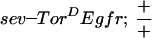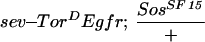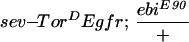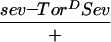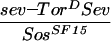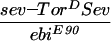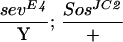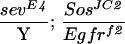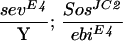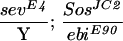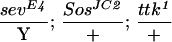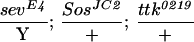Abstract
ebi regulates the epidermal growth factor receptor (EGFR) signaling pathway at multiple steps in Drosophila development. Mutations in ebi and Egfr lead to similar phenotypes and show genetic interactions. However, ebi does not show genetic interactions with other RTKs (e.g., torso) or with components of the canonical Ras/MAP kinase pathway. ebi encodes an evolutionarily conserved protein with a unique amino terminus, distantly related to F-box sequences, and six tandemly arranged carboxy-terminal WD40 repeats. The existence of closely related proteins in yeast, plants, and humans suggests that ebi functions in a highly conserved biochemical pathway. Proteins with related structures regulate protein degradation. Similarly, in the developing eye, ebi promotes EGFR-dependent down-regulation of Tramtrack88, an antagonist of neuronal development.
Keywords: ebi, EGFR, tramtrack88, F box, WD40 repeats, protein degradation
Epidermal growth factor receptors (EGFRs) have a central role in vertebrate and invertebrate development (for review, see van der Geer et al. 1994; Wassarman et al. 1995; Freeman 1998). Biochemical studies largely in mammalian systems and genetic studies in Caenorhabditis elegans and Drosophila have led to a detailed description of the signal transduction pathways elicited by activation of the EGFR (for review, see Kayne and Sternberg 1995; Schwietzer and Shilo 1997). These include the Ras/MAP kinase (MAPK), Ca2+, and phosphatidyl inositol-dependent signaling pathways (for review, see Kazlauskas 1994). In Drosophila, the Ras/MAP kinase cascade is the prominent signaling pathway triggered by the EGFR. Two other fly receptor tyrosine kinases (RTKs) that control patterning and cell fate specification, Torso (for review, see Duffy and Perrimon 1994) and Sevenless (Sev) (for review, see Zipursky and Rubin 1994), do so largely, if not exclusively, through activation of this pathway.
The development of the R7 photoreceptor neuron in the fly eye has proved to be a system amenable to detailed genetic dissection of RTK signaling pathways (Wassarman et al. 1995). Whereas the Sev RTK is required for the development of R7 only, EGFR is essential for the development of most, if not all, cells, including R7 (for review, see Freeman 1996a). This dual RTK requirement is intriguing. Constitutively active overexpressed forms of both Sev and EGFR are sufficient to induce R7 development. Furthermore, overexpression of Spitz, a ligand for EGFR, can partially rescue R7 development in a sev null mutant background. These findings have led Freeman (1996b) to propose that signals from Sev and EGFR are qualitatively equivalent. This is consistent with previous findings that overexpression of activated forms of proteins in the Ras/MAP kinase pathway induce receptor-independent R7 development (Fortini et al. 1992; Dickson et al. 1992a; Brunner et al. 1994). These observations support the view that activation of the Ras/MAPK pathway is sufficient to induce R7 development during normal development.
The activities of three transcription factors are modulated by the MAPK signaling pathway in the R7 precursor cell. Two ETS-domain-containing transcription factors, Yan and Pointed, are direct targets of MAPK phosphorylation: Pointed is activated by phosphorylation and promotes R7 induction (O’Neil et al. 1994), whereas Yan is inhibited by phosphorylation and acts as a transcriptional repressor (Rebay and Rubin 1995). Inactivation of a second repressor, Tramtrack88 (Ttk88), also is required for R7 development (Xiong and Montell 1993). Ttk88 is down-regulated via a MAPK-induced protein degradation pathway. RTK signaling induces the expression of phyllopod (phyl) (Dickson et al. 1995). Phyl and Sina, the product of the seven in absentia gene, then bind to Ttk88 and promote its ubiquitin-dependent degradation (Li et al. 1997; Tang et al. 1997).
Proteolysis has an important role in signaling downstream of activated receptors in several developmental systems. Activation of the NF-κB transcription factor and its Rel homologs in Drosophila (i.e., Dorsal and Dif) requires signal-dependent phosphorylation of the inhibitor, IκB or Cactus, followed by its degradation via a ubiquitin-dependent pathway (for review, see Verma et al. 1995). Degradation of the inhibitor allows NF-κB to enter the nucleus and directly regulate gene expression. Proteolysis also is important in transmitting signals in the Wingless (Wg), Hedgehog (Hh), and Notch (N) pathways. Slimb is required for Wg and Hh signaling and regulates the processing of intracellular components in these pathways, Cubitus interruptus and Armadillo, respectively (Jiang and Struhl 1998). On the basis of genetic and protein-binding studies, C. elegans Sel-10 was proposed to be a direct negative regulator of Notch/Lin-12 signaling (Hubbard et al. 1997). Both sel-10 and slimb encode F-box/WD40 repeat proteins homologous to Saccharomyces cerevisiae Cdc4, which itself targets a specific phosphorylated cell cycle-related substrate, Sic1, to ubiquitin-mediated protein degradation (for review, see Patton et al. 1998).
During the course of a screen for mutations affecting eye development, we identified mutations in a gene we now designate ebi (‘shrimp’ in Japanese). Genetic analysis revealed that ebi functions in EGFR pathways regulating aspects of oogenesis and embryogenesis, and in imaginal disc development. ebi is highly homologous to proteins in yeast, plants, and humans and is structurally related to Cdc4/Sel-10/Slimb. We demonstrate that ebi is required in R7 development for down-regulation of Ttk88 in response to EGFR signaling.
Results
Loss-of-function mutations in ebi and Egfr have similar phenotypes
ebi mutations were identified in a screen for enhancers of an eye mutant called roughex that plays a key role in regulating cell cycle progression in the developing eye (see Materials and Methods; Dong et al. 1997). As a consequence of cell cycle defects, photoreceptor differentiation and pattern formation in the eye are disrupted. Whereas cell cycle regulators enhance and suppress the primary cell cycle phenotype (Thomas et al. 1994; Dong et al. 1997; K.H. Zavitz and S.L. Zipursky, unpubl.), mutations in other loci, such as Star and Egfr (i.e., encoding the EGF receptor), modify the differentiation phenotype only and not the earlier cell cycle defects. Like Star and Egfr, ebi enhances the differentiation phenotype. These observations led us to consider the relationship between the EGFR signaling pathway and ebi. In this section we present evidence that ebi participates in EGFR signaling pathways. ebiE4, ebiE90, and ebiP7 are null, strong, and weak alleles, respectively (see below).
That ebi functions in the EGFR pathway was suggested initially by phenotypes of a viable heteroallelic combination of ebi (i.e., ebiP7/ebiE90). These flies exhibit phenotypes similar to weak loss-of-function Egfr alleles (i.e., Egfrtop1/Egfrf2) including partial female sterility resulting from partially ventralized eggs (Fig. 1A–C), wing vein defects, short bristles, and abnormal eyes (i.e., rough eyes) (data not shown). Further evidence that ebi participates in the EGFR pathway was provided by genetic interactions between them (also see below). For instance, flies carrying two different alleles of Egfr (Egfrtop1/Egfrf2) have a weak rough-eye phenotype, which is enhanced in flies that are heterozygous for ebi (data not shown).
Figure 1.
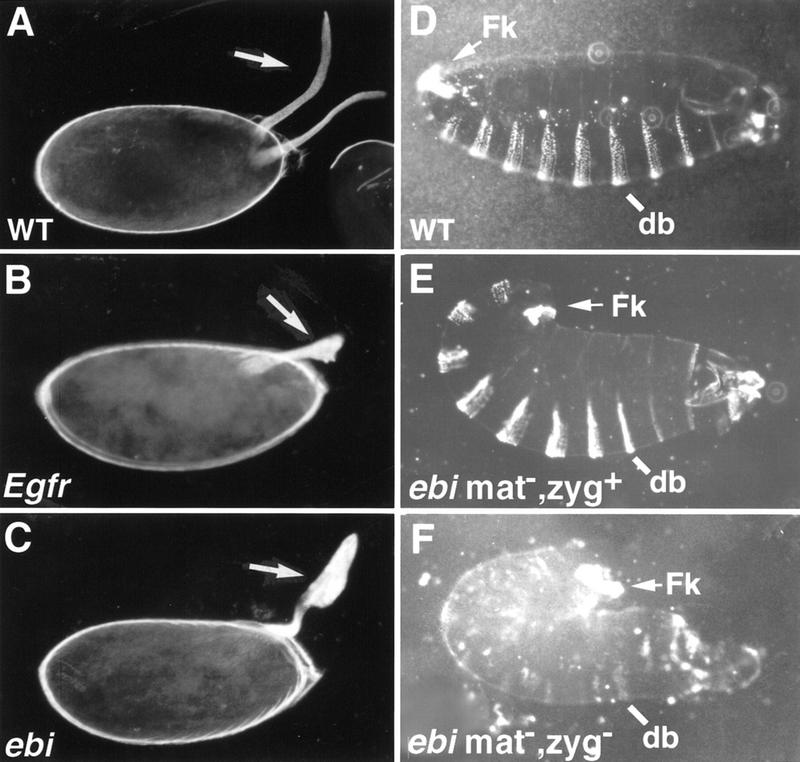
ebi mutants exhibit egg and embryonic cuticle phenotypes. (A) Two dorsal appendages (arrow) are located at the dorsal anterior region of the wild-type egg. (B) Females harboring a weak Egfr allele, Egfrtop1/Egfrtop1, produce ventralized eggs with fused dorsal appendages (arrow). (C) Eggs laid by females mutant for ebi (ebiE90/ebiP7) are also ventralized (arrow). (D) Wild-type embryos. (E) Embryos lacking maternal, but not zygotic, contribution of ebi exhibit a tail-up embryo, indicating failure of germ-band retraction. These embryos also have head defects. Embryos lacking maternal ebi were generated using the ovoD method (see Materials and Methods). (F) Embryos lacking maternal and zygotic ebi exhibit more severe phenotypes, including loss of ventral denticle belt (db) structures and a tightly curled morphology. In addition, severe head defects and malformed posterior filzkörper (Fk) were observed. Anterior is to the right.
