Abstract
MicroRNAs (miRNAs) function through the RNA-induced silencing complex (RISC), which contains an Argonaute (Ago) protein at the core. RISC assembly follows a two-step pathway: miRNA/miRNA* duplex loading into Ago, and separation of the two strands within Ago. Here we show that the 5′ phosphate of the miRNA strand is essential for duplex loading into Ago, whereas the preferred 5′ nucleotide of the miRNA strand and the base-pairing status in the seed region and the middle of the 3′ region function as additive anchors to Ago. Consequently, the miRNA authenticity is inspected at multiple steps during RISC assembly.
Keywords: Argonaute, microRNA, RISC
Introduction
MicroRNAs (miRNAs) regulate diverse biological functions by repressing expression of their target messenger RNA (mRNAs). MiRNAs function through a protein–RNA effector complex, RNA-induced silencing complex (RISC), which contains a member of the Argonaute (Ago) family of proteins as a core component. In flies, miRNAs are typically sorted into Ago1, whereas in humans miRNAs are incorporated into all four Ago proteins, Ago1–4 (Liu et al, 2004; Meister et al, 2004; Okamura et al, 2004; Tomari et al, 2007; Kawamata et al, 2009; Hafner et al, 2010; Kawamata & Tomari, 2010; Yoda et al, 2010). Most miRNAs are first transcribed as long primary miRNAs and then successively processed by two RNase III enzymes, Drosha and Dicer, into miRNA/miRNA* duplexes (Kim et al, 2009).
Assembly of miRNA/miRNA* duplexes into RISC follows an ordered pathway (Kawamata & Tomari, 2010). miRNA/miRNA* duplexes are first loaded into Ago proteins as double-stranded RNA, forming pre-RISC (Tomari et al, 2007; Kawamata et al, 2009; Yoda et al, 2010). This RISC-loading step is ATP dependent (Kawamata et al, 2009; Yoda et al, 2010) and requires the Hsc70/Hsp90 chaperone machinery (Iki et al, 2010; Iwasaki et al, 2010; Johnston et al, 2010). Subsequently, the two strands of each duplex are separated within the Ago protein. In this unwinding step, the passenger strand (typically the miRNA* strand) is discarded, and only the guide strand (typically the miRNA strand) remains in the Ago protein, forming mature RISC (Tomari et al, 2007; Kawamata et al, 2009; Yoda et al, 2010).
In both flies and humans, central mismatches direct loading of miRNA/miRNA* duplexes into pre-RISCs, whereas mismatches in the seed region (guide positions 2–8) or the middle of the 3′ region (guide positions 12–15; hereafter referred to as the 3′ region) promote unwinding (Tomari et al, 2007; Kawamata et al, 2009; Yoda et al, 2010). MiRNA/miRNA* duplexes typically bear 5′ monophosphates, 3′ hydroxyl groups and 3′ 2-nucleotide overhangs, all of which are characteristic of cleavage by RNase III enzymes (Kim et al, 2009). In addition, the guide strands of animal miRNA/miRNA* duplexes often begin with uridine (U; Lau et al, 2001; Czech et al, 2009; Okamura et al, 2009; Frank et al, 2010; Ghildiyal et al, 2010). Many of these features of miRNA/miRNA* duplexes are crucial for their functions and are recognized by specific binding pockets within Ago proteins (Ma et al, 2005; Parker et al, 2005; Wang et al, 2008; Frank et al, 2010). However, it has been poorly understood at which step in RISC assembly these features are recognized and how important they are for RISC assembly. Here, we show that 5′ phosphate of the guide strand, but not that of the passenger strand, is required for miRNA/miRNA* duplex loading into Ago proteins, whereas the preferred 5′ nucleotide of the guide strand and the base-pairing status in the seed and 3′ regions function as additive anchors to Ago proteins. Such a mechanism allows the RISC assembly pathway to assess the authenticity of miRNAs at multiple steps.
