Abstract
A recessive mutation in the Arabidopsis STERILE APETALA (SAP) causes severe aberrations in inflorescence and flower and ovule development. In sap flowers, sepals are carpelloid, petals are short and narrow or absent, and anthers are degenerated. Megasporogenesis, the process of meiotic divisions preceding the female gametophyte formation, is arrested in sap ovules during or just after the first meiotic division. More severe aberrations were observed in double mutants between sap and mutant alleles of the floral homeotic gene APETALA2 (AP2) suggesting that both genes are involved in the initiation of female gametophyte development. Together with the organ identity gene AGAMOUS (AG) SAP is required for the maintenance of floral identity acting in a manner similar to APETALA1. In contrast to the outer two floral organs in sap mutant flowers, normal sepals and petals develop in ag/sap double mutants, indicating that SAP negatively regulates AG expression in the perianth whorls. This supposed cadastral function of SAP is supported by in situ hybridization experiments showing ectopic expression of AG in the sap mutant. We have cloned the SAP gene by transposon tagging and revealed that it encodes a novel protein with sequence motifs, that are also present in plant and animal transcription regulators. Consistent with the mutant phenotype, SAP is expressed in inflorescence and floral meristems, floral organ primordia, and ovules. Taken together, we propose that SAP belongs to a new class of transcription regulators essential for a number of processes in Arabidopsis flower development.
Keywords: Arabidopsis mutant; inflorescence, flower, ovule development; transposon tagging; cadastral gene
Morphogenesis in higher plants originates from an undifferentiated group of cells, the meristem. After floral induction, the cells of a shoot apical meristem change in identity and form the inflorescence meristem that is characterized by a pattern of indeterminate growth and the production of flower meristems on its flanks. In Arabidopsis, these flower meristems are arranged in a spiral phyllotactic manner and are, unlike inflorescence meristems, determined. The flower meristems produce floral organ primordia in a precise number and pattern. Arabidopsis flowers are composed of four sepals in the first whorl, four petals in the second whorl, six stamens in the third whorl, and a pistil in the center of the flower. Inside the pistil, which is formed by the congenital fusion of two carpels, new primordia initiate and differentiate into the ovules. The initiation of primordia and the specification of their identity require a complex network of regulatory genes. A number of these regulatory genes has been isolated from Arabidopsis using molecular genetic approaches (for review, see Weigel 1995). Despite the identification of these genes and extensive studies in recent years, missing pieces in the puzzle limit our understanding of the interactions between the genes involved in flower ontogeny.
The meristem identity genes LEAFY (LFY) (Weigel et al. 1992) and APETALA1 (AP1) (Mandel et al. 1992) from Arabidopsis are involved in the establishment of the floral meristem and mutations in these two genes cause a partial conversion of a floral meristem into an inflorescence meristem. Strong ap1 mutants produce highly branched, inflorescence-like flowers with ectopic secondary flowers in the axils of the first whorl organs (Irish and Sussex 1990; Mandel et al. 1992). LFY and AP1 act synergistically together with other floral meristem identity genes, among which the APETALA2 (AP2) gene (Jofuku et al. 1994). The AP2 gene has a central role in the network of genes that control the establishment and maintenance of floral meristem and organ identity. In addition to its function as a flower meristem identity gene, AP2 specifies whorl 1 and 2 organ identity. Furthermore, it controls the temporal and spatial expression of the homeotic gene AGAMOUS (AG) in the perianth floral whorls (Bowman et al. 1991; Drews et al. 1991) and in the ovule (Modrusan et al. 1994). Finally, aberrations in the seed coat of ap2 mutants demonstrate that AP2 is also required for proper seed development (Jofuku et al. 1994). The homeotic gene AG can also be regarded as a multifunctional gene that controls at least three critical aspects of flower development (Yanofsky et al. 1990; Bowman et al. 1991; Mizukami and Ma 1997). First, it specifies stamen and carpel identity. Secondly, it is essential for floral meristem determination, as ag mutants are characterized by indeterminate floral growth, reiterating the floral program inside the flower. Third, as was proposed by the ABC model (Coen and Meyerowitz 1991), AG negatively regulates the AP2 gene activity in stamens and carpels. This regulation does not appear to be at the transcriptional level as AP2 mRNA accumulation coincides with AG expression in the inner two floral whorls (Jofuku et al. 1994).
Several genes that are functional during floral meristem determination and flower organogenesis participate in ovule and seed development as well. Sometimes their function is masked by other genes because of redundancy. Also the Arabidopsis gene SUPERMAN (SUP) acts at different stages during flower development. It specifies the boundary between whorl three and four organs (Sakai et al. 1995) as well as the growth of integuments, the cell layers that surround the nucellus of the ovule (Gaiser et al. 1995). As a final example, the AINTEGUMENTA (ANT) gene is also involved in the development of both flower organs and ovules (Elliott et al. 1996; Klucher et al. 1996).
In this report, we describe a novel Arabidopsis mutant, sterile apetala (sap), which is affected in inflorescence and flower and ovule development. The SAP gene was cloned by transposon tagging and the deduced amino acid sequence suggests that it acts most likely as a transcription regulator. Here, we demonstrate that SAP, in concert with AG, plays a role in the maintenance of floral fate in the flower. Additionally, analysis of mutant phenotypes and expression analysis revealed that SAP negatively controls AG in the inflorescence meristem and outer floral whorls. Furthermore, the observed aberrations in the ovule suggest that SAP is essential for the initiation of meiotic divisions during megasporogenesis. Together, these results indicate that SAP is a novel key regulator involved in several distinct steps of flower morphogenesis.
Results
Identification of the sap mutant
A large population of Arabidopsis plants containing the En/Spm-I/dSpm transposable element system (Aarts et al. 1995) was screened for mutants with reduced fertility. Plants with small or undeveloped siliques were selected. One mutant appeared to be male and female sterile as determined by reciprocal crosses to wild-type plants. This mutant (Fig. 1B) was designated sap because of the resemblance of its flowers to those of the homeotic Arabidopsis mutant ap2 (Komaki et al. 1988; Kunst et al. 1989; Bowman et al. 1991; Jofuku et al. 1994).
Figure 1.

Phenotype of wild-type and sap mutant plants and flowers: (A) phenotype of a wild-type Arabidopsis thaliana plant. (B) Phenotype of a unstable sap mutant plant. Plants are sterile and internodes are shorter than in wild-type plants. Revertant branch can be observed (arrow). (C) Wild-type revertant branch (left) and a branch with a sap mutant phenotype (right). Reversion to wild-type phenotype occurred by excision of the transposable element from the sap::I/dSpm allele. (D) Phenotype of a wild-type flower. (E) Phenotype of a sap early-arising flower. Sepals resemble wild-type sepals. Petals are short and narrow. Stamens are short with aberrant anthers. Petals and stamens are reduced in number. (F) Phenotype of a sap late-arising flower. First-whorl organs are sepals with occasionally stigmatic at the tips. Petals are absent. Stamens and pistil are reminiscent with whorl -3 and -4 organs in early-arising sap flowers. (G) Phenotype of a severely affected late-arising flower. First-whorl organs are carpels with stigmatic papillae (p) and ovules at the inner side. Whorl-2 and -3 organs are absent.