ebi and Egfr mutant embryos are similar also. Homozygous ebi null mutant embryos (ebiE4) exhibit a tail-up or U-shaped embryo with head defects (Fig. 1E). Embryos lacking both the zygotic and maternal contributions of ebi were created using ovoD and FRT/FLP-induced recombination (see Materials and Methods). This resulted in a more severe phenotype, including the loss of ventral denticle belt structures and a tightly curled morphology indicating a marked failure in germ-band retraction (Fig. 1F). Severe head defects were also observed. In contrast to Egfr mutants, some residual ventral cuticular structures remain in embryos lacking both the zygotic and maternal contributions of ebi (Fig. 1F).
Loss of ebi also affects Egfr-dependent expression of genes in the embryo. The EGFR ligand Spitz is expressed along the ventral midline and induces expression of different target genes, including fasciclin III (fasIII) and orthodenticle (otd), in cells located in more lateral positions. In zygotic null Egfr mutants both otd and FasIII expression are lost (Kim and Crews 1993; Raz and Shilo 1993). In wild-type stage 11/12 embryos, FasIII protein is broadly distributed in the visceral mesoderm and in a bilaterally symmetric cluster of cells flanking the midline of the ventral ectoderm (Fig. 2A). In ebi mutant embryos lacking both maternal and zygotic contribution, FasIII expression is largely abolished, although some residual patches of staining remain (Fig. 2C). Egfr-independent expression of FasIII in the anterior-most region of the embryo is unaffected in ebi mutants (data not shown). In wild-type stage 10/11 embryos, otd mRNA is expressed in the preantennal head region and in the ventral-most ectoderm (Fig. 2B). In ebi mutant embryos, otd expression was markedly reduced (Fig. 2D). These data suggested that ebi may be a component in the EGFR signal transduction pathway.
Figure 2.
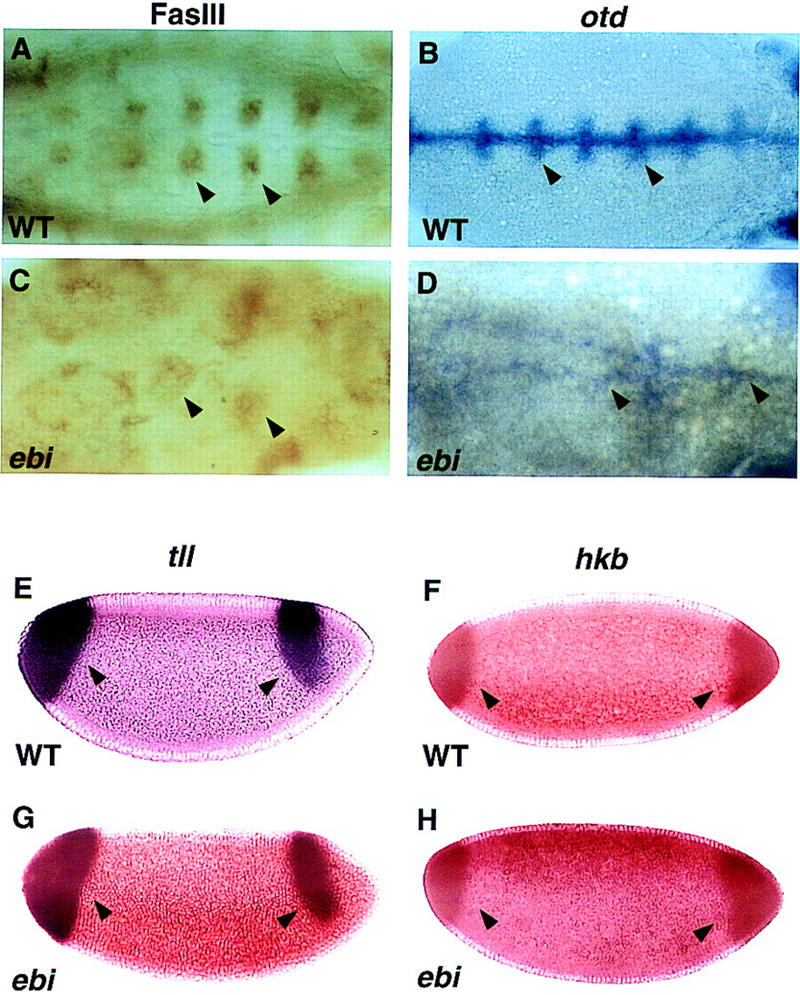
ebi is required for EGFR but not Torso receptor-dependent gene expression. RTK-dependent gene expression in wild-type embryos (A,B,E,F) and embryos lacking both maternal and zygotic ebi (C,D,G,H) were compared. The expression of otd and FasIII is EGFR dependent. Expression of tll and hkb is Torso dependent. (A,C) FasIII expression was determined by immunohistology using mAb7G10. A ventral view of stage 11/12 embryos is shown. (A) In wild-type embryos, FasIII is expressed in clusters of cells flanking the midline of the ventral ectoderm (arrowheads). (C) FasIII levels are severely reduced in ebi embryos (arrowheads). (B,D) Expression of otd mRNA was detected by in situ hybridization. A ventral view of stage 10/11 embryos is shown. (B) Wild-type embryos show strong otd staining in ventral-most ectoderm (arrowheads). (D) Although otd expression is markedly reduced in ebi mutant embryos, residual staining is observed (arrowheads). Expression of tll and hkb mRNA in ebi (G,H) is indistinguishable from wild-type (E,F, respectively). Anterior is to the right.
To assess whether ebi encoded a hitherto unidentified regulator in the Ras/MAP kinase pathway, we assessed its role in the Torso RTK pathway. Torso controls the development of the anterior and posterior termini of the embryo (for review, see Duffy and Perrimon 1994). Ras, Raf, MEK, and MAPK participate in both the EGFR and Torso RTK pathways. The expression of Torso target genes huckebein (hkb) and tailless (tll) in embryos entirely deficient in ebi (i.e., lacking both maternal and zygotic ebi) were indistinguishable from wild type (Fig. 2E–H). Although filzkörper material is present, it is malformed. It is unlikely that this cuticular defect is due to abnormal Torso signaling but, rather, arises later in development as a result of a disruption in the EGFR signaling pathway.
In summary, ebi mutant phenotypes assessed using both molecular and morphological criteria are similar to Egfr mutations. Furthermore, ebi does not function in all RTK pathways, as Torso-induced terminal development is ebi independent. These data indicate that ebi, either directly or indirectly, regulates EGFR signaling. As a step toward understanding the role of ebi in the context of a specific developmental process, we have assessed the role of ebi in R7 development in the compound eye through both genetic and molecular studies.
ebi is required for R7 development
The R7 equivalence group comprises five cells competent to become R7 neurons. They are the R7 precursor cell and the precursors to the four cone cells. Cone cell precursor cells can be induced to become R7 cells by ectopic activation of the R7 inductive pathway in these cells. Transformation of cone cells into R7 cells leads to a disorganized adult eye or a so-called rough-eye phenotype. The ability of loss-of-function ebi mutations to suppress this transformation was assessed in various genetic backgrounds. Whereas ebi dominantly suppressed R7 development induced by the activated EGFR expressed in the R7 equivalence group under the control of the sev enhancer (sev–TorDEgfr) (Fig. 3; Table 1), it did not suppress R7 development induced by the activated Sev receptor (sev–TorDSev (Fig. 3; Table 1), SevS11 (Basler et al. 1991; data not shown), or activated forms of Ras, Raf, and MAPK (data not shown). Hence, ebi is required for the transformation of cone cell precursors into R7 neurons by the activated EGFR.
Figure 3.