Results And Discussion
RISC loading requires 5′ phosphate of the guide
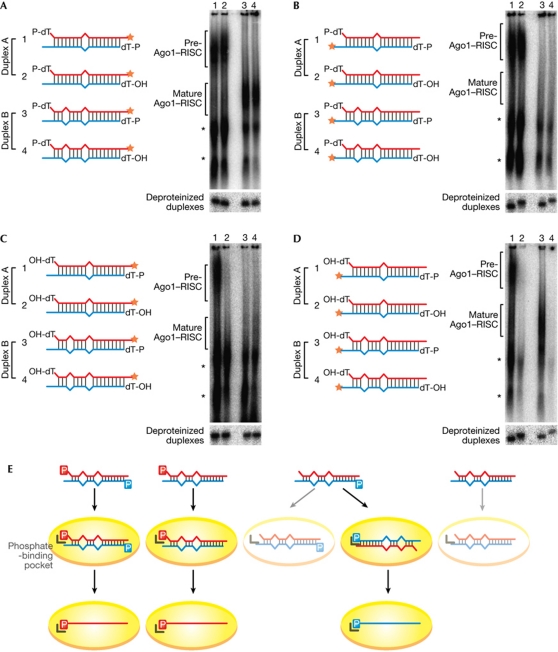
We first asked whether 5′ phosphates of the guide and passenger strands are recognized on RISC loading or unwinding of fly Ago1, a model Ago protein for the miRNA pathway that shares features with human Ago proteins (Kawamata et al, 2009; Yoda et al, 2010). We used two model duplexes, A and B, both of which contained an identical guide-strand sequence, a mismatch at guide position 1, to ensure the functional asymmetry of the duplexes (Khvorova et al, 2003; Schwarz et al, 2003), and a central mismatch at guide position 10, to promote RISC loading (Tomari et al, 2007; Kawamata et al, 2009; Yoda et al, 2010; Fig 1; supplementary Fig S1A online & supplementary Table S1 online). Duplex B bore an extra mismatch at guide position 5, which accelerates the conversion of pre-Ago1–RISC to mature Ago1–RISC; in contrast, duplex A is unwound slowly (Kawamata et al, 2009; Yoda et al, 2010). Therefore, duplex A predominantly forms pre-Ago1–RISC, whereas duplex B mostly forms mature Ago1–RISC in our standard assay condition (supplementary Fig S1A online).
Figure 1.
RISC loading requires 5′ phosphate of the guide strand. (A–D) Duplex A and duplex B with 5′ phosphate (P-dT) or hydroxyl group (OH-dT) were incubated in lysate at 25°C for 30 min, and complexes were separated on a vertical agarose native gel (upper panels). Either the guide strand (red) or the passenger strand (blue) was 3′ radiolabelled (denoted by stars). Asterisks indicate complexes previously shown to be unrelated to the Ago1–RISC assembly pathway (Kawamata et al, 2009; corresponding to complexes III and IV). Formation of these nonspecific complexes was more apparent when a mismatch at guide position 5 was absent, but showed no correlation with the presence or absence of the 5′ phosphate. Duplexes were deproteinized after incubation and their integrity was confirmed by 15% denaturing PAGE (lower panels). Note that 5′-P-dT RNAs migrate faster than 5′-OH-dT RNAs, indicating that the 3′-radiolabelled 5′-OH-dT strands remained unphosphorylated during the incubation. The sequences and structures of the duplexes are shown in supplementary Table S1 online. (E) Schematic summary of A–D. 5′ phosphate of the guide strand, but not that of the passenger strand, is strictly recognized on duplex loading by the specific binding pocket in Ago. When only the passenger strand contains 5′ phosphate, the duplex is loaded in a flipped orientation, suggesting that requirement of the 5′ phosphate of the guide strand precedes the thermodynamic asymmetry rule of small RNA duplexes. 5′-phosphate-binding pocket in Ago is depicted as an L-shaped hook. Ago, Argonaute; dT, deoxythymidine; RISC, RNA-induced silencing complex.
5′ deoxythymidine (dT) is a poor substrate for the kinase that phosphorylates 5′ hydroxyl small RNA duplexes (Nykanen et al, 2001). Indeed, in the lysate we routinely use to study Ago1–RISC assembly (overnight embryo lysate from dcr-2 null flies), 5′-dT duplexes were barely phosphorylated (supplementary Fig S1B online; Fig 1, lower panels) and behaved similarly to phosphorylation-resistant 5′-methoxy (MeO)-dT duplexes (supplementary Fig S1C online). However, when 5′-dT was prephosphorylated by T4 polynucleotide kinase, such duplexes (5′-P-dT) produced equal levels of pre- and mature Ago1–RISCs as prephosphorylated, 5′-U duplexes (5′-P-U; supplementary Fig S1A online). To assess the recognition of the 5′ phosphate of the passenger strand during RISC assembly, we blocked phosphorylation of the passenger strands by replacing the 5′ Us with dTs (5′-OH-dT). The 5′ Us of the guide strands were also replaced with dTs, but were prephosphorylated by T4 polynucleotide kinase (5′-P-dT; Fig 1A; supplementary Table S1 online). We radiolabelled the 3′ ends of the guide strands, and monitored the formation of pre-Ago1–RISC and mature Ago-RISC. The levels of pre- and mature Ago1–RISCs for duplexes A and B were not affected by the presence or absence of the 5′ phosphates of the passenger strands, indicating that the 5′ phosphate of the passenger strand is dispensable for both Ago1–RISC loading and unwinding, as long as the guide strand is 5′ phosphorylated (Fig 1A). As expected, when we radiolabelled the 3′ ends of the passenger strands we detected only pre-Ago1–RISC, because mature Ago1–RISC contains only the non-radiolabelled guide strands (Fig 1B).
