Abstract
Stable isotope dilution-multiple reaction monitoring-mass spectrometry (SID-MRM-MS) has emerged as a promising platform for verification of serological candidate biomarkers. However, cost and time needed to synthesize and evaluate stable isotope peptides, optimize spike-in assays, and generate standard curves, quickly becomes unattractive when testing many candidate biomarkers. In this study, we demonstrate that label-free multiplexed MRM-MS coupled with major protein depletion and 1-D gel separation is a time-efficient, cost-effective initial biomarker verification strategy requiring less than 100 μl serum. Furthermore, SDS gel fractionation can resolve different molecular weight forms of targeted proteins with potential diagnostic value. Because fractionation is at the protein level, consistency of peptide quantitation profiles across fractions permits rapid detection of quantitation problems for specific peptides from a given protein. Despite the lack of internal standards, the entire workflow can be highly reproducible, and long-term reproducibility of relative protein abundance can be obtained using different mass spectrometers and LC methods with external reference standards. Quantitation down to ~200 pg/mL could be achieved using this workflow. Hence, the label-free GeLC-MRM workflow enables rapid, sensitive, and economical initial screening of large numbers of candidate biomarkers prior to setting up SID-MRM assays or immunoassays for the most promising candidate biomarkers.
Keywords: Serum proteomes, serum biomarkers, biomarker verification, biomarker validation, label-free quantitation, multiple reaction monitoring (MRM)
Introduction
Mass spectrometry-based proteomics strategies often are used in the discovery of putative disease biomarkers. Extensive sample prefractionation and the use of high-performance mass spectrometers have facilitated in-depth analysis of many highly complex human proteomes, particularly serum or plasma proteomes.1–3 As a result of the sensitivity of current proteomics methods, up to a hundred or more candidate biomarkers can be identified readily for diverse diseases either using model systems or small numbers of patient samples or pools of samples.4–7 In addition, large numbers of additional biomarkers can be obtained readily by surveying the scientific literature.8
However, verification and laboratory-scale initial validation of large numbers of candidate biomarkers in human serum or plasma are quite challenging and have become rate-limiting steps in the biomarker pipeline.7, 9 Sandwich enzyme-linked immunosorbent assays (ELISA) can be highly specific and capable of detecting blood proteins at the low pg/mL range with high throughput. However, appropriate ELISA assays are not available for many candidate biomarkers and these assays can be difficult to multiplex. Furthermore, preparing specific antibodies and developing sandwich ELISAs for novel target proteins are lengthy and costly processes. Based on recent experience in the field, it is reasonable to expect that only a modest portion of candidate biomarkers discovered using proteomics ultimately will prove to be clinically useful. As a result, cost- and time-effective methods are needed to quickly screen large numbers of candidate biomarkers to identify those proteins worth investing the resources required to develop sandwich ELISAs or equivalent higher throughput assays.
In recent years, MRM-MS has emerged as an attractive targeted MS technique for biomarker verification and initial validation.10, 11 High selectivity of MRM is achieved by isolating a specific peptide parent ion and a high-yield fragment ion (an MRM transition) in a triple quadrupole mass spectrometer, and extensive multiplexing can be achieved where many peptides are monitored in a single run.10, 12 MRM assays typically are coupled with stable isotope dilution (SID) using chemically identical synthesized peptides to achieve absolute and reproducible quantitation.13–16 SID-MRM, coupled with peptide fractionation or specific enrichment, has been shown to detect proteins in plasma or serum in the low ng/mL range with a broad dynamic range of up to five orders of magnitude.12, 17–19 As expected, SID-MRM quantitation is highly reproducible, even if the measurements are carried out in different laboratories.20 Since stable labeled peptides have identical retention times as targeted peptides, unambiguous confirmation of the targeted peptides can be achieved even in the presence of closely co-eluting peptides which are commonly encountered in MRM analyses of complex proteomes such as serum or plasma. Interferences can also be easily detected by monitoring multiple MRM transitions of the stable labeled peptides and comparing the transition intensity ratios with the ratios from targeted peptides. However, the cost and lead time for synthesizing, purifying, and evaluating stable isotope standard peptides for absolute quantitation, as well as setting up spike-in experiments and standard curves, can be substantial, especially at the verification and early-stage validation steps where screening of a large number of putative candidate biomarkers may be of interest.
Alternative strategies have been explored to generate labeled peptides in a more cost-effective manner. This includes expression of a synthetic gene in medium containing labeled amino acids to generate isotope-labeled, concatenated peptides or full-length proteins that can be purified and trypsin digested to produce stable isotope standard peptides.10, 21–23 Chemical modification techniques such as mTRAQ and 18O-labeling also can be used to incorporate isotopic labels into peptides.24, 25 However, all these methods involve additional costs, time, and expertise. In addition, difficulties in expression of certain synthetic constructs have been encountered, and the release of concatenated tryptic peptides may differ from the natural protein.26
Despite the high selectivity of MRM, accurate quantitation of proteins in human serum can be compromised due to serum’s complexity and wide dynamic range of protein abundances, which span more than 10 orders of magnitude.7, 27 As a result, co-eluting ions may suppress desired peptide signals or incompletely resolved ions may interfere with specific MRM transitions.10, 17, 28 In addition, a protein can contain multiple isoforms or may undergo proteolytic processing or post-translational modifications that are physiologically relevant to a disease state but may not be apparent at the peptide level. Therefore, MRM quantitation strategies that could minimize or easily detect interferences and provide quantitation information on various forms of a protein are highly desirable.
In this study, we explored the use of label-free MRM quantitation of SDS gel-fractionated serum proteins (GeLC-MRM) as a rapid, first-level biomarker verification strategy. Label-free quantitation of ion peak areas recently has emerged as a promising strategy in discovery proteomics because it is simple to implement and allows relative quantitative comparisons across multiple samples in diverse experimental systems.29–33 However, label-free quantitation has only been used for LC-MRM assays in a few cases.10, 34, 35 More importantly, long-term reproducibility of label-free MRM analysis has not been demonstrated in previous studies. In this study, we observed that relative quantitation using label-free MRM is sufficiently reproducible and more appropriate as a first-level quantitation when screening large numbers of candidate biomarkers. By rigorously controlling experimental parameters, highly reproducible major protein immunodepletion, 1-D gel fractionation, and LC-MRM analysis could be achieved. Furthermore, GeLC-MRM could distinguish and quantitate different molecular forms of a protein, and reanalysis of the same sample on a different mass spectrometer yielded consistent results.
Materials and Methods
Reagents and Chemicals
Molecular biology-grade ethanol (200 proof) and iodoacetamide were purchased from Sigma-Aldrich (St. Louis, MO). Sodium dodecyl sulfate (SDS) and Tris were purchased from Bio-Rad (Hercules, CA). Dithiothreitol (DTT) was obtained from GE Healthcare (Piscataway, NJ). HPLC-grade acetonitrile was purchased from Thomas Scientific (Swedesboro, NJ). Sequencing-grade modified trypsin was purchased from Promega (Madison, WI). Yeast enolase MassPREP™ digestion standard was obtained from Waters (Milford, MA). All heavy label peptides, quantitated by amino acid analysis, were purchased from Sigma-Aldrich.
Abundant Protein Depletion
Human serum samples were processed as previously described.33 Briefly, samples were depleted of 20 abundant serum proteins using a ProteoPrep® 20 Immunodepletion Column (Sigma-Aldrich) on an ÄKTA, fast-performance liquid chromatography system (FPLC; GE Healthcare). Typically, 100 μL of diluted serum was filtered through a 0.22 μm microcentrifuge filter and injected onto the column. The flow-through fractions containing unbound proteins were collected, pooled, and precipitated with nine volumes of 200-proof ethanol, pre-chilled to −20 ºC. Ethanol supernatants were removed carefully, and protein pellets were frozen and stored at −20 ºC until use.
SDS-PAGE/In-Gel Trypsin Digestion
SDS-PAGE and in-gel trypsin digestion were performed as previously described.36 Briefly, depleted, ethanol-precipitated serum samples were resuspended in 50 mM Tris-Cl, 1% SDS, pH 8.5, reduced with 20 mM DTT for 1 h at 37 ºC, and alkylated with 60 mM iodoacetamide, pH 8.5 for 1 h at 37 ºC. For each sample, triplicate aliquots representing 10 μL of original serum were loaded into 10-well 12% NuPAGE mini-gels (Invitrogen, Carlsbad, CA) and separated using MES running buffer. Samples were electrophoresed until the tracking dye had migrated either 2 or 4 cm into the gel. Gels were stained with Colloidal Blue (Invitrogen), and each lane was subsequently sliced into uniform 1 mm slices. Corresponding slices from three replicate lanes for each depleted serum sample were combined in single wells of a 96-well pierced plate (Biomachines, Inc., Carrboro, NC) and digested with 0.02 μg/μL modified trypsin. This optimized trypsin digestion protocol allows a single person to easily and rapidly perform up to 384 tryptic in-gel digests (4 × 96-well plates) in parallel. Approximately 50 μL of peptide solution, which is equivalent to 30 μL of original serum, was recovered for each digest. In most cases, five μl of tryptic digests were injected for MRM analysis. In the spike-in, stable-label peptide experiment, one μl of tryptic digests were injected. In some cases, samples were pooled by combining aliquots of corresponding fractions from individual patients or controls. These pools and the remainder of individual sample digests were aliquoted and stored at −20 ºC until use.
