might function similarly in THAP1 and generated a potential model to illustrate how a dimer could be formed at this interface (Fig. 6D). The model predicts that the hydrophobic faces in THAP1 monomers pack against each other to produce a dimer, with the basic lysine and arginine residues of the bipartite NLS exposed on the outer faces of the dimer structure.
Discussion
The function of a wide range of cellular proteins may be regulated, at least in part, by dimerization, which may involve binding of identical subunits to each other (homodimers) and/or the interaction between members of the same (homotypic) or different (heterotypic) gene families (Klemm et al. 1998; Marianayagam et al. 2004; Amoutzias et al. 2008). Among the different categories of proteins known to dimerize are multiple transcription factors, including the basic-region leucine zipper (bZIP), homeodomain leucine zipper (HD-ZIP), basic-region helix-loop-helix (bHLH), nuclear receptors, MADS-box, signal transducers and activators of transcription (STATs), and nuclear factor-κB (NF-κB) families (for review, see Amoutzias et al. 2008). In the case of transcription factors, dimerization can exert both qualitative and quantitative effects on gene expression. The types of homo- and heterotypic dimers formed by a given factor can greatly expand its repertoire of compatible DNA elements. In addition, the signals which trigger dimer formation and/or dissociation by a given factor may influence its DNA binding activity and thereby regulate transcription.
Dimerization has been postulated as a mechanism regulating activity of THAP proteins (Sabogal et al. 2010), although direct demonstrations of dimer formation by specific THAP family members are relatively few. Two examples of THAP proteins confirmed to form homodimers are the D. melanogaster P-element transposase KP repressor protein (Lee et al. 1996) and the C. elegans protein, CTPB-1 (Nicholas et al. 2008), both of which display activity dependent on oligomeric status. Aside from THAP1, none of the other human THAPs have yet been shown to homodimerize, although THAP11 and THAP7 may bind each other within a transcriptional complex (Dejosez et al. 2008). The likelihood that THAP1 homodimerizes has been suggested indirectly by reports that (1) the promoters of at least some target genes contain two copies of its binding element (Cayrol et al. 2007; Kaiser et al. 2010); and (2) the affinity of monomeric THAP1 for DNA is relatively weak, indicating that it may need to form a complex with itself or other proteins to effectively bind its target element (Campagne et al. 2010). More direct evidence of THAP1 dimerization has been provided by yeast double hybrid screens, demonstrating interactions between different THAP1 constructs (Rual et al. 2005; Lanati et al. 2010). The present study, demonstrating an interaction between tagged variants in cultured cells, confirms that THAP1 associates with itself, while further identifying specific residues that modulate the interaction.
Our analysis of the Q154fs180X and Δ167–213 mutants indicates that the ability of THAP1 to bind itself is dependent on residues within a 13-aa region (154–166) of the coiled-coil domain. This sequence, together with upstream residues within the coiled-coil domain, contains elements consistent with leucine zippers in other dimerizing transcription factors. Within a leucine zipper, dimer formation is driven by interhelical interactions between hydrophobic residues at the a and d positions (a↔a′ and d↔d′ where ‘ denotes the position on the opposite monomer), as well as interhelical electrostatic interactions between differently charged residues at g and e positions (g↔e′ where e′ is five residues towards the C-terminal on the opposite monomer) (Vinson et al. 2002; Alber 1992; Ellenberger et al. 1992; Thompson et al. 1993). Thermodynamic studies have shown that the most stable dimers contain (1) isoleucine (Acharya et al. 2002) or valine (Tripet et al. 2000) at a positions; (2) leucine at d positions (Harbury et al. 1993; Moitra et al. 1997); and (3) one of four polar amino acids (glutamate, glutamine, arginine, or lysine) at g and e positions (Vinson et al. 2002), with the strongest dimers produced by interactions between a positively charged arginine and a negatively charged glutamate (Krylov et al. 1994). Consistent with these rules, THAP1 contains isoleucine and valine at a2 and a3, respectively, and leucines at d2 and d3. This scheme also accurately describes all of the g↔e′ pairs that would exist in the predicted THAP1 dimer, specifically: arginine↔glutamate (g1↔e2′); glutamine↔arginine (g2↔e3′); and lysine↔glutamine (g3↔e4′).
Leucine zippers in transcription factors such as the bZIPs are typically comprised of four or five heptads (Vinson et al. 2002), but stable oligomerization has been achieved by even a two-heptad domain provided a sufficient interface of consecutive hydrophobic residues is present (Burkhard et al. 2000; Lu and Hodges 2004). The Q154fs180X mutant most likely fails to meet these criteria, given that (1) the helical wheel analysis demonstrated no more than 2–3 continuous hydrophobic residues at any point in this region, suggesting that an amphipathic helix is unlikely to form; and (2) secondary structure analysis indicated that the novel sequence encoded by the frameshift lacks helical structure and may be disordered.
