Abstract
SR proteins are nuclear phosphoproteins with a characteristic Ser/Arg-rich domain and one or two RNA recognition motifs. They are highly conserved in animals and plants and play important roles in spliceosome assembly and alternative splicing regulation. We have now isolated and partially sequenced a plant protein, which crossreacts with antibodies to human SR proteins. The sequence of the corresponding cDNA and genomic clones from Arabidopsis revealed a protein, atSRp30, with strong similarity to the human SR protein SF2/ASF and to atSRp34/SR1, a previously identified SR protein, indicating that plants possess two SF2/ASF-like proteins. atSRp30 expresses alternatively spliced mRNA isoforms that are expressed differentially in various organs and during development. Overexpression of atSRp30 via a strong constitutive promoter resulted in changes in alternative splicing of several endogenous plant genes, including atSRp30 itself. Interestingly, atSRp30 overexpression resulted in a pronounced down-regulation of endogenous mRNA encoding full-length atSRp34/SR1 protein. Transgenic plants overexpressing atSRp30 showed morphological and developmental changes affecting mostly developmental phase transitions. atSRp30- and atSRp34/SR1-promoter–GUS constructs exhibited complementary expression patterns during early seedling development and root formation, with overlapping expression in floral tissues. The results of the structural and expression analyses of both genes suggest that atSRp34/SR1 acts as a general splicing factor, whereas atSRp30 functions as a specific splicing modulator.
Keywords: SR proteins, alternative splicing, pre-mRNA processing, plant development, overexpression, phenotype
Alternative pre-mRNA splicing is part of the expression program of a large number of genes in animals and plants. It allows the selection of different combinations of splice sites within a given pre-mRNA, generating structurally and functionally distinct protein isoforms (Breitbart et al. 1987; Manley and Tacke 1996; Cáceres and Krainer 1997). Several protein factors involved in the regulation of alternative splicing have been described, including a family of RNA-binding proteins containing arginine/serine-rich regions (SR proteins) (for reviews, see Fu 1995; Chabot 1996; Valcarcel and Green 1996; Cáceres and Krainer 1997). SR proteins are highly conserved nuclear phosphoproteins that are members of a protein family and share a serine phospho-epitope recognized by the monoclonal antibody mAb104 (Roth et al. 1991). They consist at least of one RNA-binding domain (RBD) [the typical RBD or RNA recognition motif (RRM) domain of ∼80 amino acids] (Birney et al. 1993), and possess several serine/arginine (SR) dipeptides near their carboxy termini (Zahler et al. 1992). In general, SR proteins are a defined subgroup of a large superfamily of nuclear proteins with RS-rich domains of variable sequence and position (Fu 1995).
The human SF2/ASF splicing factor, a prototype SR protein, is essential for the first cleavage step in pre-mRNA splicing (Krainer et al. 1990b; Ge et al. 1991) and can also determine in a concentration-dependent manner which 5′ splice site is selected in pre-mRNAs containing alternative sites (Ge and Manley 1990; Krainer et al. 1990a). The preferential usage of the proximal 5′ splice site at higher concentrations of SF2/ASF is counteracted by members of the hnRNP A/B family of proteins (Mayeda and Krainer 1992; Mayeda et al. 1994), suggesting that the relative abundance of these and possible other antagonistic splicing factors determines the patterns of alternative splicing in vitro and in vivo (Mayeda and Krainer 1992; Cáceres et al. 1994; Yang et al. 1994; Wang and Manley 1995; Hanamura et al. 1998). SF2/ASF and any other SR protein can complement splicing-deficient S100 extracts that essentially lack SR proteins, but individual SR proteins sometimes display distinct specificities and efficiencies in splicing different pre-mRNAs (Krainer et al. 1990b; Ge et al. 1991; Fu and Maniatis 1992; Kim et al. 1992; Mayeda et al. 1992; Fu 1993; Zahler et al. 1993; Screaton et al. 1995; Wang and Manley 1995). Furthermore, an important activity of SF2/ASF and other SR proteins is the interaction with exonic enhancer sequences, which stimulate usage of an adjacent weak splice site (Sun et al. 1993; Tian and Maniatis 1993; Staknis and Reed 1994; Ramchatesingh et al. 1995; Tacke and Manley 1995; Lou et al. 1996). The RNA-binding specificity of an SR protein is conferred by the RRM region and adjacent sequences (Cáceres and Krainer 1993; Zuo and Manley 1993; Tacke and Manley 1995; Allain and Howe 1997; Tacke et al. 1997); the various RS domains are responsible primarily for protein–protein interactions, which are modulated by the phosphorylation status of these regions (Wu and Maniatis 1993; Amrein et al. 1994; Kohtz et al. 1994; Zuo and Maniatis 1996; Xiao and Manley 1997). In addition, it has been demonstrated that RS domains modulate the RNA-binding activity and subnuclear localization of SR proteins (Li and Bingham 1991; Hedley et al. 1995; Cáceres et al. 1997). Recently, SF2/ASF, SRp20, and 9G8 have been shown to shuttle between the nucleus and the cytoplasm, and this property depends on the presence and type of RS domain (Cáceres et al. 1998).
The studies described above led to the proposal that SR proteins might function by bridging splice sites through RNA–protein and protein–protein interactions, therefore establishing early interactions for splice-site definition and for the assembly of spliceosomes. These interactions are stimulated by binding of SR proteins to nearby enhancer sequences and modulated by phosphorylation/dephosphorylation of the RS regions.
There is limited information about the regulation of SR protein expression in vivo. In the case of SF2/ASF and SRp20, significant differences in mRNA and protein levels have been observed in various cell types and tissues (Jumaa et al. 1997; Hanamura et al. 1998). SC35 expression is also highly variable in cell lines (Fu and Maniatis 1992; Vellard et al. 1992) and some SR proteins are activated by mitogens (Diamond et al. 1993; Screaton et al. 1995). Interestingly, several alternatively spliced SR protein mRNAs have been described, which code for truncated proteins of still unknown function (Ge et al. 1991; Cavaloc et al. 1994; Screaton et al. 1995; Jumaa et al. 1997). Human SRp20 autoregulates its expression at the level of splicing by binding to its own pre-mRNA, thereby preventing overexpression of the active protein (Jumaa and Nielsen 1997). It was found recently that the amounts of alternatively spliced mRNAs coding for SRp30b and SRp20 in Caenorhabditis elegans may be regulated at least in part at the level mRNA stability (Morrison et al. 1997).
Little is known about the mechanisms of intron excision in plant cells. It is generally assumed that the basic mechanism of splicing in plants is similar to that of yeast and mammals (Solymosy and Pollak 1993; Luehrsen et al. 1994; Filipowicz et al. 1995; Brown and Simpson 1998). Nevertheless, animal introns are not processed in any plant tissue so far examined (Barta et al. 1986; Van Santen and Spritz 1987; Wiebauer et al. 1988). Apparently, the process of intron recognition differs in these two kingdoms. One of the determining features of introns in plants seems to be an U- or AU-rich character, whereas the exons are more GC-rich (for review, see Brown and Simpson 1998). As SR proteins play a critical role in correct splice-site selection in mammals, it is of interest to characterize the corresponding protein family in plants. We screened for similar proteins in plants previously by using the mAb104 antibody or a specific monoclonal antibody raised against human SF2/ASF, and demonstrated that plants do possess SR proteins, including SF2/ASF-like proteins (Lopato et al. 1996a). However, the plant SR proteins are of different size and are smaller than 55 kD. We further showed that plant SR proteins are active in constitutive and alternative splicing when assayed in heterologous HeLa cell extracts.
In an effort to isolate individual plant splicing factors, we have characterized arginine/serine-rich proteins from Arabidopsis belonging to two different families (Lopato et al. 1996b; 1999). Both families show good homology in the amino-terminal RRM with animal SR proteins, but at their carboxy-terminal domains they are richer in arginine than in serine, have few SR dipeptides, and they were therefore termed RS proteins. Nevertheless, these proteins are recognized by the mAb104 antibody and can complement SR protein-deficient HeLa S100 extracts efficiently .
As described in this manuscript, we have now isolated and partially sequenced a plant protein immunoreactive with a human SF2/ASF antibody. This information was used to isolate a gene and a cDNA from Arabidopsis with significant homology to human SF2/ASF, which we termed atSRp30. Interestingly, this protein also has ∼80% similarity to a SF2/ASF-like splicing factor from Arabidopsis characterized previously (originally termed SR1, but in accordance with our nomenclature we propose renaming this protein atSRp34/SR1) (Lazar et al. 1995). We show that in spite of their extensive sequence homology atSRp30 and atSRp34/SR1 are differentially expressed in distinct types of plant cells, thus indicating their different functions. Both SR proteins are alternatively spliced, with the ratios of the different isoforms varying in different tissues and during development of the plant. Overexpression of atSRp30 under the control of a nonspecific promoter in transgenic Arabidopsis plants leads to changes in alternative splicing of its own pre-mRNA and of pre-mRNAs of several other genes, showing that atSRp30 is a modulator of splicing.
Results
Isolation and sequencing of genomic and cDNA clones of atSRp30
We initially identified plant SR proteins in magnesium chloride pellets from protein extracts of carrot and tobacco cell cultures and from Arabidopsis plants. The proteins were detected with the monoclonal antibody mAb104, specific for a shared phosphoepitope in all SR proteins, and with a monoclonal antibody specific for the human SF2/ASF splicing factor (Lopato et al. 1996a). A protein of ∼50 kD from the carrot SR preparation, which was recognized by both antibodies, was isolated and partially sequenced. Two peptides with significant homology to human SF2/ASF and atSRp34/SR1 were found. The first peptide comprised the RNP-2 submotif of the first RRM, and the second peptide had high homology to a sequence situated between RNP-2 and RNP-1 of the second RRM (see Materials and Methods; Fig. 2, below). Based on these sequences, degenerate primers were synthesized and used for PCR on purified genomic DNA from carrot, tobacco, and Arabidopsis. Sequencing of the cloned PCR products revealed two very homologous sequences from Arabidopsis and tobacco. The Arabidopsis fragment was 838 bp long and contained five introns. The fragment borders of the Arabidopsis fragment are marked with black arrows in Figure 1A. The protein sequence deduced from the PCR sequence had extensive homology to both human SF2/ASF and atSRp34/SR1 (Fig. 2). The PCR product from Arabidopsis was used as a probe to screen a λ ZAPII genomic library of A. thaliana. One genomic clone was found and designated GatSRp30 (genomic clone of Arabidopsis thaliana serine/arginine-rich protein with deduced molecular mass of ∼30 kD). It was >4.5 kb long, had 1805 bp of promoter region, but ended 12 bp upstream of the stop codon. A corresponding cDNA was obtained from an expressed sequence tag (EST) cDNA library of A. thaliana ecotype Columbia and sequenced. The mRNA corresponding to this cDNA was designated mRNA1 of atSRp30. Primers derived from the 3′ untranslated region of this cDNA were used to obtain the missing sequences of the genomic clone by PCR amplification on purified genomic DNA. The DNA fragment contained an additional intron and the rest was identical to the corresponding sequence of the cDNA1. The sequence of GatSRp30 and the deduced protein sequence are shown in Figure 1A.
Figure 2.