Western Blot
Cathepsin D (CTSD) protein levels were verified using Western blots.37 One μg of lysate from the 1205LU metastatic melanoma cell line and ~100 ng of depleted human serum derived from a pool of nine individual serum samples were separated on 12% Bis-Tris NuPAGE gels. Proteins were transferred to Immobilon-P PVDF membranes (Millipore, Billerica, MA) and blocked with non-fat dry milk or BSA prior to incubation with the anti-Cathepsin D mAb (C0715; Sigma-Aldrich) primary antibody. The antigen-antibody complex was detected with HRP (horseradish peroxidase)-labeled secondary antibodies and SuperSignal® West Pico Chemiluminescent Substrate (Pierce Biotechnology, Rockford, IL).
Label-free Multiple Reaction Monitoring
MRM experiments were performed on a 4000 QTRAP® or a 5500 QTRAP hybrid triple quadrupole/linear ion trap mass spectrometer (AB Sciex, Foster City, CA) interfaced with a nanoACQUITY UPLC® system. Tryptic digests were injected using the partial loop injection mode onto a UPLC Symmetry trap column (180 μm i.d. x 2 cm packed with 5 μm C18 resin; Waters) and then separated by RP-HPLC on a PicoFrit® column (75-μm i.d., 15-μm tip opening; New Objective, Woburn, MA) packed in-house with 25 cm of Magic C18 3-μm reversed-phase resin (Michrom Bioresources, Auburn, CA). Chromatography was performed with Solvent A (Milli-Q water with 0.1% formic acid) and Solvent B (acetonitrile with 0.1% formic acid). Three different gradients were used in the current study. For the 73 min gradient, peptides were eluted at 200 nL/min for 5–28% B over 42 min, 28–50% B over 26 min, 50–80% B over 5 min, 80% B for 4.5 min before returning to 5% B over 0.5 min. For the 20-min gradient, peptides were eluted at 300 nL/min for 5–35% B over 15 min, 35–70% B over 5 min, 70% B for 5 min before returning to 5% B in 0.5 min. For the 24-min gradient, peptides were eluted at 400 nL/min for 5–35% B over 24 min, 35% B for 3 min before returning to 5% B in 0.5 min. To minimize sample carryover, a short blank was run between each sample. For duplicate analysis, the first set of samples was completely acquired before running the duplicate set.
MRM transitions used in this study were derived from experimental MS/MS data obtained using an LTQ Orbitrap XL or the 4000 QTRAP mass spectrometer. MRM data on the 4000 QTRAP mass spectrometer were acquired with a spray voltage of 2,800 V, curtain gas of 20 p.s.i., nebulizer gas of 10 p.s.i., interface heater temperature of 150 °C, and a pause time of 5 ms. For the 5500 QTRAP mass spectrometer, spray voltage of 2,300 V, nebulizer gas of 8 p.s.i., and a pause time of 3 ms were used. Multiple MRM transitions were monitored using unit resolution in both Q1 and Q3 quadrupoles to maximize specificity. Each MRM transition had a minimum dwell time of 15 s. Scheduled MRM also was used to reduce the number of concurrent transitions and to maximize the dwell time for each transition. The detection window was set at 4 min and target scan time was set at 1 s. Data analysis and signal-to-noise (S/N) ratio were performed using MultiQuant™ version 1.1 software (AB Sciex). The most abundant transition for each peptide was used for quantitation unless interference from the matrix was observed. In these cases, another transition free of interference was chosen for quantitation.
Results and Discussion
Overview of the Label-free GeLC-MRM Strategy
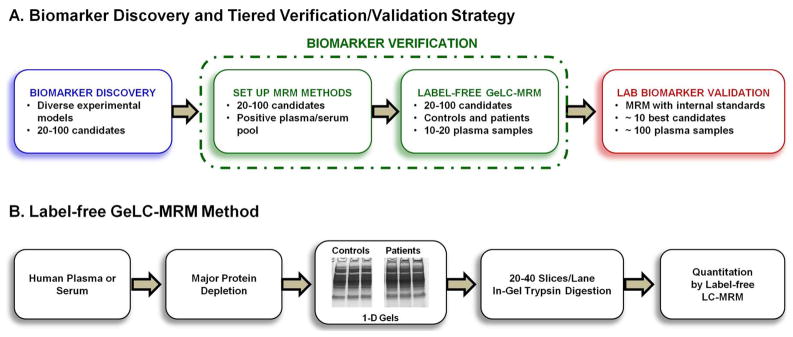
A tiered strategy for rapid verification of candidate biomarkers in human plasma or serum is shown in Figure 1A. Biomarker discovery using various proteomics techniques and experimental models often results in identification of a relatively large number of candidate biomarkers that need to be verified in human plasma or serum samples. After prioritization of these candidates based on available information, the first step in the verification process involves setting up a multiplexed MRM assay using a pool of patient samples expected to be positive for the candidate biomarkers. The serum sample is immunodepleted of abundant proteins, fractionated on 1-D gels, and digested with trypsin prior to MRM analysis (Figure 1B). Most clinically useful disease biomarkers are expected to be present in serum or plasma at low ng/mL or less,27, 38 but MRM studies using non-fractionated plasma samples were not very successful in quantifying proteins lower than the μg/mL level.10, 13 Immunodepletion of major plasma proteins allowed quantitation down to about 25 to 50 ng/mL levels.10, 17 However, this limit is still significantly higher than the mid to low pg/mL level of most well-developed ELISA assays. Limit of quantitation (LOQ) in the range of low to sub-ng/mL previously was achieved with depletion of 12 abundant proteins combined with strong cation exchange (SCX) chromatography, or by targeted enrichment.12, 17, 18 Consistent with our results, these earlier studies indicate that at least two levels of sample fractionation are necessary for reliable low ng/mL MRM quantitation of plasma proteins, unless target-specific immunoenrichment is used.
Figure 1.
Overview of the tiered label-free GeLC-MRM biomarker verification/validation strategy and method. (A) Summary of workflow for discovery, efficient verification, and laboratory-scale validation of candidate biomarkers using a tiered strategy. (B) Summary of the label-free GeLC-MRM method.
Following MRM method development, biomarker verification is completed using the abundant protein depletion/GeLC-MRM strategy to rapidly screen a modest-sized set of individual patient and control serum samples or a small number of pools of patient and control sera to determine the levels of targeted proteins. Proteins with significant differences in their expression levels between patients and controls will be carried forward to the next level analysis (laboratory-scale validation), which involves synthesis of stable isotope standard peptides followed by quantitative comparisons of larger numbers of patient and control plasma or serum. For the laboratory-scale validation, stable isotope-labeled peptides can be introduced readily into the samples after trypsin digestion using the same GeLC-MRM pipeline as used for the label-free verification step. Alternative analytical strategies could utilize in-solution trypsin digestion of the abundant protein-depleted sample followed by addition of the stable-isotope labeled internal standards and subsequent peptide fractionation by strong cation exchange, Off-Gel electrophoresis, etc. If a peptide fractionation method is used instead of SDS gels for the laboratory validation step, it will be important to ensure that a sufficient number of peptides for each protein of interest will provide reliable quantitative results in the new analysis method. This will include monitoring for potential differences in peptide recoveries between methods and new interferences in the LC-MRM analysis due to changes in sample composition resulting from peptide level rather than protein level separation. Alternatively, if an ELISA or related immunoassay is available, it can be tested on the same sera used for the initial MRM analyses and, if results are consistent, the immunoassay can be used for analyzing larger patient cohorts. We have applied this latter strategy to verify ADAM12, a disintegrin and metalloprotease domain-containing protein, as a novel biomarker for the diagnosis of ectopic pregnancy (see below).33, 39
We used SDS-PAGE instead of SCX fractionation of depleted serum because the 1-D gels offer rapid, reproducible, protein-size-based separations that can be multiplexed. SDS-PAGE also provides a convenient method for sample clean-up prior to trypsin digestion. We generally perform gel separation for 2-, 3-, or 4-cm, which takes less than 25 min for one person to simultaneously separate up to 10 samples. Gel lanes were cut precisely into 1 mm slices and efficiently digested with trypsin in batch-mode using 96-well pierced plates.36 It is important to note that only the series of gel slices bracketing the migration positions on the 1-D gels of the targeted candidate proteins need to be analyzed by MRM. The GeLC-MRM method is especially attractive if the preceding biomarker discovery strategy utilized 1-D gels as a sample prefractionation step, thereby defining the locations of candidate biomarkers on SDS gels. For example, we have observed that most cancer biomarker candidates have observed molecular weights between 20 and 90 kDa. Using a 2-cm gel separation, as described in our recent ectopic pregnancy (EP) biomarker discovery study,33 only eight gel slices need to be analyzed to cover the 25–90 kDa range; that is, eight MRM runs per serum sample. Alternatively, multiple gel slices can be combined to increase the throughput of the analysis, especially if a longer gel separation is performed. For example, we found no loss in MS detection sensitivity in comparing a 2 cm gel with 20 X 1 mm fractions versus an equivalent 4 cm gel with 20 × 2 mm fractions. Overall, the throughput of our GeLC-MRM strategy is comparable with recent sensitive SID-MRM methods for quantifying low-abundant proteins.17, 19 These SID-MRM methods utilized major protein depletion followed by SCX fractionation and pooling (n<10) from ~ 40 total collected fractions. A unique advantage of GeLC-MRM, compared with methods using peptide fractionation immediately prior to LC-MS/MS analysis, is that quantitation profiles across fractions are obtained for each peptide and all profiles for a given protein should be similar. This enables ready identification of quantitative errors due to interference and other factors, thereby reducing the need for technical replicates and, in turn, further increasing throughput (see below).