In addition to the Q154fs180X frameshift, the I149T mutation also targets a critical residue within this domain, specifically the isoleucine at a2. However, the threonine substitution did not disrupt dimerization; rather, it displayed an apparent tendency across experiments to immunoprecipitate more wtTHAP1-FLAG than was pulled out by V5-wtTHAP1. Leucine zipper proteins which normally contain threonine at a2 have been characterized (Porte et al. 1997), and a dimer involving a threonine↔isoleucine a↔a′ interaction (as would occur between I149T and wtTHAP1) is reportedly stable (Acharya et al. 2006). However, substitutions at a positions may also change the packing geometry between coils to produce aberrant trimers and even tetramers in place of dimers (Harbury et al. 1993). The behavior of the I149T mutant in the coimmunoprecipitation assay could suggest a similar effect on oligomerization by THAP1; if so, then the interaction with its target DNA element might be altered due to formation of aberrant oligomeric complexes. Nevertheless, further biophysical analyses are required to test these possibilities.
Bonetti et al. (2009) identified a DYT6-related missense mutation at position 170 (C170R) and hypothesized that it might negatively affect dimerization based on a proposed structural model of the coiled-coil domain. Our results confirm the importance of the coiled-coil domain for THAP1 self-association but suggest that residues beyond position 166 are not required. The C-terminal portion of the coiled-coil domain may instead mediate binding of THAP1 to other proteins, although this hypothesis remains to be tested. Lanati et al. (2010) also proposed a hypothetical model of THAP1 dimerization, predicting that individual monomers may associate via the zipper-like motif in the coiled-coil domain. The observed dimerization defect in the Q154fs180X mutant, which loses most of the zipper-like motif, provides direct evidence to support this prediction. In addition, we extended the previous in silico analyses of the THAP1 coiled-coil domain to predict how critical sequences within the bipartite NLS might be spatially oriented within a potential dimer structure. In cargo proteins with bipartite NLS sequences similar to THAP1, the upstream and downstream clusters of basic residues interact with a minor and major binding pocket, respectively, on carrier proteins such as importin-α that mediate nuclear entry (Conti et al. 1998; Fontes et al. 2000). We hypothesize that the translocation defect in the Q154fs180X mutant is most likely due to the loss of the downstream basic cluster, 158KLRKKLK164. Given that the fractionation/co-immunoprecipitation analysis suggested that THAP1 binds itself within the cytoplasm, it seems likely that the basic residues of the NLS must be displayed on exposed surfaces of the resulting dimer to allow interaction with the nuclear import machinery. The helical wheel and Richardson plots predict such an arrangement.
Although the I149T mutation also falls within the NLS, it did not prevent nuclear localization. The linker sequence in a bipartite NLS is believed to orient the upstream and downstream basic clusters in the proper conformation to allow binding to the two pockets on an importin protein (Lange et al. 2010). Although some mutations within NLS linkers have been shown to disrupt nuclear trafficking, others have been reported with no apparent effect (Moore et al. 1998; Lange et al. 2010). Our results demonstrate that the I149T substitution in THAP1 is an example of the latter. Among the DYT6 mutants tested, the only other variant to exhibit altered intracellular localization was C54Y, which frequently accumulated within large perinuclear inclusions that did not colocalize with two markers of the nuclear envelope (NE), lamin A/C and lamin B receptor. This finding bears some similarity to previous descriptions of the DYT1 dystonia protein, torsinA, a AAA+ chaperone protein localized to the lumen of the NE and endoplasmic reticulum (ER) (Granata and Warner 2010; Bragg et al. 2011). The DYT1 mutant form, torsinAΔE, accumulates within membrane inclusions derived from the NE and ER (Hewett et al. 2000; Kustedjo et al. 2000; Bragg et al. 2004b; Goodchild and Dauer 2004; Gonzalez-Alegre and Paulson 2004, Naismith et al. 2004), yet only when expressed at high levels in cultured cells (Bragg et al. 2004c) with only limited and controversial evidence that it may do so in brains of DYT1 individuals (Standaert 2011). It is unclear whether the immunoreactive inclusions in cells expressing the C54Y mutant are relevant to DYT6 pathogenesis, or if they bear any relationship to the aberrant membrane structures induced in cultured cells by torsinAΔE. Yet given that NE dysfunction has been postulated as a mechanism underlying DYT1 dystonia (for review, see Granata et al. 2009), this observation may suggest that there could be interactions between THAP1 and the NE that warrant further investigation.