Revertant branches were observed frequently on sap mutant plants producing wild-type flowers (Fig. 1C). This instability indicates that the mutation was caused by the insertion of an I/dSpm transposable element into the SAP gene, whose function was restored upon excision. Flowers from several revertant branches were self-pollinated and the progenies (35 plants each) were analyzed for sap mutant segregation. All progenies showed approximately a 3:1 ratio of wild-type to mutant phenotype, indicating that the parental revertant flower was heterozygous for SAP (Sap,sap::I/dSpm) and that the mutation was inherited in a Mendelian fashion as a recessive trait.
sap mutant is affected in inflorescence architecture and floral structure
In the sap mutant, inflorescences are produced with at least two to three times shorter internodes than in wild-type plants (Fig. 1C). Consequently, the flowers are positioned closer to each other. Mild-to-severe defects were observed in sap mutant flowers, depending on the age of the plant (Fig. 1E–G). The first whorl organs of sap flowers show a homeotic transformation from sepal to carpelloid with increasing severity in later arising flowers. Petals in the second whorl of the early-arising flowers are narrower and shorter than in wild-type flowers. In later-arising flowers the petals are completely absent and the four first-whorl organs are transformed into carpels bearing ovules at the inner side (Fig. 1G). These aberrations in the outer two whorls of sap flowers resemble those observed in the flowers of the Arabidopsis mutant ap2 (Komaki et. al. 1988; Kunst et. al. 1989; Jofuku et al. 1994). The number of stamens in the third whorl of sap flowers is reduced from six to four (missing the lateral stamens) or two. These stamens have shorter and thinner filaments and small degenerated anthers, which do not produce pollen. A detailed microscopic analysis showed that the tissues in the anthers did not differentiate normally (results not shown). The fourth whorl of a sap flower consists of a morphologically normal pistil with stigmatic papillae, which are longer than in wild-type plants.
Ovule development is abnormal in sap mutants
Flowers from sap plants are female sterile and never produce seeds upon pollination with wild-type pollen. To investigate whether female sterility is caused by defects in pollen–pistil interaction, we followed the pollen tube growth in sap pistils. Pollen grains were able to germinate on the stigma and the pollen tubes grew inside the pistil towards the ovary, with the formation of regular callose plugs as can be observed in wild type (Fig. 2A). However, penetration of pollen tubes into sap ovules was abolished, suggesting that the ovules were aberrant (Fig. 2B,C). Failure of pollen tube penetration was also observed by Hülskamp et al. (1995) in the Arabidopsis female sterile mutants bell1, short integuments (sin1), 47H4, and 54D12.
Figure 2.
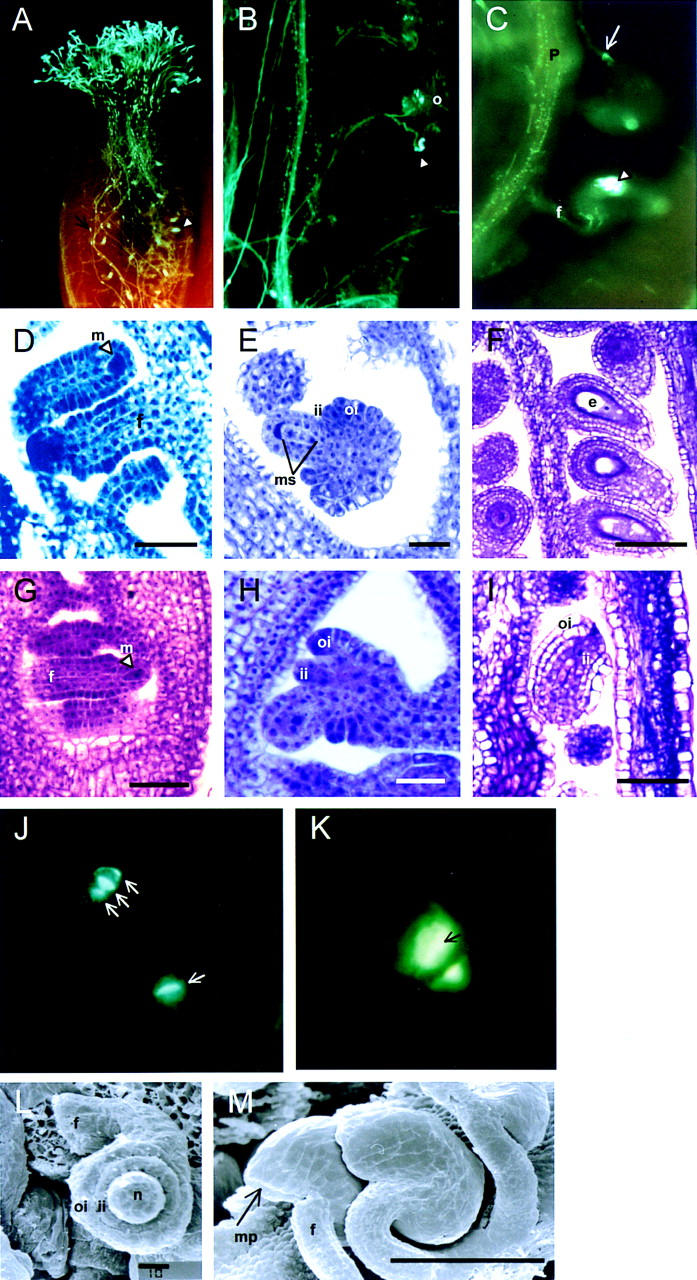
Microscopic analysis of wild-type and sap mutant ovules. (A) UV micrograph showing pollen tube growth through a mature pistil of sap. Pollen tubes are visualized by aniline blue. Regular callose plugs inside the pollen tubes (arrow) and strong staining in ovules (arrowhead) can be seen. (B) UV micrograph showing the penetration of pollen tubes into wild-type ovules. The arrowhead points to a successful penetration into the micropyle of the ovule. (C) UV micrograph of two ovules in a pollinated sap mutant plant. The upper ovule is morphologically normal and is penetrated by a pollen tube (arrow). Wild-type ovules appeared rarely in the sap mutant. They are most likely caused by somatic excision of the transposable element from the SAP gene during early stages of ovule development. The lower ovule has a typical sap phenotype with a highly stained plug of callose in the center of the ovule. An arrowhead indicates this callose deposition. (D) Light micrograph of developing wild-type ovules at stage 9 (Smyth et al. 1990). The megasporocyte is indicated by an arrowhead. (E) Light micrograph of developing wild-type ovule at stage 10 (Smyth et al. 1990). The four megaspores are formed in the nucellus after two cycles of meiotic divisions. Simultaneously, the outer and inner integuments appear. (F) Light micrograph of mature wild-type ovules. Outer and inner integuments surround embryo sacs. (G) Light micrograph of developing sap ovules at stage 9 (Smyth et al. 1990). The megasporocyte is indicated by an arrowhead. (H) Light micrograph of developing sap ovule at stage 10 (Smyth et al. 1990). Megasporogenesis is arrested and only two megaspores can be detected. (I) Light micrograph of a mature sap ovule. Embryo sac is absent, but outer and inner integuments are indistinguishable from wild-type integuments. (J) and (K) UV micrographs of aniline blue stained ovules of wild-type plants (J) and sap mutant plants (K). (J) A tetrad and dyad are visible in the wild-type ovules. Callose deposition is indicated by an arrow. (K) Stage 10 (Smyth et al. 1990) of a sap ovule with an aberrant dyad. Callose accumulates inside the megaspores as indicated by an arrow. (L) and (M) Scanning electron micrographs of stage 10 sap ovules (L) and mature sap ovules (M). The sporophytic tissues develop normally and the integuments enclose the nucellus completely. (o) Ovule; (m) megasporocyte; (ms) megaspores; (oi) outer integument; (ii) inner integument; (f) funiculus; (e) embryo sac; (p) placenta; (mp) micropyle. Bar, 10 μm (D,E,G,H,L); 50 μm (F,I) 100 μm (M).
Surprisingly large accumulation of callose was observed in mature sap ovules. To study this abnormal callose deposition and the ontogeny of sap ovules in more detail, we performed a microscopic analysis on ovules of various developmental stages. Detailed analyses of ovule development in Arabidopsis have been reported previously (Robinson-Beers et al. 1992; Modrusan et al. 1994; Schneitz et al. 1995). In wild-type Arabidopsis ovules at stage 9 (Smyth et al. 1990), a megasporocyte is differentiated in the nucellus (Fig. 2D). Shortly afterward, at stage 10, when the formation of the two integuments is initiated, the megasporocyte undergoes two cycles of meiotic divisions resulting in four megaspores (megasporogenesis, Fig. 2E). Only one out of the four megaspores is functional and survives and forms, after three cycles of nuclear division and cellularization (megagametogenesis), a mature embryo sac (Fig. 2F). During megagametogenesis, both integuments continue to grow and cover the nucellus completely by late stage 12.