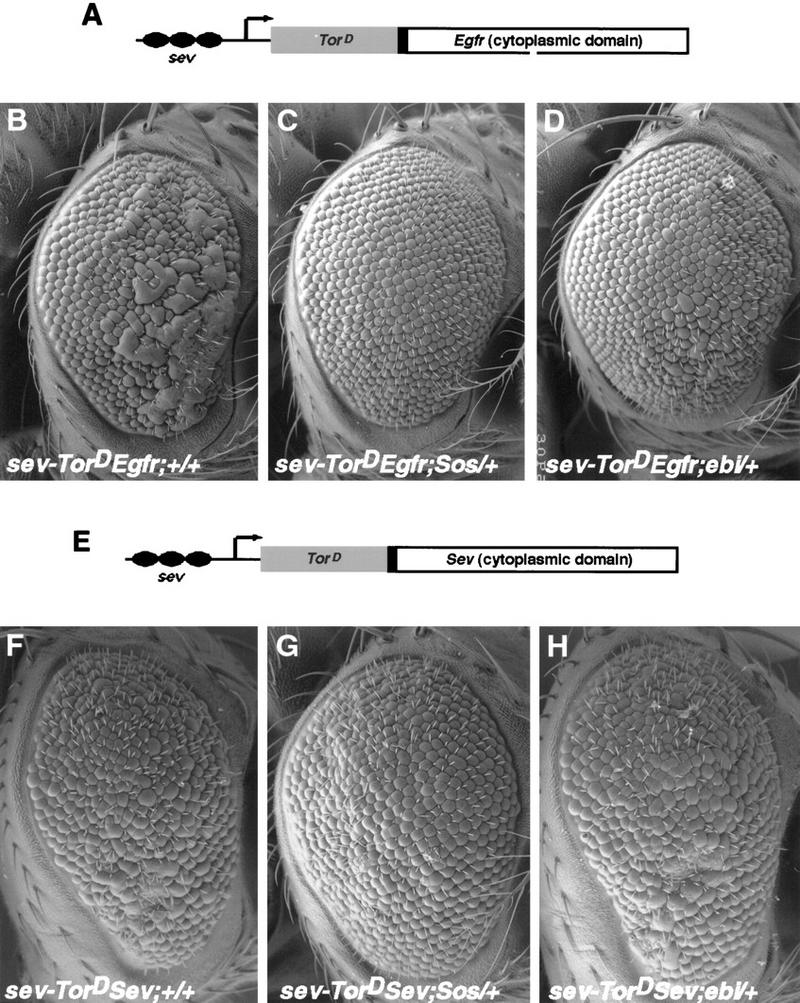
ebi is a dominant suppressor of ectopic R7 development induced by an activated EGFR. (A) A schematic representation of the sev–TorDEgfr construct (Reichman-Fried et al. 1994). (B–D) Scanning electron micrographs of adult eyes showing sev–TorDEgfr alone (B), in an SosSF15/+ background (C), and in an ebiE90/+ background. Like Sos, ebi is a dominant suppressor of sev–TorDEgfr. (E) A schematic representation of sev–TorDSev (Dickson et al. 1992b). (F–H) Scanning electron micrographs of adult eyes showing sev-TorDSev alone (F), in an SosSF15/+ background (G) and in an ebiE90/+ background (H). Although Sos suppresses the sev–TorDSev phenotype, ebi does not (see Table 1). Anterior is to the right.
Table 1.
ebi suppresses sev–TorD Egfr
| Genotype
|
Extra R cells per ommatidia
|
|
|---|---|---|
|
1.8 (n = 159) | |
|
0.5 (n = 142) | |
|
0.5 (n = 122) | |
|
2.9 (n = 89) | |
|
1.9 (n =113) | |
|
3.0 (n = 97) |
SosSF15 is a null allele (Rogge et al. 1991). (n) The number of ommatidia scored. Between four and five eyes were analyzed in semithin sections for each genotype.
To assess whether ebi participates in the induction of the R7 precursor cell into an R7 neuron, we utilized a genetically sensitized background in which only some 15%–20% of the R7 precursors become R7 neurons (Rogge et al. 1991). The R7 inductive signal is attenuated by using a strong hypomorphic allele of sev (sevE4) and a weak gain-of-function mutation in the Ras activator, encoded by the son-of-sevenless gene, SosJC2. Aside from the loss of the majority of the R7 cells, development of the eye in this genetic background is otherwise indistinguishable from wild type. ebi is a dominant enhancer of this phenotype, as are Egfr loss-of-function mutations (Table 2). These data are consistent with previous studies by Freeman (1996b), demonstrating a requirement for both the EGFR and Sev receptor in R7 induction. Hence, ebi is required for induction of the R7 precursor cell into an R7 neuron and for transformation of cone cell precursors into R7 in response to ectopic activation of EGFR. That Ttk88 down-regulation is required for R7 induction of the R7 precursor cell is supported by the finding that Ttk88 mutations are dominant suppressors of the SevE4;SosJC2/+ phenotype (Table 2).
Table 2.
ebi and ttk regulate R7 development
| Genotype
|
Ommatidia containing R7 cells (%)
|
|
|---|---|---|
|
14.7 (n = 2527) | |
|
0.1 (n = 1045) | |
|
0.2 (n = 2036) | |
|
0.3 (n = 1144) | |
|
29.6 (n = 2527) | |
|
31.1 (n = 1219) |
In ttk1, mRNA encoding p88 is missing (Xiong and Montell 1993). ttk0219 is an enhancer trap allele with a P element inserted upstream of the ttk transcription unit (Lai et al. 1996). Egfrf2 is a null allele (Clifford and Schüpbach 1994). (n) The number of ommatidia scored. Between 7 and 12 eyes were analyzed for each genotype.
To assess the role of ebi on R7 development in an otherwise wild-type background, we sought to generate homozygous null mutant clones. We were unable to generate such clones using X-ray and heat shock Flp-induced mitotic recombination. Hence, like Egfr (Xu and Rubin 1993; Dominguez et al. 1998; Kumar et al. 1998 ), ebi is required for cell proliferation and/or survival during the proliferative phase of disc development. To increase the efficiency of Flp-induced mitotic recombination, we used a Flp source driven by the eyeless (ey) promoter. The ey promoter drives expression from the earliest cell divisions in the eye primordium until the last cell division of precursor cells in the third instar. This results in the production of multiple mutant clones throughout development. Mutant clones in the eye disc were recognized by the loss of Ebi immunoreactivity. Rather small clones were observed, consistent with our observations described above with heat shock Flp and X-ray-induced mitotic recombination experiments. Clusters within these clones contained differentiating R cells. Each cluster contained a single R8 cell (i.e., stained with antibody to the Boss protein), and early clusters appeared normal. Although clusters containing eight neurons formed, disorganized clusters containing fewer differentiated neurons also were observed (data not shown).
Adult ommatidia containing homozygous mutant cells were frequently highly disorganized and showed a marked reduction in R cells (Fig. 4A,B). Mutant R cells, including R7 cells, were seen in adult mosaic ommatidia (Fig. 4C); some 80% of these cells showed an altered cellular morphology. Hence, although ebi is required for R7 development in a genetically sensitized background, R7 neurons can develop in an ebi mutant. Although we cannot rule out the formal possibility that these R7 neurons develop because of perdurance of Ebi protein in the R7 precursor cell, these data strongly suggest that R7 cells can form in an ebi-independent fashion, though less efficiently than in wild type. These data are consistent with ebi subserving a redundant function in R7 development (see below). To gain clues to the molecular pathways regulated by ebi, the gene was cloned and sequenced.
Figure 4.
ebi ommatidia are highly abnormal. ebi homozygous clones were generated using FRT-mediated mitotic recombination catalyzed by Flp recombinase expressed under the control of the ey promoter. Pigmented regions are homozygous for the ebiE4 allele; pigmentation results from two copies of a w+ transgene on the ebiE4 chromosome (see Materials and Methods). Ommatidia heterozygous for w+ transgene or lacking this transgene are lightly pigmented or appear unpigmented in the sections. (A) Wild-type ommatidia in the unpigmented region are separated by a pigmented region containing highly abnormal ommatidia. (B) Many ommatidia within the labeled regions lack R cells (see arrowheads). (C) Although many mutant ommatidia lack R cells of all different classes, some normal looking ommatidia contain both ebi mutant and wild-type cells. The arrow indicates an ommatidium containing a genotypically mutant R7 cell (i.e., containing pigment granules).
