Abstract
Background
Previous work from our laboratory demonstrated a role for the Drosophila Limonly (dLmo) gene in regulating behavioral responses to cocaine. Herein, we examined whether dLmo influences the flies’ sensitivity to ethanol’s sedating effects. We also investigated whether 1 of the mammalian homologs of dLmo, Lmo3, is involved in behavioral responses to ethanol in mice.
Methods
To examine dLmo function in ethanol-induced sedation, mutant flies with reduced or increased dLmo expression were tested using the loss of righting (LOR) assay. To determine whether mouse Lmo3 regulates behavioral responses to ethanol, we generated transgenic mice expressing a short-hairpin RNA targeting Lmo3 for RNA interference-mediated knockdown by lentiviral infection of single cell embryos. Adult founder mice, expressing varying amounts of Lmo3 in the brain, were tested using ethanol loss-of-righting-reflex (LORR) and 2-bottle choice ethanol consumption assays.
Results
We found that in flies, reduced dLmo activity increased sensitivity to ethanol-induced sedation, whereas increased expression of dLmo led to increased resistance to ethanol-induced sedation. In mice, reduced levels of Lmo3 were correlated with increased sedation time in the LORR test and decreased ethanol consumption in the 2-bottle choice protocol.
Conclusions
These data describe a novel and conserved role for Lmo genes in flies and mice in behavioral responses to ethanol. These studies also demonstrate the feasibility of rapidly translating findings from invertebrate systems to mammalian models of alcohol abuse by combining RNA interference in transgenic mice and behavioral testing.
Keywords: Ethanol Sedation, Alcohol Consumption, Drosophila Lim-Only, Lmo3, Transgenic RNA Interference
COMPLEX GENETIC AND environmental components contribute to the risk of development of alcoholuse disorders (AUDs). A variety of genes have been identified that contribute to a higher risk for AUDs (Gelernter and Kranzler, 2009). These include genes involved in ethanol metabolism, such as ALDH2 and ADH1B, as well as several receptors and signaling molecules regulating neurotransmission, such as the GABAA receptor (Ducci and Goldman, 2008). However, a complete understanding of the genetic risk factors contributing to alcoholism remains elusive. We use the fruit fly Drosophila melanogaster as a model organism to identify novel genes that regulate behavioral responses to ethanol. There are a number of similarities between behavioral responses to ethanol in flies and mammals. For instance, flies exhibit increased locomotion in response to a low dose of ethanol, and exhibit motor incoordination and sedation in response to higher doses (Guarnieri and Heberlein, 2003). Recent studies from our laboratory have demonstrated the utility of using Drosophila to identify novel genes that modify behavioral responses to ethanol and to translate these findings to mammalian models of AUDs (Corl et al., 2009).
In an unbiased genetic screen for mutants with altered sensitivity to cocaine, we identified mutations in the Drosophila LIM-only (dLmo, also known as Beadex, Bx) gene, which encodes a transcriptional regulator consisting of 2 tandem LIM domains (Kadrmas and Beckerle, 2004). Reduced dLmo function increases behavioral responsiveness to cocaine, whereas increasing dLmo levels has the opposite effect (Tsai et al., 2004). Herein, we report that dLmo also modulates sedation in response to ethanol in a bidirectional manner. To translate these findings to a mammalian system, we used RNA interference (RNAi) to reduce expression of 1 of the mammalian homologs of dLmo, Lmo3. We employed a technique to rapidly generate transgenic mice globally expressing short-hairpin RNAs (shRNAs) targeting Lmo3 by infecting single-cell embryos with lentivirus (Lois et al., 2002; Rubinson et al., 2003; Tiscornia et al., 2003). This method produced founder mice with varying degrees of Lmo3 knockdown. When tested for behavioral responses to ethanol, transgenic mice exhibited altered ethanol sedation and consumption that correlated with Lmo3 expression levels. These results suggest a role for Lmo3 in ethanol-related behaviors in mice, and demonstrate the feasibility of rapidly validating novel genes, identified in invertebrate systems, in behavioral responses to ethanol in mammals.