Next, we blocked the 5′ ends of the guide strands with 5′-OH-dT. Intriguingly, only duplex A bearing 5′-P-dT on the passenger strand formed pre-Ago1–RISC, and no mature Ago1–RISC was detected with duplex B (Fig 1C). One plausible explanation for this observation is that duplexes A and B were loaded into pre-Ago1–RISC with flipped polarity (the original 5′-P-dT passenger strands acted as the guide strands, whereas the original 5′-OH-dT guide strands acted as the passenger strands), regardless of the strong thermodynamic asymmetry determined by the mismatch at guide position 1. To examine this possibility, we radiolabelled the 3′ ends of the passenger strands and monitored RISC assembly. We detected pre- and mature Ago1–RISC for duplexes A and B when the passenger strands bore 5′-P-dTs (Fig 1D). Taking these observations together, we concluded that the 5′ phosphate of the guide strand is required on RISC loading, whereas that of the passenger strand is dispensable. Importantly, such strong requirement of the 5′ phosphate of the guide strand precedes the thermodynamic asymmetry rule of small RNA duplexes (Fig 1E). Target cleavage assays for each of the strands of duplex B, as well as a duplex of unrelated sequence, confirmed this conclusion (supplementary Fig S2 online). This is in contrast to the well-characterized fly Ago2–RISC assembly pathway, in which the 5′ phosphate of the small-interfering RNA passenger strand is checked by R2D2, a component of RISC-loading complex that also senses the small-interfering RNA thermodynamic asymmetry, before loading into Ago2 (Tomari et al, 2004).
Double check of 5′ nucleotide of the guide
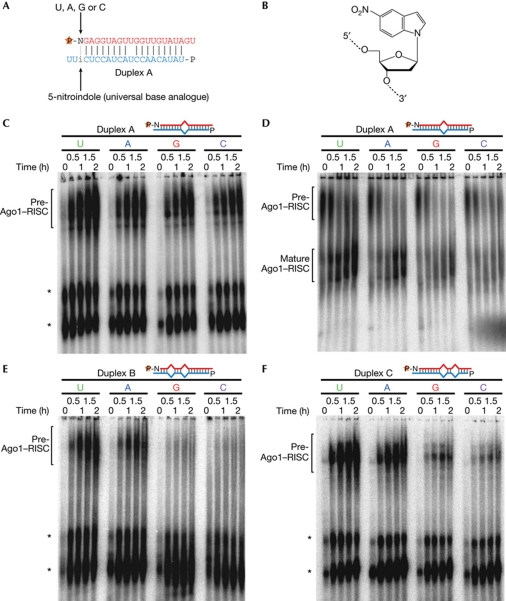
We next investigated whether the 5′ nucleotide is recognized on RISC loading or unwinding. We monitored Ago1–RISC formation by using the derivatives of duplex A bearing U, A, G or C at the 5′ end of the guide strand. We chose duplex A because its slow unwinding rate makes it easier to precisely monitor the transition from pre-Ago1–RISC to mature Ago1–RISC. However, changing the 5′ nucleotide alters the base-pairing state at this position, disturbing the thermodynamic asymmetry of the duplexes. To avoid this, we introduced a universal base, 5-nitroindole, which hybridizes equally with all four nucleotides (Liu et al, 2004), at position 19 of the passenger strand across from the 5′ end of the guide strand (Fig 2A,B). A control experiment verified that the thermodynamic asymmetry of 5-nitroindole-containing duplexes was unaffected, regardless of the 5′ nucleotide identity of the guide strand (supplementary Fig S3 online).
Figure 2.
