Abstract
The expression of the type III secretion system—a main determinant of virulence in Shigella—is controlled by regulator cascades VirF-InvE (VirB) and CpxAR two-component system. A screen for mutants that restore virulence in the cpxA background led to the isolation of a mutant of rodZ, a cytoskeletal protein that maintains the rod-shaped morphology of bacilli. InvE is normally repressed at 30 °C because of decreased messenger RNA (mRNA) stability, but rodZ mutants markedly increase invE-mRNA stability. Importantly, the inhibition of InvE production by RodZ can be genetically separated from its role in cell-shape maintenance, indicating that these functions are distinguishable. Thus, we propose that RodZ is a new membrane-bound RNA-binding protein that provides a scaffold for post-transcriptional regulation.
Keywords: bacterial cytoskeleton, post-transcriptional regulation, Shigella , type III secretion system
Introduction
Recent studies of the regulation of bacterial cell-shape have shown that a set of bacterial cytoskeletal proteins maintain the rod-shaped morphology of bacilli. Escherichia coli MreB is a cytoskeletal protein that polymerizes into filaments—similarly to eukaryotic actin—that form helical arrays within the bacterial cytosol (Jones et al, 2001). Recently, RodZ (YfgA) was identified as a putative cytoskeletal anchoring protein that colocalizes with MreB at the inner membrane. Mutants of rodZ with altered cell morphology were isolated by microscopic screening of a collection of non-essential gene deletion mutants in E. coli K-12 (Shiomi et al, 2008), analysis of a transposon library from Caulobacter crescentus (Alyahya et al, 2009) and screening of E. coli K-12 mutants that require excess amounts of the cell-division protein FtsZ for growth on rich media (Bendezu et al, 2009). MreB and RodZ interact to define the long and short axes of E. coli.
A new rodZ mutant was identified in Shigella sonnei, an organism closely related to E. coli. Clinically, Shigella invades and propagates within the epithelial cells of the human intestine, resulting in the onset of bloody diarrhoea. The pathogenesis of Shigella is mediated by the type III secretion system (TTSS) encoded on the virulence plasmid that injects effector molecules into the host epithelium during infection (Dorman & Porter, 1998). Expression of the TTSS is tightly regulated by two proteins, VirF and InvE. VirF—an AraC-type transcriptional regulator—activates the transcription of invE (virB; Adler et al, 1989; Kato et al, 1989), and InvE—a homologue of the plasmid-partitioning factor ParB (Watanabe et al, 1990)—activates transcription of mxi-spa and ipa genes that encode the components of the TTSS (Beloin & Dorman, 2003).
The TTSS supports bacterial survival by repressing unnecessary expression under adverse conditions, such as low temperature (Maurelli et al, 1984) and low osmotic-pressure (Porter & Dorman, 1994). Repression is primarily accomplished through the post-transcriptional regulation of invE messenger RNA (mRNA). At a permissive temperature of 37 °C, invE-mRNA is stable, but its stability markedly decreases at 30 °C or under low osmotic-pressure. The deletion of hfq, an important RNA chaperone that is involved in the post-transcriptional regulation of many genes, leads to the recovery of invE-mRNA stability and increased TTSS gene expression (Mitobe et al, 2008, 2009).
Expression of TTSS is also under the control of the CpxAR two-component system (Nakayama & Watanabe, 1998). Transcription of virF depends on the phosphorylated form of the response regulator CpxR, which suggests a functional hierarchy: CpxAR>VirF>InvE. In the cpxA mutant, however, transcription of invE is maintained because virF is transcribed by autophosphorylated CpxR, as is the case for other Cpx-regulated genes (Alves et al, 2003). Although invE-mRNA is transcribed, the corresponding amount of InvE protein is not synthesized in the cpxA mutant at the permissive temperature of 37 °C (Mitobe et al, 2005). To elucidate the mechanism for this, a mutant that expressed InvE-regulated TTSS genes was isolated under the cpxA mutant background. The transposon was unexpectedly found to map to the locus encoding bacterial cytoskeleton RodZ.