ebi encodes an evolutionarily conserved nuclear protein with WD40 repeats and an F-box-like sequence
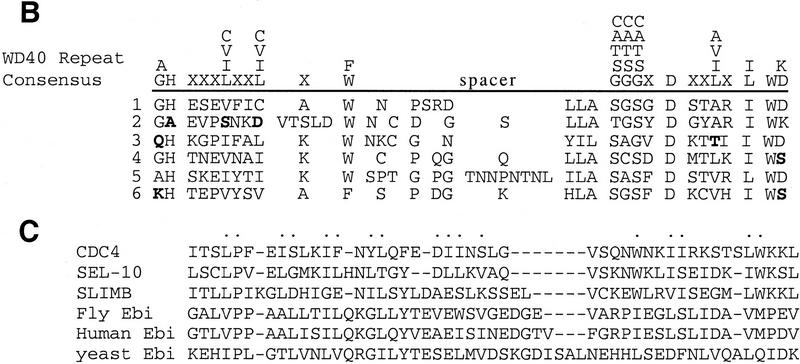
The P-element insertion in ebiP was used to clone neighboring genomic DNA (see Materials and Methods). An 11.1-kb fragment spanning the P element insertion site was used to isolate cDNA clones from an embryonic cDNA library. One cDNA encoded a transcript into which the P element had inserted within the 5′-untranslated region (UTR). Both a 7.5-kb genomic fragment encompassing this transcription unit and a 3.5-kb cDNA fused to the heat shock promoter rescued the mutant phenotype. cDNAs corresponding to this transcription unit encode a protein of 700 amino acids with a carboxy-terminal segment containing six WD40 repeats (Fig. 5A,B). The ebiE4 and ebiE90 alleles result in missense mutations. In ebiE4 the methionine encoded by codon 1 is changed to an isoleucine, and in ebiE90 a highly conserved cysteine, located at amino acid 510 between WD40 repeats 3 and 4, is changed to a tyrosine (Fig. 5A).
Figure 5.

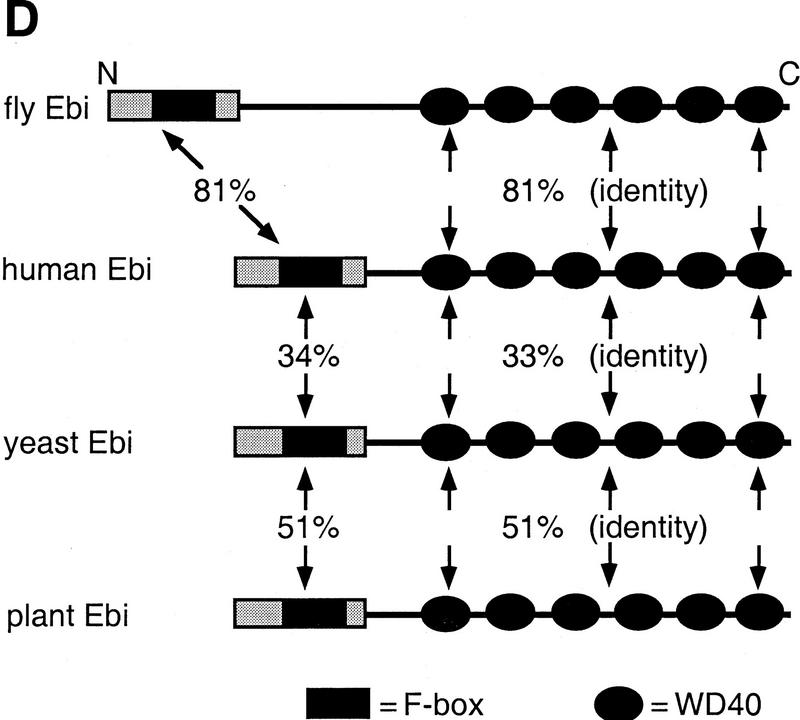
ebi encodes a protein containing WD40 repeats and a diverged F box. (A) The amino acid sequence of Ebi. The F-box-like sequence is indicated by the broken line. WD40 repeats are indicated by solid lines. Missense mutations in ebiE4 (amino acid 1) and ebiE90 (amino acid 510) alleles are indicated above the sequence. (B) Alignment of WD40 repeats. The WD40 consensus sequence according to Neer et al. (1994) is shown. Divergence from the consensus in the Ebi repeats are indicated in boldface type. (C) Alignment of F-box sequence with Ebi amino-terminal sequence (see text). Dots above the alignment indicate the conserved residues in F boxes according to Patton et al. (1998) (D) Schematic representation of fly, human, yeast, and plant Ebi proteins. The percentage of identical amino acids in each region relative to fly Ebi is indicated (see text).
ebi-related human cDNA sequences and genomic sequences from S. cerevisiae and Arabidopsis thaliana, were identified in the database. Because the initial human expressed sequence tag was not complete, additional cDNAs were isolated from adult human spleen cDNA library and sequenced. Both an amino-terminal 89-amino-acid segment and the carboxy-terminal WD40 repeats of fly ebi correspond remarkably well to these regions in the mammalian, plant, and yeast genes; the amino-terminal 89 amino acids and the WD40 repeat region share 81%, 34%, and 51% identity with the human, yeast, and plant sequences, respectively (Fig. 5D). In addition to these conserved regions the fly protein is predicted to contain an insertion of 160 amino acids between the amino terminus and the WD40 repeats.
The bipartite structure of Ebi is reminiscent of three proteins involved in protein degradation, Cdc4 from S. cerevisiae, Slimb from Drosophila melanogaster, and Sel-10 from C. elegans (see introductory section and Discussion). All three proteins contain an amino-terminal F-box and carboxy-terminal WD40 repeats; these proteins have been shown (Cdc4) or proposed (Slimb and Sel-10) to target proteins for degradation by linking them to a ubiquitin–conjugase complex. Although the amino-terminal domain of Ebi is divergent from the Cdc4 F box (as is Slimb), it shares weak sequence and structural homology (Fig. 5C). The amino-terminal half of the F box is more highly related to ebi than the carboxy-terminal region. The periodic spacing of hydrophobic residues in both Ebi and F-box sequences is consistent with these regions assuming an α-helical amphipathic conformation. Elledge and coworkers (Bai et al. 1996) demonstrated that three residues in the amino-terminal region of the Cdc4 F box were required for binding to Skp1 (a component in the E3 complex). These amino acids are conserved in Ebi, as shown in the alignment in Figure 5C, and correspond to residues P45, I52, and L57 in the Ebi sequence.
The localization of Ebi protein was determined using antibodies to three different epitopes, as well as in a transgenic line carrying an ebi genomic construct tagged with a Myc epitope. The distribution of Ebi protein determined with these different reagents was identical. Ebi is widely expressed in nuclei of the embryo and larvae (Fig. 6A–D). Staining was largely, if not exclusively, nuclear. Double staining of salivary gland nuclei with anti-Myc antibodies to detect Myc-tagged Ebi and the DNA stain DAPI demonstrated that Ebi was not associated with chromatin but, rather, was distributed in a reticular pattern throughout the nucleoplasm (Fig. 6E).
Figure 6.
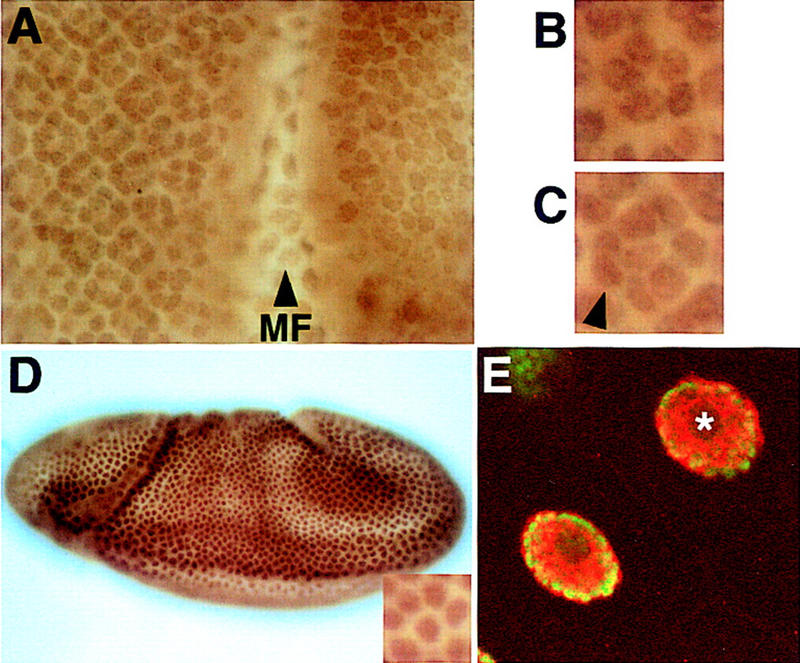
Ebi is a widely expressed nuclear protein. Third-instar eye discs and embryos were stained with an affinity purified anti-Ebi antibody (see Materials and Methods). (A) Ebi is expressed in the nuclei of all cells in the eye disc. (MF) Morphogenetic furrow. Nuclei of cells in the morphogenetic furrow are out of the focal plane shown. High magnification views of a five-cell precluster (B) and eight-cell cluster (C) are shown. The R7 precursor cell is indicated by the arrowhead in C. Anterior is to the right. (D) Ebi is widely expressed in nuclei (inset) of a stage 8 embryo, as well as throughout embryogenesis. (E) Salivary glands were stained with both anti-Myc antibodies to detect Myc-tagged Ebi (red) (see text and Materials and Methods) and the DNA stain DAPI (green). Ebi was not associated with chromosomes or the nucleolus (*) but, rather, was distributed in a reticular network pattern throughout the nucleoplasm.