MATERIALS AND METHODS
Drosophila Culture and Strains
All flies were maintained on standard cornmeal and molasses agar food at 25°C and 70% humidity. EP1306 was originally isolated from a collection of X-linked EP lines (Rorth, 1996), and showed reduced dLmo expression and increased cocaine sensitivity (Tsai et al., 2004). BxJ is a well-characterized overexpression allele of dLmo/Bx (Shoresh et al., 1998), and showed reduced behavioral responses to cocaine (Tsai et al., 2004). Both lines were out-crossed for at least 5 generations to a w1118 stock isogenic for Chromosomes II and III; these “w; iso” flies served as the genetic background control for experiments.
Drosophila Behavioral Assays and Ethanol Absorption
Ethanol sedation (loss of righting, LOR) assays were performed essentially as previously described (Corl et al., 2009; Rothenfluh et al., 2006). Samples of 25 to 30 male flies were allowed to equilibrate for approximately 10 minutes to humidified air in the booz-o-mat apparatus (Wolf et al., 2002) before starting exposure to a 100:50 mixture of ethanol vapor:humidified air (100:50 E:A). Ethanol exposure commenced at 0 minutes of the assay and was continuous thereafter. At 2-minute intervals, each tube of flies (8 tubes per assay) was twirled and the number of flies that appeared unable to right themselves was scored by an experimenter blinded to sample identity. Typical assay duration was 30 minutes. To measure ethanol absorption, samples of 25 flies were exposed to 100:50 E:A for 15 minutes and were immediately snap-frozen on dry ice and processed to determine internal ethanol concentration using a kit (Genzyme Diagnostics, Framingham, MA).
Animals
C57BL/6J mice were used for single cell embryo injections and subsequent behavioral testing. CD1 females were used for embryo implantation. Animals were group housed. Food and water were provided at all times, and animals were on a 12-hour light-dark cycle. All animal protocols were approved by the Ernest Gallo Clinic and Research Center Institutional Animal Care and Use Committee.
Design and Production of Lentivirus Expressing shRNAs
Three different 19-nucleotide small interfering RNAs were designed to target mouse Lmo3 (NM_207222) using the Wistar siRNA design tool (Levenkova et al., 2004). The three 19-nucleotide targeting sequences are listed in the Supplemental Table S1. Target sequences, including the shScr control sequence, were incorporated into hairpins and cloned into the lentiviral vector pLL3.7 as described previously (Lasek et al., 2007). The shScr sequence encodes a nonspecific hairpin, predicted to not target any gene in the mouse genome. Lentivirus was produced from shScr and shLmo3.8 plasmids in 293FT cells using the ViraPower™ packaging mix (Invitrogen, Carlsbad, CA) as described previously (Lasek et al., 2007).
Testing shRNA Efficacy in Neuro-2a Cells
Neuro-2a cells (American Type Culture Collection, Manassas, VA) were grown in DMEM plus 10% fetal bovine serum and 5% CO2. Cells were seeded into 12-well dishes, and transfected with pLL3.7 plasmids containing shLmo3 and shScr sequences using Lipofectamine™ 2000 and Opti-MEM media (Invitrogen) according to the manufacturer’s instructions. Forty-eight hours after transfection, RNA was isolated using the RNeasy® Mini Kit according to manufacturer’s instructions (Qiagen, Valencia, CA). Total RNA was treated with RNase-free DNase (Promega, Madison, WI) to remove genomic DNA contamination.
Quantitative PCR
cDNA synthesis and TaqMan® quantitative PCR (qPCR) were performed as described in Lasek and colleagues (2007). The sequences of the mouse Lmo1, Lmo2, and Lmo3 primers are listed in the Supplemental Table S1. Predesigned primers to Lmo4 (Assay ID Mm00495373) and mouse Gapdh (product number 4352932E) were purchased from Applied Biosystems (Foster City, CA).
Generation of Transgenic Mice Expressing shRNAs
C57BL/6J mice were superovulated at 3 to 4 weeks of age, and embryos were collected. High-titer lentivirus (>107 pg p24 antigen/ml) in a volume of 10 to 100 pl was injected into the perivitelline space of single-cell embryos using a pulled glass pipette, as described in Lois and colleagues (2002) and implanted into pseudopregnant CD1 females. Of 141 injected embryos, we obtained 63 pups, 39 derived from shLmo3.8 injections and 24 derived from shScr injections. To confirm genomic insertion of the lentivirus, we isolated DNA from tail clips using standard methods. PCR amplification was performed with primers against the pLL3.7 vector and green fluorescent protein (GFP) encoded by the lentiviral vector. Primer sequences are listed in the Supplemental Table S1. Of the 63 pups analyzed, 13 (6 females, 7 males) of 39 (33%) shLmo3.8-injected animals and 9 (5 females, 4 males) of 24 (37.5%) shScr-injected animals were positive for virus by genotyping. Twelve shLmo3.8 and 8 shScr animals were used for behavioral testing.