Results And Discussion
Isolation of a rodZ mutant
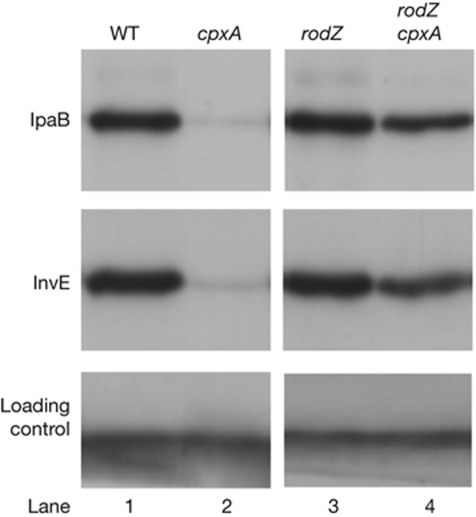
A cpxA-sensor deletion mutant showed reduced expression of InvE, as well as the TTSS effector molecule IpaB, at 37 °C (Fig 1A, lane 2). The reduction in invE expression was caused by a defect in post-transcriptional processing (Mitobe et al, 2005). To investigate this repression mechanism, secondary mutations that caused recovery of TTSS expression in the cpxA mutant background were identified. A cpxA-deletion mutant of E. coli (strain ME2824) that carried a reporter plasmid encoding a TTSS(mxiC)–lacZ fusion gene (pJM1718) was constructed and subjected to Tn5 transposon mutagenesis. After screening 2 × 104 colonies, a single Tn5-transposon-insertion mutant was isolated that exhibited enhanced β-galactosidase activity. The Tn5 insertion site was mapped to 11 base-pairs downstream from the amino-terminal end of the rodZ (yfgA) gene (see supplementary information online) that encodes a new bacterial cytoskeletal protein involved in the maintenance of cell shape. To confirm the involvement of RodZ in TTSS expression, a deletion mutant (cpxA/rodZ, ME5199) was constructed that produced wild-type levels of InvE and IpaB at 37 °C (Fig 1, lane 4). This showed that RodZ was involved in CpxA-dependent expression of TTSS-related genes. The amount of RodZ protein increased slightly in the parental cpxA mutant, and deletion of the response regulator cpxR or both of the cpxRA genes had no effect on the expression of a rodZ–lacZ reporter gene (supplementary Fig S1A,B online). Therefore, the CpxAR two-component system probably does not directly regulate transcription of the rodZ gene.
Figure 1.
Type III secretion system-encoding gene expression in Echerichia coli strains carrying a virulence plasmid from Shigella sonnei. Immunoblot analysis of IpaB and InvE expression at 37 °C. A nonspecific protein that cross-reacted with the InvE antibody (lower panel) was used as a loading control throughout the study. Lanes: 1, HW1273 (WT); 2, ME2824 (ΔcpxA); 3, ME5201 (ΔrodZ); 4, ME5199 (ΔcpxA/ΔrodZ). WT, wild type.