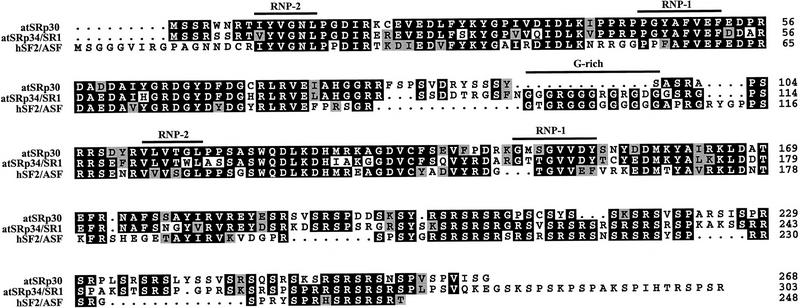
Alignment of Arabidopsis atSRp30, atSRp34/SR1, and human SF2/ASF protein sequences. (Solid area) Positions at which a single residue occurs in at least two of the sequences. (Shaded areas) Conservative substitutions such as RK, IVL, ED, FY, ST. The positions of the conserved RNP-1 and RNP-2 submotifs and the glycine-rich region are indicated.
Figure 1.
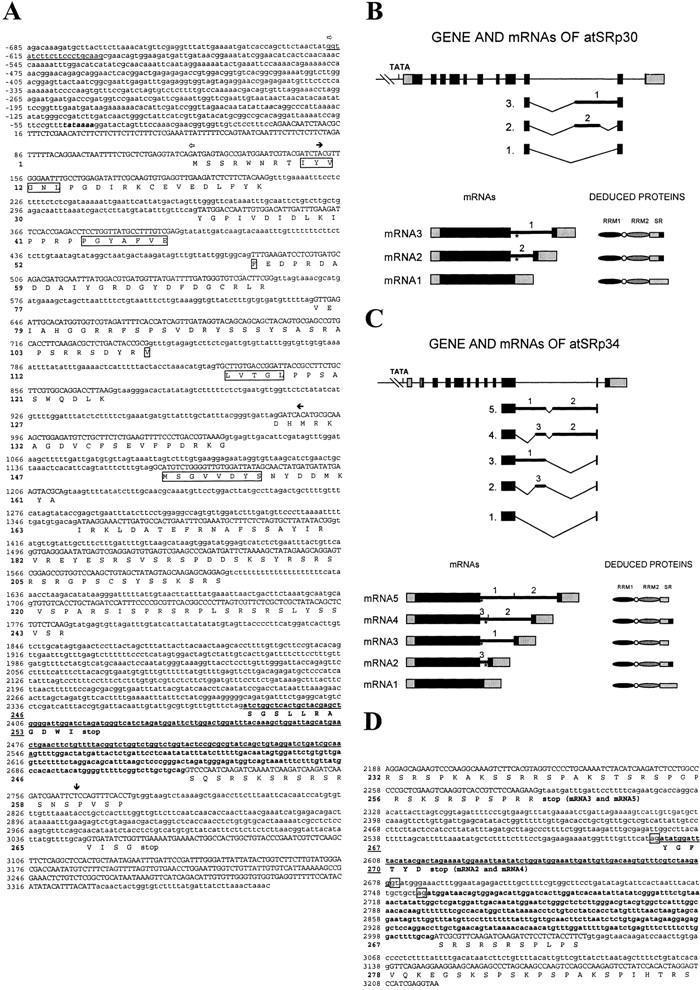
Nucleotide and deduced protein sequences of atSRp30 and of atSRp34. (A) Genomic sequence of atSRp30; promoter and intron sequences are indicated in lowercase, cDNA sequences in uppercase, and the TATA box in bold italics. The deduced protein sequence is shown below the DNA in the one-letter code. The bold sequence in the tenth intron is included in alternatively spliced mRNAs, and the underlined sequence is included when both cryptic 3′ and 5′ splice sites are used. The conserved RNP submotifs of both RRMs are boxed. The open arrows mark the ends of promoter sequences used for expression studies. Solid horizontal arrows indicate the ends of the PCR product obtained with degenerate primers and used as a probe for library screening. (↓) The end of the GatSRp30 clone. The sequence data of the atSRp30 gene was submitted to the EMBL database (accession no. AJ131214). (B) Schematic representation of GatSRp30, its mRNA isoforms, and deduced proteins. Exons are shown as boxes and introns as lines (bold lines: introns included in the alternatively spliced mRNAs). Exonic 5′ and 3′ untranslated regions are shaded, and the coding regions are black. (*) The new stop codon in the alternatively spliced products. (C) Schematic representation of GatSRp34, its mRNA isoforms, and deduced proteins. The drawings are as in B (accession no. AF001035). (D) Partial genomic sequence of atSRp34/SR1 starting from exon 10 up to the stop codon. The bold sequence in the long intron is included in alternatively spliced mRNAs; the underlined sequence is included when both cryptic 3′ and 5′ splice sites are used.
To facilitate comparison of features of atSRp30 and of the closely related Arabidopsis atSRp34/SR1 protein (Lazar et al. 1995), the genomic sequence of this protein was determined using PCR. GatSRp34 has a very similar gene structure to that of GatSRp30, except for an intron present in the 5′-untranslated region of the former. The fragment of GatSRp34 comprising the part from exon 11 to the stop codon is shown in Figure 1C, where the alternative splicing events involving intron 11 are also indicated.
Comparison of atSRp30 to other SR proteins
The deduced protein sequences of atSRp30 and atSRp34/SR1 are very homologous (80.7% similarity and 67.1% identity) to each other and both show very high similarity (75.3 and 77.8%, respectively) and identity (58.1 and 59.4%, respectively) to human SF2/ASF (Fig. 2). As atSRp30 and atSRp34/SR1 are less homologous to other animal or plant SR proteins identified to date, both proteins can be considered true orthologs of human SF2/ASF. Within their amino-terminal portions, all three proteins contain two RRMs with their conserved RNP-2 and RNP-1 submotifs, whereby the second RRM is atypical and contains the invariant SWQDLKD signature characteristic of SF2/ASF-like SR proteins (Birney et al. 1993). However, unlike atSRp34/SR1 and SF2/ASF, the RRMs of atSRp30 are not separated by a glycine-rich ‘hinge’ region but rather by a serine-rich sequence. The RS domain of atSRp30 is shorter than the one of atSRp34 because of an extension at the 3′ end of atSRp34, which includes a previously described positively charged proline/serine/lysine-rich (PSK) domain of unknown function (Lazar et al. 1995). If this unique 3′ extension is not taken into account, atSRp34/SR1 is slightly more homologous to human SF2/ASF, mainly because of their common G-rich hinge region. Taken together, these analyses suggest that in contrast to mammals, for which only one SF2/ASF protein has been described to date, two SF2/ASF homologs exist in Arabidopsis.
RNA distribution and alternative splicing forms of atSRp30 and atSRp34/SR1
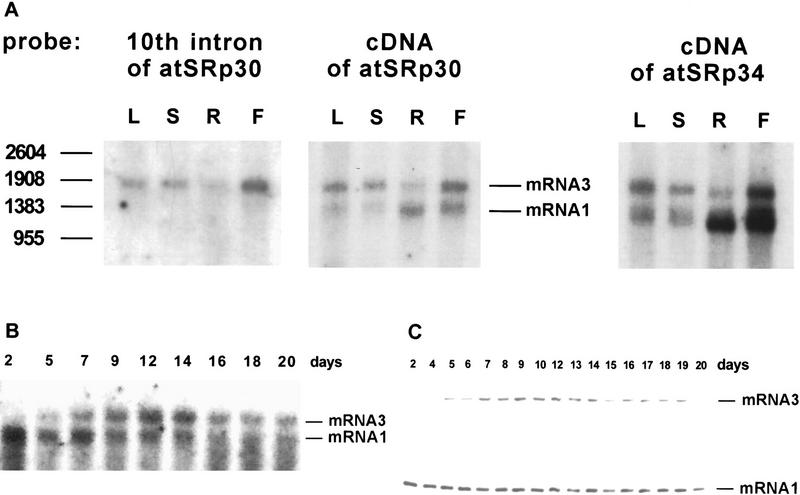
RNA blots of poly(A)+ mRNA from various tissues of wild-type Arabidopsis plants were probed with radioactive labeled atSRp30 or atSRp34/SR1 cDNAs and revealed at least two mRNA species in each case (Fig. 3A). The level of expression of each gene varied considerably in different tissues but in both cases was highest in flowers, followed by roots (Fig. 3A). In addition, the ratio of the two discernible mRNAs, mRNA3 and mRNA1, was different for each gene in the various organs. For atSRp30, mRNA3 seemed to be more abundant in leaves, stems, and flowers, whereas mRNA1 was predominant in roots. In contrast, the ratio of the two main mRNAs of atSRp34/SR1 was ∼1:1 in leaves and stems, whereas in roots and flowers mRNA1 predominated. Reprobing of the RNA blot with the long tenth intron of atSRp30 showed that mRNA3 retained sequences of this intron, indicating that alternative splicing involves this region of atSRp30 pre-mRNA.
Figure 3.
Expression of atSRp30 and atSRp34/SR1 in wild-type Arabidopsis thaliana. (A) Expression in different plant tissues. Northern blot analysis of poly(A)+ RNA from leaves (L), stems (S), roots (R), and flowers (F) is shown. One microgram of RNA was loaded per lane and the blots were either probed with a probe corresponding to the tenth intron of GatSRp30, or with the cDNAs of atSRp30 or atSRp34/SR1. (B,C) Developmental expression of atSRp30. Total RNA was isolated from whole plants on different days during development starting from the day of germination. The RNA was either used for Northern blot analysis with 10 μg/lane of total RNA and the membrane probed with atSRp30 cDNA (B), or for analysis of the RT–PCR products on a 1.2% agarose gel (C). The two primers for the PCR reaction were located in the exons adjacent to the tenth intron.
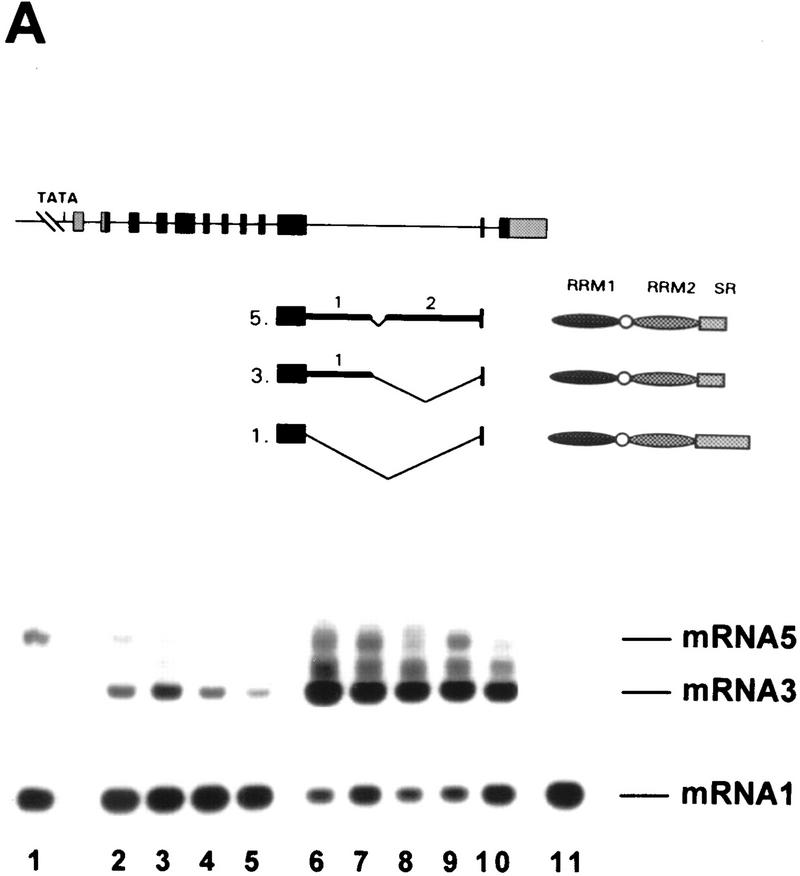
The full sequences of the various mRNA isoforms were obtained by RT–PCR cloning from total RNA using primers from the 5′ and 3′ ends of the genes (see Materials and Methods). mRNA1 corresponded to the cDNA sequence of atSRp30, whereas mRNA3 retained part of the tenth intron, because of usage of an alternative 3′ splice site (Fig. 1A,B). The mRNA2 isoform with both alternative 3′ and 5′ splice sites within intron 10 (Fig. 1A,B) was not visible in Northerns of wild-type plants (see Fig. 3A) and was only detected by RT–PCR by increasing the number of cycles (data not shown).