Ovule development in the sap mutant is indistinguishable from wild type at early stages before meiosis (Fig. 2G). All sporophytic tissues, such as nucellus, inner and outer integuments, and funiculus develop normally (Fig. 2H,I,L,M). The first aberrations became apparent during megasporogenesis, because a tetrad was never observed in sap ovules (Fig. 2H). In mature sap ovules, embryo sacs are absent completely and callose accumulates at the position normally occupied by the embryo sac (Fig. 2C,I).
To analyze the megasporogenesis in wild-type and in sap ovules in more detail, aniline blue staining was performed to visualize callose in newly formed cell walls (Rodkiewicz 1970; Schneitz et al. 1995). Figure 2J shows an ovule at the dyad and one at the tetrad stage in a wild-type ovary. In sap ovules, the megasporocyte undergoes the first meiotic division, but does not continue cell division. Frequently, callose does not only accumulate in the cell wall of the dyads but deposition was also observed inside the cells (Fig. 2K). Accumulation of callose inside megaspores is characteristic for cells that degenerate (Pimienta and Polito 1982; Vishnyakova 1991), which probably occurs in sap ovules at or immediately after the first meiotic division. At later stages, when the integuments extend towards the apex of the nucellus, still only two megaspores are present and they are filled with callose (data not shown). In mature sap ovules, callose accumulation is observed at the position where in wild-type ovules an embryo sac is formed (Fig. 2C).
Molecular cloning of the SAP gene
For the cloning of the SAP gene, the I/dSpm transposable element that cosegregates with the sap phenotype was identified. DNA flanking this I/dSpm element was amplified by an inverse polymerase chain reaction (IPCR) (Aarts 1996) and used as a probe for the screening of a wild-type Arabidopsis thaliana Landsberg erecta genomic library. A 9-kb genomic fragment was isolated. The structure of this genomic fragment and the location of the I/dSpm element insertion in SAP are illustrated in Figure 3A.
Figure 3.

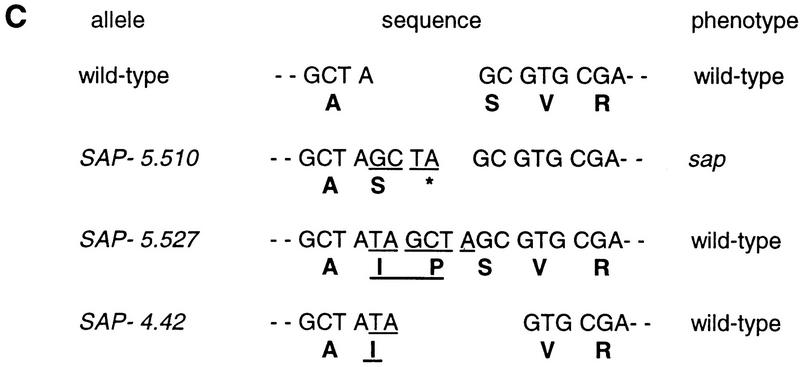
SAP genomic structure and sequences of SAP cDNA and mutant alleles. (A) Genomic structure of the SAP gene. Solid bars represent exons and the single intron of approximately 3 kb in length is indicated by an interrupted line. The locations of the start (ATG) and stop codon (GTA) are shown. The thin line represents genomic DNA outside the SAP gene. The position of the I/dSpm insertion is indicated by the large open triangle. Primer 4 was used for 5′ RACE. (E) EcoRI; (H) HinfI restriction sites. (B) Nucleotide sequence of SAP cDNA and the deduced amino acid sequence. The position of the intron is shown (▾); the position of the I/dSpm insertion is indicated ( ). The serine–glysine-rich domain is underlined. (*) The stop codon of the ORF. (C) Footprint alleles generated after excision of the I/dSpm element from the sap:I/dSpm allele. The phenotype is either as wild-type or sap mutant plants. The sequence of the duplications is underlined. In allele SAP-5.510 a duplication of four nucleotides results in the generation of a stopcodon indicated by an asterisks. In allele SAP-4.42 a duplication of two nucleotides (TA) is accompanied by a GC deletion, restoring the ORF.
). The serine–glysine-rich domain is underlined. (*) The stop codon of the ORF. (C) Footprint alleles generated after excision of the I/dSpm element from the sap:I/dSpm allele. The phenotype is either as wild-type or sap mutant plants. The sequence of the duplications is underlined. In allele SAP-5.510 a duplication of four nucleotides results in the generation of a stopcodon indicated by an asterisks. In allele SAP-4.42 a duplication of two nucleotides (TA) is accompanied by a GC deletion, restoring the ORF.
To prove that the insertion in the cloned fragment cosegregates with the sap mutant phenotype, a blot containing DNA of a segregating Sap/sap progeny was hybridized with a part of the isolated fragment as probe. All sap mutant plants (6 plants) were homozygous for the specific I/dSpm element insertion, whereas all plants (21 plants) with a wild-type phenotype exhibited either two hybridizing fragments or only one band representing the wild-type allele (data not shown).
Excision of an I/dSpm element often leads to footprints in the form of short DNA deletions or additions (Aarts et al. 1995). Therefore, a few stable sap mutants and Sap revertant plants with a wild-type phenotype were chosen for transposon-footprint analysis. The DNA sequences of the derived alleles are shown in Figure 3C. The stable sap plant (SAP-5.510) carries a 4-bp addition, resulting in the generation of a stop codon, whereas the footprints in the Sap plants leave the open reading frame intact (SAP-5.527 and SAP-4.42).
Taken together these data show that transposon insertion and excisions in the cloned genomic fragment correlate fully with the sap mutant phenotype. Thus, this cloned fragment contains part of the SAP gene.
The map position of the SAP gene was determined by RFLP using a population of recombinant inbred line (RIL) markers (Lister and Dean 1993). It is localized on chromosome 5, between genetic markers um579D and m247 (data not shown). This position demonstrates that the sap mutant is a novel homeotic mutant and not an allele of one of the known homeotic mutants.
Sequence of the SAP gene
The 3-kb EcoRI genomic fragment, that hybridizes with the IPCR clone, was sequenced completely. This sequence analysis combined with RT–PCR analysis (data not shown) revealed that the fragment contains the 3′-terminal exon and part of the intron of the SAP gene. Attempts to isolate a cDNA clone corresponding to the SAP gene from an Arabidopsis flower-specific cDNA library (Weigel et al. 1992) were not successful, probably because of a very low and specific SAP gene expression in flowers (see below). Therefore, the 5′ terminal coding part of SAP was identified by 5′ rapid amplification of cDNA ends (RACE)–PCR using poly(A)+ RNA isolated from young flower buds. In this way, the 5′ terminal part of the SAP cDNA including a putative 5′-untranslated leader sequence of 81 bp was cloned. Using this 5′-terminal part of the SAP cDNA as probe, another EcoRI genomic fragment of 1081 bp was isolated. Sequence analysis showed that this genomic fragment contains upstream sequences, the complete 5′-exon and part of the single SAP intron (Fig. 3A).efficacy
As shown in Figure 3B, the SAP cDNA has a length of 1811 bp and contains an open reading frame for a deduced protein of 446 amino acid residues with a predicted molecular mass of 49 kD. The coding region is followed by a 3′ untranslated region of 390 bp. Based on the comparison of the genomic and cDNA sequence, it is concluded that the SAP gene contains a single intron of ∼3 kb. This was confirmed by comparison of fragments obtained by RT–PCR and PCR on genomic DNA using SAP-specific primers (data not shown).
A computer search of the available sequence databases using the BLAST program (Altschul et al. 1990) revealed no significant similarities of the predicted SAP protein sequence to known protein sequences. When we compared the SAP cDNA sequence with the EST (expressed sequence tag) database of Arabidopsis, no clear homologies were detected either. However, a further examination of the SAP protein revealed small sequence motifs that are characteristic for transcriptional regulators. A long serine-rich stretch (STSSSSS) is present at the amino terminus of the protein. Similar domains have been found in a number of eukaryotic transcription factors and it has been implicated that these sites are targets for phosphorylation (Mitchell and Tjian 1989). This amino-terminal serine-rich region is adjacent to a glycine-rich region (DNGAGGSGG). Together these two stretches compose a serine/glycine-rich domain, which is a motif often found in eukaryotic transcriptional regulators (Janssen-Timmen et al. 1989; Gashler et al. 1993). In conclusion, SAP encodes a novel protein containing a few hallmarks of a transcriptional regulator.