Increased Throughput and Sensitivity with Short LC Gradients and Scheduled MRM
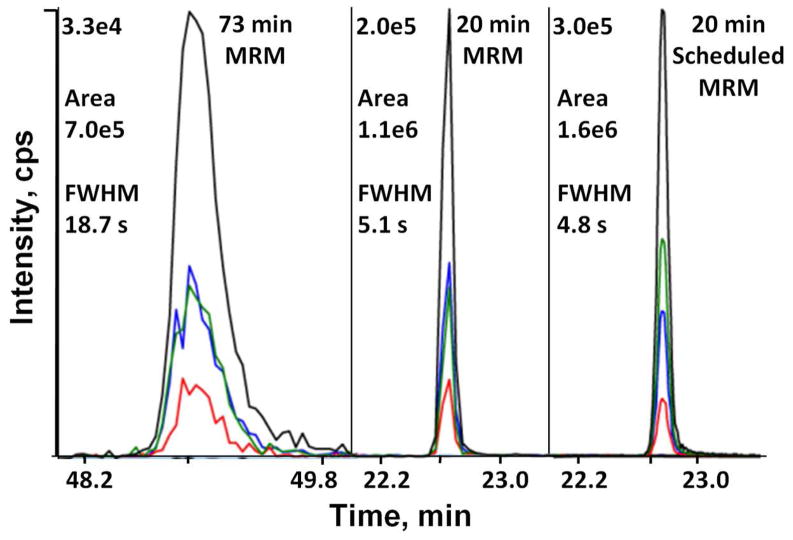
Discovery-based proteomics strategies typically use long, shallow LC gradients (more than 60 min) during MS/MS analyses to maximize peptide separation and therefore achieve greater proteome coverage. However, shorter gradients will increase throughput and generally should increase peak heights in MRM analyses, and long gradients may not be needed if interference from the greater complexity is not limiting. Here, we investigated the use of a shorter, steeper LC-MRM gradient and its effect on peptide detection and quantitation. Comparison of an enolase digest analyzed using a 73 min vs. a 20 min gradient showed that the shorter gradient produced more intense peaks with improved overall sensitivity of the MRM analysis (Supplemental Figure 1A). For example, the extracted ion chromatogram (XIC) of the 400.7/637.3 transition for the peptide YDLDFK is 6.0-fold more intense in the short gradient while displaying only a 1.4-fold increase in area (Figure 2). Examination of 15 enolase peptides showed an average intensity increase of 2.9-fold, with only minor changes in peak area (average 1.2-fold increase). The gain in intensity is due to narrower peak widths with the short gradient (average full-width at half-maximum [FWHM] of 4.6 sec) compared to an average FWHM of 11.0 sec for the long gradient, which improves the S/N for most peaks and results in lower LOQ for most peptides. However, the very narrow peak widths resulted in less data points across the peak, which can result in less reliable quantitation. With a dwell time of 15 msec and 134 transitions, the cycle time of 2.7 sec resulted in less than eight data points for most of the peaks. Scheduled MRM,12 which measures MRM transitions over specific time windows, was therefore used to increase sampling across the narrower peaks and to permit analysis of more transitions when shorter gradients were used. With scheduled MRM, most peaks have more than 12 data points resulting in adequate data for smooth peak shapes and more reliable quantitation (Figure 2). In addition to smoother peak shapes, we also observed some changes in relative intensities for some peptide transitions when scheduled MRM was used. For example in the peptide YDLDFK, the relative intensities of the y3 and y4 transitions were reversed when analyzed using scheduled MRM (Figure 2). This is most likely due to the difference in the dwell time used. The dwell time for the unscheduled MRM analysis is fixed at 15 msec, but the dwell time used by the scheduled MRM is dependent on the number of concurrent transitions. Due to a low number of concurrent transitions, the estimated dwell time for peptide YDLDFK is 200 msec for a scheduled MRM using a target scan time of 1 sec. Even though relatively minor differences in transition intensity ratios were occasionally observed when comparing unscheduled and scheduled MRM analyses, these intensity ratios appeared to be consistent within each method, and therefore will not affect the reliability of quantitation as long as a consistent MRM method is used.
Figure 2.
Comparison of UPLC gradients for LC-MRM assays. XIC of MRM transitions for the enolase peptide YDLDFK from the 73 and 20 min gradients, and a 20 min gradient with scheduled MRM enabled. For scheduled MRM, the detection window was set at 2 min, and target scan time was 1 s. The intensities, areas, and peak widths of the most intense y5 transition (400.69/637.32, black line) are indicated. Other transitions are y4 (400.69/522.29, green line), y3 (400.69/409.21, blue line), and b4 (400.69/507.21, red line). The time axes are shown using the same scale.
While the use of a shorter gradient increases throughput and sensitivity of the MRM assay, it could potentially increase matrix interference due to the shorter separation distance. However, the immunodepletion of 20 abundant proteins and the 1-D gel separation have greatly reduced sample complexity and lowered the possibility of matrix interference. For example, GeLC-MRM analysis of an intrauterine pregnancy (IUP) pooled serum sample for 20 peptides (117 transitions) from ADAM12, CGA, CGB, and PAEP did not show any significant co-eluting interference when analyzed using the 20 min gradient (Supplemental Figure 1B). In some of the XICs, additional peaks were observed at a different retention time but were recognized easily as spurious based on greater-than-expected retention time shifts and/or changes in the pattern of co-eluting transitions for a given peptide and/or variations in relative yields across gel slices for multiple peptides derived from the same protein. However, it is possible that interference may become a significant problem at low detection limits for some peptides when short gradients are used. To test this, we recently compared the quantitation of EP candidate biomarkers in a small series of IUP and EP patient samples using label-free LC-MS/MS performed with the 73 min gradient on an Orbitrap versus LC-MRM quantitation on a 4000 QTRAP with the 20 min gradient.33 Both quantitation methods yielded similar relative amounts of the candidate biomarkers across the analyzed samples, indicating that shortening the gradient time to 20 min had no deleterious effect on the quantitation. In cases where greater increases in sample complexity/interference are encountered, the gradient slope can be adjusted to achieve greater separation without increasing the run time. In many cases, it also is possible to use another interference-free MRM transition should an interfering peak be encountered with appreciable frequency. Finally, if interference persists, a longer gradient can be used with the trade-off that throughput will be somewhat decreased.
GeLC-MRM Quantitation Profile
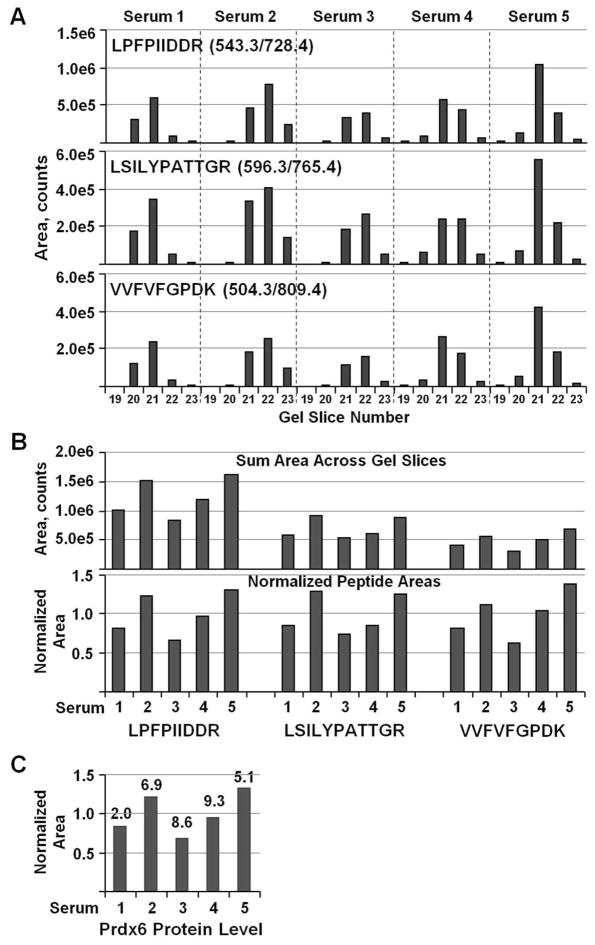
The utility of GeLC-MRM is demonstrated by the quantitation of Prdx6 in a series of human serum samples. Prdx6 is a bifunctional 1-Cys peroxiredoxin that plays important physiological roles in antioxidant defense and phospholipid turnover.40 Prdx6 also has been hypothesized to promote cancer growth and invasiveness, and increased expression has been observed in various malignancies.40–42 In these analyses, serum samples from five individuals were immunodepleted and subjected to 1-D gel separation for 4 cm. Scheduled MRM was set up to monitor three Prdx6 peptides with three transitions per peptide. The most intense transition was used for the quantitation of each peptide. Analysis of tryptic digests from a series of gel slices corresponding to the 24 to 40 kDa regions of the gel showed that Prdx6 was primarily in gel slices 21 and 22 (~25 to 30 kDa), which agrees well with the calculated size of 25 kDa (Figure 3A). Slight variations in gel slicing or gel separation cause minor, non-detrimental shifts in the distribution across gel slices between samples. Most importantly, all three Prdx6 peptides showed identical gel slice distributions for a given specimen. This is an extremely useful indicator that confirms the three peptides used for quantitation are derived from the same protein, and shows that quantitation is not affected by any matrix-interfering signal or spray instability during chromatographic separation. Any substantial problem during the chromatographic separation, such as spray instability, would likely perturb the quantitation profile of only a single peptide due to the different retention times of each peptide. Similarly, matrix interference experienced by a peptide will result in an easily noticeable change to the quantitation profile across gel slices. This ability to easily recognize quantitation problems with the GeLC-MRM method should reduce the need for technical replicates, as problematic runs can be accurately identified and reanalyzed. Peptide-level sample fractionation methods, such as the commonly used SCX, do not have this advantage because the peptides of a protein will not track together. Of course, acquiring multiple transitions per peptide and monitoring their ratios is a good method that is usually used to identify matrix interferences,17 but the gel fraction protein distribution profiles provide an independent check. Furthermore, multiple strong transitions are not always obtained for every peptide, particularly low-abundance peptides, thereby limiting the utility of using transition ratios as the sole method for identifying interference.