Expression of invE in rodZ mutants
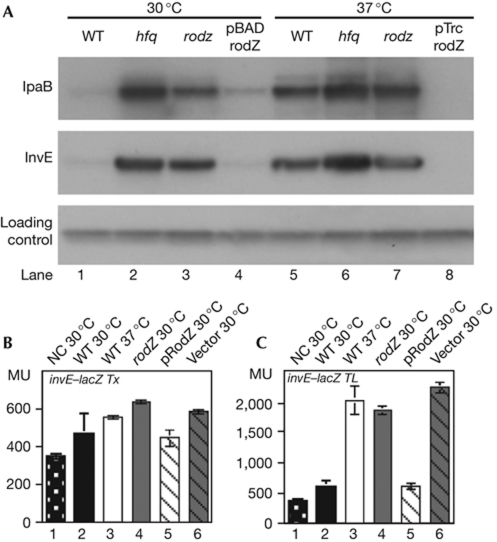
To determine the regulatory role of RodZ, the rodZ deletion mutation was introduced into wild-type S. sonnei. The expression of invE is normally repressed at 30 °C (Fig 2A, lane 1), but repression is abolished in an hfq deletion mutant (Fig 2A, lane 2) that is involved in the post-transcriptional control of invE-mRNA (Mitobe et al, 2008). Similarly to the hfq mutant, robust expression of InvE and IpaB proteins was detected in the rodZ mutant strain (MS5201) at 30 °C (Fig 2A, lane 3). Expression of Hfq and RodZ were mutually independent because the amounts of the corresponding proteins were not affected in the respective rodZ and hfq mutants (supplementary Fig 1C online). The copy number of the virulence plasmid—which was measured by real-time PCR to detect the invE gene—was unaffected in either mutant (see supplementary information online). Expression of RodZ in the rodZ mutant with the pBAD-rodZ plasmid repressed expression of InvE and IpaB at 30 °C (Fig 2A, lane 4), but not at 37 °C (data not shown). Overexpression of RodZ with pTrc-rodZ repressed InvE and IpaB expression, even at 37 °C (Fig 2A, lane 8).
Figure 2.
Type III secretion system-encoding gene expression in Shigella sonnei. (A) Immunoblot analysis of IpaB and InvE at 30 °C (lanes 1–4) and 37 °C (lanes 5–8). Lanes: 1 and 5, MS390 (WT); 2 and 6, MS4831 (Δhfq); 3 and 7, MS5201 (ΔrodZ); lane 4, MS5201 carrying pBAD-rodZ; lane 8, MS5201 carrying pTrc-rodZ. (B) β-galactosidase activity of an invETx–lacZ transcriptional fusion reporter protein (pJM4320). (C) β-galactosidase activity of an invETL–lacZ translational fusion reporter protein (pJM4321). (B,C) Bars: 1, MS506 (NC, avirulent S. sonnei); 2, MS390 (WT S. sonnei); 3, MS390 at 37 °C; 4, MS5201 (ΔrodZ); 5, MS5201 (ΔrodZ) MS5201 carrying pBAD-rodZ; 6, MS5201 carrying pBAD18Kan. Error bars represent s.d. of four independent experiments. MU, Miller units; NC, negative control; WT, wild type.
Transcription of the invE gene was measured using the lacZ reporter plasmid. In wild-type S. sonnei, the β-galactosidase activity directed by the invE–lacZ transcriptional fusion was almost the same at the non-permissive and permissive temperatures, 30 and 37 °C, respectively. This is different from the activity measured using the invE–lacZ translational fusion, in which β-galactosidase activity was lower at 30 °C (Fig 2B,C). Thus, the temperature-dependent repression of invE expression in wild-type S. sonnei occurs at the post-transcriptional level.
In the rodZ mutant, the β-galactosidase activity driven by the invE–lacZ transcriptional fusion was similar to that of the wild-type strain or the rodZ mutant strain carrying rodZ expression plasmid pBAD-rodZ (Fig 2B). The difference between the wild-type and the rodZ mutants at 30 °C was greater for the invE–lacZ translational-fusion construct (Fig 2C). Consistently, the introduction of pBAD-rodZ into the rodZ mutant decreased β-galactosidase activity to the level of the wild-type strain. The results suggest that mutation of rodZ affects the expression of virulence genes at a post-transcriptional step.