SAP is expressed predominantly in inflorescence and floral meristems and ovules
Phenotypic analysis of sap mutant plants indicates that the SAP gene product is required for inflorescence and flower and ovule development. To determine how the expression pattern of SAP correlates with the sap mutant phenotype, we studied the SAP mRNA distribution in wild-type Arabidopsis plants by in situ hybridization.
SAP transcripts first become detectable in the inflorescence meristem, and SAP remains uniformly expressed in the floral meristems throughout stages 1 and 2 of flower development (Fig. 4A,B). After stage 3, when sepal primordia have started to arise, SAP expression becomes restricted to floral whorls 2, 3, and 4, whereas no hybridizing signal was detected in whorl 1 (Fig. 4B). Expression is maintained at approximately the same level in developing petals, stamens, and carpel primordia until stage 6. After stage 6, when the organ primordia emerge further, the level of SAP expression diminishes and no SAP mRNA is detectable at stage 8 (Fig. 4C). At later developmental stages SAP transcript appears again in ovule primordia in the nucellus region before the initiation of integuments. When integument primordia start to arise (stage 10), the expression of SAP increases and is predominantly detectable in the developing inner and outer integuments (Fig. 4F). In expanding integuments, SAP expression seems to be maintained at the same level, but it decreases at mid-stage 12 and becomes restricted to the basal part of the integuments. No expression was detected in mature ovules (data not shown).
Figure 4.
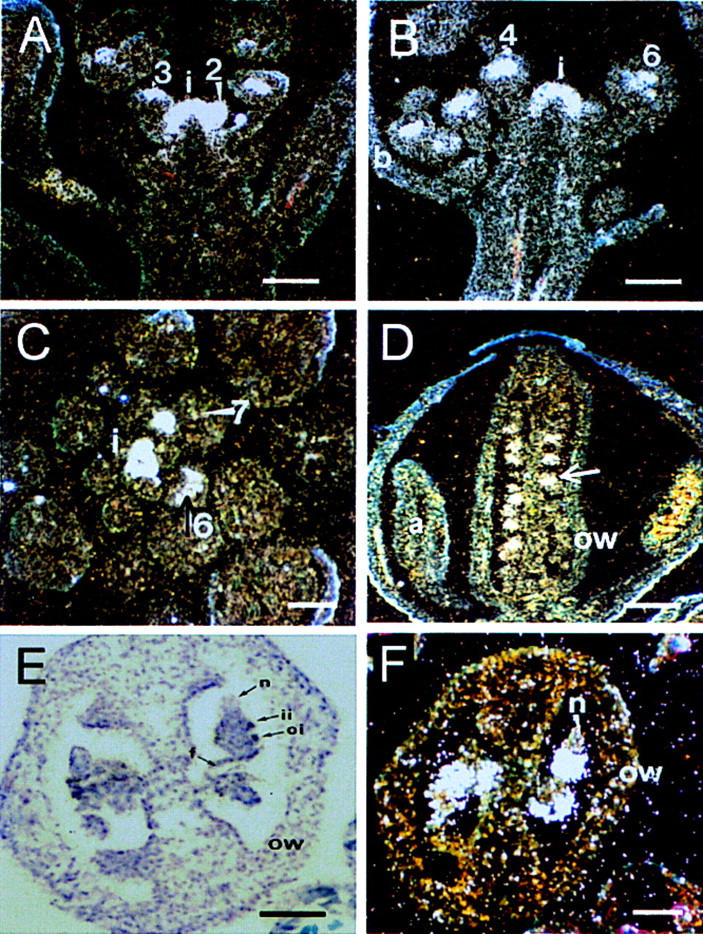
Expression of SAP mRNA in wild-type Landsberg erecta inflorescence and flowers. Numbers refer to the stage of flower development according to Smyth et al. (1990). Expression of SAP was determined by in situ hybridization with 35S-labeled antisense RNA corresponding to the SAP cDNA. Hybridization signals were viewed using dark-field microscopy. (i) Inflorescence meristem; (b) bract; (p) petal; (a) anther; (s) stigma; (ow) ovary wall; (oi) outer integuments; (ii) inner integument; (f) funiculus; (n) nucellus. (A,B) Longitudinal sections through inflorescence. Strong expression is detected in the inflorescence meristem and at the flanks of the apex, corresponding to the initiation of a floral primordium and young floral buds (stages 1 and 2). At flower stages 3–4 expression is detectable in the central region between the sepal primordia. At stage 6, hybridization signals are visible in the primordia of the petals, the stamens and the central gynoecium but not in the sepals. (C) Cross section through an inflorescence apex. The developing floral buds surround the inflorescence meristem in a spiral pattern. Expression is maintained throughout stages 2–5 and decreases during stage 6, whereas expression is abolished after stage 7. (D) Longitudinal section of a stage-12 flower bud. Strong SAP expression is present in ovules (arrow), but not in other floral organs. (E,F) Cross sections through a pistil of stage 11 imaged under bright field (E) or dark-field (F). Strong hybridization signals are present in the developing outer and inner integuments, and weaker signals are detectable in the nucellus. Bars, 100 μm (A–D); 50 μm (E,F).
The in situ hybridization experiments demonstrate that the location of SAP expression in wild-type Arabidopsis plants is correlated with the phenotypic aberrations observed in the sap mutant. The homeotic transformations in the first whorl of sap flowers appear to be initiated at a very early stage, because at later developmental stages when sepal primordia arise, SAP expression is not detectable anymore.
To determine whether residual SAP mRNA is present in sap mutant plants, Northern blot analysis was performed using poly(A)+ RNA isolated from inflorescences with young flower buds at various developmental stages (0–6). Although the expression of SAP is weak, accumulation of SAP mRNA was detectable in wild-type inflorescences, but was abolished completely in inflorescences of the sap mutant (data not shown). Because there is a strong resemblance between the phenotypes of sap and the ap2 mutant, we reprobed the Northern blot with AP2 to analyze whether the sap phenotype was partly caused by altered AP2 gene expression. However, AP2 is expressed in the sap mutant at the same level as in wild-type inflorescences (data not shown). A reciprocal Northern blot carrying mRNA from three ap2 mutant alleles (ap2-1, ap2-5, ap2-6), and probed with SAP, did not show difference in expression either (data not shown), indicating that there is no direct correlation between AP2 and SAP expression.
Interaction between SAP and homeotic genes
Some features of the sap mutant resemble characteristics of ap2 mutants, which may suggest a possible genetic interaction between these two genes. To test this hypothesis we produced and analyzed double-mutant plants. Because it was demonstrated previously (Drews et al. 1991) that AP2 is a negative regulator of AG, we also raised double mutants between sap mutant plants and the ag-1 mutant allele.
sap/ap2 double mutants
Several ap2 mutant alleles were used to raise double mutants with sap. We used the weak ap2-1 allele in our double-mutant studies to monitor a possible synergistic effect of SAP in an ap2 mutant background. In ap2-1 flowers, leaves develop in whorl 1, a reduced number of staminoid petals are formed in the second whorl, and normal stamens and carpels develop in whorls 3 and 4, respectively (Bowman et al. 1991, Fig. 5A). As is shown in Figure 5B, sap/ap2-1 flowers possess features of both sap and ap2-1 single mutants. Whorl one organs are leaf-like as in ap2-1 single mutants implying that AP2 is epistatic to SAP in the first whorl. With respect to whorl two organs, the genes seem to act synergistically because sap/ap2-1 double mutants never produce second whorl organs. The morphology of the stamens resembles those of the sap single mutant. The pistil is normal morphologically though the size is somewhat reduced, resulting in a smaller number of ovules.
Figure 5.