Processing of Transgenic Mouse Brains and GFP Fluorescence Imaging
After the completion of behavioral tests on transgenic mice, animals were euthanized with CO2 and decapitated. Brains were rapidly removed and cut in half through the midline. Half of the brain was placed in 4% paraformaldehyde for 24 hours to post-fix for GFP imaging and then stored in phosphate buffer. The other half of the brain was dissected into forebrain, midbrain, and cerebellum sections, and snap-frozen in liquid nitrogen for subsequent RNA isolation using Trizol® (Invitrogen) and qPCR. To image GFP fluorescence, 40-μm sagittal brain sections were cut using a vibrating blade microtome (Leica Microsystems, Bannockburn, IL) and mounted on slides. Images were acquired using a Zeiss confocal microscope, and visualized using Zeiss LSM software (Carl Zeiss MicroImaging, LLC, Thornwood, NY).
Ethanol Loss-of-Righting-Reflex and 2-Bottle Choice Consumption Assays
Mice aged 9 to 16 weeks were tested first for ethanol-induced loss-of-righting-reflex (LORR) at 3 doses of ethanol (3.2, 3.6, and 4.0 g/kg) in a random-order balanced design with a 1 week interval between each dose. The same mice were used for the 2-bottle-choice ethanol consumption experiment, and were allowed to rest for 2 weeks after the final LORR test. LORR and 2-bottle choice ethanol consumption experiments were performed as described in Kapfhamer and colleagues (2008). Both males and females were tested; we did not detect a significant difference in Lmo3 expression between males and females, so data from males and females were combined in the analysis.
RESULTS
Mutations in Drosophila dLmo Affect Ethanol Sedation
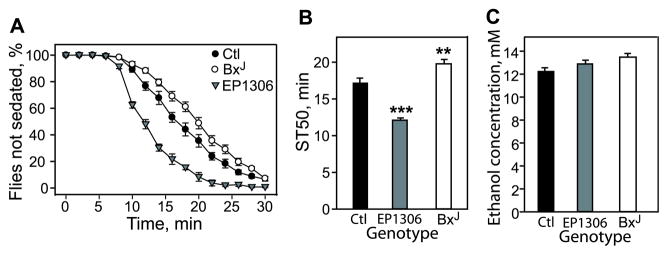
To investigate whether dLmo might play a role in ethanolinduced sedation, the LOR assay was used to test control flies and 2 dLmo mutants: BxJ, with increased levels of dLmo, and EP1306, with reduced dLmo (Tsai et al., 2004). When exposed to ethanol, EP1306 mutant flies sedated more quickly, showing a significantly reduced ST50 (time to 50% sedation) of 12.1 minutes compared with 17.1 minutes for the w;iso control flies. BxJ flies, by contrast, exhibited increased resistance to sedation, with an ST50 of 19.8 minutes (Fig. 1A,B). Ethanol absorption was not altered in the mutants (Fig. 1C). Thus, dLmo negatively regulates the flies’ sensitivity to ethanol-induced sedation, and this regulation is bidirectional.
Fig. 1.
dLmo/Bx mutants display altered resistance to ethanol sedation and no change in ethanol absorption. (A) Ethanol sedation curves indicating that EP1306 flies show increased sensitivity and BxJ flies show decreased sensitivity to ethanol sedation in the loss of righting (LOR) assay when compared with control (Ctl) flies. Ctl flies were in the w;iso genetic background. (B) The median sedation time (ST50)—the time required for half of the ethanol-exposed flies to show LOR—was calculated by linear interpolation. Error bars represent SEM, and asterisks denote statistical significance by one-way ANOVA followed by post hoc Holm-Sidak testing (**p < 0.01; ***p < 0.001; n = 9 to 12, where n is the number of samples, not the number of flies). (C) Samples of mutant and control flies were exposed to ethanol vapor under identical conditions as for behavioral assays, and were snap-frozen after 15 minutes and processed for internal ethanol absorption. No significant difference was seen between mutant and control flies (n = 10 or 11).