5′ nucleotide of the guide strand and base pairing in the seed and 3′ regions function as additive anchors to Argonaute proteins. (A) Schematic representation of 5′ nucleotide of the guide strand and 5-nitroindole at position 19 of the passenger strand. (B) Structure of 5-nitroindole. (C) Duplex A derivatives with 5′ U, A, G or C were incubated in lysate at 10°C and separated by agarose native gel. (D) Duplex A derivatives with 5′ U, A, G or C were incubated in lysate at 10°C for 2 h, 20-fold excess of non-radiolabelled duplexes were added (time=0), and then ‘chase’ conversion of pre-Ago1–RISC into mature Ago1–RISC was monitored at 30°C. (E) The same experiment as in C with duplex B derivatives bearing a seed mismatch. (F) The same experiment as in C with duplex C derivatives bearing a 3′ mismatch. Asterisks indicate complexes previously shown to be unrelated to the Ago1–RISC assembly pathway (Kawamata et al, 2009; corresponding to complexes III and IV). Note that these nonspecific complexes are barely observed in D, because of long preincubation followed by the addition of excess non-radiolabelled duplexes. The sequences and structures of duplexes are shown in supplementary Table S2 online. Ago, Argonaute; RISC, RNA-induced silencing complex.
By using these duplexes, we monitored Ago1–RISC loading at 10°C at which temperature unwinding is suppressed. We found that, although 5′ U was preferred, changing the 5′ nucleotide did not affect pre-Ago1–RISC formation, suggesting that the identity of the 5′ nucleotide is not crucial for RISC loading of duplex A derivatives (Fig 2C; supplementary Fig S4A online). We next analysed the role of 5′ nucleotides in unwinding. To this end, we incubated the duplexes in lysate for 2 h at 10°C to nearly saturate the formation of pre-Ago1–RISC, added excess non-radiolabelled duplexes and then monitored the ‘chase’ conversion from pre-Ago1–RISC to mature Ago1–RISC at 30°C, at which temperature unwinding is efficient. Remarkably, although the initial amounts of pre-Ago1–RISCs before unwinding (at time 0) were nearly equal, accumulation of mature Ago1–RISCs showed a pronounced preference for duplexes with 5′ U>A>G>C (Fig 2D; supplementary Fig S4B online). Thus, the 5′ nucleotide identity of the guide strand is checked not only on RISC loading, but also on or after unwinding, thereby amplifying the preference for 5′ U through the RISC assembly pathway. Frank et al (2010) have recently shown that a rigid loop in the isolated MID domain of human Ago2 forms a specific binding pocket for 5′ U and A, and human Ago2 and fly Ago1 have the same amino-acid sequence for this rigid loop. Our data indicate that the specificity of the 5′-nucleotide-binding pocket in the context of full-length Ago proteins can be influenced during the course of RISC assembly.
Additive anchors for Ago proteins
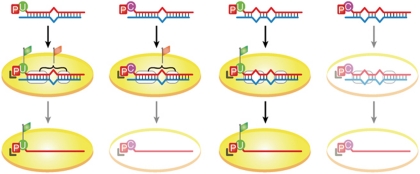
Why are unfavourable 5′ nucleotides (that is, G and C) of duplex A derivatives tolerated in pre-Ago1–RISC, but forbidden in mature Ago1–RISC? We hypothesized that, compared with mature Ago1–RISC, pre-Ago1–RISC uses an extra anchor to accommodate unfavourable 5′ nucleotides. As mentioned above, mismatches in the seed or the 3′ regions promote separation of miRNA/miRNA* duplexes within Ago proteins (Kawamata et al, 2009; Yoda et al, 2010); mismatches in these regions obstruct Ago proteins, to hold miRNAs in their double-stranded state. If so, base pairing in these regions might act as an anchor for Ago proteins. To test this possibility, we used derivatives of duplex B, which harbour a mismatch at guide position 5 as well as a central mismatch, bearing U, A, G or C at the 5′ end of the guide strand. In contrast to duplex-A derivatives, formation of pre-Ago1–RISC was much less efficient for duplex-B derivatives with 5′ G or C, compared to those with 5′ U or A (Fig 2E; supplementary Fig S4C online). Duplex C, which harbours a 3′ mismatch at position 13 and a central mismatch, also yielded similar results (Fig 2F; supplementary Fig S4D online). These observations indicate that, when seed and 3′ base pairing is absent, unfavourable nucleotides are not allowed in pre-Ago1–RISC. Taken together, we concluded that favourable 5′ nucleotide and base pairing in the seed and 3′ regions function as additive anchors for Ago1, although it is difficult to quantify the weighing of each anchor (Fig 3).
Figure 3.