The same approach was used to sequence the mRNA isoforms of atSRp34/SR1 (Fig. 1C). The longest mRNA species, which is also the most abundant in wild-type plants, was named mRNA5 and retained most of intron 11 (which is in the position corresponding to intron 10 of atSRp30) except for an internal intron of ∼80 nucleotides. Two other minor alternative mRNAs could be detected during early plant development, but only by RT–PCR (data not shown). mRNA4 uses an additional alternative 3′ splice site; mRNA2 uses alternative 5′ and 3′ splice sites (Fig. 1C,D); and mRNA3, which is the main alternatively spliced mRNA in plants when atSRp30 is overexpressed from the 35SCaMV promoter (Fig. 5, below), uses one alternative 5′ splice site.
Figure 5.
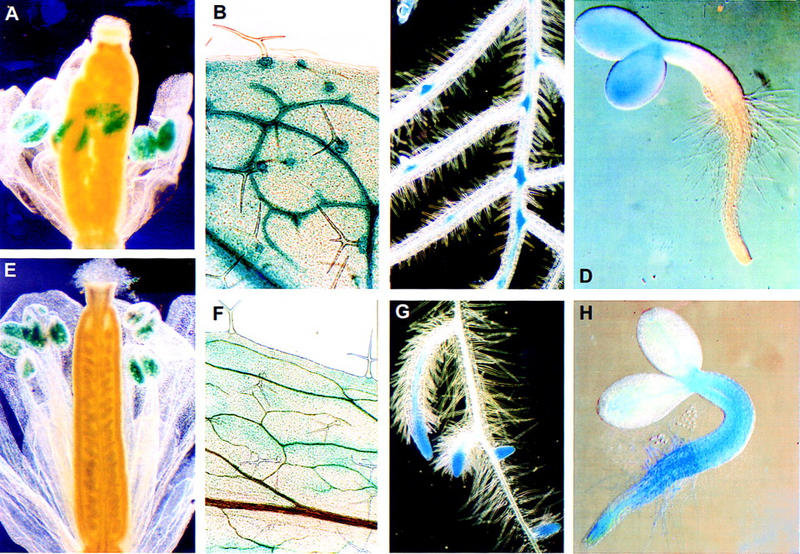
Transcriptional analysis of atSRp30 (A–D) and atSRp34/SR1 (E–H) promoter–GUS fusions in transgenic Arabidopsis. GUS staining of (A,E) flowers at the postanthesis stage; (B,F) staining patterns in the leaves; (C,G) primary and developing lateral roots; (D,H) seedlings (2 days after germination).
The above alternatively spliced mRNAs of atSRp30 and atSRp34/SR1 all had an in-frame stop codon near the 5′ end of the retained intronic sequences. The hypothetical shorter proteins, designated atSRp30s and atSRp34s, lack the carboxy-terminal part of the RS domain and instead have other sequences that are shown in bold in Figure 1A for atSRp30s and in Figure 1C for atSRp34s.
To obtain information about changes in the transcriptional and alternative splicing patterns of atSRp30 during plant development, total RNA was isolated from whole plants of different ages and used for RNA blot hybridization and RT–PCR analysis (Fig. 3B,C). Primers derived from exons adjacent to the 10th intron were used for RT–PCR. The results from both methods were in good agreement and showed that the expression of mRNA1 is highest in younger plants and starts to decline around day 12, whereas the expression of mRNA3 is extremely low in young seedlings, peaks between days 9–14, and declines slowly afterward. Although we do not know how the ratio of these two transcripts is regulated, these regulatory events might determine the quantity of genuine atSRp30 protein in individual cells.
Antibody detection of Arabidopsis SR proteins
Polyclonal antibodies were raised in chickens against purified recombinant atSRp30 and atSRp34/SR1 and used to identify antigens in different ammonium sulfate fractions of Arabidopsis extracts. The extracts were fractionated by sequential precipitation with ammonium sulfate and magnesium chloride, to enrich for SR proteins (Roth et al. 1990). The anti-p30 and anti-p34 antibodies recognized proteins in the 60%–90% ammonium sulfate cut. The immunoreactive proteins in the dialyzed 60%–90% cut precipitated quantitatively in the presence of 20 mm magnesium chloride. No crossreaction of plant proteins with the pre-immune immunoglobulin fraction was found (data not shown).
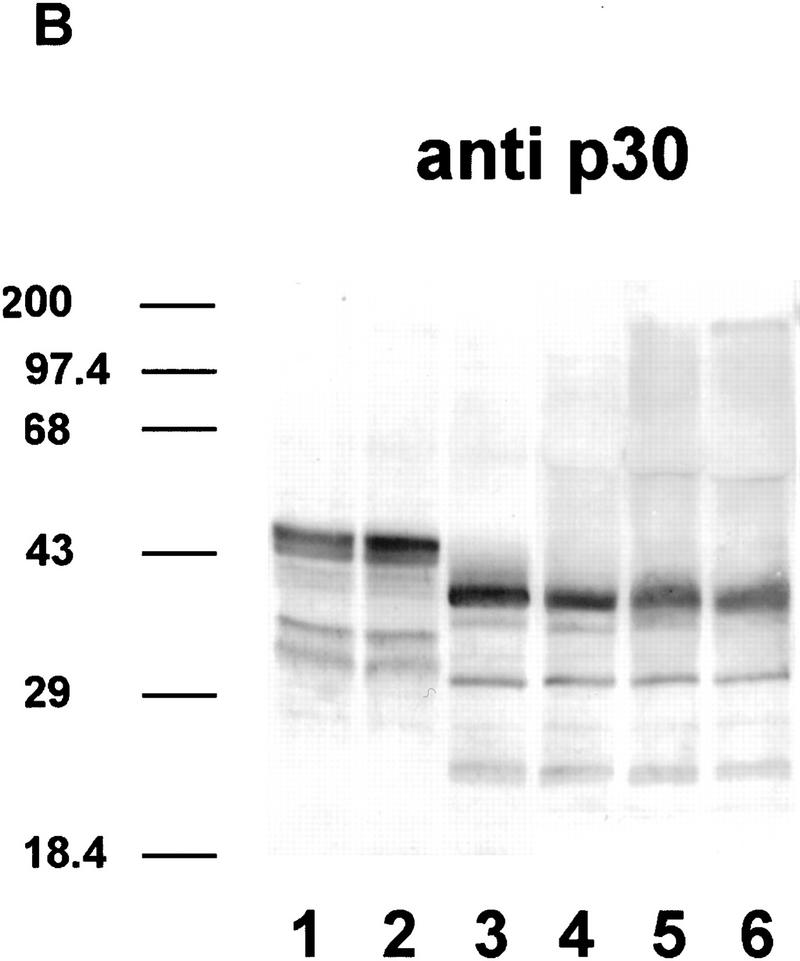
The magnesium precipitate was immunoblotted and probed with four different antibodies. With anti-p30, six protein bands were detected as three doublets, with the 43- to 46-kD doublet showing the highest intensity. The two other doublets migrated with apparent molecular masses of 38–40 and 31–34 kD (Fig. 4A, lane 2). The immunostaining pattern with the monoclonal antibody mAb104 (Fig. 4A, lane1), which is specific for a serine phosphoepitope common to SR proteins (Roth et al. 1990) was very similar, although the smallest band was more pronounced than with anti-p30 (lane 2). This could be indicative of an additional SR protein of ∼30 kD, or it may reflect the presence of multiple or stronger phosphoepitopes within p30. We did not detect high molecular mass proteins with the mAb104 antibody, although (Lazar et al. 1995), have reported immunoreactive proteins up to 120 kD.
Figure 4.