The similarity of Ebi to F-box/WD40 repeat-containing proteins and its nuclear localization suggested that Ebi may regulate EGFR signaling through degradation of nuclear proteins. Recent studies revealed an important role for both EGFR (Freeman 1996b) and degradation of a specific transcription factor, Ttk88 (Li et al. 1997; Tang et al. 1997), for R7 development. In the next section, we consider the relationship of ebi, Egfr, and ttk88.
ebi regulates Ttk88 protein levels in the developing eye imaginal disc
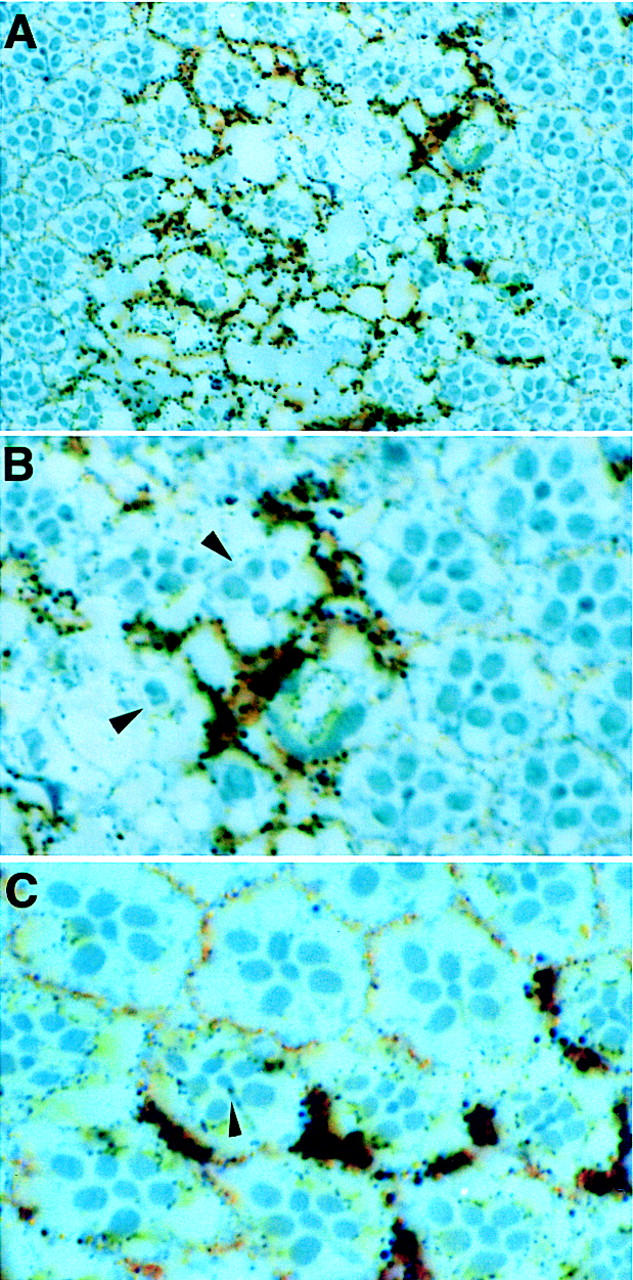
The structural similarity between Ebi and F-box/WD40-repeat proteins involved in protein degradation prompted us to explore the relationship between ebi and Ttk88 protein levels in the developing eye (Fig. 7). Ttk88 is expressed at very low levels in undifferentiated cells in the developing eye disc and at high levels in developing cone cell nuclei; it is not expressed in developing photoreceptor cells. It was shown previously (Li et al. 1997; Tang et al. 1997) that transformation of cone cells into R7 by misexpression of phyl under the sev promoter led to Ttk88 degradation. Ectopic R7 induction by TorDEgfr driven by the sev promoter also leads to marked degradation of Ttk88. sev–TorDEgfr-induced Ttk88 degradation is dominantly suppressed by ebi. Similarly, ebi dominantly suppressed the pGMR–phyl-induced decrease in Ttk88, as well as the pGMR–phyl-induced eye phenotype (data not shown).
Figure 7.
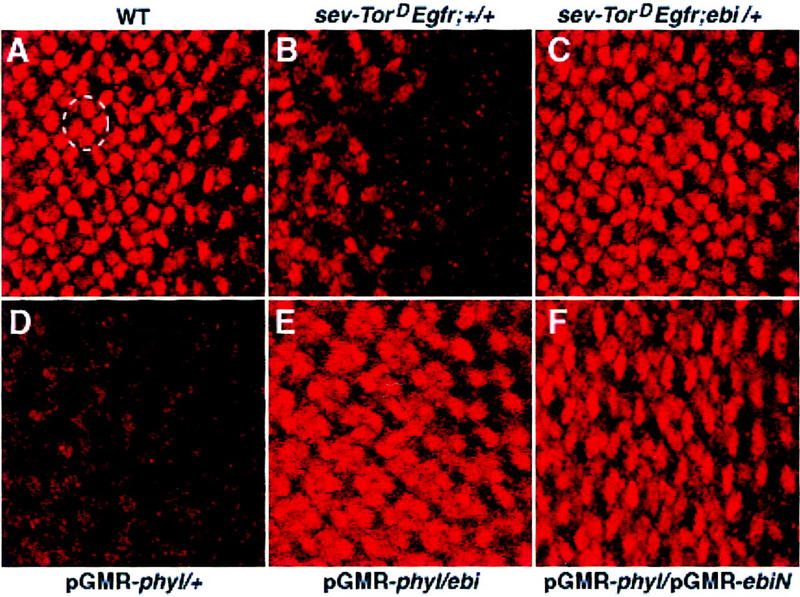
ebi promotes Ttk88 protein degradation. Eye discs labeled with anti-Ttk88 (A–F). (A) In wild-type, Ttk88 is expressed in cone cell nuclei. A single ommatidium is circled to show four labeled cone cells. (B) Ttk88 staining in cone cells is severely reduced in eye discs expressing sev–TorDEgfr. Residual labeling was seen in the posterior region of eye discs, which correlates with the less severe phenotype seen in the posterior part of the adult eye (see Fig. 5B). (C) Ttk88 levels are restored in eye discs heterozygous for ebiE90. pGMR–phyl also induces Ttk88 degradation (D). (E) ebiE4 dominantly suppresses phyl-induced Ttk88 degradation. (F) pGMR-driven expression of a dominant negative ebi transgene, pGMR–ebiN, also restores Ttk88 protein levels in discs expressing pGMR–phyl.
We sought to examine the role of ebi in regulating Ttk88 levels in an otherwise wild-type eye disc. Analysis of Ttk88 levels on the small mutant clones generated with ey-Flp revealed no obvious differences. To explore this issue further, reduction in ebi was achieved by expressing the dominant-negative form of ebi in all cells posterior to the morphogenetic furrow in an ebi heterozygous background (Fig. 8). Dominant-negative ebi contained the amino-terminal half of the protein from amino acids 1–334 (see Materials and Methods for evidence that this dominant-functions as a negative allele) expressed under the control of the pGMR promoter. In wild-type eye discs, Ttk88 staining was not observed in a focal plane in which photoreceptor cell nuclei are located. In contrast, in mutant discs, an average of 36 ± 6 (n = 9 discs) Ttk88-positive nuclei were observed in this region. Most Ttk88-positive nuclei were found 8–10 rows posterior to the morphogenetic furrow.
Figure 8.
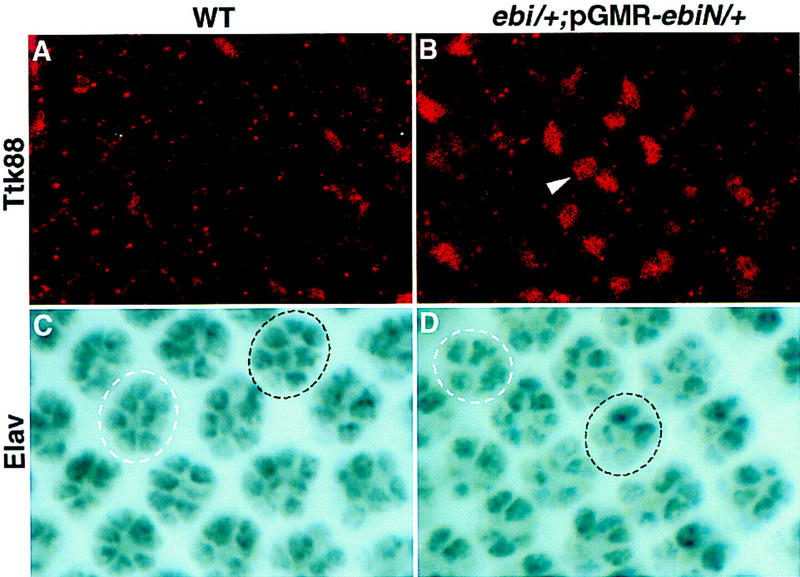
Ttk88 is up-regulated in ebiP25/+; pGMR–ebiN/+ eye discs. (A,B) Anti-Ttk88 staining; (C,D) anti-Elav staining. (A) In wild type, Ttk88 is not expressed in a focal plane containing differentiating R cell nuclei. (B) In ebiP25/+; pGMR–ebiN/+ eye discs, Ttk88 is up-regulated in this region. (C) Wild-type eye disc. An eight-cell cluster, 11 rows posterior to the morphogenetic furrow, is encircled by a broken black line. The R7 cell is slightly out of the focal plane and appears only lightly stained. A more mature cluster, three rows posterior, is encircled by a broken white line. (D) ebiP25/+; pGMR–ebiN/+ eye disc. Two ommatidia containing less than eight Elav-stained R cells at equivalent stages of development as those shown in C are encircled.