Phenotypes of sap/ap2 double mutants. (A) Phenotypes of wild-type (1), ap2-1 (2), and sap (3) mutant flowers. (B) Phenotype of sap/ap2-1 double mutant flower. Sepals are leaf-like as in ap2-1 single mutants and petals are completely absent. The number of stamens is reduced and anthers are malformed or carpelloid. Pistil is smaller than in ap2-1 and sap single mutants. (C) Longitudinal section through mature wild-type ovules. Outer and inner integuments surround the embryo sacs. (D) Longitudinal section through a mature sap/ap2-1 ovule. These ovules exhibit protruding nucellus (n) and the outer integument (oi) does not enclose the ovule completely. Bar, 50 μm. (E) UV micrograph of cleared wild-type ovule at stage 9 (Smyth et al. 1990) stained with aniline blue to visualize callose deposition. A tetrad is visible and the arrows indicate callose deposition. (F) UV micrograph of a cleared sap/ap2-1 ovule at stage 9 (Smyth et al. 1990) stained with aniline blue to visualize callose deposition. Callose accumulates (arrow) in the nucellus before megaspore formation. (G) Phenotype of an early-arising sap flower. (H) Phenotype of an ap2-5 mutant flower. First whorl organs show carpelloid features with occasionally stigmatic tissue (arrow). Petals are reduced in size. Stamens are reduced in number but develop normally. The pistil is not affected. (I) Phenotype of an ap2-6 mutant flower. Whorl-one organs are transformed into carpels, petals are absent and the number of stamens is reduced. (J) Phenotype of an early-arising ap2-5/sap double mutant flower. Four carpels are formed in whorl one, which are partially fused to a gynoecium. Petals and stamens are completely absent. The pistil contains ovules with similar aberrations as is observed in sap/ap2-1 double mutants. (K) Inflorescence of an ap2-5/sap double mutant. (L) Phenotype of a late-arising ap2-5/sap double mutant flower. Four completely fused carpels are formed in whorl one, which covers the central gynoecium. (M) Light micrograph of a cross section through a wild-type flower. The floral whorls are indicated. The central gynoecium is composed of two fused carpels. (N) Light micrograph of a cross section through a sap/ap2-5 flower. The four carpels in whorl 1 are fused to a tube. Ovules in the outer and inner gynoecium are indicated with arrows. Bars, M and N, 100 μm.
Like sap single mutants sap/ap2-1 plants are completely sterile. Defects in ovule development appear to be enhanced in sap/ap2-1 mutants. Microscopic analysis (Fig. 5, cf. C and D) reveals that sap/ap2-1 ovules not only lack an embryo sac, as in sap single mutants, but in addition the outer integument does not cover the inner integument and nucellus completely. This strong synergistic effect suggests that both genes are essential for normal outer integument development, in accordance with the expression patterns of SAP and AP2 (Jofuku et al. 1994).
The sap single mutant shows defects in megasporogenesis, therefore we examined whole mount of sap/ap2-1 ovules to visualize callose deposition during cell division. In these ovules, callose becomes apparent as a group of intensely fluorescing cells at the chalazal region of the nucellus at stage 9 (Fig. 5, cf.E and F), when differentiation of a megasporocyte occurs in wild-type plants. In contrast to sap single mutants, megaspores, either as dyad or tetrad, were never observed in sap/ap2-1 ovules, indicating that mutations in both genes lead to an arrest in megasporogenesis at earlier stages than in sap single mutants. The fact that ap2-1 ovules display a wild-type phenotype indicates that AP2 is redundant during megasporogenesis and acts in a different pathway to control ovule development other than SAP.
We also raised double mutants between sap and stronger alleles of ap2. The ap2-5 mutant allele (Kunst et al. 1989) has carpelloid sepals, which produce occasionally stigmatic tissue (Fig. 5H). The petals are reduced in size and the number of fertile stamens is reduced to four or two. No morphological abnormalities are observed in ap2-5 pistils. The ap2-6 (Kunst et al. 1989) is a strong allele with whorl-one organs transformed into unfused carpels and petals completely missing. The reproductive organs are fertile, although the number of stamens is reduced dramatically (Fig. 5I). sap/ap2-5 and sap/ap2-6 double mutants are very similar indicating that the maximum synergistic level is reached (Fig. 5J–L). In both types of double mutants, the four sepals are transformed completely into carpels, which are fused to a gynoecium-like structure. In side this structure, only the whorl-four pistil is present, whereas petals and stamens are absent. Cross sections through these flowers show that the outer pistil is composed of four carpels bearing ovules at the inner side (Fig. 5M,N). Analysis of the ovules from these double mutants using aniline blue staining for callose deposition revealed a similar abnormality as shown in Figure 5F for the sap/ap2-1 double mutant. In conclusion, these double-mutant studies demonstrate that the sap mutant is synergistic to ap2 mutant alleles.
sap/ag-1 double mutants
Double mutants between sap and the ag-1 mutant allele were generated and displayed a remarkable phenotype (Fig. 6A,D–F). Early-arising sap/ag-1 flowers resemble ag-1 single mutants exhibiting an indeterminate number of whorls of sepals and petals (Fig. 6B,C). The first-whorl organs are not carpel-like as in sap flowers but more sepalloid implying that AG is epistatic to SAP with respect to first-whorl organ identity. The petals in the second whorl of the double mutant are shorter and narrower than the petals in both ag-1 and wild-type flowers (Fig. 6D). In contrast to the sap mutant the number of organs in the second and third whorl are as in wild-type flowers. Later-arising sap ag-1 flowers (from approximately seventh flower) produce secondary flower buds in the axils of the second and all subsequent floral whorl organs (Fig. 6D–F). This pattern of floral bud formation reiterates in axillary flowers giving rise to a highly branched inflorescence-like structure (Fig. 6F). When the axillary flowers develop, the regions between the floral whorls elongate forming floral internodes in such a way that the floral organs and secondary flowers are arranged in a spiral phyllotaxis rather than a whorled mode as in wild type. These indeterminate flowers display several features of an inflorescence, such as elongated internodes and spiral phyllotaxis. This can be interpreted as a partial reversion from floral to an inflorescence identity.
Figure 6.