Figure 3.
GeLC-MRM quantitation profiles of Prdx6 across gel slices. (A) MRM quantitation of three Prdx6 peptides in tryptic digests of gel slices #19 to 23 (24–40 kDa region) from five depleted serum samples. The peptide sequences together with the MRM transition used for quantitation are indicated. (B) The amount of peptide in each serum sample was determined by summing the signal of the strongest transition across all gel slices. The bottom chart shows peptide amounts after normalizing against the average value for each peptide. (C) The relative Prdx6 protein level in each sample calculated from the average of the normalized peptide values. The %CV was determined from the three normalized peptide values and is indicated on the top of each bar.
To compare Prdx6 levels across serum samples, the total amount of protein in each sample was determined by first summing each peptide’s peak area across all gel slices analyzed (Figure 3B, top panel). The summed area for each sample then was normalized by dividing it by the average value for that peptide in all samples (Figure 3B, bottom panel). Finally, the protein amount in each sample was determined by taking the average of the three normalized peptide values (Figure 3C). The reproducibility of the quantitation is apparent from the similarity in the relative abundance among different samples for each peptide (Figure 3B, bottom panel), with a coefficient of variation (CV) of less than 10% for the normalized peptide values. Therefore, the GeLC-MRM method provides quantitation profiles at both the gel-slice level and the peptide level that help ensure that the correct peptides for a targeted protein have been quantitated and the observed changes in abundance are consistent.
Importance of Gel Fractionation
There are a number of alternative methods of fractionating serum proteins after major protein depletion, including peptide SCX, peptide Off-Gel electrophoresis, protein Off-Gel electrophoresis, and protein solution IEF. However, fractionation of intact proteins by 1-D SDS gels is simplest to perform and preserves information about protein size, thereby providing insights into some size changes associated with proteolytic processing, extensive post-translational modification, or alternatively spliced isoforms that are more likely to be missed by alternative fractionation methods.
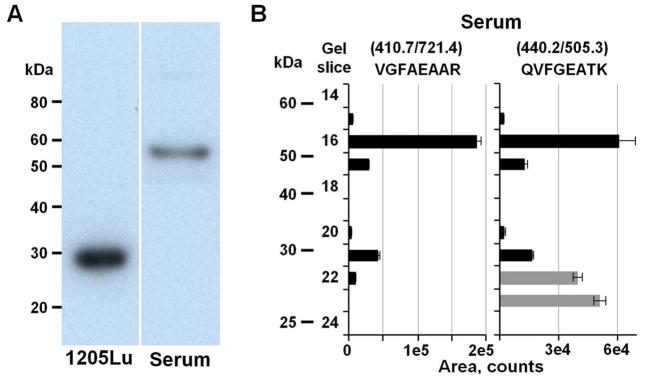
The advantages of gel separation for MRM analysis is illustrated by quantitation of CTSD, a multifunctional lysosomal aspartic endopeptidase. CTSD is synthesized as an inactive pre-proenzyme that is cleaved and glycosylated into a 52 kDa procathepsin D. The N-terminal propeptide subsequently is removed to produce a 48 kDa single-chain, intermediate, active form, which is processed further to yield a mature active form comprised of a 34 kDa heavy chain and a 14 kDa light chain.43 We previously identified CTSD as one of the proteins implicated in melanoma metastatic potential37 and therefore set up MRM assays to quantitate CTSD in human sera. Western blot using C0715 monoclonal anti-CTSD antibody that is specific to the heavy chain showed a band at ~30 kDa in the metastatic melanoma 1205LU cell extract (Figure 4A, left lane). However, GeLC-MRM analysis of CTSD in a major protein-depleted human serum sample revealed the presence of two forms: a dominant form at ~ 52 kDa and a minor form at ~ 30 kDa (Figure 4B) that was not readily detected in the Western blot of the serum sample. The two targeted CTSD peptides showed a similar relative abundance level for each gel slice analyzed except for slices 22 and 23, indicating a problem with the MRM-based quantitation in these two fractions. Further investigation revealed that the MRM transition pattern for the QVFGEATK peptide is altered in slices 22 and 23 compared to the rest of the slices analyzed, indicating the presence of co-eluting interferences in these two fractions (Supplemental Figure 2). However, accurate quantitation based on the QVFGEATK peptide was not significantly affected because the CTSD amount in gel slice 22 is only 3.7% of the total amount based on the quantitation profile of the VGFAEAAR peptide. These results clearly demonstrate the utility of GeLC-MRM in identifying quantitation errors in selected fractions due to interference by using peptide quantitation profiles across gel slices.
Figure 4.
Detection and quantitation of different forms of CTSD. (A) Western blot of CTSD in a melanoma 1205LU cell extract and major protein depleted human serum using C0715 monoclonal anti-cathepsin D antibody. (B) GeLC-MRM quantitation of CTSD in the major protein depleted human serum. MRM assays were run on the 20–70 kDa region of the gels. The CTSD peptide sequences and the MRM transitions are indicated at the top of the panel. Grey bars represent quantitation values from a co-eluting interference with similar ratios for two of the three monitored MRM transitions as the targeted peptide (see Supplemental Figure 2). Error bars indicate standard deviations from duplicate injections.
Subsequent Western blot analysis of the identical depleted human serum confirmed the presence of the ~52 kDa species (Figure 4A, right lane), which is consistent with prior reports that the 52 kDa procathepsin D is indeed the major form present in human serum.44 Interestingly, the 52 kDa proenzyme has been reported to be over-expressed and secreted by numerous cancer cell lines.43, 45 In addition, a serological immunoassay specific for procathepsin D was found to be superior to immunoassays for total cathepsin D in detecting increased levels of the protein in serum of patients with metastatic breast carcinoma.46
It is not uncommon for serum proteins to be present in multiple forms or to be variably proteolytically processed, and it is likely that, at least in some cases, specific forms of a protein will provide a better biomarker assay than assays that do not distinguish proteolytic fragments or homologous proteins or spliceforms. In addition to CTSD, we previously detected and quantitated two forms of ADAM12, a novel EP candidate biomarker, in serum using the GeLC-MRM strategy.33 With prior detailed knowledge of any targeted protein or protein fragment, appropriate MRM assays could be designed to target most forms of a protein, although isoform-specific peptides may not always be proteotypic and detectable.47 Knowing the presence of and having the ability to quantitate various forms of a protein as provided by the GeLC-MRM analysis are invaluable for accurate correlation of expression levels with a disease condition, because a disease-relevant change may be molecular-form specific, which might not be apparent from quantitation of the total protein level.