shRNAs Reduce Lmo3 Expression in Cell Culture
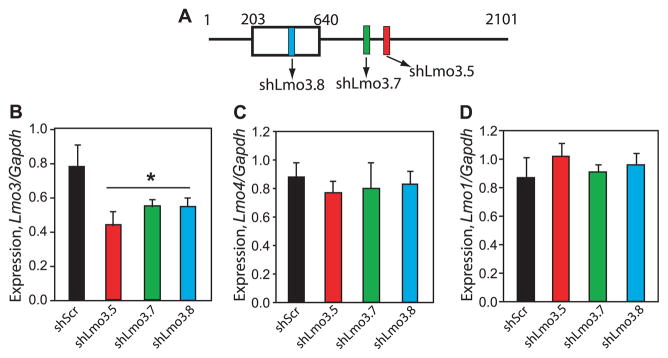
To begin the investigation of whether the mammalian Lmo genes are involved in behavioral responses to ethanol, we targeted Lmo3 in the mouse using RNAi. Lmo3 is 1 of 4 mammalian homologs of dLmo, and is expressed at high levels in brain regions important for behavioral responses to ethanol, particularly in the forebrain (Bulchand et al., 2003; Hinks et al., 1997). We designed 3 independent shRNAs specific for Lmo3 (Fig. 2A). To test their ability to reduce Lmo3 expression, lentiviral plasmids encoding each Lmo3 shRNA (shLmo3) were transfected into mouse Neuro-2a cells in parallel to the control shScr plasmid, which encodes a scrambled shRNA sequence that does not target any mouse gene (Lasek et al., 2007). Two days after transfection, RNA was isolated and subjected to first-strand cDNA synthesis and real-time qPCR. We found that Lmo3 levels were significantly reduced by 30 to 43% in cells transfected with each of the shLmo3 plasmids compared with the shScr control (Fig. 2B). Transfection of shLmo3 plasmids had no detectable effect on the expression of the closely related Lmo family members Lmo4 (Fig. 2C), Lmo1 (Fig. 2D), and Lmo2 (data not shown). These results indicate that these shRNAs specifically attenuated expression of Lmo3 without affecting expression of the other Lmo genes.
Fig. 2.
Efficacy of short-hairpin RNAs (shRNAs) targeting Lmo3 for RNA interference in Neuro-2a cells. (A) Schematic illustrating the position of shRNAs targeting the Lmo3 transcript. Colored boxes show shRNA location, open box illustrates the protein coding region, and numbers indicate the nucleotide position along the Lmo3 mRNA. (B) Expression of Lmo3 in Neuro-2a cells transfected with lentiviral plasmids expressing shRNAs targeting Lmo3 (shLmo3.5, shLmo3.7, and shLmo3.8) or the control shScr. RNA was isolated 2 days after transfection and subjected to qPCR. Lmo3 expression is normalized to expression of the housekeeping gene, Gapdh. Asterisk indicates a significant difference between the shLmo3 constructs and shScr by one-way ANOVA (*p = 0.007; n = 3). (C, D) Lmo4 (C) and Lmo1 (D) expression in transfected Neuro-2a samples described in (B).