The DYT6 mutants tested here are associated with a range of clinical phenotypes, both in terms of disease onset (early vs. late) and somatic distribution of symptoms (generalized vs. focal). The clinical spectrum of DYT6 dystonia has grown more heterogeneous as additional THAP1 mutations have been identified, and it has been difficult to account for the phenotypic variability at the level of genotype. Among the descriptions of DYT6 mutations reported to date, there does not appear to be a consistent genotype:phenotype relationship in terms of symptom distribution, but there is a potential trend related to disease onset. Early onset dystonia has typically been documented for THAP1 mutations within the N-terminal DNA binding module and in C-terminal mutations which disrupt the NLS; whereas the mutations associated with late onset disease have generally been C-terminal variants not predicted to alter nuclear localization (Fuchs et al. 2009; Bressman et al. 2009; Djarmati et al. 2009; Bonetti et al. 2009; Paisan-Ruiz et al. 2009; Xiao et al. 2010; Groen et al. 2010; Houlden et al. 2010; Söhn et al. 2010; de Carvalho Aguiar et al. 2010; Cheng et al. 2011; Zittel et al. 2010; Clot et al. 2011; Schneider et al. 2011; Jech et al. 2011). In that regard, our observation of normal nuclear localization by the I149T mutation in the NLS appears consistent with this trend, as this mutation has been linked to late onset dystonia. A potential exception is the ΔF132 mutation which falls outside the N-terminal binding domain and NLS yet is associated with early onset generalized dystonia. However, unlike most of the known DYT6 mutations, the ΔF132 deletion was detected on both alleles, which may account for the severity of the phenotype.
In summary, this study performed a phenotypic analysis of wtTHAP1 and a collection of DYT6 mutant forms in cultured cells, with an emphasis on probing the effects of different mutations on the protein’s ability to bind itself. The demonstration that the Q154fs180X mutant is defective in both dimerization and nuclear translocation confirmed the importance of specific residues within the coiled-coil domain for both of these functions. Given that other DYT6 mutations have been identified within this region, these data may ultimately guide further structure:function analyses of this domain, which could prove useful in understanding the heterogeneous clinical phenotypes associated with different DYT6 disease variants.
Supplementary Material
Purified recombinant wtTHAP1-FLAG was captured on anti-FLAG agarose beads and used as bait to pull down V5-THAP1 variants in lysates from transfected HEK-293T cells. Consistent with the coimmunoprecipitation analysis, all V5-THAP1 variants were pulled down by wtTHAP1-FLAG, except for the Q154fs180X mutant. No V5-wtTHAP1 was pulled down by anti-FLAG beads in the absence of immobilized wtTHAP1-FLAG (control).
Immunofluorescence of U2OS cells expressing V5-tagged THAP1 variants, stained with anti-V5 monoclonal antibody (red), wheatgerm agglutinin-Alexa488 (membranes, green), and TO-PRO®-3-iodide (nuclei; blue). Several of the DYT6 mutants tested produced staining patterns which could not be distinguished from that of wtTHAP1. (A–D) V5-wtTHAP1 was detected primarily in the nucleus and, in many cells, also in the cytosol. Similar distributions were observed for the F81L (E–H), ΔF132 (I–L), T142A (M–P), and A166T (Q–T) mutants. Images captured via laser confocal microscopy at final magnification of 100X under oil immersion. Scale bar = 10 μM.
Double label immunofluorescence in transfected U20S cells demonstrating localizations of V5-wtTHAP1 (A–D) or V5-C54Y mutant (E–H), the nuclear envelope marker, lamin A/C, and TO-PRO®-3-iodide (nuclei). Lamin A/C immunoreactivity (green) consisted of prominent labeling in and around the nucleus which overlapped to some extent with intranuclear V5 staining (red), particularly for wtTHAP1 (D). However, the perinuclear inclusions produced by the C54Y mutant were not immunoreactive for lamin A/C (H). Images shown were captured by laser confocal microscopy at 100X final magnification under oil immersion. Scale bars = 10 μM.
Double label immunofluorescence in transfected U20S cells demonstrating localizations of V5-wtTHAP1 (A–D) or V5-C54Y mutant (E–H), the nuclear envelope marker, lamin B receptor, and TO-PRO®-3-iodide (nuclei). Similar to Lamin A/C, staining for lamin B receptor (green) showed strong labeling around the nucleus with diffuse immunoreactivity also within the nucleoplasm. Although some colocalization with V5 (red) was detected in the nucleus, the C54Y+ inclusions did not colocalize with lamin B receptor (E). Images shown were captured by laser confocal microscopy at 100X final magnification under oil immersion. Scale bars = 10 μM.