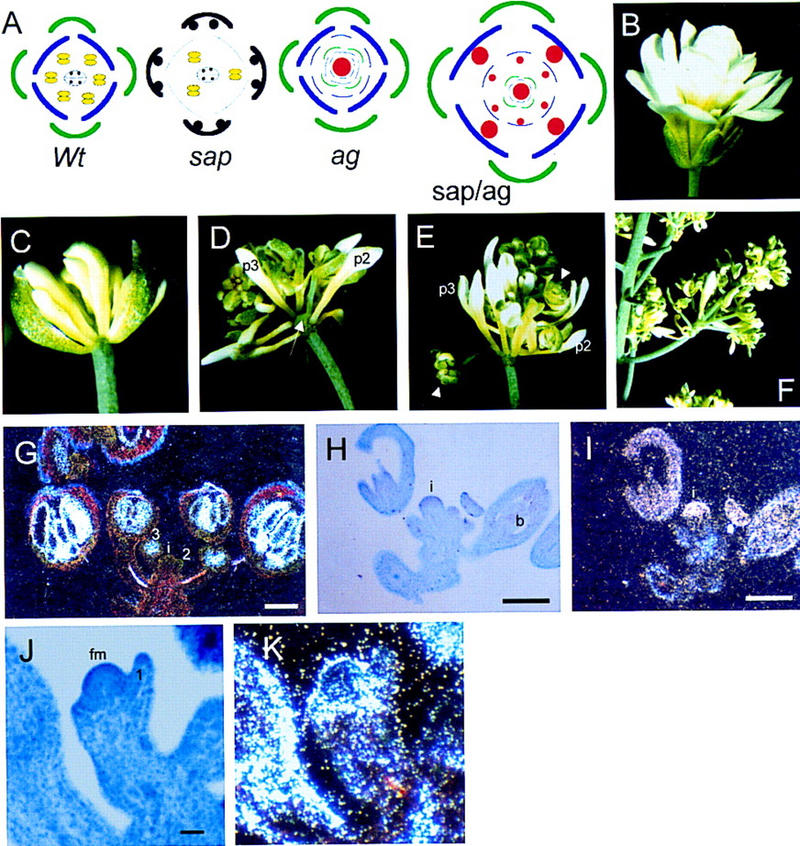
Phenotypes of sap/ag double mutants and expression pattern of AG in wild-type and sap inflorescences. (A) Schematic representation of position and identity of the floral organs in wild-type (wt), sap, ag-1, and sap/ag-1 mutant flowers. Organs are marked as follows: sepals (green); petals (blue); degenerated petals in sap (thin blue lines); stamens (yellow), carpels with ovules (black); new floral buds (red). (B) Phenotype an ag-1 mutant flower. (C) Phenotype of an early-arising sap/ag-1 double mutant flower. One sepal was removed to show the inner floral organs. sap/ag-1 flowers are indeterminate with a repeated pattern of sepals-petals-petals from the outermost whorl to the center as in ag-1 single mutants. Compared to wild-type petals, petals of sap/ag-1 are shorter and narrower as in sap. (D–E) Developing sap/ag-1 flowers resembling inflorescences. Secondary flowers, indicated by an arrowhead, develop in axils of the second (p2) and third (p3) whorl petals and all internal floral organs. Internodes are formed between the floral whorls (arrow). (F) Old sap/ag-1 flower producing an inflorescence-like branch. (G–K) Expression of AG was determined by in situ hybridization with 35S-labeled antisense RNA. Hybridization signals were viewed using dark-field microscopy. (i) Inflorescence meristem; (b) bud; (fm) floral meristem. (G) Distribution of AG mRNA in a wild-type inflorescence. Signal appears in the center of buds at early stage 3 and in older buds only in stamens and carpels. (H–I) Longitudinal section through a sap inflorescence imaged under bright field (H) or dark field (I). Strong hybridizing signal is visible in the inflorescence meristem and in the entire bud (b). These buds are composed of only carpelloid sepals and a gynoecium in the fourth whorl. (J–K) Longitudinal section through a sap floral meristem at early stage 3 of flower development imaged under bright field (J) or dark field (K). Strong hybridizing signal is visible in a ring around the floral apex. The first-whorl primordia just emerge at the flanks of the meristem.
The phenotype of sap/ag-1 double mutants demonstrates that characteristics of the sap mutant depend on the presence of an active AG gene, suggesting that SAP repress the expression of AG in the outer two floral whorls. To investigate this in more detail, the distribution of AG mRNA in sap flowers was analyzed. In wild-type flowers, AG is not expressed in inflorescence meristems and stage 1–2 floral buds (Fig. 6G). At early stage 3, when the sepal primordia just emerge, AG mRNA is first detectable in the floral apex and not in the sepal primordia. At later developmental stages, hybridization signals were observed in stamen and carpel primordia only. In contrast, high expression levels were detectable in inflorescence meristems of the sap mutant. AG mRNA is also present in all whorls of the highly affected floral buds, which are composed of carpelloid sepals and a developing gynoecium (Fig. 6H,I). In sap flower buds at a stage when the first-whorl organs just emerge at the flanks of the floral meristem (Fig. 6J,K), hybridization signal is detectable in a ring around the floral apex and at a lower level in the first whorl primordia and apex. These data indicate that, unlike wild-type flowers, AG is expressed in inflorescence meristems and at the earliest stages of sap flower formation.
Discussion
Upon floral induction, the inflorescence meristem normally produces flowers with whorls of distinct organs. We have demonstrated that the Arabidopsis SAP gene, consistent with its mRNA expression pattern, is involved in several floral developmental programs. In concert with AG it is required to maintain floral meristem identity, it controls floral organ identity by suppressing AG activity in the outer whorls and it interacts with AP2 to initiate megasporogenesis in ovules. Sequence comparison revealed that the SAP gene encodes a novel protein with no substantial sequence similarities to other known proteins. However, small domains inside the SAP protein share sequences present in transcriptional regulators, suggesting that SAP may play a role in controlling several developmental programs at the transcriptional level.
SAP is required for megasporogenesis
The sap mutant is female sterile, which trait is inherited in a sporophytic manner. Microscopic analysis of the development of sap ovules revealed no obvious aberrations in the ontogeny of the sporophytic tissues of the female reproductive organ, whereas megasporogenesis is arrested after the first meiotic division. These observations imply that SAP, as a sporophytically controlled gene, is involved directly in female gametophyte development and presumably plays a crucial role during meiotic cell division. No information is available about the role of SAP in microsporogenesis, because anthers degenerate prematurely in sap mutant plants.
Several mutants that affect female gametophyte development in Arabidopsis have been described (for review, see Schneitz et al. 1997), though the majority of these mutants display aberrations in sporophytic as well as gametophytic tissues of the ovule. Often, defects in integument development are accompanied by partial or complete absence of magasporogenesis, as it is observed in the sin1, ant and huellenlos (hll) mutants (Robinson-Beers et al. 1992; Lang et al. 1994; Elliott et al. 1996; Klucher et al. 1996; Schneitz et al. 1997). These mutants are referred to as sporophytic and megagametogenesis-defective (smd) mutants by Schneitz et al. (1997). Another class of female defective mutants, from which only a few members have been described (Schneitz et al. 1997) are characterized by defects in gametogenesis and are designated embryo sac-defective (emd) mutants. According to this classification, the sap mutant belongs to the class of megasporogenesis-defective (msd) mutants, from which only SAP has been cloned. The emd and msd mutants provide valuable knowledge about the interdependency between sporophytic and gametophytic tissues. Based on the mutants, we conclude that defects in sporophytic tissues often lead to aberrant gametophyte development, however, in contrast sporophyte development is independent from megasporogenesis and megagametogenesis.
SAP genetically interacts with AP2 in various developmental programs
The SAP gene is strongly expressed in the inflorescence meristem and the emerging floral primordia in wild-type Arabidopsis plants. This pattern of expression overlaps with that of AP2 and it also coincides with AP2 expression at later developmental stages, during floral organogenesis and ovule development. Combined with the resemblance of the phenotypes of the single mutants, we hypothesized that SAP and AP2 interact genetically. Expression analysis revealed that AP2 expression is not affected in the sap mutant and, vice versa, demonstrating that they do not regulate each other at the transcriptional level.
Although both genes are expressed in the inflorescence meristem, the phenotype of the single mutants revealed that only SAP is essential for establishing the inflorescence structure. The complete transformation of sepals into carpels and the absence of petals in the ap2-5/sap and ap2-6/sap double mutants indicate that the genes are synergistic.
Another intriguing aspect of the interaction between SAP and AP2 is the more dramatic defect observed during ovule development. Megasporogenesis in all double mutants analyzed appears to be arrested at even an earlier stage than in the sap single mutant, whereas the ap2 mutant alleles are normally fertile. This new phenotype indicates that both genes SAP and AP2 are involved in the initiation of megasporogenesis and are able to complement each other at this stage. This ascribes a new function for AP2, which is normally masked by redundancy of genes also involved in this process. Because in sap ovules the development proceeds until the second meiotic division, SAP is dispensable for the initial stage of megasporogenesis but absolutely required for later processes. The fact that the aberrations in the sap mutant are inherited as a recessive trait indicates that SAP is a sporophytically expressed gene controlling the reduction to haploid spore cells.
SAP genetically interacts with AG to maintain floral meristem identity
sap/ag-1 double mutants display several interesting transformations with respect to floral architecture as well as meristem identities. First, the restoration of the first-whorl organ identity to sepals and the formation of petals in the second and third whorls demonstrate that the role of SAP in these whorls depends on AG expression.