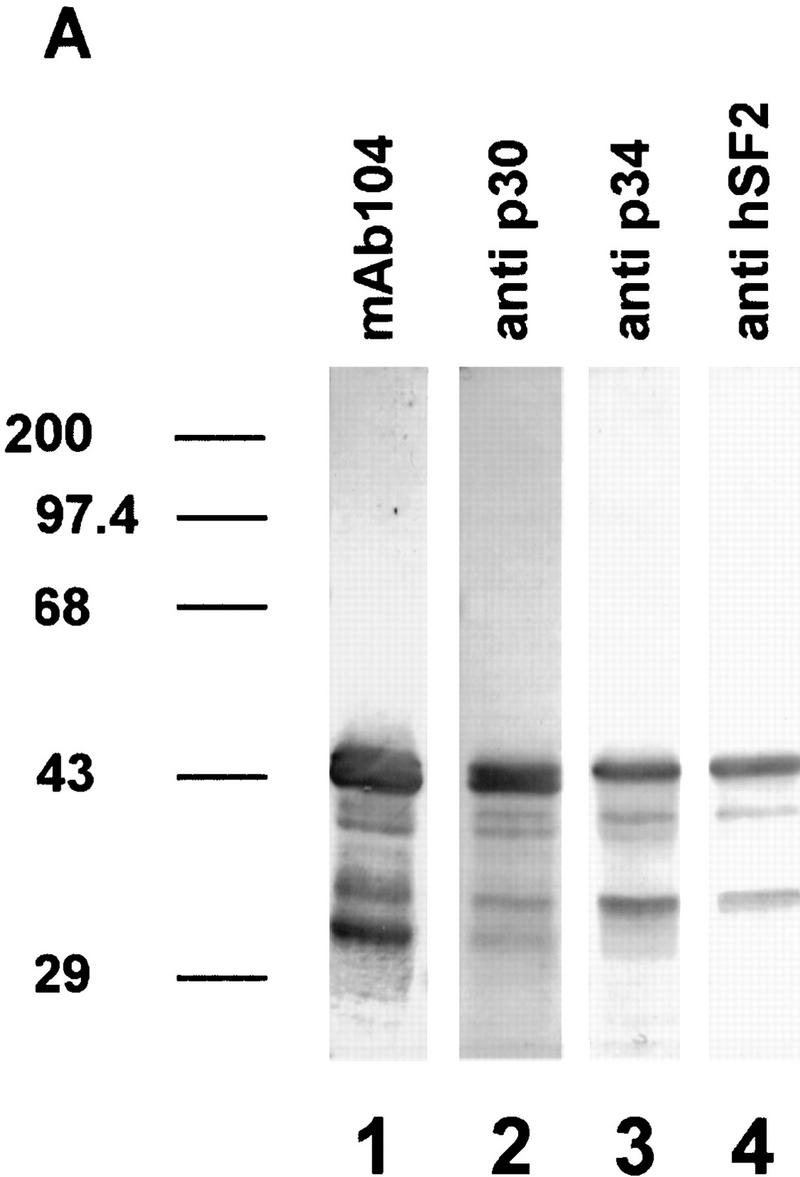
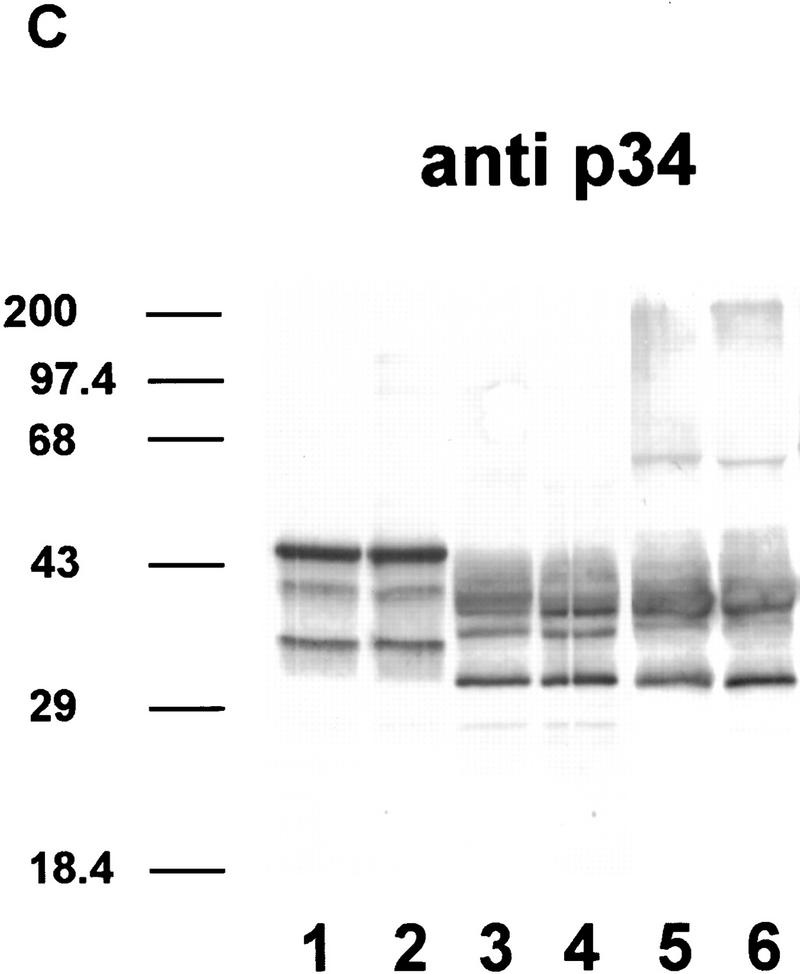
Immunodetection of phosphorylated and dephosphorylated SR proteins from cell cultures of Arabidopsis. (A) Immunodetection of proteins in SR protein preparations (lanes 1–4). (Lane 1) Monoclonal antibody mAb104, which is specific for a common phosphoepitope of SR proteins; (lane 2) polyclonal antibody raised against recombinant atSRp30; (lane 3) polyclonal antibody raised against recombinant atSRp34/SR1; (lane 4) monoclonal antibody specific for human SF2/ASF. Protein markers are indicated at the left. (B,C) Dephosphorylation of SR proteins with alkaline phosphatase. (Lanes 1,2) SR proteins incubated at 37°C without enzyme for 0 and 24 hr, respectively; (lanes 3,4) SR proteins incubated with 0.2 and 1.0 U/μl of enzyme for 3 hr; (lanes 5,6) SR proteins incubated with 1.0 and 2.0 U/μl of enzyme for 24 hr. Proteins were probed either with anti-p30 (B) or anti-p34 (C).
The anti-p34 antibodies recognize mainly three proteins of ∼46–47, 40, and 34 kD. As the same antibody shows minimal cross-reaction with recombinant atSRp30 (data not shown), these bands probably represent atSRp34/SR1-related proteins and correlate well with published overexpression data in which the protein was immunodetected with a size of 47–48 kD (Lazar et al. 1995). The 40-kD protein comigrates with an alternatively spliced isoform of atSRp34/SR1 (see below), whereas the nature of the 34-kD polypeptide remains to be determined. Interestingly, the immunoblot with anti-p34 is very similar to that obtained with a monoclonal antibody specific for human SF2/ASF (Fig. 4A, cf. lanes 3 and 4). Therefore, anti-hSF2, which recognizes a discontinuous epitope within RRM1 of hSF2/ASF but does not recognize other human SR proteins (Hanamura et al. 1998) cross-reacts with the same set of Arabidopsis proteins as anti-p34, supporting the notion that hSF2/ASF resembles atSRp34/SR1 in this region more closely than it does atSRp30.
The fact that atSRp30 and atSRp34/SR1 are present in the SR protein preparation and comigrate with specific bands in the mAb104 immunoblot suggests that both proteins are phosphorylated. To test this suggestion, we treated the SR protein preparation with increasing concentrations of alkaline phosphatase and for different times (Fig. 4B,C). Compared to the untreated controls (lanes 1,2), these treatments resulted in an increase in the mobility of all immunostained bands. Under these conditions, anti-p30 recognized predominantly three proteins (27, 31, and 38 kD), whereas anti-p34 recognized two proteins (31 and 38 kD). As expected, all these proteins were no longer stained with mAb104, except for the 38-kD band, which probably represents a partially dephosphorylated intermediate resistant to further dephosphorylation (data not shown). Interestingly, dephosphorylation led to the appearance of new bands of higher apparent molecular mass, consistent with our observation that unphosphorylated recombinant atSRp30 and atSRp34/SR1 are very insoluble and have a strong tendency to aggregate.
Transcriptional activity of SR protein promoters in Arabidopsis
It is striking that in contrast to mammals, Arabidopsis possesses two SF2/ASF-like proteins. As revealed by the Northern blot analysis, the genes of both proteins are transcribed in all plant organs. It was therefore of interest to investigate if both genes are generally transcribed in all tissues or specifically in different tissues, or to which extent they have overlapping activities. To this end, the promoters of GatSRp30 and GatSRp34, including the 5′-untranslated regions up to the start codon, were fused to the coding region of a reporter β-glucuronidase gene and transferred into A. thaliana plants by Agrobacterium tumefaciens-mediated root transformation. β-Glucouronidase (GUS) activity was never observed in any tested nontransgenic Arabidopsis tissues, as well as in transgenic plants harboring a promoterless GUS gene. Control plants containing a 35S cauliflower mosaic virus (CaMV) promoter–GUS fusion were easily stained, demonstrating that substrate accessibility was not limiting GUS activity. To exclude an influence of the transgene position on promoter activity, the analyses were carried out with several independent transgenic lines.
Using detailed histochemical analyses, we found that both genes had distinct but also overlapping patterns of transcriptional activities. Flowers in the postanthesis stage (Fig. 5A,E) showed essentially the same staining patterns for both genes, with the most pronounced expression in the pollen grains and weak staining of sepals and style. Additionally, weak activity was observed in the abscission zone of the flower in atSRp30–GUS transgenic plants (Fig. 5A). The leaves of atSRp30–GUS plants showed strongest staining in vascular bundles and in the support cells of each trichome (Fig. 5B), whereas the atSRp34–GUS fusion was weakly active only in secondary veins and in the cells around them, but activity was never observed in trichomes or support cells (Fig. 5F). The differences in the GUS-staining patterns of these genes were especially evident during the different stages of lateral root development. Both atSRp30 and atSRp34–GUS fusions were active at the very first stages of lateral root initiation, when pericycle cells are stimulated, dedifferentiate, and proliferate. Then, at the later stages, when redifferentiation occurs to form the lateral root meristem, atSRp30–GUS was expressed in the basal enlarged cells of the forming lateral root (Fig. 5C), whereas atSRp34–GUS expression was characteristic of the activated root meristem (Fig. 5G). The staining patterns during early seedling development provided additional evidence for the distinct transcriptional activities of atSRp30 and atSRp34/SR1. Each of the genes was expressed in specific regions along the apical–basal axis of the seedling. In atSRp30–GUS seedlings, expression was restricted to the cotyledons (Fig. 5D), whereas in atSRp34–GUS seedlings, expression was observed in hypocotyl and in root (Fig. 5H), suggesting that these genes can be transcriptionally active in regions with different patterns of cell division. This suggestion was confirmed by differential expression of atSRp30 and atSRp34/SR1 during lateral root formation, as well as in trichome support cells.
Overexpression of atSRp30 in transgenic plants
In Drosophila, ectopic overexpression of SRp55/B52 resulted in various developmental abnormalities, although the identity of affected transcripts remains unknown (Kraus and Lis 1994). It was therefore of interest to study the effect of overexpression of atSRp30 on plant development and on the alternative splicing patterns of individual plant transcripts. The constructs used contained either a complete gene (pG30) or a cDNA (pC30) encoding atSRp30 under the control of the strong constitutive promoter of the 35S RNA from CaMV. The 35S CaMV promoter is strong and constitutive in all plant tissues studied, which was confirmed in control experiments using GUS as a reporter gene. To control for the conditions of the regeneration and transformation procedure in the RNA analysis of transformants, negative controls were either transformed with a 35SRNA promoter–GUS control (pBI121) or were transformants with the same construct, but which for unknown reasons did not show overexpression.
Both fusion constructs, pG30 and pC30, were used for Agrobacterium-mediated transformation of Arabidopsis roots. Forty independent transgenic lines were regenerated for each construct; eight of the lines transformed with pG30 and twelve transformed with pC30 were used in further work. Some of these transgenic lines were used for RNA blot and RT–PCR analyses as shown in Figure 6, A and B. Surprisingly, all pG30 transformants (Fig. 6A,B, lines 1–4) expressed mainly mRNA3, which possesses an alternative 3′ splice site within the long intron (Fig. 1B); nevertheless, mRNA1 was still more abundant in these plants than in the control plant (Fig. 6A, lane 10), as judged by the RNA blot analysis (four times more product was loaded in lane 10 in Fig. 6B). Although mRNA1 levels in both types of transformants are quite different (Fig. 6A), the RT–PCR shows similar yields (Fig. 6B) because a high number of cycles was employed to detect all alternatively spliced products. As expected, all transformants containing pC30 overexpressed only mRNA1 (Fig. 6A, lanes 5–9). In both types of transformants, overexpressed mRNAs were >100-fold more abundant than in wild-type plants. Interestingly, when pC30 transformants were analyzed by RT–PCR, only mRNA2, which uses cryptic 3′ and 5′ splice sites in intron 10 (Figs. 6B and 1B), was present, whereas a small amount of mRNA3 was visible in wild-type plants. This result shows that an excess of atSRp30 changes 5′ splice-site selection of the endogenous atSRp30 pre-mRNA.
Figure 6.