Reproducibility of the Depletion and GeLC-MRM Method
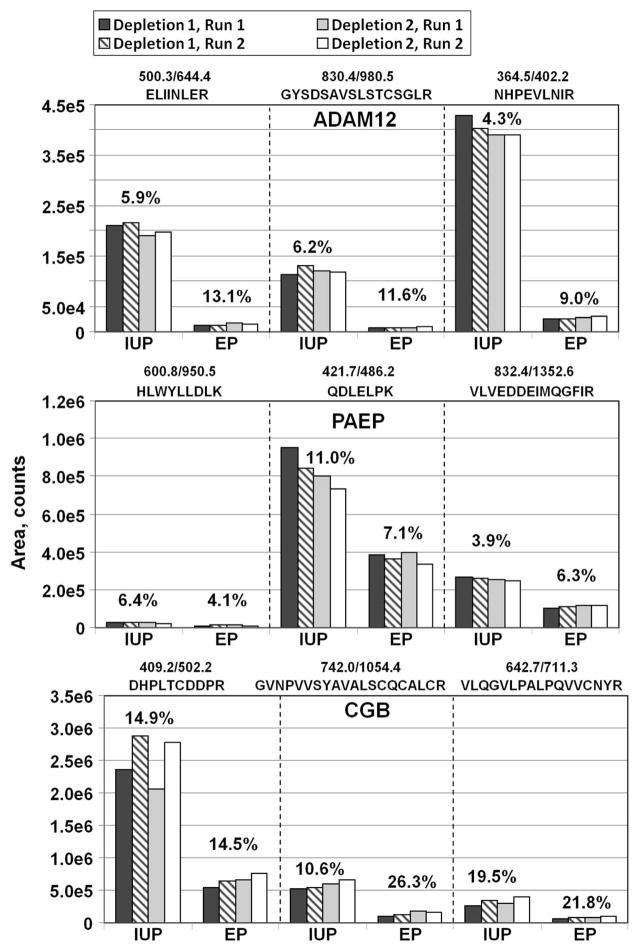
As noted above, due to current limitations of MS instrument sensitivity and dynamic range, multiple sample fractionation steps are required to permit quantitation of serum proteins in the low ng/mL range.17, 19 However, fractionation steps prior to LC-MRM have the potential of introducing noise and reducing the reproducibility of label-free quantitation. In the analysis strategy used here, variability could be introduced during any of the steps from major protein depletion to MS analysis. To assess the reproducibility of the overall label-free GeLC-MRM method, replicate fractionations pooled EP and IUP sera were performed and three putative EP biomarkers, ADAM12, PAEP, and CGB,33 were quantified using label-free MRM. Separate pooled serum samples were prepared by combining serum from nine individuals with EP and nine individuals with IUP. Replicate 100 μl aliquots of pooled serum samples were immunodepleted on separate days. The depleted IUP and EP samples were run on short 1-D gels and digested with trypsin (Figure 5; Depletions 1 and 2). Tryptic digests for gel slices 12 to 15 then were analyzed using duplicate injections of each tryptic digest on a 4000 QTRAP mass spectrometer for each depletion. To maximize any potential variability, the replicate injections (Run 2) were performed only after all fractions from the first set (Run 1) of samples were analyzed. Quantitative comparison of all the peptides showed good reproducibility of the overall analytical pipeline, including major protein depletions, gels, trypsin digests, and the MRM analyses with consistent recovery of the proteins in technical replicates (Figure 5). At the peptide level, most of the replicates had CVs of less than 15%, while three replicates had CVs between 20 and 26% for CGB. At the protein level, the CVs for CGB were 12.9% (IUP) and 16.2% (EP), whereas the protein level CVs for ADAM12 and PAEP were ≤ 10% for all replicates of both samples. In addition, the observed fold change between IUP and EP was similar for the three peptides in their respective protein group, that is, 14.2±0.8 fold for ADAM12, 2.3±0.1 fold for PAEP, and 4.0±0.1 fold for CGB, indicating high technical reproducibility for multiple samples at both the peptide and protein levels.
Figure 5.
Reproducibility of the label-free MRM analysis using major protein depletion and 1-D SDS fractionation. MRM data for ADAM12, PAEP, and CGB are shown. Three peptides for each protein were quantitated. Depletions 1 and 2 are replicate Top-20 depletions of EP and IUP serum samples performed on different days. Run 1 and Run 2 are duplicate injections of tryptic digests onto a 4000 QTRAP mass spectrometer. Bars indicate areas of the strongest MRM transition for each peptide, summed for gel slices 12–15. The % CV of all replicate runs is indicated above each sample.
In agreement with our results, good reproducibility in replicate Top-6 protein depletions and in-solution trypsin digestions have previously been reported in the quantitation of plasma proteins.10 In a related study, we used dissociation-enhanced lanthanide fluoroimmunoassay (DELFIA) to determine the level of ADAM12 in a larger group of 199 patients and found that median ADAM12 concentration was 18.6 ng/mL in IUP versus 2.5 ng/mL in EP.39 Considering that the immunoassay used a much larger, different cohort of patients from the smaller pools analyzed above, the 7.4-fold change in IUP/EP level as measured by DELFIA is in good agreement with our 14.2-fold change determined above using MRM. Furthermore, the ADAM12 MRM assay was more sensitive than the DELFIA immunoassay. In the latter assay, the majority (69%) of EP patients had ADAM12 levels below the 2.5 ng/mL lower limit of detection by DELFIA,39 and this minimum value was therefore assigned to these patients rather than zero, indicating the IUP/EP fold change measured by DELFIA would actually be higher if the DELFIA assay were more sensitive. Taken together, these data show that GeLC-MRM with major protein depletion can reproducibly quantitate serum ADAM12 in the low ng/mL or lower range.
Previously, a comparison of Top-7 versus Top-12 protein depletion showed that better S/N, LOQ, and CVs were obtained when a larger number of major proteins were depleted.17 Hence, it is very likely that our depletion of 20 major proteins would expose more low-abundance targets for quantitation and improve quantitation of target proteins due to the removal of potential co-eluting interferences. However, there is always the potential of non-specific antibody binding or complex formation between the depleted major proteins and some low-abundant proteins, resulting in the loss or variable recovery of some desired low-abundant proteins. So far, we have not obtained clear evidence of this potential complication for any candidate biomarkers tested, which suggests that such occurrences are likely to be relatively rare.
Reproducibility of Label-free GeLC-MRM Quantitation over Extended Time Periods
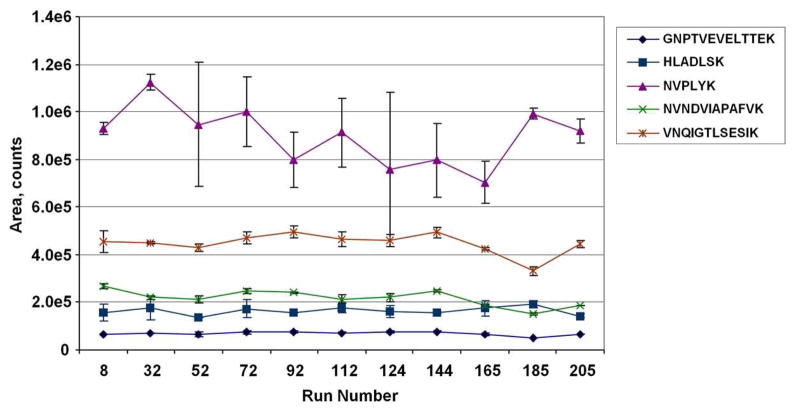
Major benefits of using stable-isotope internal standard peptides for MRM quantitation include greater assay consistency over long time periods and portability of assays between laboratories. To evaluate the long-term reproducibility of the label-free GeLC-MRM assay, we monitored the quantitation of an external quality control reference interspersed between experimental samples. Ten femtomoles of an enolase standard digest were injected in duplicate at various intervals during a label-free GeLC-MRM analysis of a series of depleted serum samples. A total of 206 injections, including 22 enolase runs, were performed over a period of 12 days on a 4000 QTRAP using the 20 min UPLC gradient. The values for five representative enolase peptides over the entire experiment are shown in Figure 6 and the CVs from these standard runs were determined for 16 enolase peptides that had average intensities ranging from 1.7E+3 to 2.4E+5 cps (Table 1). The majority of the peptides (75%) have a CV of < 16%, demonstrating very good long-term reproducibility for label-free LC-MRM quantitation. Only three peptides, eluting either at the beginning or end of the gradient, displayed a CV greater than 20%.
Figure 6.
Reproducibility of label-free LC-MRM quantitation over a large number of runs. Quantitation of five representative enolase peptides from duplicate injections of 10 femtomoles of enolase tryptic peptides interspersed over a total of 206 MRM runs. MRM transitions used are listed in Table 1. Error bars indicate standard deviations from duplicate injections.
Table 1.
Reproducibility of MRM Quantitation of 10 Femtomol Enolase Standard Peptides
| Sequence | Q1 m/z | Q3 m/z | Retention Time (Min)a | Intensity (cps)a | Area (counts)a | %CVb |
|---|---|---|---|---|---|---|
| SVYDSR | 363.67 | 540.24 | 17.3±0.1 | 5.88E+04 | 4.60E+05 | 21.5 |
| GVLHAVK | 362.23 | 567.36 | 17.6±0.1 | 3.05E+03 | 2.51E+04 | 10.5 |
| ANIDVK | 330.19 | 361.21 | 17.6±0.1 | 2.32E+04 | 1.90E+05 | 12.9 |
| HLADLSK | 392.22 | 437.21 | 17.6±0.1 | 2.01E+04 | 1.64E+05 | 14.8 |
| IATAIEK | 373.23 | 561.32 | 18.0±0.1 | 5.74E+04 | 4.55E+05 | 11.6 |
| IGSEVYHNLK | 580.31 | 674.36 | 19.0±0.1 | 5.30E+03 | 4.14E+04 | 14.4 |
| NVPLYK | 367.22 | 520.31 | 19.5±0.1 | 1.01E+05 | 8.94E+05 | 18.4 |
| LNQLLR | 378.74 | 401.29 | 20.4±0.1 | 8.20E+04 | 6.30E+05 | 11.0 |
| TFAEALR | 404.22 | 559.32 | 20.9±0.1 | 2.40E+05 | 1.69E+06 | 12.1 |
| GNPTVEVELTTEK | 708.86 | 948.49 | 21.3±0.1 | 9.34E+03 | 6.88E+04 | 11.8 |
| VNQIGTLSESIK | 644.86 | 834.46 | 21.7±0.1 | 6.44E+04 | 4.49E+05 | 10.3 |
| AADALLLK | 407.76 | 743.47 | 22.0±0.1 | 4.71E+04 | 3.45E+05 | 15.6 |
| YDLDFK | 400.69 | 637.32 | 22.2±0.1 | 9.94E+04 | 7.39E+05 | 12.9 |
| NVNDVIAPAFVK | 643.86 | 1073.60 | 23.3±0.1 | 2.82E+04 | 2.19E+05 | 15.3 |
| TAGIQIVADDLTVTNPK | 878.48 | 1073.55 | 24.0±0.1 | 3.24E+03 | 2.21E+04 | 29.5 |
| AVDDFLISLDGTANK | 789.90 | 918.49 | 25.7±0.1 | 1.72E+03 | 1.19E+04 | 32.3 |
Average values from 22 enolase standard runs spread over a total of 206 MRM analyses spanning 12 days. Peptides with average intensities less than 200 cps were excluded from this table. Cps, counts/second.