Transgenic Mice Display a Range of Lmo3 Expression in the Brain
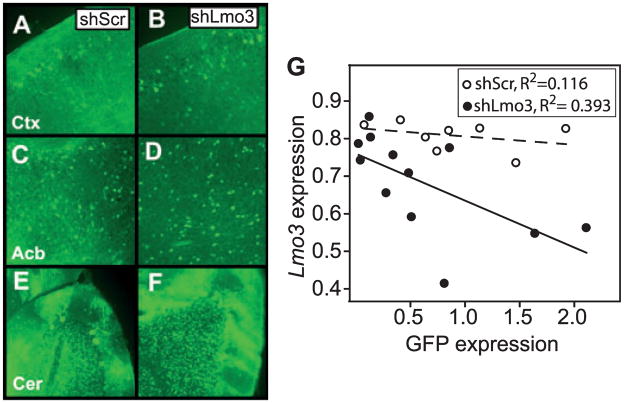
To knock down Lmo3 expression in mice, we injected single- cell embryos with lentivirus expressing either shLmo3.8 or shScr. We reasoned that infection of individual mice at the embryo stage with shLmo3 lentivirus would yield animals with a range of Lmo3 expression due to variation in the number of viruses integrated into the genome of each founder (Lois et al., 2002) as well as potential effects of the integration site, generating an allelic series for behavioral testing. After behavioral testing (see below), the brains of founder animals were analyzed for expression of GFP, which is encoded by the integrated virus. We found that GFP was widely expressed in the brains of animals containing either integrated shScr or shLmo3.8 lentivirus. GFP fluorescence was visible as bright spots in cell bodies and processes of neurons in many brain regions, including cells in the cortex, nucleus accumbens, and cerebellum (Fig. 3A–F). No gross differences were observed in the levels and patterns of GFP fluorescence between groups of animals that had been injected with shScr or shLmo3.8 lentivirus. However, the levels of GFP fluorescence varied among individual animals within each group. To quantify the amount of GFP and Lmo3 transcript in individual animals, we performed qPCR using RNA isolated from the forebrain of each founder animal (Fig. 3G). Within the shScr group, GFP expression (normalized to Gapdh expression as a control for input RNA) ranged from 0.08 to 1.92; comparable levels of GFP expression were observed in the shLmo3.8 group (range: 0.03 to 1.64). Infection with shScr did not affect expression of Lmo3, as Lmo3 expression varied from 0.74 to 0.85 in these animals, similar to the levels of Lmo3 observed in animals that did not contain integrated virus (data not shown). In contrast, expression of Lmo3 ranged from 0.41 to 0.86 in shLmo3.8-infected animals, indicating that shLmo3.8 was, at least in some animals, effective in reducing levels of Lmo3 in vivo. Importantly, GFP expression was inversely correlated with Lmo3 expression in shLmo3.8- infected mice (Fig. 3G, R = −0.628, p = 0.029), indicating that higher levels of integrated shLmo3.8 virus correlated with decreased Lmo3 expression in the brains of these mice.
Fig. 3.
Characterization of transgenic mice expressing shLmo3.8 or shScr. (A–F) Green fluorescent protein (GFP) fluorescence in representative sagittal adult brain sections of transgenic mice infected at the single-cell embryo stage with lentivirus encoding shLmo3.8 (B, D, F) or shScr (A, C, E). GFP expression from the viral vector is visible in cell bodies (bright spots) and processes of neurons throughout the brain, including the cortex (A, B, Ctx), nucleus accumbens (C, D, Acb), and cerebellum (E, F, Cer). Hazy green areas represent background autofluorescence from the tissue, rather than infection per se. (G) Expression of GFP and Lmo3 transcript levels were negatively correlated in the forebrains of transgenic shLmo3.8 (closed circles, n = 12), but not shScr (open circles, n = 8) mice. GFP (x-axis) and Lmo3 (y-axis) expression values were normalized relative to expression of the housekeeping gene Gapdh.
Lmo3 Expression Correlates with Levels of Behavioral Responses to Ethanol
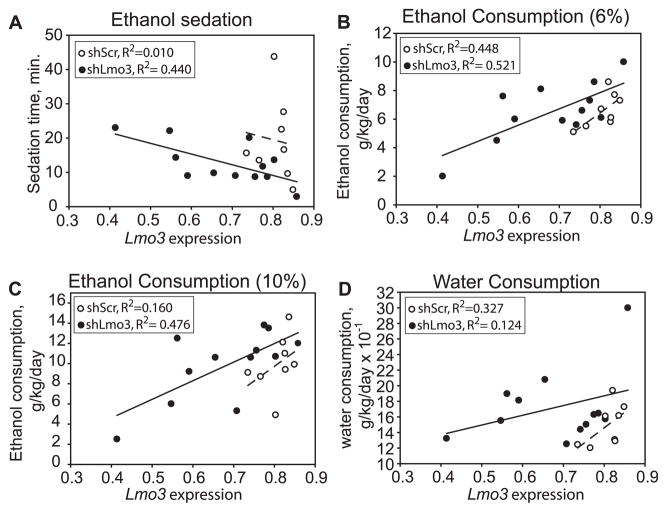
To determine if Lmo3 is involved in behavioral responses to ethanol, we tested transgenic founder mice expressing shScr or shLmo3.8 for their response to the acute sedative /hypnotic effect of ethanol in the LORR test. We observed a significant negative correlation between expression levels of Lmo3 and sedation time at a dose of 3.2 g/kg ethanol (Fig. 4A, R = −0.663, p = 0.019), suggesting that reduced expression of Lmo3 leads to increased sensitivity to the sedating effect of this dose of ethanol. Sedation time at higher doses of ethanol (3.6 and 4 g/kg) was not significantly correlated with Lmo3 expression (data not shown). The same animals were tested 2 weeks after the completion of the LORR test for voluntary ethanol consumption using a 2-bottle choice protocol. Significant positive correlations were observed between Lmo3 expression and ethanol consumption at 6% (Fig. 4B, R = 0.719, p = 0.008), 10% (Fig. 4C, R = 0.691, p = 0.013), and 14% ethanol (R = 0.591, p = 0.043, data not shown). Although not significant, we observed a trend correlating Lmo3 expression with consumption at 2 additional ethanol concentrations, 3% (R = 0.540, p = 0.07) and 20% (R = 0.527, p = 0.078). To determine if Lmo3 levels were specifically correlated with ethanol consumption or were more generally associated with drinking behavior, we examined the correlation between Lmo3 expression and water consumption, and observed no significant relationship (Fig. 4D, R = 0.353, p = 0.261). Together, these results indicate that lower levels of Lmo3 are associated with decreased voluntary ethanol consumption in mice.