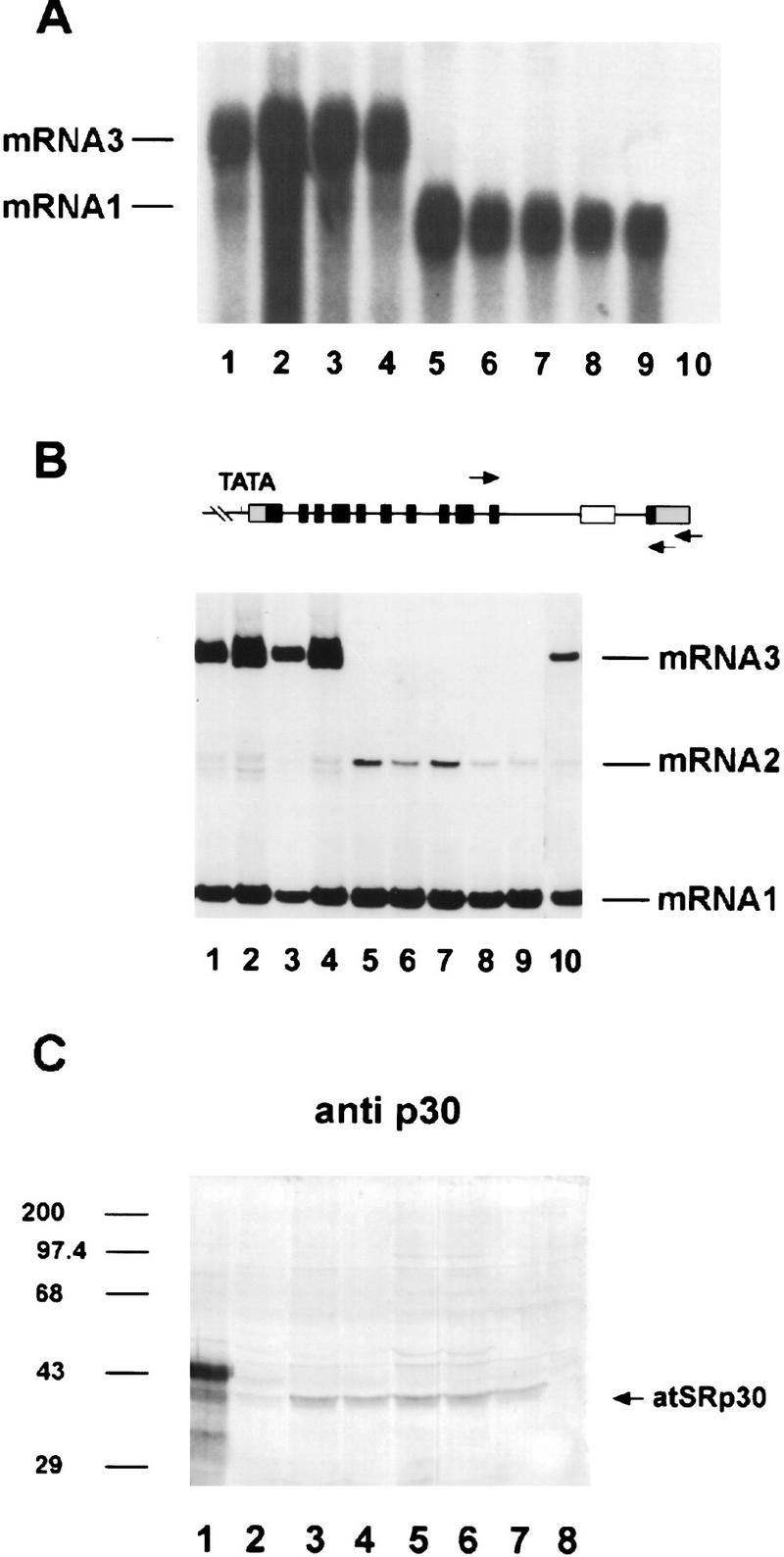
Overexpression of atSRp30 in transgenic Arabidopsis plants. The cDNA and the genomic sequences of atSRp30 were cloned under the control of the strong 35S CaMV promoter (constructs pC30 and pG30, respectively) and used to transform Arabidopsis plants. (A) Northern blot analysis of total RNA isolated from independent transgenic lines probed with atSRp30 cDNA. (Lanes 1–4) Plants transformed with the pG30 construct; (lanes 5–9) plants transformed with pC30; (lane 10) control plant transformed with pBI121 (35S CaMV–GUS construct). mRNA isoforms are indicated. (B) RT–PCR analysis of the same transgenic lines. The primers used are indicated with arrows in the diagram of GatSRp30. (Lane 10) Four times more material was loaded in this control lane. (C) Immunodetection of atSRp30 in total protein extracts of overexpressing transgenic plants. (Lane 1) SR protein preparation from Arabidopsis cell culture (wild type); (lane 2) line transformed with pG30; (lanes 3–7) lines transformed with pC30; (lane 8) pC30 transgenic line that does not overexpress mRNA1. Antibody raised against atSRp30 (anti-p30) was used for immunodetection.
In light of the above results, it was of interest to measure overexpression of atSRp30 at the protein level. Total soluble protein extracts of transgenic plants were analyzed by Western blotting using anti-p30 for immunodetection (Fig. 6C) and compared to a control plant (lane 8, a transformed plant with no overexpression of atSRp30) and to an SR protein preparation from an Arabidopsis suspension culture (lane 1). The SR protein preparation had the characteristic pattern for anti-p30 (cf. Figs. 6C, lane 1, and 4A, lane 2), whereas no specific proteins were immunostained in the total protein extract of the control plant (Fig. 6C, lane 8), because of the low abundance of atSRp30 in plants. In contrast, a specific protein band was visible in the pC30 transformants (lanes 3–7), comigrating with a protein band in the SR protein preparation (lane 1). In contrast, in the pG30 transformant (lane 2), only a faint protein band was detected. These results correlate well with the RNA expression pattern of mRNA1 in both types of transformants (Fig. 6A). It is interesting that overexpression of atSRp30 yielded an immunostained protein of 38 kD and not a band comigrating with the more abundant 43- to 46-kD doublet seen in the SR protein preparation (Fig. 6C, lane 1). One likely explanation is that overexpressed atSRp30 is only partially phosphorylated, which may be related to our observation that even exhaustive dephosphorylation results in a 38-kD atSRp30 protein that retains the mAb104 phosphoepitope (Fig. 4B, lanes 3–6). However, no shorter protein product (atSRp30s) of the highly abundant mRNA3 in pG30 transformants could be detected with any antibody used (see also Fig. 6C, lane 2).
Phenotypic changes in plants overexpressing atSRp30
Overexpression of atSRp30 resulted in strong phenotypes with pleiotropic changes both in morphology and development of the transgenic plants. No significant differences were observed between plants transformed with pG30 and pC30 constructs, although the levels of atSRp30 protein were different (Fig. 6C, cf. lane 2 and lanes 3–7). The observations were reproducible in T2 and subsequent generations of independent transgenic lines, and cosegregated with antibiotic resistance. In transgenic plants, the transition from vegetative to flowering stage was delayed greatly under short day conditions. The time from germination to the formation of the first mature silique was 65–78 days in overexpressing plants, compared to 42–47 days in control plants grown under the same conditions. In addition, adult plants showed reduced apical dominance, resulting in a ‘bushy’ phenotype with an increased number of secondary inflorescences and a shortened primary inflorescence (Fig. 7C). Growing of the transgenic plants under long-day conditions resulted in partial reversion to a normal phenotype. But despite of the partial recovery of apical dominance, the transition to flowering and the time to senescence remained delayed. Under long-day conditions, the majority of plants from overexpressing lines started to flower between 35 and 39 days after germination, whereas control plants flowered between days 25 and 28. The initial inflorescences were very short and did not exceed 0.5–1 cm when the first flower opened. The size of flowers in transgenic plants was ∼30% larger than in control plants (4 and 3 mm, respectively). Transgenic plants also had larger rosette leaves (Fig. 7A, cf. to wild type in B) with trichomes having predominantly four to five 5 branches, compared to wild-type leaves, which had mainly three-branched trichomes. In strong overexpressing lines, the primary inflorescence produced numerous secondary branches with a vegetative rosette-like appearance. These branches developed from 7–15 vegetative-like leaves and finally formed inflorescences. Under long-day conditions, the rosette leaf numbers determined at the time of flowering were somewhat lower in transgenic lines compared to the wild type (at average 12 and 16 leaves, respectively). Independent transgenic lines overexpressing atSRp30 displayed the described characteristics to various extents, with the main impact on the time for transition from the vegetative to the reproductive phase.
Figure 7.

Phenotypic changes in plants overexpressing atSRp30. (A) Plant overexpressing atSRp30; (B) wild-type plant; (C) plant overexpressing atSRp30 grown under short day conditions displays no apical dominance, resulting in a bushy phenotype.
Overexpressed atSRp30 modulates alternative splicing in vivo
The influence of overexpression of atSRp30 on its own alternative splicing pattern and the high homology of atSRp30 to human alternative splicing factor SF2/ASF suggested that this protein could be a modulator of plant splicing. This assumption cannot be tested in vitro, as no plant splicing extracts are available. We therefore used total RNA preparations from pG30 transformants (lower level of atSRp30 overexpression) and pC30 transformants (higher level of atSRp30 overexpression) for RT–PCR analysis of several plant introns that are known to be processed alternatively under wild-type conditions.
Among several genes tested, such as FCA (Macknight et al. 1997), rubisco activase (Werneke et al. 1989), agamous (Yanofsky et al. 1990), and LSD1 (Dietrich et al. 1997), no changes in the splicing patterns were observed in plants overexpressing atSRp30. However, pre-mRNA splicing of three plant genes was found to be affected by overexpression, as described below.
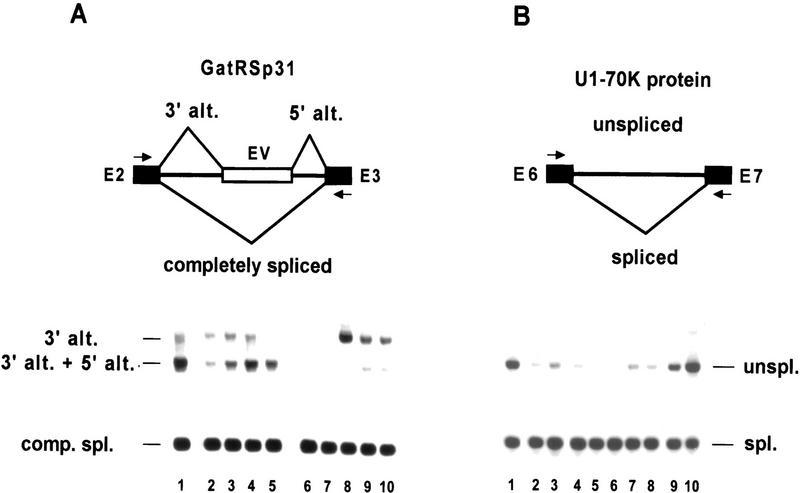
The splicing factor atRSp31 belongs to a novel family of plant RS proteins and possesses an alternatively spliced second intron (792 nucleotides) (Lopato et al. 1996b). In the wild-type plant, either a cryptic 3′ splice site or both alternative 3′and 5′ splice sites were used in this intron (Fig. 8A), whereas the latter form prevailed in control lines (lane 1) and pG30 transformed lines (lanes (2–5). In contrast, pC30 lines (lanes 6–10) expressed mainly the alternative 3′ splice site form, although its abundance was variable (in lanes 6 and 7 this form is only visible in the original photograph). These transcripts are probably subject to nonsense-mediated decay and potentially code for the same truncated protein.