This increase in Ttk88-positive cells also parallels a concomitant decrease in the number of cells stained with the pan-neuronal stain, anti-Elav. In wild-type eye discs (n = 10), all ommatidia 8–10 rows posterior to the morphogenetic furrow have at least seven Elav-positive cells (i.e., R1–R6 and R8). However, in mutant discs, many ommatidia (213 of 398 ommatidia; n = 9 discs) in this region contained less than seven stained cells. Interestingly, a considerably smaller fraction of ommatidia (64 of 602 ommatidia) in rows 11–13 contain less than eight Elav-positive R cells (Fig. 8C,D); in wild-type discs, all clusters contain eight Elav-positive cells in this region. Hence, a reduction in ebi activity delays neuronal development and this is correlated with persistent nuclear expression of the Ttk88 protein. In summary, both ebi and Egfr promote Ttk88 down-regulation, thereby promoting neuronal development. Further work is required to assess the relationship between ebi and ttk in Egfr signaling in other developmental contexts.
Discussion
EGFR plays multiple roles in Drosophila development. A complex molecular circuitry modulates the activity of this pathway. Components include proteins in the Ras/MAPK pathway, various EGF-like ligands, receptor antagonists, transcription factors, and modulators of receptor or ligand function (for review, see Perrimon and Perkins 1997). In this paper we describe the ebi gene, encoding an evolutionarily conserved protein that regulates the EGFR signaling pathway in Drosophila. We propose that it plays a role in modulating EGFR signaling during R7 development by promoting degradation of an antagonist of neuronal development, Ttk88.
Ebi is required for EGFR signaling
Several lines of genetic evidence support the view that ebi participates in the EGFR signaling pathway. Strong loss-of-function ebi and Egfr mutations lead to similar embryonic defects, whereas weak loss-of-function mutations result in similar wing, eye, and oocyte abnormalities. Furthermore, the weak Egfr phenotypes in the wing and eye are dominantly enhanced in genetic backgrounds heterozygous for strong ebi loss-of-function mutations. Flies doubly heterozygous for strong ebi and Egfr mutations also show wing and eggshell patterning defects similar to those seen in weak homozygous Egfr mutants (L. Tsuda and S.L. Zipursky, unpubl.). Like Egfr, ebi is a dominant enhancer of a weak sev phenotype associated with sevE4; SosJC2/+. Conversely, the dominant phenotype induced by ectopic expression of a constitutively active chimeric EGFR is dominantly suppressed by ebi.
Genetic evidence indicates that in the developing embryo ebi specifically regulates the EGFR pathway. In embryos lacking both maternal and zygotic ebi, expression of Torso RTK target genes hkb and tll are indistinguishable from wild type. That ebi functions in a pathway distinct from the canonical Ras/MAPK signaling cascade is supported further by the finding that the phenotypes induced in the eye by activated forms of Ras, Raf, MEK, and MAPK are not dominantly suppressed by ebi. Nevertheless, the lack of a dosage interaction does not preclude a role for ebi downstream from activated MAP kinase; it simply states that ebi is not a limiting factor in the Ras/MAPK pathway. Genetic studies in the embryo demonstrated that activated MAP kinase kinase (Dsorsu4) suppressed an Egfr null (Lim et al. 1997) but not an ebi null mutant phenotype (L. Tsuda and S.L. Zipursky, unpubl.). Hence, ebi lies downstream or in parallel to MAPK.
Surprisingly, whereas ebi suppresses dominant Egfr eye phenotype, it does not suppress dominant phenotypes induced by two different activated forms of the Sev receptor, a truncated form in which only the cytoplasmic domain is tethered to the membrane (SevS11) and a second construct, sev–TorDSev, which is analagous to sev–TorDEgfr. This is particularly perplexing because the dominant forms of EGFR and the two different forms of activated Sev drive the same developmental transformation, namely cone cell precursors into R7 neurons, and have been assumed to share the downstream signaling cascade. Although it is possible that this reflects an intrinsic difference between the signaling pathways downstream from the two different receptors, it is also possible that this simply reflects differences in the level of signal or the kinetics of signaling from these two receptors. It is clear that under the conditions used to assay the genetic requirements for Sev and EGFR-mediated cone cell transformation into R7 that the level of inductive signal for EGFR was closer to threshold than for Sev (see Table 1). As the level of signaling approaches threshold, it is more likely that a gene will show a dosage-dependent interaction.
ebi regulates Ttk88 levels
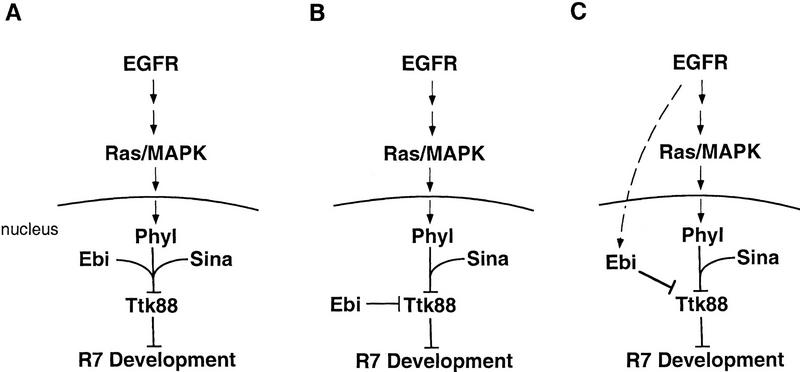
ebi is a dominant suppressor of the transformation of cone cell precursor cells into R7 neurons induced by expression of the activated EGFR or by a downstream target, phyl. It was demonstrated previously (Li et al. 1997; Tang et al. 1997) that phyl overexpression down-regulated Ttk88 protein levels in a sina-dependent fashion. Genetic and biochemical studies support a model in which Sina and Phyl interact directly with Ttk88 and promote its ubiquitin-dependent protein degradation. Here, we demonstrate that down-regulation of Ttk88 in these cells, mediated either by overexpression of Phyl or the activated EGFR, is ebi-dependent. That ebi also functions in the R7 precursor cell was demonstrated using a sensitized system (i.e., sevE4; SosJC2/+) in which the level of the R7 inductive signal hovers near threshold; removing a single copy of ebi in this background prevents R7 development. Conversely, removing a single copy of ttk88 suppresses the phenotype. The structural similarity of ebi to other proteins that directly regulate protein stability (see below) suggests that, like Sina and Phyl, Ebi may play a direct role in controlling Ttk levels in response to an R7 inductive signal. Sina/Phyl and Ebi may be related to each other in several different ways (see Fig. 9). They may define components in the same pathway or two parallel pathways downstream from the EGFR inductive signal. Alternatively, ebi may not contribute to signal-dependent down-regulation of Ttk88 protein directly but, rather, may set a threshold level of Ttk88 by contributing to constitutive protein down-regulation. That some R7 neurons can develop in an ebi mutant background suggests that Ebi and Sina/Phyl may act in a redundant fashion to control Ttk88 stability. Studies in other systems support the view that multiple degradation pathways frequently contribute to the stability of specific proteins. For instance, two distinct ubiquitin-dependent pathways have been shown to regulate Matα2 protein levels (Chen et al. 1993), and both signal-dependent and signal-independent proteolytic pathways regulate stability of IκB (Chen et al. 1995). As we have been unable to detect ttk88 mRNA in either wild-type discs or in ebi heterozygous discs in which dominant-negative ebi is expressed uniformly posterior to the morphogenetic furrow, we cannot exclude a role for ebi in controlling Ttk88 transcription or mRNA stability.
Figure 9.