A second feature of the double mutant is the formation of axillary flower buds in the axils of the organs in the second and all subsequent whorls. The organs in these whorls and the axillary flowers are arranged in a spiral phyllotaxy, which is typical for inflorescences. Similar inflorescence-like characteristics and reversions to inflorescence shoot meristem are present in ap1 and lfy single and ag/ap1, ag/lfy double mutants (Bowman et al. 1993; Weigel et al. 1992; Schultz and Haughn 1991). Even in ag single mutants grown under short day conditions, reversions of the indeterminate floral meristem to the indeterminate inflorescence meristem occurs independently from the early acting genes AP1 and LFY (Mizukami and Ma 1997). Because single sap and ag mutant flowers grown under our conditions do not exhibit transformations into inflorescence-like structures, the phenotype of the double mutant indicates that SAP acts locally in concert with AG either to maintain or to promote floral meristem fate. The residual floral character of the transformed flowers is revealed by the presence of sepal and petal-like organs and implies that SAP and AG interact in a local manner similar to AP1. Most likely, the early acting meristem identity genes LFY and AP1 initiate a determinate floral meristem, whereas several other genes are required for the final definition of floral meristem identity. Here, we demonstrated that at least two genes, SAP and AG, facilitate in part the final definition of floral meristem identity in Arabidopsis.
SAP is a negative regulator of AG
One of the functions of AP2 is to prevent ectopic expression of the AG gene in the perianth whorls, but AP2 seems not to be capable of suppressing AG on its own (Drews et al. 1991; Okamuro et al. 1993; Jofuku et al. 1994). Recently, it was shown that two other genes, LEUNIG (LUG) and CURLY LEAF (CLF) may play a role in concert with AP2 to repress AG in the first- and second-whorl organs of Arabidopsis flowers (Liu and Meyerowitz 1995; Goodrich et al. 1997). lug mutants are characterized by narrow leaves and floral organs and display an enhanced ap2 phenotype with respect to floral-organ identity. Mutations in CLF cause severe effects on leaf morphology and on flowering time. In addition, the flowers of clf are very similar to ap2 flowers, though the petals are smaller and narrower compared to petals of weak ap2 alleles. Double-mutant analysis and misexpression of AG in ap2, lug, and clf mutants have demonstrated that these genes act as negative regulators of AG at different stages of development (Drews et al. 1991; Liu and Meyerowitz 1991; Goodrich et al. 1997). AP2 and LUG are both required for the initial specification of AG expression pattern from stage 2 onwards. In clf mutants, AG is expressed ectopically in hypocotyls, cotelydons, leaves, and at later floral stages (from stage 9) in the petals. This suggests that CLF is needed to maintain the suppression at later developmental stages.
Several observations provide evidence that SAP, like AP2, LUG, and CLF, is a cadastral gene negatively regulating AG expression. First, the phenotype of sap single and ap2/sap double mutants show striking similarities to that of plants in which AG was constitutively expressed (Mizukami and Ma 1992). The double mutants exhibit more severe homeotic transformation in flowers than either single mutant. This corroborates our suggestion that SAP is one of the partners of AP2 to suppress AG. Secondly, in ag/sap double mutants sepals and petals develop, which demonstrates that for SAP action in the perianth organs AG expression is needed. Furthermore, it shows that SAP is not a floral organ identity gene required to specify sepal and petal identity. Finally, our in situ hybridization experiments reveal that AG is misexpressed in the inflorescence meristem and during the initial stages of sap floral organ development. We did not detect SAP expression in sepal primordia and in flower buds after developmental stage 7, suggesting that another regulator causes the carpelloidy in the first whorl of sap mutants. Alternatively, the carpelloidy is already established before the sepal primordia emerge (before stage 2), when SAP mRNA is present and AG is ectopically expressed in a sap mutant background. The role of SAP in specifying the spatial expression of AG is additive to that of the other cadastral genes AP2, LUG, and CLF, because none of these regulators control AG in the inflorescence meristem.
This report and earlier observations with AP2, LUG, and CLF, demonstrates that a complex pattern of spatially restricted genes, each acting at different developmental stages is required to co-ordinatively regulate the expression of the homeotic gene AG.
Materials and methods
Plant material
A. thaliana ecotype Landsberg erecta was used in this work. All plants were grown under standard greenhouse conditions (16 hr light; 20–22 °C). T3 seeds of two En/Spm-I/dSpm transposon-containing lines were used for the screening for sterile mutants (Aarts et al. 1995). The original sap::I/dSpm transposon- tagged mutant was found in the line H32.3 containing ∼10 different I/dSpm elements.
Seeds of ag-1 and ap2 mutant alleles were obtained from the Arabidopsis Biological Resource Center (Columbus, OH).
Construction of double mutants
sap/ap2
As sap mutant plants are completely sterile we pollinated Sap,sap::I/dSpm heterozygous flowers with ap2 pollen. F1 progeny of those crosses were all phenotypically normal. Seeds obtained from a few self-fertilized F1 plants were harvested and the progenies were analyzed. As the ap2-5 and ap2-6 mutants were in the Columbia genetic background the appearance of the F2 plants varied slightly with respect to size and internode length depending on the genetic background. Among the F2 progeny four flower phenotypes were observed consistent with a 9:3:3:1 ratio, as listed below. For sap/ap2-1, the distribution of F2 plants was 173 wild-type, 51 ap2-1, 54 sap, and 18 plants with a novel phenotype (ratio 9:3:3:1; χ2 = 0.013; P > 0.9). For sap/ap2-5, the distribution of F2 plants was 71 wild-type, 21 ap2-5 19 sap, and 8 plants with a novel phenotype (ratio 9:3:3:1; χ2 = 0.75; P > 0.9). For sap/ap2-6, the distribution of F2 plants was 57 wild type, 20 ap2-5, 16 sap, and 6 plants with a novel phenotype (ratio 9:3:3:1; χ,2 = 0.5; P > 0.9). Some of the plants with the novel phenotype produced revertant branches with a normal length of the internodes (in F2 sap/ap2-1) and with fertile ap2 flowers indicating that they represented sap/ap2 double mutants.
sap/ag-1
To obtain sap/ag-1 double mutants stable, heterozygous Sap,sap::I/dSpm plants were crossed with the Ag,ag-1 plants. Among the F2 progenies obtained from Ag,ag-1/Sap, sap::I/dSpm self-pollinated plants four different phenotypes were found represented by 159 wild-type, 55 ag-1, 50 sap, and 17 plants with a novel phenotype (ratio 9:3:3:1; χ2 = 0.26; P > 0.90). A few plants with the novel phenotype had revertant branches with ag-1-like flowers, indicating that the new phenotype belonged to sap/ag-1 double mutants.
Inverse PCR analysis
DNA flanking the I/dSpm element was obtained by IPCR as described by Aarts (1996). The resulting IPCR product was cloned in pMOSBlueT-vector (Amersham) and sequenced with the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer).
Mapping of the SAP locus
For the mapping of SAP, a PCR fragment obtained by using the SAP gene-specific primer 1 (5′-TCGACTCATTTAAGGCTTC-3′) and primer 2 (5′-CAGTGCACCGAAATCCCATAGATG-3′), was used as RFLP probe. This probe was hybridized to a blot containing the SstI-digested DNAs of 98 RILs (Lister and Dean 1993). Recombination frequencies and map distances were calculated with the JoinMap program (Stam 1993), using mapping data provided by C. Lister and C. Dean (Cambridge Laboratory, John Innes Institute, Norwich, UK).
Genomic library screening
A Landsberg erecta genomic library, obtained through the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH) and the European DNA Resource Centre (Max-Delbrück Laboratory, Köln, Germany), was screened with the SAP-specific IPCR fragment as a probe. Hybridizations were done as described previously (Angenent et al. 1992).
DNA and RNA analysis
Genomic DNA from Arabidopsis leaves was isolated as described by Aarts (1996), and 400 ng was digested, electrophorized, and blotted onto Hybond N+ membranes. Blots were hybridized according to Angenent et al. (1992) and washed with 0.1× SSC at 65°C.
Poly(A)+ RNA from different Arabidopsis tissues was isolated using a QuickPrep Micro mRNA Purification Kit as described by manufacturer (Pharmacia Biotech), and 5 μg was denaturated by glyoxal (1.5 m) prior to electrophoresis. Hybridization was performed according to Angenent et al. (1992).