Figure 8.
Regulation of the alternative splicing pattern of atRSp31 and U1–70K in plants overexpressing atSRp30. The relevant alternative splicing events are shown in the diagrams at the top. RT–PCR was carried out using primers (marked with arrows) derived from the adjacent exons. (A) Changes in splicing in the second intron of GatRSp31. (Lane 1) Control line transformed with pBI121 (35SCaMV–GUS construct), (lanes 2–5) lines transformed with the pG30 construct; (lanes 6–10) lines transformed with pC30. (B) More correctly spliced mRNA encoding U1–70K protein is produced upon overexpression of atSRp30. (Lane 1) control plant transformed with pBI121 (35SCaMV–GUS construct); (lanes 2,3) lines transformed with the pG30 construct; (lanes 4–8) plants transformed with pC30; (lane 9) control pC30 transgenic plant that does not overexpress mRNA1; (lane 10) 19-day-old wild-type plant.
Another well-described alternative splicing event occurs in the case of the U1–70K gene of Arabidopsis, in which transcripts retaining intron 6 are more abundant in most tissues (except roots) than the correctly spliced transcript (Golovkin and Reddy 1996). The RT–PCR results from control plants (Fig. 8B, lanes 1,9,10) confirm this observation. However, in both atSRp30 transformants, either the lower expressing pG30 lines (lanes 2 and 3) or the higher expressing pC30 lines (lanes 4–8), most of the transcripts were spliced correctly and only a low level of intron retention was observed. This regulatory event could potentially influence the level of active protein.
The most interesting case of alternative splicing modulation was found in the case of the long intron (intron 11) of GatSRp34, which is a close homolog of GatSRp30 (Fig. 9). In wild-type plants (Fig. 9, lane 1), part of this intron is alternatively spliced to generate mRNA5. However, in pG30 transformants (lanes 2–5), mRNA 3 was the main alternatively spliced form, whereas the level of mRNA1 appeared unchanged. Surprisingly, when atSRp30 was highly overexpressed, the alternatively spliced mRNA3 became the main transcript, whereas the amount of mRNA1 was considerably decreased. Assuming that shorter PCR products are probably overrepresented, the reduction of mRNA1 may be even more drastic. This assumption was confirmed by immunoblot analysis of total protein extracts: atSRp34/SR1 was detected in control plants (Fig. 9B, lane 3) and in a pG30 transformant (lane 5), but not in a pC30 transformant (lane 4). However, a smaller specific protein band (atSRp34s, 38 kD) became visible in this case, indicating that the protein product of mRNA3 predominates in this transformant. These results clearly show that overexpression of atSRp30 strongly influences gene expression of its close homolog, atSRp34/SR1.
Figure 9.
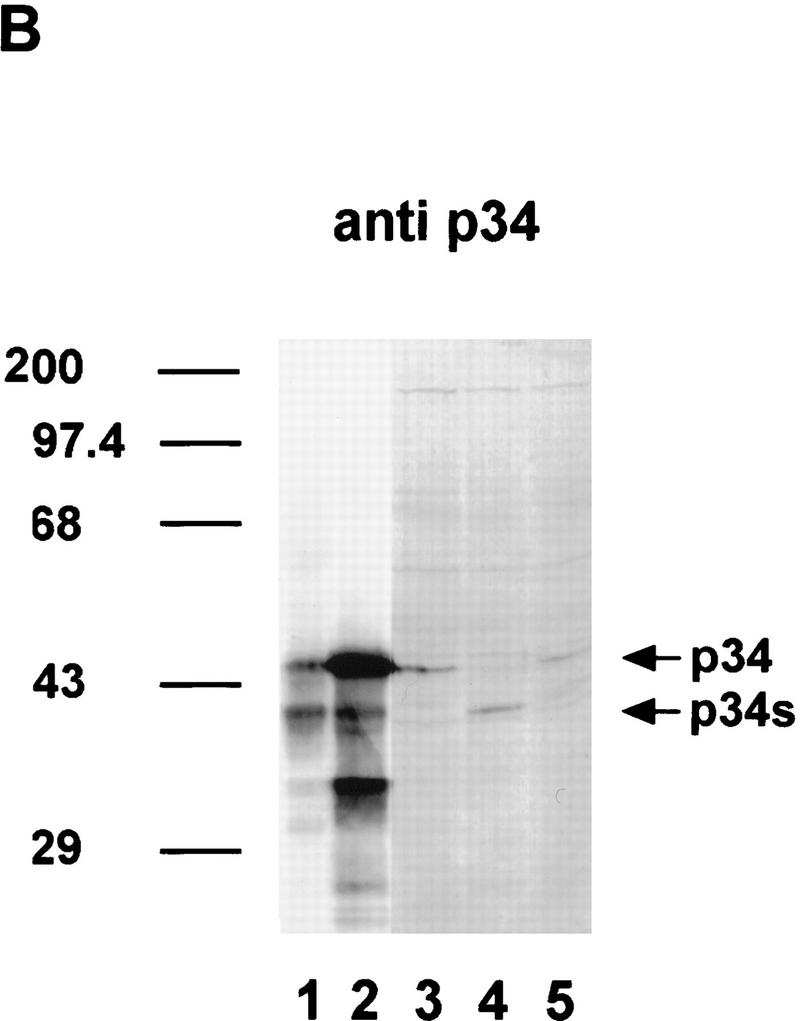
Changes in the expression pattern of atSRp34/SR1 in plants overexpressing atSRp30. (A) Preferential usage of an alternative 5′ splice site in the eleventh intron of atSRp34/SR1. The gene, the relevant alternative splicing events, and the deduced proteins are depicted above the gel showing the RT–PCR products obtained with primers from the adjacent exons. (Lane 1) Control, untransformed wild-type plant; (lanes 2–5) lines transformed with the pG30 construct; (lanes 6–11) plants transformed with pC30; (lane 11) control pC30 transgenic plant that does not overexpress mRNA1. (B) Immunodetection of atSRp34 and atSRp34s in total protein extracts from transgenic plants. (Lanes 1,2) SR protein preparations isolated from Arabidopsis plants and cell culture, respectively; (lane 3) control plant transformed with pC30 that does not overexpress mRNA1; (lane 4) line transformed with pC30; (lane 5) line transformed with the pG30 construct. Anti-p34 antibodies were used for immunodetection.
Discussion
atSRp30 is a member of the SR protein family
In addition to their characteristic domains, which include one or two RRMs and a carboxy-terminal RS domain with multiple SR dipeptides, SR proteins possess phosphoepitope(s) recognized by mAb104 and they are insoluble in the presence of millimolar concentrations of magnesium salts. The antibody to atSRp30 recognized a complex pattern of bands in the Mg precipitate, whereas in total protein fractions no specific band could be detected. The failure to detect atSRp30 in crude lysates reflects the low abundance of the protein and/or the limited sensitivity of the antibody. However, when atSRp30 was overexpressed, the antibody detected a specific protein with an apparent mobility of 40 kD. In contrast, atSRp34/SR could be detected by anti-p34 in total protein fractions. Our immunoblot data confirm that atSRp30 is a true SR protein, as it is present in SR protein preparations and shows corresponding immunostained bands with mAb104. However, as complete dephosporylation could not be achieved, we do not know how many of these proteins represent modified forms of atSRp30 or closely related proteins.
Is the expression of atSRp30 autoregulated?
When atSRp30 pre-mRNA was overexpressed, mRNA3, which is generated from use of an alternative 3′ splice site in the tenth intron, was the main transcript detected, whereas the level of mRNA1 was enhanced only moderately (Fig. 6). There are several possible explanations for this phenomenon. First, there may be a limiting splicing factor that is titrated out because of overexpression of the atSRp30 gene. Such a factor would have to be specific for the tenth intron, as all the other introns in atSRp30 are spliced properly. Second, mRNA3 and mRNA1 may be synthesized for the most part in different cells, via cell-type-specific alternative splicing. As the constitutive 35S CaMV promoter used in the overexpression experiments is active in most tissues, the large amount of mRNA3 may reflect inappropriate expression in cell types in which a factor required for correct processing of the tenth intron is absent. To clarify this issue, experiments aimed at determining the cells in which the alternative splicing event takes place will be necessary.
The fact that the ratio of mRNA 3 to mRNA 1 is constant in all transformants overexpressing GatSRp30 (Fig. 6A) is consistent with a potential autoregulatory mechanism involving atSRp30. Overproduced atSRp30 protein may stimulate the alternative splicing event, thus down-regulating its own expression, as has been shown to be the case for several animal splicing factors, such as Sxl (Bell et al. 1991), tra (Mattox and Baker 1991), and SWAP (Zachar et al. 1987) from Drosophila, as well as mouse SRp20 (Jumaa and Nielsen 1997). In agreement with this model, we could not detect a protein product from mRNA3 (atSRp30s). We obtained evidence for an influence of atSRp30 on alternative splicing of transcripts from the endogenous gene in plants overexpressing the atSRp30 cDNA (Fig 6B, lanes 5–9), resulting in preferential use of an alternative 5′ splice site. Overexpression of atSRp30 might have a different effect on the large amount of exogenous pre-mRNA present in cells where this protein is not expressed normally, resulting in preferential use of an alternative 3′ splice site (Fig. 6A,B, lanes 1–4). Unfortunately, this hypothesis cannot be tested in vitro because no plant splicing extracts are available. Future experiments should address whether atSRp30 can indeed autoregulate its own production.
atSRp30 overexpression influences splice-site choice in several plant pre-mRNAs and induces changes in plant development
Many in vivo and in vitro experiments have shown that human SF2/ASF is involved in the selection of 5′ splice sites by binding cooperatively to this site (Zuo and Manley 1994) and to U1 snRNP (Kohtz et al. 1994; Jamison et al. 1995; Zahler and Roth 1995). Furthermore, like other SR proteins, SF2/ASF influences splice-site selection in part by binding to enhancer sequences that are present frequently in intronic or exonic regions. Upon binding to these elements, the protein activates splicing by recruiting the splicing machinery to an adjacent splice site through protein–protein interactions (Wu and Maniatis 1993; Amrein et al. 1994; Zuo and Manley 1994). Arabidopsis atSRp30 was a good candidate for being a splicing modulator, given its similarity to human SF2/ASF and its peculiar expression pattern. The ability to stably overexpress atSRp30 in whole plants that remained viable allowed us to determine, for the first time, the effects of increased SR protein levels on alternative splicing of specific endogenous transcripts. Some but not all endogenous transcripts tested were affected. In the case of atSRp30 pre-mRNA itself, and of atSRp34/SR1 pre-mRNA, intronic alternative 5′ splice sites were activated, whereas in atRSp31 and U1 70K pre-mRNAs, the usage of the normal splice sites was generally enhanced but variable. We have no explanation for this variability in individual transformants (see Fig. 8), as the atSRp30 protein levels in several pC30 transformants were similar (Fig. 6C). The most pronounced effect of atSRp30 overexpression was on the splicing pattern of the close homolog atSRp34/SR1, in which preferential use of the alternative 5′ splice site gave rise mostly to a truncated mRNA isoform, atSRp34s, and virtually no atSRp34/SR1 protein. Although we have no information to date concerning the activity of the smaller atSRp34s, these results suggest that normal expression of atSRp30 protein in the same cells that transcribe atSRp34/SR1 is important for the synthesis of authentic atSRp34/SR1 protein. This regulatory loop might explain the different expression patterns of both factors in root tissue and young seedlings. It remains to be determined whether there is a reciprocal effect of atSRp34/SR1 overexpression on alternative splicing of atSRp30 pre-mRNA.