Models for Ebi function in R7 induction. Data presented in this paper are consistent with the following models for Ebi function in R7 development. (A) Ebi functions in a common biochemical pathway with Phyl and Sina to down-regulate Ttk88; (B) Ebi is a signal-independent constitutive negative regulator of Ttk88; (C) Ebi participates in a parallel pathway regulating Ttk88 downregulation activated by the EGFR.
ebi is highly conserved in evolution and may play a direct role in protein degradation
ebi encodes a protein predicted to contain an amino-terminal F-box-like sequence and six carboxy-terminal WD40 repeats. Biochemical and genetic studies have demonstrated that Cdc4, the founding member of the F-box/WD40 repeat-containing protein family, plays a central role in regulating cell cycle in S. cerevisiae through phosphorylation-dependent down-regulation of the cyclin-dependent kinase inhibitor Sic1 (Feldman et al. 1997; Skowyra et al. 1997). In Cdc4, the WD40 repeats bind to the phosphorylated form of Sic1, and the F-box promotes binding through Skp1 to a complex containing Cdc53 and Cdc34 which, in turn, promotes transfer of ubiquitin from Cdc34 to Sic1 (for review, see Patton et al. 1998). Although the F-box-like sequence in Ebi is divergent from Cdc4, the striking effects of reduction of ebi function on Ttk protein levels in cone cell precursors expressing phyl or activated forms of the EGFR raise the intriguing possibility that Ebi plays an analogous role in signal-dependent degradation of Ttk88.
Two other F-box/WD-repeat-containing proteins have been implicated in signaling pathways regulating development in Drosophila and C. elegans. Jiang and Struhl (1998) have recently shown that the Drosophila slimb gene encodes a six WD40-repeat-containing protein with a diverged F box, which has a role in both the Wingless and Hedgehog signaling pathways by controlling the levels of Armadillo and Cubitus interruptus, respectively. Greenwald and colleagues (Hubbard et al. 1997) have demonstrated that the C. elegans gene, sel-10, is a negative regulator of lin-12 signaling. Although it is not known whether Slimb interacts directly with components in the Wingless or Hedgehog pathways, Sel-10 directly interacts with cytoplasmic domains of Lin-12, and the related mammalian Notch3 receptor. Comparison of sel-10 mutant phenotypes in an otherwise wild-type background and the phenotype in backgrounds sensitized for lin-12 function suggests that Sel-10 function is compensated either by sel-10 homologs or by alternative pathways regulating lin-12 levels or both. Slimb and Sel-10 homologs have been identified in vertebrates, suggesting that they may be conserved elements in the Hh/Wg and Lin-12 (i.e., Notch) signaling pathways, respectively.
Yeast, plant, and human genes have been identified that encode proteins which share remarkable sequence conservation with Ebi. This suggests that Ebi plays a role in a biochemical pathway conserved from fungi to humans. Identification of proteins with which Ebi interacts directly in vivo will provide insights into its precise biochemical function.
Materials and methods
Genetics
Genetic markers and chromosomes are described in Lindsley and Zimm (1992). Two EMS alleles of ebi, ebiE4, and ebiE90, were isolated as dominant enhancers of rux3 from a genetic screen described in Dong et al. (1997). ebi mutations were mapped to bands 21B–21D as they failed to complement Deficiency(2L)al. Testing of available P-element lethal insertions in the region from the Kiss collection led to the identification of a third allele, l(2)k16213, designated ebiP. ebiP7 is a weaker allele and was isolated by imprecise excision of this P insertion. Overexpression of the amino-terminal 334 amino acids (i.e., lacking the WD40 repeats) induced a weak rough eye phenotype. Several lines of evidence indicate that this represents a dominant-negative phenotype: This phenotype was dominantly enhanced by removing a single copy of ebi (i.e., ebi/+). Conversely, the phenotype was suppressed by adding additional copies of a genomic wild-type ebi transgene.
Production of mosiac clones
ebi germ-line mosaic clones were generated using the FLIP-DFS technique (Chou and Perrimon 1992). In brief, virgin females carrying the ebiE4 FRT40A/CyO were crossed with hs-FLP/Y; ovoD1, FRT40A/Elp1, Bc, Gla males. Progeny were heat-shocked for 1 hr at 37°C during larval stages, and females of genotypes hs-FLP/+; ebiE4 FRT40A/ovoD1, FRT40A were examined for the presence of germ-line clones. Both null (ebiE4/ebiE4) and paternally rescued (ebiE4/+) animals, derived from females lacking maternal ebi activity during oogenesis, die during embryogenesis. To distinguish between null and paternally rescued embryos, mosaic females possessing ebi germ-line clones were crossed with males carrying ebiE4/CyO, ftz-lacZ, a balancer chromosome that contains a lacZ gene under the control of the fushi tarazu promoter. The genotypes of embryos were determined by following the expression pattern of the lacZ gene, which was detected by β-galactosidase activity. Embryos without the lacZ marker are referred to as maternal−/zygotic−. Their siblings, which express the lacZ gene, are referred to as maternal−/zygotic+. Larval cuticles were prepared in Hoyer’s mount. Cuticles were examined using dark-field optics. Clones in the eye were generated in flies heterozygous for ebiE4 using FRT-mediated recombination catalyzed by Flp recombinase (provided by expression of flp under the ey promoter. The genotype of these flies was: y, w P[ry, eyflp]/w; ebiE4, P[w+]27F, FRT40A /FRT40A. Mosaic eyes contained patchy pigmentation with regions of dark red (mutant clones) in an otherwise light yellow eye. Analysis of clones was as described previously (Reinke and Zipursky 1988).
In situ hybridization and immunocytochemistry
In situ hybrydization on whole-mount Drosophila embryos was performed according to Tautz and Pfeifle (1989). Single-stranded antisense digoxigenin-containing RNA probes were prepared using the Genius kit (Boehringer Mannheim). Probes were prepared from plasmids containing the tll (Pignoni et al. 1990), hkb (Bronner et al. 1994), and otd (Wieschaus et al. 1992) cDNAs. For visualization, embryos were mounted in glycerol and were analyzed and photographed with a Zeiss Axiophot equipped with Nomarski optics.
Immunocytochemistry was performed as described in Patel et al. (1987). Antibody was used at dilutions 1:50. The FasIII monoclonal antibody (7G10) was obtained from C. Goodman (University of California, Berkeley). Confocal microscopy was done using a Bio-Rad MRC 1024.
SEM and plastic eye section were carried out as described previously (Cheyette et al. 1994). In Table 1 the number of ommatidia containing R7 cells was determined using the pseudopupil method (Banerjee et al. 1987). To determine the ebi expression pattern, a Myc epitope tag sequence was inserted in-frame with the start codon of the ebi genomic rescue construct. Anti-Myc antibody (mAb1-9E10.2 from the ATCC, final dilution 1:25) staining was performed in lines carrying this construct.
Molecular biology
A 5.5-kb BamHI genomic fragment flanking the P-element site of ebiP was isolated by plasmid rescue and subsequently used as a probe to screen a Drosophila genomic library in λEMBL3 (Tamkun et al. 1992). An 11.1-kb DNA flanking the P-element insertion site was isolated and used to screen a 0- to 24-hr embryo cDNA library in λEXLX(+) (Palazzolo et al. 1989). Fourteen cDNAs corresponding to four classes of transcription units were isolated. The P element was inserted into the 5′ UTR of one of the four transcripts. The longest cDNA corresponding to this transcript is 3.5 kb. It has two small introns (65 and 79 bp). ebi cDNAs were sequenced by ABI automated sequencing. ebiE4 and ebiE90 mutant chromosomes were amplified by PCR and sequenced to determine the molecular lesions. In ebiE4 the ATG start codon is changed to ATC (codon for isoleucine), and in ebiE90 the codon TGC encoding cysteine 510 is changed to TAC encoding tyrosine. ebiE4 is likely to be a null allele based on sequencing. DNA and protein databases were searched for homologous sequences using the BLAST program (Altschul et al. 1990).
Isolation of ebi homologs
A human EST clone (114411), which encodes a partial ORF with significant similarity to fly ebi, was obtained from Research Genetics, Inc. The 1.0-kb EST clone was used to screen human adult spleen cDNA library (a gift of Dr. Owen Witte, UCLA). A 2.6-kb human cDNA was isolated and sequenced.
Rescue and expression
Germ-line transformation of Drosophila was performed using standard methods (Spradling and Rubin 1982). A 7.5-kb genomic fragment encompassing the ebi transcription unit and a 3.5-kb ebi cDNA were inserted into pCaspeR4 and CaspeR-hs transformation vectors, respectively. Heat shock at 37°C was administered for 30 min every 12 hr. A DNA fragment encoding the first 334 amino acids of Ebi was inserted into the pGMR transformation vector. This functioned as a dominant-negative construct (see above). More than three independent lines of each construct were generated.
Generation of antibodies
Three peptides corresponding to amino-terminal (TEVEWSVGEDGEVA), carboxy-terminal (WNSKGTKVGASASDG), and middle (SQKKSQNSNEAGSSS) regions of Ebi were synthesized and injected into rabbits to raise antibodies by Chiron Technologies Ltd. Antisera were purified using the peptides as affinity reagents. Ebi staining was performed using one of the purified antisera, 2903, which was raised against the peptide corresponding to middle region of Ebi. The final dilution for the antibodies was 1:300.