Acknowledgments
We gratefully acknowledge Dr. John R. Engen for advice on structural modeling of THAP1, the MGH Neuroscience Center Image and Analysis Core (supported by NIH P30NS045776) for use of the Nikon scanner; and Ms. Suzanne McDavitt for skilled editorial assistance in the preparation of the manuscript. Support for this work was provided by: NIH NS064450 (DCB), NS069973 (DCB), NS071025 (DCB), RR026123 (LJO), and NS037409 (LJO, NS); the Benign Essential Blepharospasm Research Foundation (DCB); and Tyler’s Hope for a Dystonia Cure, Inc. (DCB). The authors declare no financial conflicts of interest.
Abbreviations
- aa
amino acid
- AAA+
ATPases Associated with a variety of cellular Activities superfamily
- bZIP
basic region leucine zipper transcription factors
- bHLH
basic region helix-loop-helix transcription factors
- DMEM
Dulbecco’s Modified Eagle Medium
- ER
endoplasmic reticulum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HDAC1
histone deacetylase-1
- HEK-293
human embryonic kidney-293 cells
- HD-ZIP
homeodomain leucine zipper transcription factors
- HRP
horseradish peroxidase
- IgG
immunoglobulin
- MADS-box
transcription factor family named for four members (MCM1, Agamous, Deficiens, SRF)
- ORFs
open reading frames
- NE
nuclear envelope
- NFκB
nuclear factor κB family
- NLS
nuclear localization signal
- PBS
phosphate buffered saline
- PDB
Protein Databank
- pRb/E2F
retinoblastoma tumor suppressor protein/E2F transcription factor family pathway
- PSIPRED
Protein Structure Prediction Server
- RRM1
ribonucleotide reductase M1
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- STATs
signal transducers and activators of transcription
- TBS-T
tris-buffered saline-Tween
- THAP1
thanatos-associated [THAP] domain-containing apoptosis-associated protein 1
- U2OS
human osteosarcoma cells
- WGA
wheatgerm agglutinin
- wt
wild-type
References
- Acharya A, Ruvinov SB, Gal J, Moll JR, Vinson C. A heterodimerizing leucine zipper coiled coil system for examining the specificity of a position interactions: amino acids I, V, L, N, A, and K. Biochemistry. 2002;41(48):14122–14131. doi: 10.1021/bi020486r. [DOI] [PubMed] [Google Scholar]
- Acharya A, Rishi V, Vinson C. Stability of 100 homo and heterotypic coiled-coil a-a′ pairs for ten amino acids (A, L, I, V, N, K, S, T, E, and R) Biochemistry. 2006;45(38):11324–11332. doi: 10.1021/bi060822u. [DOI] [PubMed] [Google Scholar]
- Alber T. Structure of the leucine zipper. Curr Opin Genet Dev. 1992;2(2):205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- Amoutzias GD, Robertson DL, Van de Peer Y, Oliver SG. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem Sci. 2008;33(5):220–229. doi: 10.1016/j.tibs.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Bessière D, Lacroix C, Campagne S, Ecochard V, Guillet V, Mourey L, Lopez F, Czaplicki J, Demange P, Milon A, Girard JP, Gervais V. Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways. J Biol Chem. 2008;283(7):4352–4363. doi: 10.1074/jbc.M707537200. [DOI] [PubMed] [Google Scholar]
- Bonetti M, Barzaghi C, Brancati F, Ferraris A, Bellacchio E, Giovanetti A, Ialongo T, Zorzi G, Piano C, Petracca M, Albanese A, Nardocci N, Dallapiccola B, Bentivoglio AR, Garavaglia B, Valente EM. Mutation screening of the DYT6/THAP1 gene in Italy. Mov Disord. 2009;24(16):2424–2427. doi: 10.1002/mds.22861. [DOI] [PubMed] [Google Scholar]
- Bragg DC, Kaufman CA, Kock N, Breakefield XO. Inhibition of N-linked glycosylation prevents inclusion formation by the dystonia-related mutant form of torsinA. Mol Cell Neurosci. 2004a;27(4):417–426. doi: 10.1016/j.mcn.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Bragg DC, Slater DJ, Breakefield XO. TorsinA and early-onset torsion dystonia. Adv Neurol. 2004b;94:87–93. [PubMed] [Google Scholar]
- Bragg DC, Camp SM, Kaufman CA, Wilbur JD, Boston H, Schuback DE, Hanson PI, Sena-Esteves M, Breakefield XO. Perinuclear biogenesis of mutant torsin-A inclusions in cultured cells infected with tetracycline-regulated herpes simplex virus type 1 amplicon vectors. Neuroscience. 2004c;125(3):651–661. doi: 10.1016/j.neuroscience.2004.01.053. [DOI] [PubMed] [Google Scholar]