Plants overexpressing atSRp30 constitutively and ubiquitously showed interesting changes in morphology, as well as in several aspects of development. Although some of the details will have to be investigated more thoroughly, many of the differences can be explained by changes in phase transitions and in the definition of cell fates. In this work, we have demonstrated that atSRp30 can change the expression of other genes drastically by influencing their splicing patterns. As the expression pattern of atSRp30 is normally very tissue specific, some of the observed effects of overexpression might simply be caused by its expression in inappropriate tissues, where it influences the expression of genes that are not the natural targets of this splicing factor. In addition, the observed decrease in the expression of the splicing factor atSRp34 upon overexpressing atSRp30 may in turn affect the expression of other genes, leading to the observed phenotypic changes. On the other hand, the changes in trichome development resulting in additional branching may reflect overexpression of atSRp30 in the supporting cell of the trichomes, which is a normal site of atSRp30 expression. Therefore, atSRp30 may be a determinant of trichome development. Finding the natural targets of regulation by atSRp30 remains a critical goal to explain the observed changes in phenotype, as well as to promote our understanding of developmental pathways in plants.
Two SF2/ASF-like splicing factors in Arabidopsis
Immunoblot analysis with mAb104 suggested previously that the complexity of the SR protein family is higher in animals than in plants (Lopato et al. 1996a), as several higher molecular mass proteins have been identified in SR preparations from mammals but not from those in plants. The existence of two SF2/ASF-like factors in Arabidopsis may compensate for the absence of orthologs of other members of the SR protein family. The similar gene structure of atSRp30 and atSRp34/SR1 is indicative of an ancestral gene duplication event. Interestingly, the penultimate long intron is conserved in both genes and is involved in alternative splicing events. Judging from Northern blot analysis of RNAs from different plant organs, the ratio of the various SR protein mRNAs is quite variable and is probably reflected in the abundance of the resulting proteins. In both genes, the alternative splicing event results in predicted proteins with a truncated RS region and a few new additional carboxy-terminal amino acids. Whereas a shorter protein of atSRp34/SR1 could be detected clearly in plants overexpressing atSRp30, we never observed atSRp30s in these plants, although the corresponding alternatively spliced mRNA is abundant. Expression of atSRp30s cDNA in Escherichia coli yielded stable protein (data not shown). Therefore, either mRNA3 is not translated efficiently, or it is degraded through a premature nonsense codon-mediated decay pathway, or atSRp30s is preferentially recognized by one of the plant protein-degradation systems.
Comparative analysis of atSRp30 with all known plant and animal SR proteins revealed that it is a paralog of atSRp34/SR1 and that both Arabidopsis proteins are closest to human SF2/ASF. The main structural difference is that atSRp30 lacks a G-rich region between the two RRMs, which may influence the flexibility and perhaps the specificity of the RNA-binding region. The notion that the two proteins might have distinct activities is strengthened by the observation that in many cases their expression patterns are quite different, as shown in Figure 5. In general, atSRp30 expression is more confined to specialized cell types and tissues, like trichomes, cotyledons, or lateral root primordia, suggesting a special role for this protein in the initiation of organ formation, whereas atSRp34/SR1 is more strongly expressed in meristematic tissue. Furthermore, our sequence and immunological data indicate that atSRp34/SR1 is more similar to hSF2/ASF than is atSRp30. Taken together, these data suggest that atSRp34/SR1 is a more general splicing factor, similar to hSF2/ASF, whereas atSRp30 might have more specialized functions, perhaps acting as a regulatory splicing factor that modulates alternative splicing and gene expression in specific cell types.
Materials and methods
Purification of SR proteins and protein sequencing
The SR protein purification was described previously (Lopato et al. 1996a). The proteins were separated by SDS-PAGE. After Coomassie G staining, the protein bands were excised and subjected to in-gel digestion with Lys-C endopeptidase. The resulting peptides were separated by HPLC and sequenced by automated Edman degradation as described previously (Wang et al. 1996).
Isolation and sequencing of genomic DNA and cDNA of atSRp30 and atSRp34/SR1
Total DNA of carrot, tobacco, and Arabidopsis was isolated by the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson 1980). The most conserved part of the sequenced peptide YVGNLPGDI was used to design two degenerate forward primers. Two reverse degenerate primers were derived from the peptide sequence SWQDLKDHM, which was also part of a sequenced peptide. All four combinations of degenerate primers were used in PCR reactions with genomic DNA from different plants as a template. PCR products were subcloned and sequenced. The PCR product from Arabidopsis was used as a probe for screening a genomic λZAPII library of A. thaliana var. Columbia (Stratagene). One positive clone, GatSRp30, was found, mapped, subcloned, and sequenced (EMBL accession no. AJ131214). A partial cDNA sequence was found by a BLAST search through the EST databases with the accession no. R65514, 4018 Arabidopsis thaliana cDNA clone 17J1T7, and was kindly provided by the Arabidopsis Biological Resource Center at Ohio State University. Sequence homology was analyzed using Genetics Computer Group (GCG, University of Madison, Madison, WI) sequence analysis software package version 7.1-UNIX. Sequencing of the cDNA and the genomic clones was done using an A.L.F. DNA Sequencer and Auto Read Sequencing kit (Pharmacia). To obtain a cDNA clone of atSRp34/SR1, the EST clone with the accession no. ACT76795, 11573 atcDNA clone 151I9T7, was sequenced. The genomic sequences for atSRp34/SR1 (accession no. AF001035) were isolated by PCR and sequenced.
Analysis of alternatively spliced isoforms
RNA blot analyses were done as described (Lopato et al. 1996b). cDNAs encoding alternatively spliced isoforms were obtained by RT–PCR using total RNA preparations from A. thaliana plants at different stages of development and primers derived from the 3′ untranslated region (for reverse transcription). (1) 5′-AAATGAGCTCAAATGTATATGTATGGAAAAACC-3′ (atSRp30) and (2) 5′-AATGAGCTCGAAACGATATCTTCAAAAAAAAAC-3′ (atSRp34/SR1) (the underlined nucleotides correspond to restriction sites) and from the beginning of the 5′ untranslated region and the end of coding region (for PCR); (3) 5′-AAACTGGATCCAGAACAATCTAACGCTTTCTCG-3′ (atSRp30) and (4) 5′-ATATAGGATCCTCAACCAGAUAUCACAGGTG-3′ (atSRp30); (5) 5′-AAATATCTAGAGATCTCAAATCGACGACC-3′ (atSRp34/SR1) and (6) 5′-ATATAGGATCCCATTTTACCTCGATGGAC-3′ (atSRp34/SR1). All PCR products were sequenced. Alternative splicing of the long introns was studied using primers derived from adjacent exons; (7) 5′-AATGAGCTCTGTGTCACCTGCTAGATCC-3′ (atSRp30) and (8) 5′-ATATAGGATCCAGATATCACAGGTGAAAC-3′ (atSRp30); (9) 5′-ATAGGATCCAGGAGCAGAAGTCCCAAGGCAAAG-3′ (atSRp34/SR1) and (10) 5′-AAAGTCGACAGAAGGTAGAGGAGATCTTGATC-3′ (atSRp34/SR1). The fragments were analyzed on a 1.2 % agarose gel.
Constructs for promoter analysis and overexpression
Aproximately 1 kb of promoter sequences of atSRp30 and atSRp34/SR1 plus their complete 5′ untranslated regions (including the first intron in the case of atSRp34/SR1) were obtained by PCR from genomic clone GatSRp30 and genomic DNA of A. thaliana, respectively, as a template, using primers (11) 5′-AAACTAAGCTTGGTATCTTCTTCCCTGCAAG-3′ (atSRp30); (12) 5′-AAACCTAGGCGGCTACTCAGCTGATACCTCAGAGCAG-3′ (atSRp30); (13) 5′-AAACTAAGCTTAAATATTGAACCGGCCTCGGTTC-3′ (atSRp34/SR1); (14) 5′-AAACTGGATCCTCTTCCTGTTGGTCGTCGACGATTTG-3′ (atSRp34/SR1); and (15) 5′-AAACTGGATCCTCTTCCTTTATCAAATCC-3′ (atSRp34/SR1) containing HindIII and BamHI restriction sites. These fragments were digested with HindIII and BamHI and fused to the GUS reporter gene (Jefferson 1987) in pBI101 (Clontech).
The cDNA and genomic sequences of atSRp30 were amplified from cDNA and genomic DNA from A. thaliana using primers 3 and 1, containing BamHI and SacI restriction sites, respectively. The DNA fragments were sequenced and inserted into the BamHI–SacI restriction sites of the pBI121 (Clontech) vector (with the GUS gene deleted) under the control of the 35S CaMV promoter, giving rise to constructs named pC30 and pG30, respectively.
Cultivation of plants and suspension cultures and plant transformation procedure
Conditions for maintaining carrot and tobacco BY2 suspension cultures were as described earlier (Lopato et al. 1996a). The suspension culture of A. thaliana ecotype Columbia was a gift from Czaba Koncz (Max-Planck-Institut für Züchtungsforschung, Köln, Germany) and was maintained as described in (Lopato et al. 1999). A. thaliana ecotype Columbia was used in all transfer experiments. Seeds were surface-sterilized by a 30-min imbibition with water, followed by a 1-min incubation in 70% ethanol, 10 min in 10% bleach and five rinses with sterile water. Sterilized seeds were grown in MS medium (Murashige and Skoog 1962), supplemented with 1% sucrose. Plants were usually maintained in 16-hr light/8-hr dark cycle at 23°C, except as otherwise indicated. The cell culture was grown in medium containing Murashige and Skoog salts, 2× Gamborg’s vitamins (Gamborg et al. 1968), 1 mg/liter 2,4-dichlorophenoxyacetic acid (2,4-D), and 3% (wt/vol) sucrose. Cell cultures were incubated in 50 ml of medium in 250-ml conical flasks on a rotary shaker at 110 rpm at 23°C in dim light. Cell suspensions were subcultured every 7 days and were diluted threefold with fresh medium at each subculture.
All constructs were introduced into Agrobacterium tumefaciens LBA4404 (Hoekema et al. 1983) using a triparental mating procedure with the helper plasmid pRK2013 (Ditta et al. 1980). Root explants of A. thaliana were transformed using the method described by (Valvekens et al. 1988) with one modification: Root explants were taken from 15-day-old seedlings grown on vertically placed agar plates (Huang and Ma 1992). Seeds of transgenic plants were germinated in the presence of 50 μg/ml of kanamycin.
Histochemical assay of GUS expression
Histochemical assays of GUS expression were carried out with 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc, Duchefa) as a substrate and were performed on intact seedlings or excised organs of mature plants grown in vivo as described by Jefferson (1987). The samples were treated with 70% ethanol for 2–6 hr to remove chlorophyll from tissues where necessary.
Purification of SR proteins from Arabidopsis and immunoblotting
SR proteins were purified from 3-week-old Arabidopsis plants or 5-day-old suspension cultures using a two-step salt precipitation method as described (Lopato et al. 1996a). Total protein extracts were prepared with the buffer for SR protein isolation (Zahler et al. 1992). Proteins from the magnesium precipitate and total protein extracts were separated on a 12.5% SDS gel. Protocols for immunoblotting and detection were described earlier (Lopato et al. 1999). Protein dephosphorylation was carried out with alkaline phosphatase from calf intestine (Biolabs).