Acknowledgments
We thank Gerry Rubin and Corey Goodman for anti-Elav, and anti-FasIII antibodies, respectively, Owen Witte for a human spleen cDNA library, Trudi Schüpbach for Egfr alleles, and Barry Dickson for ey-Flp. We also thank Drs. Owen Witte, Utpal Banerjee, Mike Tyers, Tom Clandinin, and Chi-Hon Lee, as well as members of the Zipursky laboratory, for helpful discussions and for reading various versions of this manuscript. This work was supported by a National Research Service Award (NRSA) training grant (GM07185) (X.D.), a Human Frontiers Research Fellowship (L.T.), a senior research fellowship from the Leukemia Society of America (K.Z.), a Medical Scientist Training Program training grant (M.L.), and a grant from the National Institutes of Health (S.L.Z.). S.L.Z. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL zipursky@hhmi.ucla.edu; FAX (310) 206-3800.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofman K, Ma L, Goebl M, Harper JW, Elledge S. Skp1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Banerjee U, Renfranz PJ, Pollock JA, Benzer S. Molecular characterization and expression of sevenless, a gene involved in neuronal pattern formation in the Drosophila eye. Cell. 1987;49:281–291. doi: 10.1016/0092-8674(87)90569-1. [DOI] [PubMed] [Google Scholar]
- Basler K, Christen B, Hafen E. Ligand-independent activation of the sevenless Receptor tyrosine kinase changes the fate of cells in the developing Drosophila eye. Cell. 1991;64:1069–1081. doi: 10.1016/0092-8674(91)90262-w. [DOI] [PubMed] [Google Scholar]
- Bronner G, Chu-LaGraff Q, Doe CQ, Cohen B, Weigel D, Taubert H, Jäckle H. Sp1/egr-like zinc-finger protein required for endoderm specification and germ-layer formation in Drosophila. Nature. 1994;369:664–668. doi: 10.1038/369664a0. [DOI] [PubMed] [Google Scholar]
- Brunner D, Oellers N, Szabad J, Biggs WH, Zipursky SL, Hafen E. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT α 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes & Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131:643–653. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Schüpbach T. Molecular analysis of the Drosophila EGF receptor homolog reveals that several genetically defined classes of alleles cluster in subdomains of the receptor protein. Genetics. 1994;137:531–550. doi: 10.1093/genetics/137.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson B, Sprenger F, Morrison D, Hafen E. Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature. 1992a;360:600–603. doi: 10.1038/360600a0. [DOI] [PubMed] [Google Scholar]
- Dickson B, Sprenger F, Hafen E. Prepattern in the developing Drosophila eye revealed by an activated torso-sevenless chimeric receptor. Genes & Dev. 1992b;6:2327–2339. doi: 10.1101/gad.6.12a.2327. [DOI] [PubMed] [Google Scholar]
- Dickson BJ, Dominguez M, van der Straten A, Hafen E. Control of Drosophila photoreceptor cell fates by Phollopod, a novel nuclear protein acting downstream of the Raf kinase. Cell. 1995;80:453–462. doi: 10.1016/0092-8674(95)90496-4. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Wasserman JD, Freeman M. Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol. 1998;8:1039–1048. doi: 10.1016/s0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- Dong X, Zavitz KH, Thomas BJ, Lin M, Campbell S, Zipursky SL. Control of G1 in the developing Drosophila eye: rca1 regulates Cyclin A. Genes & Dev. 1997;11:94–105. doi: 10.1101/gad.11.1.94. [DOI] [PubMed] [Google Scholar]
- Duffy JB, Perrimon N. The torso pathway in Drosophila: Lessons on receptor tyrosine kinase signaling and pattern formation. Dev Biol. 1994;166:380–395. doi: 10.1006/dbio.1994.1324. [DOI] [PubMed] [Google Scholar]
- Feldman RMR, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Simon MA, Rubin GM. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature. 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- Freeman M. Cell determination strategies in the Drosophila eye. Development. 1996a;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- ————— Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996b;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- ————— Complexity of EGF receptor signalling revealed in Drosophila. Curr Opin Genet Dev. 1998;8:407–411. doi: 10.1016/s0959-437x(98)80110-x. [DOI] [PubMed] [Google Scholar]
- Hubbard EJA, Wu G, Kitajewski J, Greenwald I. sel-10 a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes & Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signaling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Kazlauskas A. Receptor tyrosine kinases and their targets. Curr Opin Genet Dev. 1994;4:5–14. doi: 10.1016/0959-437x(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Kayne PS, Sternberg PW. Ras pathways in Caenorhabditis elegans. Curr Opin Genet Dev. 1995;5:38–43. doi: 10.1016/s0959-437x(95)90051-9. [DOI] [PubMed] [Google Scholar]
- Kim SH, Crews ST. Influence of Drosophila ventral epidermal development by the CNS midline cell and spitz class genes. Development. 1993;118:893–901. doi: 10.1242/dev.118.3.893. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo B-Z, Moses K. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Harrison SD, Karim F, Li Y, Rubin GM. Loss of tramtrack gene activity results in ectopic R7 cell formation, even in a sina mutant background. Proc Natl Acad Sci. 1996;93:5025–5030. doi: 10.1073/pnas.93.10.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li Y, Carthew RW, Lai Z. Photoreceptor cell differentiation requires regulated proteolysis of the transcription repressor Tramtrack. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- Lim Y, Tsuda L, Inoue YH, Irie K, Yamada TA, Hata M, Nishi Y, Matsumoto K, Nishida Y. Dominant mutations of Drosophila MAP Kinase Kinase and their acitivities in Drosophila and yeast MAP Kinase cascades. Genetics. 1997;146:263–273. doi: 10.1093/genetics/146.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:298–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Palazzolo MJ, Hyde DR, VijayRaghavan K, Mecklenburg K, Benzer S, Meyerowitz E. Use of a new strategy to isolate and characterize 436 Drosophila cDNA clones corresponding to RNAs detected in adult heads but not in early embryos. Neuron. 1989;3:527–539. doi: 10.1016/0896-6273(89)90211-0. [DOI] [PubMed] [Google Scholar]
- Patel NH, Snow PM, Goodman CS. Characterization and cloning of fasciclin III: A glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinational control in ubiquitin-dependent proteolysis: Don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Perkins LA. There must be 50 ways to rule the signal: The case of the Drosophila EGF receptor. Cell. 1997;89:13–16. doi: 10.1016/s0092-8674(00)80177-4. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, Merriam JR, Lengyel JA. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell. 1990;62:151–163. doi: 10.1016/0092-8674(90)90249-e. [DOI] [PubMed] [Google Scholar]
- Raz E, Shilo B. Establishment of ventral cell fates in the Drosophila embryonic ectoderm requires DER, the EGF receptor homolog. Genes & Dev. 1993;7:1937–1948. doi: 10.1101/gad.7.10.1937. [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Reichman-Fried M, Dickson B, Hafen E, Shilo B. Elucidation of the role of breathless, a Drosophila FGF receptor homolog, in tracheal cell migration. Gene & Dev. 1994;8:428–439. doi: 10.1101/gad.8.4.428. [DOI] [PubMed] [Google Scholar]
- Reinke R, Zipursky SL. Cell-cell interaction in the Drosophila retina: The bride of sevenless gene is required in photoreceptor R8 for R7 development. Cell. 1988;55:321–330. doi: 10.1016/0092-8674(88)90055-4. [DOI] [PubMed] [Google Scholar]
- Rogge RD, Karlovich CA, Banerjee U. Genetic dissection of a neurodevelopmental pathway: Son of sevenless functions downstream of the sevenless and EGF receptor tyrosine kinases. Cell. 1991;64:39–48. doi: 10.1016/0092-8674(91)90207-f. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Shio B. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 1997;13:191–196. doi: 10.1016/s0168-9525(97)01091-3. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosophorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Gunning DA, Cho J, Zipursky SL. Cell-cycle progression in the Developing Drosophila Eve: roughex encodes a novel protein required for the establishment of G1. Cell. 1994;77:1003–1014. doi: 10.1016/0092-8674(94)90440-5. [DOI] [PubMed] [Google Scholar]
- van der Geer P, Hunter T, Lindberg PA. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- Verma IM, Stevenson JK, Schwarz EM, Van Antwep D, Miyamoto S. Rel/NF-kB/IkB family: intimate tales of association and dissociation. Genes & Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- Wassarman DA, Therrien M, Rubin GM. The Ras signaling pathway in Drosophila. Curr Opin Genet Dev. 1995;5:44–50. doi: 10.1016/s0959-437x(95)90052-7. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Perrimon N, Finkelstein R. Orthodenticle activity is required for the development of medial structures in the larval and adult epidermis of Drosophila. Development. 1992;115:801–811. doi: 10.1242/dev.115.3.801. [DOI] [PubMed] [Google Scholar]
- Xiong W, Montell C. tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes & Dev. 1993;7:1085–1096. doi: 10.1101/gad.7.6.1085. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Rubin GM. Determination of neuronal cell fate: Lessons from the R7 neuron of Drosophila. Annu Rev Neurosci. 1994;17:373–397. doi: 10.1146/annurev.ne.17.030194.002105. [DOI] [PubMed] [Google Scholar]