In situ hybridization
The plant material was fixed at 40°C for 2 days in 4% formaldehyde, 5% acetic acid, 50% ethanol, and stepwise dehydrated until 100% ethanol prior to infiltration with xylene and subsequent embedding in paraffin and sectioning. Sections treated with paraffin and rehydrated were taken according to the in situ hybridization procedure essentially as described by Angerer and Angerer (1991). The complete SAP cDNA was used for generating the antisense probe. The AG antisense RNA contained nucleotides 240–977 of the AG sequence published in Yanofsky et al. (1990). Hybridization was done at 42°C in 2.25× SSPE (1× SSPE: 0.18 m NaCl, 0.001 m NaPO4 at pH 7.7, 0.001 m EDTA), 50% formamide, and washed at 42°C in 0.1× SSC, 50% formamide. Sections were exposed for 2–4 weeks (AG probe) and 8 weeks (SAP probe). Photographs were taken with a Diaplan microscope equipped with dark-field optics (Leitz, Wetzlar, Germany).
5′ RACE
5′ RACE was performed according to the protocol of the Marathon cDNA Amplification Kit (Clontech). Briefly, 1 μg of polyadenylated RNA from young flower buds (stages 0–6) was used as a template for first-strand cDNA synthesis. The first PCR was performed using adaptor primer 1 and the SAP gene-specific primer 3 (5′-GTGTGACCAACACCGTCTATTTCCG-3′), which corresponds to nucleotides 713–739 of SAP cDNA. A second round of PCR was done using adaptor primer 2 and a nested gene-specific primer 4 (5′-ATCCGTCGGCGAACCCTCCG-3′), corresponding to nucleotides 537–555 of SAP cDNA. The resulting 5′-RACE–PCR product of 555 bp in length was cloned and sequenced as described before. The accession number of the SAP cDNA sequence is AJ223125.
Callose and pollen tube staining in ovaries
Pollen tube growth and callose deposition were visualized by staining with aniline blue as described previously (Angenent et al. 1993).
Acknowledgments
We thank Ruth De Groodt for technical assistance with in situ hybridization experiments and Andy Perreira and Arjen van Tunen for kindly providing the transposon lines and helpful discussions. Gerrit Stunnenberg is acknowledged for taking care of the plants. Ap2 and ag mutant alleles were derived from the Arabidopsis Biological Research Center (Columbus, OH). This research was supported by the Graduate School of Experimental Plant Sciences (EPS) with a fellowship awarded to M.V.B. This project was partially financed by the Dutch Organization for Scientific Research (NWO).
Footnotes
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL G.C.Angenent@CPRO.DLO.NL; FAX 31-317-41-80-94.
References
- Aarts MGM. ‘An En/Spm based transposable element system for gene isolation in Arabidopsis thaliana.’ PhD thesis. Wageningen, The Netherlands: Wageningen Agricultural University; 1996. [Google Scholar]
- Aarts MGM, Corzaan P, Stiekema WJ, Pereira A. A two-element Enhancer-Inhibitor transposon system in Arabidopsis thaliana. Mol Gen Genet. 1995;247:555–564. doi: 10.1007/BF00290346. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Giish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Angenent GC, Busscher M, Franken J, Mol JNM, van Tunen AJ. Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell. 1992;4:983–993. doi: 10.1105/tpc.4.8.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, Colombo L, van Tunen AJ. Petal and stamen formation in petunia is regulated by the homeotic gene FBP1. Plant J. 1993;3:101–112. doi: 10.1046/j.1365-313x.1993.04010101.x. [DOI] [PubMed] [Google Scholar]
- Angerer LM, Angerer RC. Localization of mRNAs by in situ hybridization. Methods Cell Biol. 1991;35:37–71. doi: 10.1016/s0091-679x(08)60568-3. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvazer J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743. [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: Genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis gene AGAMOUS by the APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQJ, Gerentes D, Perez P, Smyth DR. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser JC, Robinson-Beers K, Gasser CS. The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell. 1995;7:333–345. doi: 10.1105/tpc.7.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gashler AL, Swaminathan S, Sukhatme VP. A novel repression module, an extensive activation domain, and a bipartite nuclear localisation signal defined in the immediate-early transcription factor Egr-1. Mol Cell Biol. 1993;13:4556–4571. doi: 10.1128/mcb.13.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- Gubler U, Hoffman BJ. A simple and very efficient method for generating complimentary DNA libraries. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Huala E, Sussex IM. LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development. Plant Cell. 1992;4:901–913. doi: 10.1105/tpc.4.8.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M, Schneitz K, Pruitt RE. Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell. 1995;7:57–64. doi: 10.1105/tpc.7.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. Function of the APETALA1 gene during Arabidopsis floral development. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen-Timmen U, Lemaire P, Mattei M-G, Revelant O, Charnay P. Structure, chromosome mapping and regulation of the mouse zinc-finger gene Krox-24; evidence for a common regulatory pathway for immediate-early serum-response genes. Gene. 1989;80:325–336. doi: 10.1016/0378-1119(89)90296-5. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin SA, Savidge B, Yanofsky M. Molecular basis of the cauliflower phenotype in Arabidopsis. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki MK, Okada K, Nishino E, Shimura Y. Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development. 1988;104:195–203. [Google Scholar]
- Kunst L, Klenz JE, Martinez-Zapater J, Haughn GW. AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell. 1989;1:1195–1208. doi: 10.1105/tpc.1.12.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JD, Ray S, Ray A. sin1, a mutation affecting female fertility in Arabidopsis, interacts with mod1, its recessive modifier. Genetics. 1994;137:1101–1110. doi: 10.1093/genetics/137.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Keijzer CJ, Koornneef M. A seed shape mutant of Arabidopsis that is affected in integument development. Plant Cell. 1994;6:385–392. doi: 10.1105/tpc.6.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development. 1995;121:975–991. doi: 10.1242/dev.121.4.975. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF. A gene triggering flower formation in Arabidopsis. Nature. 1995;377:522–524. doi: 10.1038/377522a0. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- ————— Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol Biol. 1995;28:767–784. doi: 10.1007/BF00042064. [DOI] [PubMed] [Google Scholar]
- ————— Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell. 1997;9:393–408. doi: 10.1105/tpc.9.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrusan Z, Reiser L, Feldmann KA, Fischer RL, Haughn GW. Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell. 1994;6:333–349. doi: 10.1105/tpc.6.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, den Boer BGW, Jofuku KD. Regulation of Arabidopsis flower development. Plant Cell. 1993;5:1183–1193. doi: 10.1105/tpc.5.10.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimienta E, Polito VS. Ovule abortion in ‘Nonpareil’ almond (Prunus dulcis [Mill.] D.A. Webb) Am J Bot. 1982;69:913–920. [Google Scholar]
- Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haughn GW, Fischer RL. The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell. 1995;83:735–742. doi: 10.1016/0092-8674(95)90186-8. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell. 1992;4:1237–1249. doi: 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodkiewicz B. Callose in cell walls during megasporogenesis in angiosperms. Planta. 1970;93:39–47. doi: 10.1007/BF00387650. [DOI] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature. 1995;378:199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- Schneitz K, Hülskamp M, Pruitt RE. Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 1995;7:731–749. [Google Scholar]
- Schneitz K, Hülskamp M, Kopczak SD, Pruitt RE. Dissection of sexual organ ontogenesis: A genetic analysis of ovule development in Arabidopsis thaliana. Development. 1997;124:1367–1376. doi: 10.1242/dev.124.7.1367. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Development. 1991;119:745–765. doi: 10.1105/tpc.3.8.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam P. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 1993;3:739–744. [Google Scholar]
- Vishnyakova MA. Callose as an indicator of sterile ovules. Phytomorphology. 1991;41:245–252. [Google Scholar]
- Weigel D. The genetics of flower development: From floral induction to ovule morphogenesis. Annu Rev Genetics. 1995;29:19–39. doi: 10.1146/annurev.ge.29.120195.000315. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcriptional factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Yanofsky M. Floral meristems to floral organs: Genes controlling early events in Arabidopsis flower development. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:167–188. [Google Scholar]