- Bragg DC, Armata IA, Nery FC, Breakefield XO, Sharma N. Molecular pathways in dystonia. Neurobiol Dis. 2011;42(2):136–147. doi: 10.1016/j.nbd.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressman SB, Raymond D, Fuchs T, Heiman GA, Ozelius LJ, Saunders-Pullman R. Mutations in THAP1 (DYT6) in early-onset dystonia: a genetic screening study. Lancet Neurol. 2009;8(5):441–446. doi: 10.1016/S1474-4422(09)70081-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P, Meier M, Lustig A. Design of a minimal protein oligomerization domain by a structural approach. Protein Sci. 2000;9:2294–2301. doi: 10.1110/ps.9.12.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11(2):82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- Campagne S, Saurel O, Gervais V, Milon A. Structural determinants of specific DNA-recognition by the THAP zinc finger. Nucleic Acids Res. 2010;38(10):3466–3476. doi: 10.1093/nar/gkq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, Girard JP. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109(2):584–594. doi: 10.1182/blood-2006-03-012013. [DOI] [PubMed] [Google Scholar]
- Cheng FB, Wan XH, Feng JC, Wang L, Yang YM, Cui LY. Clinical and genetic evaluation of DYT1 and DYT6 primary dystonia in China. Eur J Neurol. 2011;18(3):497–503. doi: 10.1111/j.1468-1331.2010.03192.x. [DOI] [PubMed] [Google Scholar]
- Clot F, Grabli D, Burbaud P, Aya M, Derkinderen P, Defebvre L, Damier P, Krystkowiak P, Pollak P, Leguern E, San C, Camuzat A, Roze E, Vidailhet M, Durr A, Brice A. Screening of the THAP1 gene in patients with early-onset dystonia: myoclonic jerks are part of the dystonia 6 phenotype. Neurogenetics. 2011;12(1):87–89. doi: 10.1007/s10048-010-0264-3. [DOI] [PubMed] [Google Scholar]
- Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, Girard JP. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci U S A. 2005;102(19):6907–6912. doi: 10.1073/pnas.0406882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94(2):193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Conway JF, Parry DA. Structural features in the heptad substructure and longer range repeats of two-stranded alpha-fibrous proteins. Int J Biol Macromol. 1990;12(5):328–334. doi: 10.1016/0141-8130(90)90023-4. [DOI] [PubMed] [Google Scholar]
- De Carvalho Aguiar P, Fuchs T, Borges V, Lamar KM, Silva SM, Ferraz HB, Ozelius L. Screening of Brazilian families with primary dystonia reveals a novel THAP1 mutation and a de novo TOR1A GAG deletion. Mov Disord. 2010;25(16):2854–2857. doi: 10.1002/mds.23133. [DOI] [PubMed] [Google Scholar]
- Dejosez M, Levine SS, Frampton GM, Whyte WA, Stratton SA, Barton MC, Gunaratne PH, Young RA, Zwaka TP. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 2010;24(14):1479–1484. doi: 10.1101/gad.1935210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djarmati A, Schneider SA, Lohmann K, Winkler S, Pawlack H, Hagenah J, Brüggemann N, Zittel S, Fuchs T, Raković A, Schmidt A, Jabusch HC, Wilcox R, Kostić VS, Siebner H, Altenmüller E, Münchau A, Ozelius LJ, Klein C. Mutations in THAP1 (DYT6) and generalised dystonia with prominent spasmodic dysphonia: a genetic screening study. Lancet Neurol. 2009;8(5):447–52. doi: 10.1016/S1474-4422(09)70083-3. [DOI] [PubMed] [Google Scholar]
- Elhiti M, Stasolla C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal Behav. 2009;4(2):86–88. doi: 10.4161/psb.4.2.7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol. 2000;297(5):1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- Fuchs T, Gavarini S, Saunders-Pullman R, Raymond D, Ehrlich ME, Bressman SB, Ozelius LJ. Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nat Genet. 2009;41(3):286–288. doi: 10.1038/ng.304. [DOI] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 2008;24(18):2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- Gavarini S, Cayrol C, Fuchs T, Lyons N, Ehrlich ME, Girard JP, Ozelius LJ. Direct interaction between causative genes of DYT1 and DYT6 primary dystonia. Ann Neurol. 2010;68:549–553. doi: 10.1002/ana.22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Alegre P, Paulson HL. Aberrant cellular behavior of mutant torsinA implicates nuclear envelope dysfunction in DYT1 dystonia. J Neurosci. 2004;24:2593–2601. doi: 10.1523/JNEUROSCI.4461-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci U S A. 2004;101:847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79(5):767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Granata A, Schiavo G, Warner TT. TorsinA and dystonia: from nuclear envelope to synapse. J Neurochem. 2009;109(6):1596–1609. doi: 10.1111/j.1471-4159.2009.06095.x. [DOI] [PubMed] [Google Scholar]