Stability of invE-mRNA in the rodZ mutant
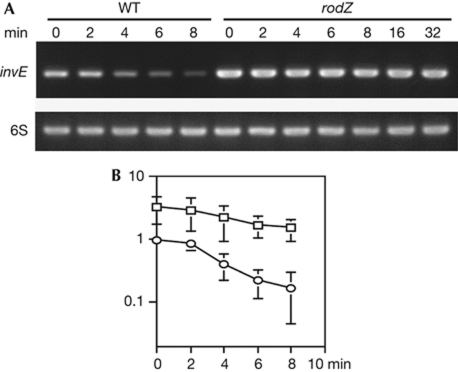
To confirm the prediction that RodZ has a function similar to that of the RNA chaperone Hfq, the stability of invE-mRNA in the rodZ mutant was measured after addition of rifampicin, a potent inhibitor of transcription initiation. In the wild-type strain MS390, invE-mRNA was degraded rapidly at 30 °C. By contrast, the basal level of invE-mRNA in the rodZ mutant was elevated, compared with the wild-type strain. Moreover, invE-mRNA remained stable for at least 30 min (Fig 3A). As determined by reverse transcription-PCR, the half-life of invE-mRNA was estimated to be more than 20 min in the rodZ mutant (Fig 3A), whereas in the hfq mutant this was approximately 5.8 min (Mitobe et al, 2008). Thus, the mutation of rodZ enhanced invE-mRNA stability more than the mutation of hfq. These results indicate a functional relationship between RodZ expression and invE-mRNA stability, which might suggest that the cytoskeletal protein RodZ has RNA-binding activity.
Figure 3.
Stability of invE-messenger RNA. (A) Reverse transcription-PCR of invE-mRNA using 6S RNA as a control. Minutes indicate the time after rifampicin treatment. WT, Shigella sonnei strain MS390; rodZ, strain MS5201 (ΔrodZ). (B) Real-time PCR of invE-mRNA in wild-type S. sonnei (MS390, circles) and ΔrodZ (MS5201, squares). mRNA was normalized to 6S RNA. Values relative to the wild type at time 0 are plotted on the semilog plot. Error bars represent s.d. of four independent experiments. mRNA, messenger RNA; WT, wild type.
RodZ-invE RNA interaction in vitro
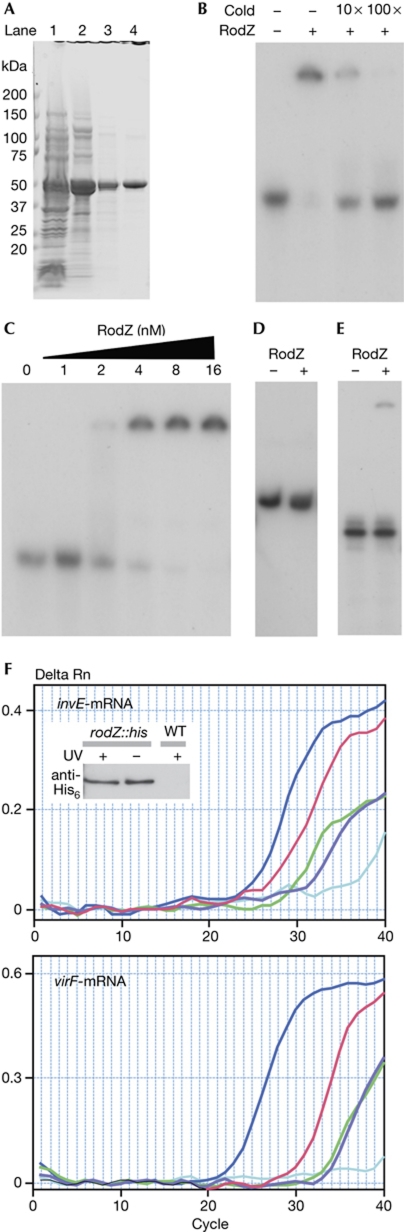
To determine whether RodZ interacts directly with invE-mRNA, a recombinant His-tagged S. sonnei RodZ fusion protein was purified to remove trace amounts of nuclease activity. Purified RodZ-His6 formed a single band when analysed by 5–15% SDS–polyacrylamide gel electrophoresis (Fig 4A) and bound to an invE RNA probe in a gel-shift assay (Fig 4B). To assess the strength of the RodZ–invE RNA interaction, the apparent dissociation constant (Kd) was determined by measuring the disappearance of the RNA probe in a gel-shift assay. The apparent Kd for the formation of the RodZ–invE RNA complex was 3.5 nM (Fig 4C), which was significantly higher than that found for the Hfq–invE RNA (Kd=19.2 nM for the monomer under similar experimental conditions; Mitobe et al, 2008). The interaction of RodZ–invE with RNA was not detected in buffer with 300 mM potassium glutamate (data not shown), which suggests that the affinity could be lower than the estimated values under physiological conditions.