Fig. 4.
Correlations between Lmo3 expression and behavioral responses to ethanol in transgenic shLmo3.8 and shScr mice. In all graphs, normalized Lmo3 expression is plotted on the x-axis and the behavioral response on the y-axis. Mice expressing shLmo3.8 (n = 12) are represented by closed circles and shScr (n = 8) by open circles. (A) Negative correlation between Lmo3 expression and ethanol sedation in the loss-of-righting-reflex test with a 3.2 g/kg dose of ethanol. (B, C) Positive correlation between Lmo3 expression and 2-bottle choice ethanol consumption of 6% (B) and 10% (C) ethanol. (D) Water consumption and Lmo3 expression were not significantly correlated.
DISCUSSION
The work presented here demonstrates that the Drosophila Lmo gene, dLmo, and its mouse homolog Lmo3 regulate behavioral responses to alcohol. We found that responses in the fly broadly paralleled to those in the mouse. Reduced dLmo expression (observed in the EP1306 mutant) caused increased sensitivity to the sedating effect of ethanol. Similarly, in the mouse, reduced Lmo3 expression correlated with increased sedation time in the LORR assay. In addition to its effect on ethanol sedation, reducing Lmo3 levels correlated with decreased ethanol consumption, suggesting that Lmo3 may play a role in alcohol preference in mammals.
Lmo3 is 1 of 4 Lmo genes in the mouse. We have studied another member of this family, Lmo4, in behavioral responses to cocaine and ethanol. We found that mice with a 50% reduction in Lmo4 levels exhibit enhanced locomotor sensitization to cocaine (Lasek et al., 2010). However, these mice did not show any differences in several behavioral assays related to alcohol, including locomotor stimulation, LORR, and 2-bottle choice ethanol consumption (David Kapfhamer, personal communication). Interestingly, however, Drosophila dLmo, which is the only Lmo gene in flies, regulates behavioral responses to both cocaine and ethanol. These findings suggest that inmammals, the role of the Lmo genes in regulating responses to abused drugs has diverged such that different homologs function in pathways that are drug-specific.