The coefficients of variation were determined from the peak area for the peptides in the 22 standard runs.
One of the major challenges in MRM-MS using nanospray ionization is maintaining good spray stability throughout the chromatographic gradient. On our system, we noticed greater spray instability near the beginning and the end of the chromatographic gradient. Therefore, the quantitation of very hydrophilic and hydrophobic peptides is most likely to be affected by the unstable spray, and this is probably the major factor in the observed greater variability of the early- and late-eluting enolase peptides. Switching to higher flow rates or modifying the gradient usually will improve quantitation of such peptides affected by spray instability, but unless such peptides are indispensible for the quantitation of desired proteins, it is easier to select alternate peptides eluting at regions not affected by spray instability.
The results in this experiment indicate that most of the tested enolase peptides are suitable for monitoring long-term reproducibility of label-free MRM assays as an external standard. However, aside from the very hydrophilic and hydrophobic peptides, a small number of peptides such as the NVPLYK peptide were found to have variable ion intensity (CV=18.4%) even between duplicate injections (Figure 6). This variability could be caused by spray instability at the elution time, influence of co-eluting ions or ionization characteristics of the peptide. This result indicates that, as expected, not all peptides of a protein will behave well in MRM analysis. Similar observations have been reported for a number of peptides from more abundant plasma proteins.10 Therefore, peptide hydrophobicity and reproducibility across multiple replicate runs are two additional parameters that should be taken into consideration when selecting the best peptides for MRM quantitation of a protein of interest.
Long-term reproducibility of quantitation relies on a properly maintained MS instrument. With regular cleaning of the nanospray interface and Q0, the 4000 QTRAP used in this experiment could be run continuously for at least five months before degradation of signal intensity due to charging or contamination of Q1 was observed. With the more sensitive 5500 QTRAP, a higher incidence of Q1 charging was observed, presumably due to its larger orifice. Hence, when using label-free MRM, it is important to periodically evaluate an external standard such as the enolase digest as a quality control for the entire LC-MRM system.
Reproducible quantitation also requires a stable HPLC system at nL/min flow rates. The Waters nanoACQUITY UPLC system used in this study was observed to be quite robust and yielded reproducible chromatographic elution times for all the enolase peptides (Table 1). A standard deviation of ± 0.1 min, which is equivalent to CVs of 0.4 to 0.6%, was observed for elution times of all the peptides. This reproducibility is especially important when performing scheduled MRM, because use of narrower time segments will permit monitoring of a larger number of MRM transitions.
Reproducibility of Label-free Quantitation on Different MS Instruments
To simulate portability of label-free quantitation between laboratories and to further evaluate the long-term reproducibility of label-free MRM quantitation, GeLC-MRM quantitation of ADAM12 in 18 depleted serum samples using a 4000 QTRAP mass spectrometer and the 20 min gradient33 was repeated on a 5500 QTRAP mass spectrometer. The four gel slices (numbers 12 to 15) containing ADAM12 were analyzed for each sample. To further challenge the label-free quantitation, the reanalysis was performed on a different column with a shallower 24 min LC gradient, as described in Materials and Methods. In addition, the reanalysis was performed 16 months later and the samples have undergone one additional freeze-thaw cycle.
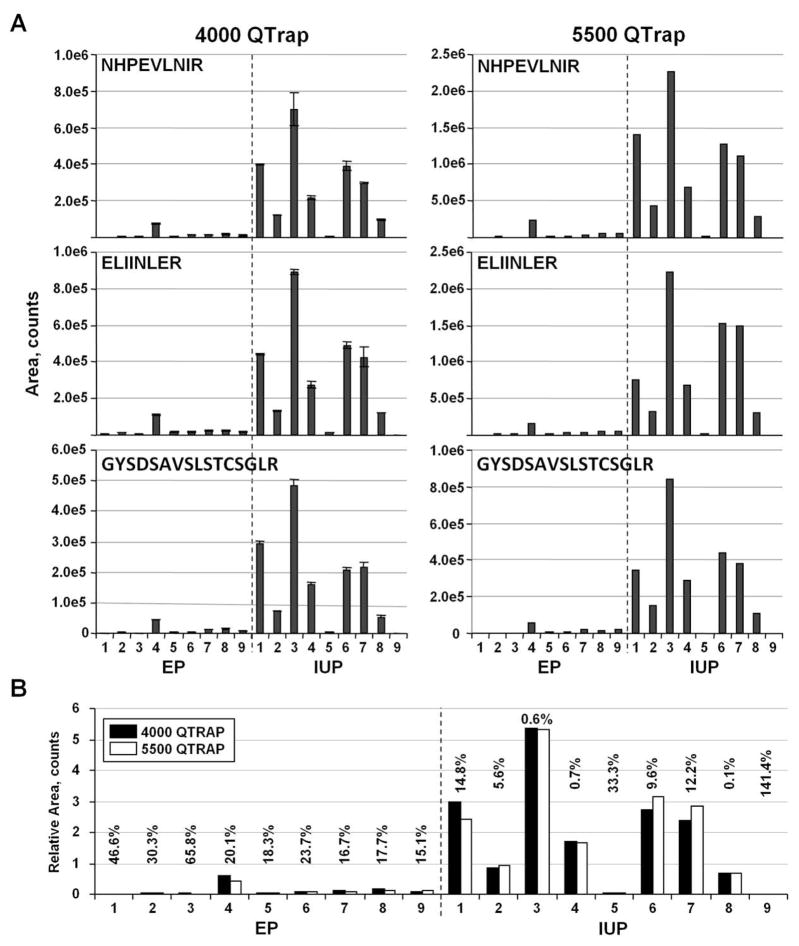
Duplicate runs were performed in the 4000 QTRAP analysis, where the first set of runs (72 total sample injections, excluding standards) was completed before starting the replicate runs (Figure 7A, left panels). The duplicate quantitation of each peptide in all the samples produced an average CV of 8% (n=54). Most of the duplicate measurements (70%) have a CV of < 10%, further demonstrating the reproducibility of label-free MRM quantitation. When compared to the 5500 QTRAP reanalysis, very similar relative quantitative patterns for the three targeted ADAM12 peptides were obtained (Figure 7A, right panels). As expected, stronger signals generally were observed for the 5500 QTRAP analysis due to its greater sensitivity. Generally, an approximately five-fold increase in sensitivity was observed on the 5500 QTRAP instrument.
Figure 7.
Reproducibility of the label-free MRM quantitation on different MS instruments. (A) Label-free GeLC-MRM quantitation of three ADAM12 peptides from nine EP and nine IUP serum samples, initially analyzed in duplicate on a 4000 QTRAP (left panels) and reanalyzed 16 months later on a 5500 QTRAP using a shallower 24 min UPLC gradient (right panels). Error bars indicate standard deviations. (B) Comparison of normalized ADAM12 protein quantitation from the two MS instruments for the 18 serum samples. The %CV for the instrument comparisons is indicated above the bars for each sample.
Due to the different sensitivities of the two instruments, the data were normalized to allow direct comparison of the two datasets. The peptide area for each sample was first normalized by dividing by the averaged area of each peptide group. This normalization was performed separately for the 4000 QTRAP and 5500 QTRAP datasets. The protein amount in each sample was then determined from the average of the three normalized peptide values for each dataset (Figure 7B). Comparison of the normalized datasets showed good reproducibility for most samples. The median CV for all 18 samples was 17.2%. Five samples: EP1, EP2, EP3, IUP5, and IUP9, had CVs greater than 20% between instruments due to the low levels of ADAM12 that resulted in very low quantifiable signals (< 5,000 peak area counts) for at least one of the three peptides in both instruments (Figure 7A). Exclusion of these five, low-level samples resulted in an average CV of 12%, which is comparable to commonly reported CVs using SID-MRM.10, 17, 19 However as noted above, SID-MRM has the additional benefit of clearly identifying targeted peptides from potential interferences, and provides a more direct method for comparing results obtained from different MS instruments.
We also explored two other methods of normalizing the datasets. In the first alternate method, the 5500 QTRAP dataset was normalized to the average of all IUP samples at the peptide level. A 4000 QTRAP/5500 QTRAP response ratio then was obtained for each peptide and applied to all the 5500 QTRAP samples. The second alternate method utilized an IUP/EP pooled reference sample that also was analyzed as a quality control interspersed among the sample runs. For each peptide, an average area was determined from five IUP/EP pooled reference runs in each dataset. A 4000 QTRAP/5500 QTRAP response ratio was determined for each peptide and applied to each 5500 QTRAP sample. These two alternate methods of normalization resulted in a median CV of 18.9% and 17.2%, respectively, for all 18 samples. Hence, all three normalization methods produced very similar results with no clear advantage for any specific method. These results demonstrate that samples processed at different times, and even on different instruments using label-free GeLC-MRM, can be compared after normalization to either an external reference or a common set of samples. The ability to compare label-free datasets generated at different times with different instruments will permit inclusion of additional samples for expanded comparison, as long as a common reference sample is available for normalization.