- Granata A, Warner TT. The role of torsinA in dystonia. Eur J Neurol. 2010;17(Suppl 1):81–87. doi: 10.1111/j.1468-1331.2010.03057.x. [DOI] [PubMed] [Google Scholar]
- Groen JL, Ritz K, Contarino MF, van de Warrenburg BP, Aramideh M, Foncke EM, van Hilten JJ, Schuurman PR, Speelman JD, Koelman JH, de Bie RM, Baas F, Tijssen MA. DYT6 dystonia: mutation screening, phenotype, and response to deep brain stimulation. Mov Disord. 2010;25(14):2420–2427. doi: 10.1002/mds.23285. [DOI] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Hewett J, Gonzalez-Agosti C, Slater D, Ziefer P, Li S, Bergeron D, Jacoby DJ, Ozelius LJ, Ramesh V, Breakefield XO. Mutant torsinA, responsible for early-onset torsion dystonia, forms membrane inclusions in cultured neural cells. Hum Mol Genet. 2000;9(9):1403–1413. doi: 10.1093/hmg/9.9.1403. [DOI] [PubMed] [Google Scholar]
- Houlden H, Schneider SA, Paudel R, Melchers A, Schwingenschuh P, Edwards M, Hardy J, Bhatia KP. THAP1 mutations (DYT6) are an additional cause of early-onset dystonia. Neurology. 2010;74(10):846–850. doi: 10.1212/WNL.0b013e3181d5276d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jech R, Bareš M, Křepelová A, Urgošík D, Havránková P, Růžička E. DYT6- A novel THAP1 mutation with excellent effect on pallidal DBS. Mov Disord. 2011 Mar 21; doi: 10.1002/mds.23599. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kaiser FJ, Osmanoric A, Rakovic A, Erogullari A, Uflacker N, Braunholz D, Lohnau T, Orolicki S, Albrecht M, Gillessen-Kaesbach G, Klein C, Lohmann K. The dystonia gene DYT1 is repressed by the transcription factor THAP1 (DYT6) Ann Neurol. 2010;68(4):554–559. doi: 10.1002/ana.22157. [DOI] [PubMed] [Google Scholar]
- Klemm JD, Schreiber SL, Crabtree GR. Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol. 1998;16:569–592. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- Kohler JJ, Schepartz A. Kinetic studies of Fos. Jun. DNA complex formation: DNA binding prior to dimerization. Biochemistry. 2001;40(1):130–42. doi: 10.1021/bi001881p. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem. 2009a;284(1):478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009b;106(25):10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustedjo K, Bracey MH, Cravatt BF. Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem. 2000;275(36):27933–27939. doi: 10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- Krylov D, Mikhailenko I, Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 1994;13(12):2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanati S, Dunn DB, Roussigne M, Emmett MS, Carriere V, Jullien D, Fryer J, Erard M, Cailler F, Girard JP, Bates DO. Chemotrap-1: an engineered soluble receptor that blocks chemokine-induced migration of metastatic cancer cells in vivo. Cancer Res. 2010;70(20):8138–8148. doi: 10.1158/0008-5472.CAN-10-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282(8):5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, McLane LM, Mills RE, Devine SE, Corbett AH. Expanding the definition of the classical bipartite nuclear localization signal. Traffic. 2010;11:311–323. doi: 10.1111/j.1600-0854.2009.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Mul YM, Rio DC. The Drosophila P-element KP repressor protein dimerizes and interacts with multiple sites on P-element DNA. Mol Cell Biol. 1996;16(10):5616–5622. doi: 10.1128/mcb.16.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SM, Hodges RS. Defining the minimum size of a hydrophobic cluster in two-stranded alpha-helical coiled-coils: effects on protein stability. Protein Sci. 2004;13(3):714–726. doi: 10.1110/ps.03443204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting Coiled Coils from Protein Sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lupas A. Predicting coiled-coil regions in proteins. Curr Opin Struct Biol. 1997;7(3):388–393. doi: 10.1016/s0959-440x(97)80056-5. [DOI] [PubMed] [Google Scholar]