How might the Lmo genes regulate behavioral responses to ethanol? dLmo and Lmo3 are expressed in several brain regions in flies and in mice. Gross abnormalities in the brain have not been observed in either dLmo mutant flies, the shLmo3 transgenic mice described here, or Lmo3 knockout mice, which are viable, fertile, and exhibit normal organ development (Tsai et al., 2004; Tse et al., 2004). dLMO and LMO3 are members of the LIM-only family of proteins, whose function is to modulate transcription by binding to DNA-binding proteins through their 2 tandem LIM domains (Kadrmas and Beckerle, 2004). In vertebrates, LMO proteins are thought to play important roles in specifying cell fate, particularly in the developing nervous system (Bulchand et al., 2003; Joshi et al., 2009; Lee et al., 2008; Remedios et al., 2004). One possibility is that LMO3 affects the patterning of specific brain structures during development, such as the cortex or amygdala (Bulchand et al., 2003; Remedios et al., 2004), through its ability to regulate transcription. Subtle changes in patterning during development might later affect behavioral responses to ethanol in adults. Little is known regarding the molecular mechanism of LMO3 function or its transcriptional targets, although binding partners of LMO3 include the neuronal transcription factor HEN2 (Aoyama et al., 2005), p53 (Larsen et al., 2010), and the calcium and integrin binding protein, CIB (Hui et al., 2009). In the adult animal, Lmo3 expression is increased by seizure activity in the hippocampus (Hinks et al., 1997). Lmo3 expression is also increased by dopamine treatment of glial cell cultures (Shi et al., 2001). The mechanism of LMO3 function in behavioral responses to ethanol has not been studied. It is tempting to speculate that Lmo3 expression/and or function is regulated by ethanol in the adult brain, and that Lmo3 in turn modulates behavioral responses to ethanol in the adult animal. As an attempt to address this possibility, we examined Lmo3 expression in response to ethanol in the cortex, striatum, hippocampus, cerebellum, and olfactory bulb by qPCR, and observed no significant changes following single or repeated ethanol injections (Amy W. Lasek, Ulrike Heberlein, and David Kapfhamer, unpublished results), suggesting that Lmo3 expression is not regulated by ethanol, at least under these conditions. Future studies will examine the function of LMO3 in ethanol-related behaviors in more detail.
Invertebrate model systems, such as Drosophila and Caenorhabditis elegans, provide an opportunity to identify novel genes that modulate acute responses to alcohol (Wolf and Heberlein, 2003). However, validation of these genes in mammalian systems, in which more complex aspects of alcoholrelated behaviors can be modeled, is necessary to provide relevance to human AUDs. Herein, we have used a technique to rapidly generate transgenic gene knockdown animals for behavioral testing. In theory, founder animals can be bred to maintain gene knockdown (Lois et al., 2002), although we found by breeding 1 of our founders that GFP fluorescence was maintained only through the first generation and appeared to be silenced in the second and third generations (data not shown). This may be due to silencing at the lentiviral integration site, perhaps due to toxicity from high shRNA expression (Cao et al., 2005; Grimm et al., 2006).
Traditional methods for testing gene function in mice have relied on the use of constitutive and conditional gene knockouts and classical transgenic animals. Producing these mice is expensive and time-consuming, and may not yield an easily interpretable behavioral phenotype due to potential developmental compensations from the complete loss of gene function. In addition to the relative ease and speed of their generation, mice with gene knockdown by shRNA may be advantageous, as gene function is not completely eliminated, thus circumventing potential lethality associated with null alleles as well as the above-mentioned compensatory mechanisms. By utilizing transgenic mice expressing lentiviral-delivered shRNAs, animals with different degrees of gene knockdown can be produced rapidly as a screening tool to complement the generation of traditional gene knockout animals. As we have done here, founder mice may be examined for correlations in behavioral responses to ethanol as a first determinant of whether a gene may play a role in alcoholrelated behaviors.
Supplementary Material
Primer and Probe Sequences
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Acknowledgments
We thank Shirley Pease for providing details on injection of mouse embryos with lentivirus and David Kapfhamer for helpful discussions. This work was funded by the Department of Defense through the U.S. Army Medical Research and Materiel Command (grant no. W81XWH-06-1-0156; the U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick, MD 21702-5014, is the awarding and administering acquisition office. The content of the information represented does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred), the State of California for medical research on alcohol and substance abuse through the University of California at San Francisco, and the National Institute on Alcohol Abuse and Alcoholism (AA16654-INIA).