Expression of atSRp30 and atSR34/SR1 in bacteria
The coding region of atSRp30 cDNA was amplified by PCR using the primers 5′-ATATACCATGGGTAGCCGATGGAATCGTAC-3′ and 4 (listed above).
The coding region of atSRp34/SR1 cDNA was amplified by PCR using the primers 5′-ATATACCATGGGCAGTCGTTCGAG-3′ and 6 (above). The primers contain NcoI and BamHI restriction sites. To obtain the NcoI restriction site, the fourth nucleotide of the coding region in both cases was changed to G. Thus expressed atSRp30 and atSRp34/SR1 have Ser2-Arg and Ser2-Gly substitutions, respectively. The fragments were cloned into the bacterial expression vector pET-3d (Novagen) and transformed into the E. coli strain BL21(DE3)pLysS (Novagen).
Single colonies were grown in 100 ml of LB medium to a density of 0.4–0.5 OD550, transferred to 900 ml of fresh prewarmed medium and incubated for 1 hr. The cultures were induced with 0.4 mm IPTG and were grown for a further 5 hr. The bacterial pellet was washed twice in 500 ml of ice-cold washing buffer (100 mm NaCl, 10 mm Tris-HCl at pH 7.5) and used for inclusion body isolation: 2 grams of bacterial pellet was resuspended in 18 ml of ice-cold buffer A (10 mm Tris-HCl at pH 7.9, 100 mm KCl, 2 mm DTT, 35% sucrose). A total of 4.5 ml of ice-cold buffer B (333 mm Tris-HCl at pH 8.0, 100 mm EDTA at pH 8.0, 40 mg of lysozyme) was added and incubated on ice for at least 10 min. Lysis buffer (22.5 ml, 1 m LiCl, 20 mm EDTA, 0.5% NP-40) was added and the solution was sonicated for a total of 2 min at full power (15-sec bursts with cooling periods of 1 min in between). Following centrifugation for 10 min (Sorvall, SS34, 13000 rpm), the pellet was washed twice in 25 ml of buffer C (10 mm Tris-HCl at pH 8.0, 0.1 mm EDTA, 0.5 m LiCl, 0.05% NP-40, 1 mm DTT), and then twice with the same buffer lacking LiCl. The isolated inclusion bodies contained >95% pure protein and were used for polyclonal antibody preparation (Lopato et al. 1999).
Acknowledgments
We thank L. Waigmann, F. Kragler, T. Skern, and Z. Rattler for invaluable discussions. The EST clones were provided by the Arabidopsis Biological Resource Centre (Ohio State University, Columbus). This work was supported by a grant (P12364-GEN) from the Österreichischer Fonds zur Förderung der wissenschaftlichen Forschung, from the Österreichische Nationalbank (P6633), and from the International Association for the Promotion of Cooperation with Scientists from the New Independent States of the former Soviet Union (INTAS) (UA-95-17) to A.B. A.R.K. and R.K. were funded in part by grant CA13106 from the National Cancer Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Andrea@bch.univie.ac.at; FAX 43-1-4277 9616.
References
- Allain FHT, Howe PWA. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J. 1997;16:5764–5774. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein H, Hedley ML, Maniatis T. The role of specific protein-RNA and protein–protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- Barta A, Sommergruber K, Thompson D, Hartmuth K, Matzke MA, Matzke AJM. The expression of a nopaline synthase—human growth hormone chimaeric gene in transformed tobacco and sunflower callus tissue. Plant Mol Biol. 1986;6:347–357. doi: 10.1007/BF00034942. [DOI] [PubMed] [Google Scholar]
- Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart RE, Andreadis A, Nadal Ginard B. Alternative splicing: A ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Brown JWS, Simpson CG. Splice site selection in plant pre-mRNA splicing. Ann Rev Plant Physiol Plant Mol Biol. 1998;49:77–95. doi: 10.1146/annurev.arplant.49.1.77. [DOI] [PubMed] [Google Scholar]
- Caćeres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— . Mammalian pre-mRNA splicing factors. In: Krainer AR, editor. Eukaryotic mRNA processing. Oxford, UK: IRL Press; 1997. pp. 174–212. [Google Scholar]
- Cáceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes & Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Popielarz M, Fuchs JP, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: A novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B. Directing alternative splicing: Cast and scenarios. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- Diamond RH, Du K, Lee VM, Mohn KL, Haber BA, Tewari DS, Taub R. Novel delayed-early and highly insulin-induced growth response genes. Identification of HRS, a potential regulator of alternative pre-mRNA splicing. J Biol Chem. 1993;268:15185–15192. [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell. 1997;88:685–694. doi: 10.1016/s0092-8674(00)81911-x. [DOI] [PubMed] [Google Scholar]
- Ditta G, Stanfield S, Corbin D, Helinski DR. Broad host range DNA cloning system for gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Gniadkowski M, Klahre U, Liu H-X. Pre-mRNA splicing in plants. In: Lamond A, editor. Pre-mRNA processing. Georgetown, TX: R.G. Landes Publishers; 1995. pp. 66–77. [Google Scholar]
- Fu XD. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- ————— The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Ge H, Zuo P, Manley JL. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Golovkin M, Reddy AS. Structure and expression of a plant U1 snRNP 70K gene: Alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell. 1996;8:1421–1435. doi: 10.1105/tpc.8.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- Hedley ML, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc Natl Acad Sci. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA. A binary plant vector strategy based on separation of vir and T-region of the A. tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Huang H, Ma H. An improved procedure for transforming Arabidopsis thaliana root explant. Plant Mol Biol Rep. 1992;10:372–383. [Google Scholar]
- Jamison SF, Pasman Z, Wang J, Will C, Lührmann R, Manley JL, Garcia-Blanco MA. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: Characterization of required elements. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Guenet JL, Nielsen PJ. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Mol Cell Biol. 1997;17:3116–3124. doi: 10.1128/mcb.17.6.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zuo P, Manley JL, Baker BS. The Drosophila RNA-binding protein RBP1 is localized to transcriptionally active sites of chromosomes and shows a functional similarity to human splicing factor ASF/SF2. Genes & Dev. 1992;6:2569–2579. doi: 10.1101/gad.6.12b.2569. [DOI] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia Blanco MA, Manley JL. Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990a;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- ————— Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes & Dev. 1990b;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Kraus ME, Lis JT. The concentration of B52, an essential splicing factor and regulator of splice site choice in vitro, is critical for Drosophila development. Mol Cell Biol. 1994;14:5360–5370. doi: 10.1128/mcb.14.8.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G, Schaal T, Maniatis T, Goodman HM. Identification of a plant serine-arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc Natl Acad Sci. 1995;92:7672–7676. doi: 10.1073/pnas.92.17.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bingham PM. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- Lopato S, Mayeda A, Krainer A, Barta A. Pre-mRNA splicing in plants: Characterization of SR splicing factors. Proc Natl Acad Sci. 1996a;93:3074–3079. doi: 10.1073/pnas.93.7.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S, Waigmann E, Barta A. Characterization of a novel arginine/serine-rich splicing factor in Arabidopsis. Plant Cell. 1996b;8:2255–2264. doi: 10.1105/tpc.8.12.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S, Gattoni R, Fabini G, Stevenin J, Barta A. A novel family of plant splicing factors with a Zn knuckle motif: Examination of RNA binding and splicing activities. Plant Mol Biol. 1999;39:761–773. doi: 10.1023/a:1006129615846. [DOI] [PubMed] [Google Scholar]
- Lou H, Gagel RF, Berget SM. An intron enhancer recognized by splicing factors activates polyadenylation. Genes & Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Taha S, Walbot V. Nuclear pre-mRNA processing in higher plants. Prog Nucleic Acid Res MolBiol. 1994;47:149–193. doi: 10.1016/s0079-6603(08)60252-4. [DOI] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes & Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Mattox W, Baker BS. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes & Dev. 1991;5:786–796. doi: 10.1101/gad.5.5.786. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Zahler AM, Krainer AR, Roth MB. Two members of a conserved family of nuclear phosphoproteins are involved in pre-mRNA splicing. Proc Natl Acad Sci. 1992;89:1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Caceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M, Harris KS, Roth MB. smg mutants affect the expression of alternatively spliced Sr protein mRNAs in Caenorhabditis elegans. Proc Natl Acad Sci. 1997;94:9782–9785. doi: 10.1073/pnas.94.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 1962;15:473–497. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchatesingh J, Zahler AM, Neugebauer KM, Roth MB, Cooper TA. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton GR, Caceres JF, Mayeda A, Bell MV, Plebanski M, Jackson DG, Bell JI, Krainer AR. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solymosy F, Pollak T. Uridylate-rich small nuclear RNAs (UsnRNAs), their genes and pseudogenes, und UsnRNPs in plants: Structure and function. A comparative approach. Crit Rev Plant Sci. 1993;12:275–369. [Google Scholar]
- Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Mayeda A, Hampson RK, Krainer AR, Rottman FM. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes & Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: Creation of an SRp40-specific splicing enhancer. BioEssays. 1997;19:189–192. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Green MR. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem. 1996;21:296–301. [PubMed] [Google Scholar]
- Valvekens D, Van Montague M, Van Lijsebettens M. A. tumefaciens-mediated transformation of Arabidopsis thaliana root explants using kanamycin selection. Proc Natl Acad Sci. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Santen VL, Spritz RA. Splicing of plant pre-mRNAs in animal systems and vice versa. Gene. 1987;56:253–265. doi: 10.1016/0378-1119(87)90142-9. [DOI] [PubMed] [Google Scholar]
- Vellard M, Sureau A, Soret J, Martinerie C, Perbal B. A potential splicing factor is encoded by the opposite strand of the trans-spliced c-myb exon. Proc Natl Acad Sci. 1992;89:2511–2515. doi: 10.1073/pnas.89.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Manley JL. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- Wang R, Kobayashi R, Bishop JM. Cellular adherence elicits ligand-independent activation of the Met cell-surface receptor. Proc Natl Acad Sci. 1996;93:8425–8430. doi: 10.1073/pnas.93.16.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneke JM, Chatfield JM, Ogren WL. Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell. 1989;1:815–825. doi: 10.1105/tpc.1.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebauer K, Herrero J-J, Filipowicz W. Nuclear pre-mRNA processing in plants: Distinct modes of 3′-splice-site selection in plants and animals. Mol Cell Biol. 1988;8:2042–2051. doi: 10.1128/mcb.8.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Proc Natl Acad Sci. 1997;94:1148–1153. [Google Scholar]
- Yang X, Bani MR, Lu SJ, Rowan S, Ben David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Zachar Z, Garza D, Chou TB, Goland J, Bingham PM. Molecular cloning and genetic analysis of the suppressor-of-white-apricot locus from Drosophila melanogaster. Mol Cell Biol. 1987;7:2498–2505. doi: 10.1128/mcb.7.7.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Roth MB. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: A conserved family of pre-mRNA splicing factors. Genes & Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neugebauer KM, Lane WS, Roth MB. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes & Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- Zuo P, Manley JL. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The human splicing factor ASF/SF2 can specifically recognize pre-mRNA 5′ splice sites. Proc Natl Acad Sci. 1994;91:3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]