Figure 4.
RodZ–invE RNA binding. (A) Purified RodZ protein. Lane: 1, crude extract; 2, SP Sephadex; 3, P-11 phosphocellulose; 4, Ni-TED. (B) Gel-shift analysis of RNA binding using invE RNA probe (2 nM) and RodZ (16 nM). Cold: 10- and 100-fold unlabelled invE RNA. (C) Determination of the apparent Kd of the RodZ–invE RNA-binding interaction. A constant amount of RNA (2 nM) was mixed with the indicated amounts of RodZ. (D) Gel-shift analysis of the RodZ–invE DNA interaction using 2 nM invE PCR product and RodZ (16 nM). (E) Gel-shift analysis of the RodZ–bla–RNA interaction using 2 nM RNA probe and RodZ (16 nM). (F) Detection of in vivo invE and virF mRNA binding at 37 °C by real-time PCR. Blue, positive control by 10 ng total RNA; red, invE–mRNA–RodZ∷His6 complex from crosslinked MS5401 (rodZ∷his6) strain; purple, negative control from uncrosslinked MS5401 strain; green, negative control from crosslinked wild-type strain; cyan, RNaseA-treated sample from crosslinked MS5401 strain. Tethered RNA–protein complex for the complementary DNA synthesis was detected by His6 antibody. mRNA, messenger RNA; UV+, ultraviolet-irradiated samples; UV−, no treatment; WT, wild type.
To examine the specificity of RodZ binding to RNA, an invE DNA probe with the same sequence as the RNA probe was constructed and subjected to a gel-shift assay. The invE DNA probe did not form complexes with 2 nM RodZ (Fig 4D). Low concentrations of unlabelled invE RNA interfered with the formation of RodZ–invE RNA complexes (Fig 4B), and the inhibition of the RodZ–invE RNA interaction required high concentrations of calf thymus DNA, yeast transfer-RNA or polyphosphate (0.1–1 mg/ml; supplementary Fig S2 online). Binding of RodZ to RNA with an unrelated sequence, such as the 149-nucleotide bla RNA probe, was weak (Fig 4E). These data suggest that RodZ has high specificity for its substrate and nucleotide sequence.
RodZ–invE RNA interaction in vivo
To detect in vivo interactions between RNA and RodZ, MS5401 cells expressing His-tagged RodZ were exposed to ultraviolet light in the mid-logarithmic phase of growth, to crosslink RodZ to its target RNA. His-tagged RodZ–RNA complexes were purified from the cell lysate using magnetic beads. After washing with a high-salt buffer, the complexes were dissociated from the beads, and the crosslinked RodZ–RNA complexes were subjected to protease digestion. After subsequent treatment with RNase-free DNaseI, invE and virF mRNAs in the samples were detected by complementary-DNA synthesis and then by real-time PCR using specific photosequencing probes. The results shown in Fig 4F indicate that the level of invE-mRNA (red line) in the ultraviolet-treated sample was higher than that found in samples from cells without ultraviolet treatment (purple line) or from cells that did not express Hig-tagged RodZ (green line). Binding specificity was examined by measuring virF mRNA as an unrelated mRNA species within the same sample. The relative level of virF mRNA was 24- to 25-fold lower than that of invE-mRNA, as determined by the deviation of amplification plots against positive controls using an intact total RNA (blue line).