Sensitivity of Label-free GeLC-MRM Quantitation
A major challenge of MRM is the quantitation of low-abundant proteins that are potential clinically relevant and are usually present in serum or plasma in the low ng/mL range or below. Two lines of evidence indicate that the GeLC-MRM analysis of depleted serum samples is able to quantify serum proteins in this range. First, as mentioned above, ADAM12 could not be quantitated in 69% of EP patients using DELFIA with a 2.5 ng/mL lower limit of detection.39 In contrast, MRM performed on the 4000 QTRAP could not detect ADAM12 in only two of the nine EP samples (22%). In all other EP samples, at least one ADAM12 peptide could be detected with a minimum peak area of 11,000 counts. With the more sensitive 5500 QTRAP, only one EP sample did not have any ADAM12 peptide with a minimum peak area of 11,000 counts (Figure 7A).
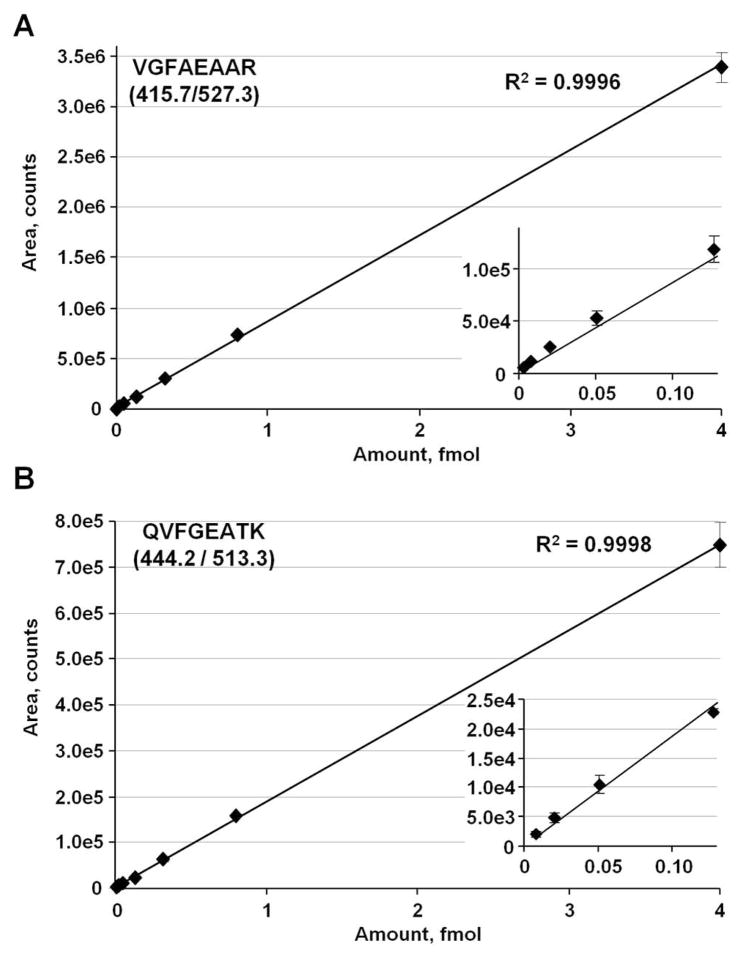
Second, GeLC-MRM using the 4000 QTRAP easily could detect CTSD that has a reported concentration of ~16 ng/mL in serum.48, 49 To further examine the sensitivity of the GeLC-MRM method, we synthesized two heavy isotope-labeled internal standard peptides for CTSD and spiked them at 3 amol to 4 fmol (equivalent to 0.2 to 288 ng/mL proCTSD) into a representative 25–60 kDa pool of tryptic digests of depleted and gel fractionated human serum. Duplicate runs were performed for each concentration point using the more sensitive 5500 QTRAP mass spectrometer, with the replicate measurement only acquired after completion of the first set of runs. As shown in Figure 8, a linear response was observed for both spiked peptides from at least 8 amol to 4 fmol. The heavy QVFGEATK peptide was not reliably detected at 3 amol, with a signal detected only in one of the duplicate set of runs at ~ 250 cps. However, the more intense response from the heavy VGFAEAAR peptide resulted in reliable quantitation at 3 amol level. Therefore, a LOQ of 0.2 ng/mL can be achieved for serum CTSD using the GeLC-MRM method described here.
Figure 8.
GeLC-MRM quantitation of two, heavy, isotope-labeled CTSD peptides using a 5500 QTRAP mass spectrometer. Heavy peptides were spiked at 3, 8, 20, 51, 128, 320, 800, and 4000 amol into a tryptic digest of a fraction of depleted human serum after 1-D gel separation. (A) Standard curve of transition 415.7/527.3 for the (13C6 15N4) VGFAEAAR peptide. (B) Standard curve of transition 444.2/513.3 for the (13C6 15N2) QVFGEATK peptide. Inserts, expanded views of the lower regions of the standard curves. Error bars are standard deviations from duplicate injections. The most intense transition of each peptide was used in the standard curve plots. XICs for the peptides are shown in Supplemental Figure 3.
Since an identical depleted serum fraction was used as matrix background in the spike-in experiment, the endogenous levels of VGFAEAAR and QVFGEATK should have been constant in all 16 runs (data not shown). The peak area of unlabeled VGFAEAAR (410.7/517.3) showed an average of 6.9E+4 counts with a CV of 6.1%, whereas the peak area of unlabeled QVFGEATK (440.2/505.3) exhibited an average of 3.2E+4 counts with a CV of 6.3% (n=16). The CVs described here represent an example of the reproducibility that can be achieved with short-term, label-free MRM quantitation. Similar reproducibility has been reported for short-term, label-free MRM quantitation, where 60% of 137 MRMs targeting high- and medium-abundant plasma proteins had CVs less than 10%.10
The LOQ for a protein is dependent on the peptide being quantitated and the specific measured transitions. Peptides that do not ionize well will have higher LOQs, whereas peptides with strong response will achieve lower LOQs. For example, a spike-in experiment using heavy isotope-labeled versions of the three Prdx6 peptides described in Figure 3 yielded intensities of around 4E+3 cps, with S/N ranging from 50 to 105 at the 3 amol level, which corresponded to a serum concentration of 100 pm/mL. These intensities were at least four-fold more intense than the CTSD peptides at the same level. Therefore, with a limit of quantitation at the low attomole level for most peptides, the GeLC-MRM fractionation and analysis strategy is able to quantitate proteins in serum down to the 100–200 pg/mL level.
Conclusion
Technological advances in proteomics and other omics discovery strategies have led to a flood of candidate biomarkers for cancers, other diseases, and clinical disorders such as EP. However, the success rate for translating candidates into clinical assays has been extremely low. To improve the probability of success, we have developed a tiered verification/validation strategy that allows large numbers of candidates to be quickly screened at low cost and with reasonable throughput in the first stage of screening. This first-tier analysis uses label-free GeLC-MRM quantitation, which we demonstrate is a robust method that provides relative quantitation of biomarkers in the low ng/mL to sub ng/ml range from less than 100 μl of serum or plasma. It has the added capability of distinguishing various molecular weight-forms of proteins that often are not distinguished using alternative proteome fractionation strategies and may not be distinguished by some immunoassays. The throughput of the method is comparable to alternative existing methods capable of quantitating low-abundant proteins without immunoenrichment of the targeted proteins.17, 19 But in contrast to these alternative methods, expensive, stable-isotope standards or specific antibodies are not required. We have successfully applied the GeLC-MRM strategy to validate a number of EP candidate biomarkers and subsequently confirmed one of the low-abundant proteins using higher throughput immunoassays on a larger patient sample set.33, 39 Therefore, the method described here offers a cost-effective strategy to rapidly distinguish the most promising candidate biomarkers as a prelude to subsequent larger-scale SID-MRM or immunoassays.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants CA120393 and CA131582 to D.W.S. and HD036455 to K.T.B., as well as an institutional grant to the Wistar Institute (NCI Cancer Core Grant CA10815). We thank Ms. Mea Fuller for her assistance preparing the manuscript and the Wistar Institute Proteomics Core for assistance with the project.