- Marianayagam NJ, Sunde M, Matthews JM. The power of two: protein dimerization in biology. Trends Biochem Sci. 2004;29(11):618–625. doi: 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- McDonnell AV, Jiang T, Keating AE, Berger B. Parcoil2: Improved prediction of coiled coils from sequence. Bioinformatics. 2006;22(3):356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- McLachlan AD, Stewart M, Smillie LB. Sequence repeats in alpha-tropomyosin. J Mol Biol. 1975;98(2):281–291. doi: 10.1016/s0022-2836(75)80118-5. [DOI] [PubMed] [Google Scholar]
- Metallo SJ, Schepartz A. Certain bZIP peptides bind DNA sequentially as monomers and dimerize on the DNA. Nat Struct Biol. 1997;4(2):115–7. doi: 10.1038/nsb0297-115. [DOI] [PubMed] [Google Scholar]
- Moitra J, Szilák L, Krylov D, Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry. 1997;36(41):12567–12573. doi: 10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- Murre C, McCawb PS, Vaessinc H, Caudyc M, Janc LY, Janc YN, Cabrerad CV, Buskine JN, Hauschkae SD, Lassarf AB, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nair R, Carter P, Rost B. NLSdb: database of nuclear localization signals. Nucleic Acids Res. 2003;31(1):397–399. doi: 10.1093/nar/gkg001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith TV, Heuser JE, Breakefield XO, Hanson PI. TorsinA in the nuclear envelope. Proc Natl Acad Sci U S A. 2004;101:7612–7617. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas HR, Lowry JA, Wu T, Crossley M. The Caenorhabditis elegans protein CTBP-1 defines a new group of THAP domain-containing CtBP corepressors. J Mol Biol. 2008;375(1):1–11. doi: 10.1016/j.jmb.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Osmanovic A, Dendorfer A, Erogullari A, Uflacker N, Braunholz D, Rakovic A, Vierke G, Gil-Rodríguez C, Münchau A, Albrecht M, Brüggemann N, Gillessen-Kaesbach G, Klein C, Lohmann K, Kaiser FJ. Truncating mutations in THAP1 define the nuclear localization signal. Mov Disord. 2011 Apr 14; doi: 10.1002/mds.23611. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Ozelius LJ, Bressman SB. Genetic and clinical features of primary torsion dystonia. Neurobiol Dis. 2010;42:127–135. doi: 10.1016/j.nbd.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisán-Ruiz C, Ruiz-Martinez J, Ruibal M, Mok KY, Indakoetxea B, Gorostidi A, Massó JF. Identification of a novel THAP1 mutation at R29 amino-acid residue in sporadic patients with early-onset dystonia. Mov Disord. 2009;24(16):2428–2429. doi: 10.1002/mds.22849. [DOI] [PubMed] [Google Scholar]
- Parry DA, Fraser RD, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J Struct Biol. 2008;163(3):258–269. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Porte D, Oertel-Buchheit P, John M, Granger-Schnarr M, Schnarr M. DNA binding and transactivation properties of Fos variants with homodimerization capacity. Nucleic Acids Res. 1997;25(15):3026–3033. doi: 10.1093/nar/25.15.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussigne M, Cayrol C, Clouaire T, Amalric F, Girard JP. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene. 2003a;22(16):2432–2442. doi: 10.1038/sj.onc.1206271. [DOI] [PubMed] [Google Scholar]
- Roussigne M, Kossida S, Lavigne AC, Clouaire T, Ecochard V, Glories A, Amalric F, Girard JP. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci. 2003b;28(2):66–69. doi: 10.1016/S0968-0004(02)00013-0. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC. THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nat Struct Mol Biol. 2010;17(1):117–123. doi: 10.1038/nsmb.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SA, Ramirez A, Shafiee K, Kaiser FJ, Erogullari A, Brüggemann N, Winkler S, Bahman I, Osmanovic A, Shafa MA, Bhatia KP, Najmabadi H, Klein C, Lohmann K. Homozygous THAP1 mutations as cause of early-onset generalized dystonia. Mov Disord. 2011 Mar 21; doi: 10.1002/mds.23561. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Söhn AS, Glöckle N, Doetzer AD, Deuschl G, Felbor U, Topka HR, Schöls L, Riess O, Bauer P, Müller U, Grundmann K. Prevalence of THAP1 sequence variants in German patients with primary dystonia. Mov Disord. 2010;25(12):1982–1986. doi: 10.1002/mds.23207. [DOI] [PubMed] [Google Scholar]