Schematic summary for multilayer checkpoints of microRNA authenticity on RISC loading and unwinding. 5′ phosphate of the guide strand is required for RISC loading. Favourable 5′ nucleotide and base pairing in the seed and 3′ regions function as additive anchors for Ago1 (represented as flags; at least one flag is necessary for Ago1 to retain the small RNA, although the weighing of each flag is unknown). Ago, Argonaute; RISC, RNA-induced silencing complex.
Given that base pairing in the seed and 3′ regions are important for miRNAs to recognize their target mRNAs (Bartel, 2009), Ago proteins generally favour stable base pairing in these regions. However, natural miRNA/miRNA* duplexes tend to have mismatches in the seed and/or 3′ regions, in addition to the central region, underlining the idea that unwinding of miRNAs is the mirror image of target recognition (Kawamata et al, 2009; Kawamata & Tomari, 2010). In this regard, seed or 3′ mismatches in miRNA/miRNA* duplexes are a ‘necessary evil’ for RISC assembly; they are naturally disfavoured by Ago proteins, hindering RISC loading of miRNA/miRNA* duplexes, and yet they are required later for efficient unwinding. Circumventing this dilemma, the strong preference of the 5′ nucleotide of the miRNA guide strand, especially 5′ U, functions as an additive anchor and helps Ago proteins to load mismatch-containing miRNA/miRNA* duplexes. Our study highlights a strategy of miRNAs and Ago proteins for assembling RISC, with miRNA authenticity being checked at multiple steps.
Methods
Preparation of Drosophila melanogaster overnight embryo was described previously in detail (Haley et al, 2003). In vitro RISC assembly assay, target cleavage assay, deproteinization, denaturing PAGE, 5′ radiolabelling with T4 polynucleotide kinase (Takara) and 32P-ATP (MP Biomedicals), 3′ radiolabelling with poly (A) polymerase (USB) and α-32P cordycepin-5′-triphosphate (PerkinElmer) and native gel analysis were performed as described previously (Nykanen et al, 2001; Haley et al, 2003; Tomari et al, 2007; Kawamata et al, 2009).
Small RNA duplexes and target RNAs for native gel analysis. The sequences and structures of the small RNA duplexes used in this study are shown in supplementary Tables S1–3 online. Oligonucleotides were synthesized by Sigma Genosys, Hokkaido System Science, IDT or GeneDesign. For experiments in Fig 1A,C supplementary Fig S1A,C online and supplementary Fig S3B online (right), RL 4 × mRNA was used as a target, as described previously (Kawamata et al, 2009). For experiments in Fig 1B,D; supplementary Fig S3B online (left), either RL 1 × mm1 mRNA or RL 1 × mm5 mRNA was used as a target, as described previously (Kawamata et al, 2009). For Fig 2, 2′-O-methyl antisense oligonucleotide containing a 5-nitroindole was used as the target: 5′-mUmCmUmUmCmAmCmUmAmUmAmCmAmAmCmCmUmAmCmUmAmCmCmUmC/5-nitroindole/mAmCmCmUmU-3′. This antisense oligonucleotide was complementary to the guide-strand sequence of duplex A and duplex B, except for one central mismatch.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank R. Carthew (Northwestern University, USA) for dcr-2 flies. We are also grateful to H. Seitz and P. Zamore for sharing unpublished results, and members of the Tomari laboratory for stimulating discussions and critical comments on the manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas (‘Functional machinery for non-coding RNAs’), a Grant-in-Aid for Young Scientists from the Japan Ministry of Education, Culture, Sports, Science and Technology, as well as a Carrier Development Award from The International Human Frontier Science Program Organization.
Author contributions: T.K. and Y.T. designed the experiments and wrote the manuscript; T.K. and M.Y. conducted the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ (2009) Hierarchical rules for Argonaute loading in Drosophila. Mol Cell 36: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B (2010) Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465: 818–822 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD (2010) Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16: 43–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M et al. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR–CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Tang G, Zamore PD (2003) In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30: 330–336 [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M (2010) In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y (2010) Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39: 292–299 [DOI] [PubMed] [Google Scholar]
- Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G (2010) HSP90 protein stabilizes unloaded Argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell 21: 1462–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y (2010) Making RISC. Trends Biochem Sci 35: 368–376 [DOI] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y (2009) Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol 16: 953–960 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ (2005) Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321 [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC (2009) Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D (2005) Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD (2004) A protein sensor for siRNA asymmetry. Science 306: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD (2007) Sorting of Drosophila small silencing RNAs. Cell 130: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ (2008) Structure of the guide-strand-containing Argonaute silencing complex. Nature 456: 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y (2010) ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol 17: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.