Footnotes
Supporting Information Available: Comparison of long and short UPLC gradients for LC-MRM assays showing gradients and transition signals; XIC of the three MRM transitions for the CTSD peptide QVFGEATK showing interfering signal in larger fractions; and XIC for the three transitions of the two heavy isotope-labeled CTSD peptides that illustrate low attomole sensitivity on a 5500 QTRAP mass spectrometer. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Veenstra TD, Conrads TP, Hood BL, Avellino AM, Ellenbogen RG, Morrison RS. Biomarkers: mining the biofluid proteome. Mol Cell Proteomics. 2005;4(4):409–18. doi: 10.1074/mcp.M500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman SA, Joo WA, Echan LA, Speicher DW. Higher dimensional (Hi-D) separation strategies dramatically improve the potential for cancer biomarker detection in serum and plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849(1–2):43–52. doi: 10.1016/j.jchromb.2006.10.069. [DOI] [PubMed] [Google Scholar]
- 3.Teng PN, Bateman NW, Hood BL, Conrads TP. Advances in proximal fluid proteomics for disease biomarker discovery. J Proteome Res. 2010;9(12):6091–100. doi: 10.1021/pr100904q. [DOI] [PubMed] [Google Scholar]
- 4.Jimenez CR, Knol JC, Meijer GA, Fijneman RJ. Proteomics of colorectal cancer: overview of discovery studies and identification of commonly identified cancer-associated proteins and candidate CRC serum markers. J Proteomics. 2010;73(10):1873–95. doi: 10.1016/j.jprot.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Makridakis M, Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J Proteomics. 2010;73(12):2291–305. doi: 10.1016/j.jprot.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Goo YA, Goodlett DR. Advances in proteomic prostate cancer biomarker discovery. J Proteomics. 2010;73(10):1839–50. doi: 10.1016/j.jprot.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24(8):971–83. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 8.Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005;563(Pt 1):23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surinova S, Schiess R, Huttenhain R, Cerciello F, Wollscheid B, Aebersold R. On the development of plasma protein biomarkers. J Proteome Res. 2011;10(1):5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 10.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–88. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol. 2010;28(7):710–21. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 12.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6(10):1809–17. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-Ms/MS using protein cleavage and isotope dilution mass spectrometry. J Proteome Res. 2004;3(3):644–52. doi: 10.1021/pr049963d. [DOI] [PubMed] [Google Scholar]
- 14.Barr JR, Maggio VL, Patterson DG, Jr, Cooper GR, Henderson LO, Turner WE, Smith SJ, Hannon WH, Needham LL, Sampson EJ. Isotope dilution--mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem. 1996;42(10):1676–82. [PubMed] [Google Scholar]
- 15.Barnidge DR, Dratz EA, Martin T, Bonilla LE, Moran LB, Lindall A. Absolute quantification of the G protein-coupled receptor rhodopsin by LC/MS/MS using proteolysis product peptides and synthetic peptide standards. Anal Chem. 2003;75(3):445–51. doi: 10.1021/ac026154+. [DOI] [PubMed] [Google Scholar]
- 16.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100(12):6940–5. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6(12):2212–29. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54(11):1796–804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keshishian H, Addona T, Burgess M, Mani DR, Shi X, Kuhn E, Sabatine MS, Gerszten RE, Carr SA. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8(10):2339–49. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27(7):633–41. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brun V, Dupuis A, Adrait A, Marcellin M, Thomas D, Court M, Vandenesch F, Garin J. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol Cell Proteomics. 2007;6(12):2139–49. doi: 10.1074/mcp.M700163-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Kito K, Ota K, Fujita T, Ito T. A synthetic protein approach toward accurate mass spectrometric quantification of component stoichiometry of multiprotein complexes. J Proteome Res. 2007;6(2):792–800. doi: 10.1021/pr060447s. [DOI] [PubMed] [Google Scholar]
- 23.Pratt JM, Simpson DM, Doherty MK, Rivers J, Gaskell SJ, Beynon RJ. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat Protoc. 2006;1(2):1029–43. doi: 10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- 24.DeSouza LV, Taylor AM, Li W, Minkoff MS, Romaschin AD, Colgan TJ, Siu KW. Multiple reaction monitoring of mTRAQ-labeled peptides enables absolute quantification of endogenous levels of a potential cancer marker in cancerous and normal endometrial tissues. J Proteome Res. 2008;7(8):3525–34. doi: 10.1021/pr800312m. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Jia W, Sun W, Jin W, Guo L, Wei J, Ying W, Zhang Y, Xie Y, Jiang Y, He F, Qian X. Combination of improved (18)O incorporation and multiple reaction monitoring: a universal strategy for absolute quantitative verification of serum candidate biomarkers of liver cancer. J Proteome Res. 2010;9(6):3319–27. doi: 10.1021/pr9011969. [DOI] [PubMed] [Google Scholar]
- 26.Mirzaei H, McBee JK, Watts J, Aebersold R. Comparative evaluation of current peptide production platforms used in absolute quantification in proteomics. Mol Cell Proteomics. 2008;7(4):813–23. doi: 10.1074/mcp.M700495-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 28.Sherman J, McKay MJ, Ashman K, Molloy MP. How specific is my SRM?: The issue of precursor and product ion redundancy. Proteomics. 2009;9(5):1120–3. doi: 10.1002/pmic.200800577. [DOI] [PubMed] [Google Scholar]
- 29.Lee AY, Paweletz CP, Pollock RM, Settlage RE, Cruz JC, Secrist JP, Miller TA, Stanton MG, Kral AM, Ozerova ND, Meng F, Yates NA, Richon V, Hendrickson RC. Quantitative analysis of histone deacetylase-1 selective histone modifications by differential mass spectrometry. J Proteome Res. 2008;7(12):5177–86. doi: 10.1021/pr800510p. [DOI] [PubMed] [Google Scholar]
- 30.Nittis T, Guittat L, LeDuc RD, Dao B, Duxin JP, Rohrs H, Townsend RR, Stewart SA. Revealing novel telomere proteins using in vivo cross-linking, tandem affinity purification, and label-free quantitative LC-FTICR-MS. Mol Cell Proteomics. 2010;9(6):1144–56. doi: 10.1074/mcp.M900490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paweletz CP, Wiener MC, Bondarenko AY, Yates NA, Song Q, Liaw A, Lee AY, Hunt BT, Henle ES, Meng F, Sleph HF, Holahan M, Sankaranarayanan S, Simon AJ, Settlage RE, Sachs JR, Shearman M, Sachs AB, Cook JJ, Hendrickson RC. Application of an end-to-end biomarker discovery platform to identify target engagement markers in cerebrospinal fluid by high resolution differential mass spectrometry. J Proteome Res. 2010;9(3):1392–401. doi: 10.1021/pr900925d. [DOI] [PubMed] [Google Scholar]
- 32.Nagaraj N, Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res. 2011;10(2):637–45. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 33.Beer LA, Tang HY, Sriswasdi S, Barnhart KT, Speicher DW. Systematic discovery of ectopic pregnancy serum biomarkers using 3-D protein profiling coupled with label-free quantitation. J Proteome Res. 2011;10(3):1126–38. doi: 10.1021/pr1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteaker JR, Zhang H, Zhao L, Wang P, Kelly-Spratt KS, Ivey RG, Piening BD, Feng LC, Kasarda E, Gurley KE, Eng JK, Chodosh LA, Kemp CJ, McIntosh MW, Paulovich AG. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007;6(10):3962–75. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 35.Choi S, Kim J, Yea K, Suh PG, Ryu SH. Targeted label-free quantitative analysis of secretory proteins from adipocytes in response to oxidative stress. Anal Biochem. 2010;401(2):196–202. doi: 10.1016/j.ab.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Tang HY, Beer LA, Speicher DW. In-depth analysis of a plasma or serum proteome using a 4D protein profiling method. Methods Mol Biol. 2011;728:47–67. doi: 10.1007/978-1-61779-068-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han MJ, Wang H, Beer LA, Tang HY, Herlyn M, Speicher DW. A systems biology analysis of metastatic melanoma using in-depth three-dimensional protein profiling. Proteomics. 2010;10(24):4450–62. doi: 10.1002/pmic.200900549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem. 2010;56(2):177–85. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 39.Rausch ME, Beer L, Sammel MD, Takacs P, Chung K, Shaunik A, Speicher D, Barnhart KT. A disintegrin and metalloprotease protein-12 as a novel marker for the diagnosis of ectopic pregnancy. Fertil Steril. 2011;95(4):1373–8. doi: 10.1016/j.fertnstert.2010.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manevich Y, Fisher AB. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005;38(11):1422–32. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Lee SB, Ho JN, Yoon SH, Kang GY, Hwang SG, Um HD. Peroxiredoxin 6 promotes lung cancer cell invasion by inducing urokinase-type plasminogen activator via p38 kinase, phosphoinositide 3-kinase, and Akt. Mol Cells. 2009;28(6):583–8. doi: 10.1007/s10059-009-0152-6. [DOI] [PubMed] [Google Scholar]
- 42.Ho JN, Lee SB, Lee SS, Yoon SH, Kang GY, Hwang SG, Um HD. Phospholipase A2 activity of peroxiredoxin 6 promotes invasion and metastasis of lung cancer cells. Mol Cancer Ther. 2010;9(4):825–32. doi: 10.1158/1535-7163.MCT-09-0904. [DOI] [PubMed] [Google Scholar]
- 43.Benes P, Vetvicka V, Fusek M. Cathepsin D--many functions of one aspartic protease. Crit Rev Oncol Hematol. 2008;68(1):12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuhlsdorf M, Imort M, Hasilik A, von Figura K. Molecular forms of beta-hexosaminidase and cathepsin D in serum and urine of healthy subjects and patients with elevated activity of lysosomal enzymes. Biochem J. 1983;213(3):733–40. doi: 10.1042/bj2130733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leto G, Tumminello FM, Crescimanno M, Flandina C, Gebbia N. Cathepsin D expression levels in nongynecological solid tumors: clinical and therapeutic implications. Clin Exp Metastasis. 2004;21(2):91–106. doi: 10.1023/b:clin.0000024740.44602.b7. [DOI] [PubMed] [Google Scholar]
- 46.Brouillet JP, Dufour F, Lemamy G, Garcia M, Schlup N, Grenier J, Mani JC, Rochefort H. Increased cathepsin D level in the serum of patients with metastatic breast carcinoma detected with a specific pro-cathepsin D immunoassay. Cancer. 1997;79(11):2132–6. [PubMed] [Google Scholar]
- 47.Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25(1):125–31. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 48.Merseburger AS, Hennenlotter J, Stenzl A, Beger G, Rinnab L, Kuczyk MA, Kuefer R. Cathepsin D serum levels are not a valid serum marker in renal cell carcinoma. Urol Int. 2007;79(1):41–3. doi: 10.1159/000102912. [DOI] [PubMed] [Google Scholar]
- 49.Fukuda ME, Iwadate Y, Machida T, Hiwasa T, Nimura Y, Nagai Y, Takiguchi M, Tanzawa H, Yamaura A, Seki N. Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res. 2005;65(12):5190–4. doi: 10.1158/0008-5472.CAN-04-4134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.