Signals were similarly detected in samples grown at 30 °C. The difference to that obtained at 37 °C was a rightward shift of total plots including the positive control, which is in agreement with the fact that little difference was observed in RNA-binding activity in vitro at either temperature (data not shown). This suggests that the temperature dependency of the invE-mRNA degradation might depend on an as yet unidentified factors such as Hfq, which shows temperature-dependent RNA-binding activity in vitro (Mitobe et al, 2008).
Functional mapping of RodZ
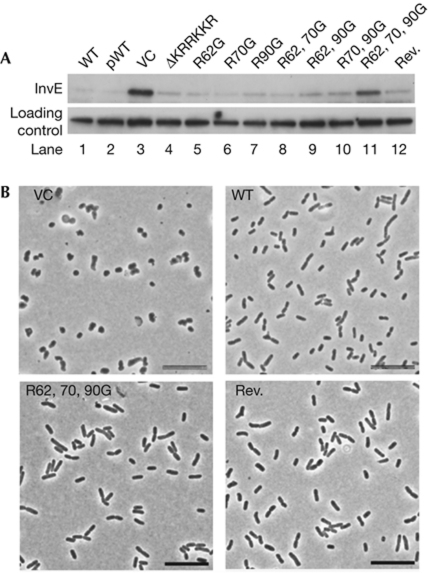
These results indicated that RodZ has RNA-binding activity with significant specificity in addition to its role as a cytoskeletal protein in rod-shaped bacteria. RodZ contains a helix-turn-helix (HTH) motif in the N-terminal region and a short basic region (KRRKKR) near the transmembrane domain (Shiomi et al, 2008; Bendezu et al, 2009). To identify the region necessary for RNA binding, a protein variant with an internal deletion of the short basic region (ΔKRRKKR) was constructed. This mutation resulted in the loss of RodZ RNA-binding activity in vitro, which shows that this region is essential for RNA binding (supplementary Fig S3B online). However, when a plasmid expressing ΔKRRKKR RodZ was introduced into a rodZ mutant, InvE expression was repressed as much as in the transformant with the wild-type rodZ expression plasmid (Fig 5A, lanes 2 and 4). Therefore, the deletion of only KRRKKR was not sufficient to interfere with RodZ–invE-mRNA processing in vivo.
Figure 5.
Functional domain mapping. (A) Immunoblot analysis of InvE protein at 30 °C in MS5201 (ΔrodZ) strains carrying the indicated expression plasmids. Lanes: 1, WT MS390; 2, ΔrodZ strain carrying pBAD-rodZ; 3, VC; 4, ΔKRRKKR; 5, R62G; 6, R70G; 7, R90G; 8, R62G, R70G; 9, R62G, R90G; 10, R70G, R90G; 11, R62G, R70G, R90G; 12, R62G, R70G, R90G with KRRKKR (Rev.). (B) Phase contrast images of strain MS5201 (ΔrodZ) expressing the indicated RodZ variants. Scale bars, 10 μm. Rev., reversion of the short basic region; VC, vector control; WT, wild type.
RodZ also has several conserved, basic amino-acid residues in a short segment between the HTH domain and KRRKKR (NCBI Conserved Domain no. PRK10856). By using the rodZ ΔKRRKKR mutant, each of these three Arg residues (R62, R70 and R90) was substituted with Gly (supplementary Fig S3A online). The rodZ mutants carrying expression plasmids with one or two Arg substitutions repressed expression of InvE in a manner that was similar to the rodZ mutant carrying the wild-type rodZ expression plasmid (Fig 5A, lane 5–10). However, the expression of a rodZ mutant with all three Arg substitutions (R62G, R70G and R90G) did not repress InvE expression (Fig 5A, lane 11). This increase in production of InvE protein was accompanied by increased stability of invE-mRNA (supplementary Fig S3C online). In addition, a plasmid encoding an R62G, R70G and R90G rodZ allele without deletion of KRRKKR repressed InvE synthesis (Fig 5A, lane 12). Taken together, these results suggest that the short basic region (KRRKKR) and the three additional Arg residues have important roles in the repression of InvE synthesis in vivo.