- Standaert DG. Update on the pathology of dystonia. Neurobiol Dis. 2011;42(2):148–151. doi: 10.1016/j.nbd.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiya G. Transcriptional dysregulation: a cause of dystonia? Lancet Neurol. 2009;8(5):416–418. doi: 10.1016/S1474-4422(09)70087-0. [DOI] [PubMed] [Google Scholar]
- Thompson KS, Vinson CR, Freire E. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry. 1993;32(21):5491–5496. doi: 10.1021/bi00072a001. [DOI] [PubMed] [Google Scholar]
- Tripet B, Wagschal K, Lavigne P, Mant CT, Hodges RS. Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled-coil: 20 amino acid substitutions in position “d”. J Mol Biol. 2000;300(2):377–402. doi: 10.1006/jmbi.2000.3866. [DOI] [PubMed] [Google Scholar]
- Van Gerpen JA, Ledoux MS, Wszolek ZK. Adult-onset leg dystonia due to a missense mutation in THAP1. Mov Disord. 2010;25(9):1306–7. doi: 10.1002/mds.23086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22(18):6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius CM, Wagner S, Green MR. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol Cell. 1999;4(2):219–28. doi: 10.1016/s1097-2765(00)80369-x. [DOI] [PubMed] [Google Scholar]
- Xiao J, Zhao Y, Bastian RW, Perlmutter JS, Racette BA, Tabbal SD, Karimi M, Paniello RC, Wszolek ZK, Uitti RJ, Van Gerpen JA, Simon DK, Tarsy D, Hedera P, Truong DD, Frei KP, Dev Batish S, Blitzer A, Pfeiffer RF, Gong S, LeDoux MS. Novel THAP1 sequence variants in primary dystonia. Neurology. 2010;74(3):229–238. doi: 10.1212/WNL.0b013e3181ca00ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zittel S, Moll CK, Brüggemann N, Tadic V, Hamel W, Kasten M, Lohmann K, Lohnau T, Winkler S, Gerloff C, Schönweiler R, Hagenah J, Klein C, Münchau A, Schneider SA. Clinical neuroimaging and electrophysiological assessment of three DYT6 dystonia families. Mov Disord. 2010;25(14):2405–2412. doi: 10.1002/mds.23279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purified recombinant wtTHAP1-FLAG was captured on anti-FLAG agarose beads and used as bait to pull down V5-THAP1 variants in lysates from transfected HEK-293T cells. Consistent with the coimmunoprecipitation analysis, all V5-THAP1 variants were pulled down by wtTHAP1-FLAG, except for the Q154fs180X mutant. No V5-wtTHAP1 was pulled down by anti-FLAG beads in the absence of immobilized wtTHAP1-FLAG (control).
Immunofluorescence of U2OS cells expressing V5-tagged THAP1 variants, stained with anti-V5 monoclonal antibody (red), wheatgerm agglutinin-Alexa488 (membranes, green), and TO-PRO®-3-iodide (nuclei; blue). Several of the DYT6 mutants tested produced staining patterns which could not be distinguished from that of wtTHAP1. (A–D) V5-wtTHAP1 was detected primarily in the nucleus and, in many cells, also in the cytosol. Similar distributions were observed for the F81L (E–H), ΔF132 (I–L), T142A (M–P), and A166T (Q–T) mutants. Images captured via laser confocal microscopy at final magnification of 100X under oil immersion. Scale bar = 10 μM.
Double label immunofluorescence in transfected U20S cells demonstrating localizations of V5-wtTHAP1 (A–D) or V5-C54Y mutant (E–H), the nuclear envelope marker, lamin A/C, and TO-PRO®-3-iodide (nuclei). Lamin A/C immunoreactivity (green) consisted of prominent labeling in and around the nucleus which overlapped to some extent with intranuclear V5 staining (red), particularly for wtTHAP1 (D). However, the perinuclear inclusions produced by the C54Y mutant were not immunoreactive for lamin A/C (H). Images shown were captured by laser confocal microscopy at 100X final magnification under oil immersion. Scale bars = 10 μM.
Double label immunofluorescence in transfected U20S cells demonstrating localizations of V5-wtTHAP1 (A–D) or V5-C54Y mutant (E–H), the nuclear envelope marker, lamin B receptor, and TO-PRO®-3-iodide (nuclei). Similar to Lamin A/C, staining for lamin B receptor (green) showed strong labeling around the nucleus with diffuse immunoreactivity also within the nucleoplasm. Although some colocalization with V5 (red) was detected in the nucleus, the C54Y+ inclusions did not colocalize with lamin B receptor (E). Images shown were captured by laser confocal microscopy at 100X final magnification under oil immersion. Scale bars = 10 μM.