References
- Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, Tokita H, Ohira M, Nakagawara A. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Subramanian L, Tole S. Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn. 2003;226:460–469. doi: 10.1002/dvdy.10235. [DOI] [PubMed] [Google Scholar]
- Cao W, Hunter R, Strnatka D, McQueen CA, Erickson RP. DNA constructs designed to produce short hairpin, interfering RNAs in transgenic mice sometimes show early lethality and an interferon response. J Appl Genet. 2005;46:217–225. [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE, Heberlein U. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Ducci F, Goldman D. Genetic approaches to addiction: genes and alcohol. Addiction. 2008;103:1414–1428. doi: 10.1111/j.1360-0443.2008.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet. 2009;126:91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Guarnieri DJ, Heberlein U. Drosophila melanogaster, a genetic model system for alcohol research. Int Rev Neurobiol. 2003;54:199–228. doi: 10.1016/s0074-7742(03)54006-5. [DOI] [PubMed] [Google Scholar]
- Hinks GL, Shah B, French SJ, Campos LS, Staley K, Hughes J, Sofroniew MV. Expression of LIM protein genes Lmo1, Lmo2, and Lmo3 in adult mouse hippocampus and other forebrain regions: differential regulation by seizure activity. J Neurosci. 1997;17:5549–5559. doi: 10.1523/JNEUROSCI.17-14-05549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L, Ji C, Hui B, Lv T, Ha X, Yang J, Cai W. The oncoprotein LMO3 interacts with calcium- and integrin-binding protein CIB. Brain Res. 2009;1265:24–29. doi: 10.1016/j.brainres.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Joshi K, Lee S, Lee B, Lee JW, Lee SK. LMO4 controls the balance between excitatory and inhibitory spinal V2 interneurons. Neuron. 2009;61:839–851. doi: 10.1016/j.neuron.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat RevMol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kapfhamer D, Bettinger JC, Davies AG, Eastman CL, Smail EA, Heberlein U, McIntire SL. Loss of RAB-3/A in Caenorhabditis elegans and the mouse affects behavioral response to ethanol. Genes Brain Behav. 2008;7:669–676. doi: 10.1111/j.1601-183X.2008.00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S, Yokochi T, Isogai E, Nakamura Y, Ozaki T, Nakagawara A. LMO3 interacts with p53 and inhibits its transcriptional activity. Biochem Biophys Res Commun. 2010;392:252–257. doi: 10.1016/j.bbrc.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Lasek AW, Janak PH, He L, Whistler JL, Heberlein U. Downregulation of mu opioid receptor by RNA interference in the ventral tegmental area reduces ethanol consumption in mice. Genes Brain Behav. 2007;6:728–735. doi: 10.1111/j.1601-183X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- Lasek AW, Kapfhamer D, Kharazia V, Gesch J, Giorgetti F, Heberlein U. Lmo4 in the nucleus accumbens regulates cocaine sensitivity. Genes Brain Behav. 2010;9:817–824. doi: 10.1111/j.1601-183X.2010.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee B, Joshi K, Pfaff SL, Lee JW, Lee SK. A regulatory network to segregate the identity of neuronal subtypes. Dev Cell. 2008;14:877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenkova N, Gu Q, Rux JJ. Gene specific siRNA selector. Bioinformatics. 2004;20:430–432. doi: 10.1093/bioinformatics/btg437. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Remedios R, Subramanian L, Tole S. LIM genes parcellate the embryonic amygdala and regulate its development. J Neurosci. 2004;24:6986–6990. doi: 10.1523/JNEUROSCI.0001-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfluh A, Threlkeld RJ, Bainton RJ, Tsai LT, Lasek AW, Heberlein U. Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell. 2006;127:199–211. doi: 10.1016/j.cell.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Shi J, Cai W, Chen X, Ying K, Zhang K, Xie Y. Identification of dopamine responsive mRNAs in glial cells by suppression subtractive hybridization. Brain Res. 2001;910:29–37. doi: 10.1016/s0006-8993(01)02393-9. [DOI] [PubMed] [Google Scholar]
- Shoresh M, Orgad S, Shmueli O, Werczberger R, Gelbaum D, Abiri S, Segal D. Overexpression Beadex mutations and loss-of-function heldup-a mutations in Drosophila affect the 3′ regulatory and coding components, respectively, of the Dlmo gene. Genetics. 1998;150:283–299. doi: 10.1093/genetics/150.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LT, Bainton RJ, Blau J, Heberlein U. Lmo mutants reveal a novel role for circadian pacemaker neurons in cocaine-induced behaviors. PLoS Biol. 2004;2:e408. doi: 10.1371/journal.pbio.0020408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse E, Smith AJ, Hunt S, Lavenir I, Forster A, Warren AJ, Grutz G, Foroni L, Carlton MB, Colledge WH, Boehm T, Rabbitts TH. Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol Cell Biol. 2004;24:2063–2073. doi: 10.1128/MCB.24.5.2063-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FW, Heberlein U. Invertebrate models of drug abuse. J Neurobiol. 2003;54:161–178. doi: 10.1002/neu.10166. [DOI] [PubMed] [Google Scholar]
- Wolf FW, Rodan AR, Tsai LT, Heberlein U. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J Neurosci. 2002;22:11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer and Probe Sequences
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.