Finally, the influence of rodZ mutations on cell shape was investigated. The rodZ mutant with a plasmid encoding all three Arg substitutions, with or without the short basic region (KRRKKR), adopted the same cell shape as a rodZ mutant that harboured the wild-type rodZ plasmid (Fig 5B). This shows that neither the substitution of the three Arg residues nor the deletion of the short KRRKKR basic region of RodZ influences the cell shape. Thus, the RodZ protein might have two functions: an architectural role in the cytoskeleton and a regulatory role for processing mRNA.
Our experimental results indicate that RodZ and invE-mRNA interact directly in living cells, leading to post-transcriptional repression of InvE synthesis due to the decreased stability of invE-mRNA. However, repression of InvE synthesis was observed in the rodZ mutant expressing ΔKRRKKR RodZ (Fig 5A, lane 4) even though this mutant RodZ had lost its RNA binding in vitro (supplementary Fig S3B online). One explanation for this apparent contradiction is that for effective interaction of RodZ with invE-mRNA, an auxiliary factor is involved that restores the invE-repression activity in mutants lacking one of the two elements, the KRRKKR sequence or the three Arg residues (Fig 5A, lanes 4 and 12). However, this putative cofactor was unable to restore the activity of the mutant lacking both elements (Fig 5A, lane 11). Taken together, we speculate that either of the elements is sufficient for completion of post-transcriptional repression. RodZ might mediate spatial coincidence between the processed mRNA and putative factors for post-transcriptional regulation. Further analysis of RodZ immunoprecipitates—in which we have already observed more than 12 unidentified bands following SDS–polyacrylamide gel electrophoresis—should resolve this issue.
The precise role of the three Arg residues, which were initially suggested to be additional RNA-binding sites, remains to be elucidated. Crystal structure of the cytoplasmic domain of RodZ in Thermotoga maritime shows that the Arg residues are predicted to exist in the fourth and fifth helices. Proper conformation of the fourth helix is important because amino-acid substitutions F60A and Y64A in a GFP-RodZ1−138-RFP are unable to interact with MreB, and result in round cells (van den Ent et al, 2010). Dissimilarly to aromatic amino-acids, substitution of the three Arg residues might not cause drastic changes in the conformation of the helix motif, as the shape of bacteria expressing those variants did not change (Fig 5B). RodZ has an additional Arg residue at the 66th amino acid. Addition of the further R66G substitution to the three Arg substitutions with ΔKRRKKR, however, resulted in a round cell-shape, but not by a single R66G substitution (data not shown). These results suggest that a subtle change of protein structure, which could disrupt putative interaction with the cofactor or additional RNA-binding sites, had already occurred in the mutant of three Arg substitutions with ΔKRRKKR.
Our results support a hypothesis that there are two fundamental roles for RodZ in rod-shaped bacteria: to maintain proper cytoskeletal architecture and to function in mRNA processing. As RodZ is localized to the membrane, it might function as an anchor to position nascent mRNA molecules. The recent discovery of an RNaseE complex that also localizes to the membrane (Taghbalout & Rothfield, 2008) supports the idea that post-transcriptional regulation occurs in spaces adjacent to the membrane. Thus, membrane-bound RodZ might provide a platform for post-transcriptional regulation.
Methods
Bacterial strains and plasmids. Bacterial strains and plasmids used in this study are listed in supplementary Table SI online.
Genetic screening. The EZ-Tn5<KAN-2> transposome (Epicenture) was introduced by electroporation into E. coli strain ME2824, which carries the mxiC(TTSS)–lacZ fusion plasmid, pJM1718. Transformants were plated on LB X-gal agar with kanamycin and chloramphenicol and incubated overnight at 37 °C.
Purification of RodZ and gel-shift assays. Recombinant RodZ-His6 protein was expressed in E. coli BL21(DE3) (Novagen) carrying pET22b-RodZ plasmid, purified by Hi-prep SP (GE Healthcare) and P-11 phosphocellulose (Whatman), followed by Ni-TED column (Macherey Nagel). For gel-shift assay, RodZ was mixed with 20 femtomoles of labelled RNA probe consisting of the first 140 nucleotides of the invE gene in a 10 μl of RNA-binding buffer, as described previously (Mitobe et al, 2008).
In vivo crosslinking of mRNA. A volume of 12 ml of MS5401 culture (OD600=1.0 at 37 °C) was subjected to ultraviolet crosslinking under a fluence of 0.2 J/cm2 ultraviolet-B (45 s, Funa UV linker FS800, Funakoshi) at 4 °C. The cells were lysed in 1 ml of binding buffer and mixed with 12.5 μl of Dynabeads TARON (Dynal), which was pretreated with a lysate of avirulent MS506 strain. After 30 min binding at 4 °C, the beads were collected and extensively washed by saturated ammonium sulphate (pH 8.0). RNA–protein complexes were released from beads, and treated by Turbo DNase kit (Ambion). The sample was subjected to complementary DNA synthesis and real-time PCR by ABI PRISM 7000 using photoquencing probes against invE and virF genes.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Y. Horiuchi from the Life Science Research Institute, Kinki University, for assistance with electron microscopy; M. Ino from Saitama Prefectural University for her kind assistance with biochemical analyses; and Dr M. Wachi from the Tokyo Institute of Technology for providing the A22 reagent. This research was supported by a grant-in-aid from the Ministry of Health, Labor and Welfare of the Japanese Government (H21·kokusai-igaku).
Footnotes
The authors declare that they have no conflict of interest.
References
- Adler B, Sasakawa C, Tobe T, Makino S, Komatsu K, Yoshikawa M (1989) A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol Microbiol 3: 627–635 [DOI] [PubMed] [Google Scholar]
- Alves R, Savageau MA (2003) Comparative analysis of prototype two-component systems with either bifunctional or monofunctional sensors: differences in molecular structure and physiological function. Mol Microbiol 48: 25–51 [DOI] [PubMed] [Google Scholar]
- Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C (2009) RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci USA 106: 1239–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin C, Dorman CJ (2003) An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol Microbiol 47: 825–838 [DOI] [PubMed] [Google Scholar]
- Bendezu FO, Hale CA, Bernhardt TG, de Boer PA (2009) RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ, Porter ME (1998) The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol Microbiol 29: 677–684 [DOI] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104: 913–922 [DOI] [PubMed] [Google Scholar]
- Kato J, Ito K, Nakamura A, Watanabe H (1989) Cloning of regions required for contact hemolysis and entry into LLC-MK2 cells from Shigella sonnei form I plasmid: virF is a positive regulator gene for these phenotypes. Infect Immun 57: 1391–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli AT, Blackmon B, Curtiss R III (1984) Temperature-dependent expression of virulence genes in Shigella species. Infect Immun 43: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitobe J, Arakawa E, Watanabe H (2005) A sensor of the two-component system CpxA affects expression of the type III secretion system through posttranscriptional processing of InvE. J Bacteriol 187: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitobe J, Morita-Ishihara T, Ishihama A, Watanabe H (2008) Involvement of RNA-binding protein Hfq in the post-transcriptional regulation of invE gene expression in Shigella sonnei. J Biol Chem 283: 5738–5747 [DOI] [PubMed] [Google Scholar]
- Mitobe J, Morita-Ishihara T, Ishihama A, Watanabe H (2009) Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiol 9: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama S, Watanabe H (1998) Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J Bacteriol 180: 3522–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Dorman CJ (1994) A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J Bacteriol 176: 4187–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H (2008) Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J 27: 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghbalout A, Rothfield L (2008) New insights into the cellular organization of the RNA processing and degradation machinery of Escherichia coli. Mol Microbiol 70: 780–782 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Johnson CM, Persons L, de Boer P, Lowe J (2010) Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J 29: 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Arakawa E, Ito K, Kato J, Nakamura A (1990) Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of invE with ParB of plasmid P1. J Bacteriol 172: 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.