Abstract
Ubiquitin-specific protease 22 (USP22) edits the histone code by deubiquitinating H2A and H2B as part of the mammalian SAGA (Spt-Ada-Gcn5) complex, and is required for transcriptional regulation and normal cell-cycle progression. Here, we show that USP22 affects the expression of p21 by altering far upstream element (FUSE)-binding protein 1 (FBP1) ubiquitination, as ablation of USP22 leads to increased FBP1 ubiquitination and decreased FBP1 protein occupancy at the p21 gene. Surprisingly, increased polyubiquitination of FBP1 does not alter its protein stability, but instead modulates the stable recruitment of FBP1 to target loci. Our results indicate a mechanism by which USP22 regulates cell proliferation and tumorigenesis.
Keywords: USP22, cell cycle, FBP1, p21, ubiquitin
Introduction
Ubiquitin-specific protease 22 (USP22) is able to remove ubiquitin moieties from histones H2A and H2B in vitro (Zhang et al, 2008; Zhao et al, 2008). In higher eukaryotes, monoubiquitination of histone H2A on Lys 119 is mediated by the polycomb repressive complex 1 and is associated with gene silencing (Wang et al, 2004). Removal of this mark is needed for the reactivation of gene expression and proper cell-cycle progression (Joo et al, 2007). Given the ability of USP22 to deubiquitinate histones, its function was initially linked with regulation of gene transcription. USP22 was shown to affect the expression of c-Myc target genes, and its depletion leads to the accumulation of cells in the G1 phase of the cell cycle (Zhang et al, 2008). The deubiquitinating activity of USP22 was also shown to be important for androgen receptor-mediated transactivation in vivo (Zhao et al, 2008). Furthermore, USP22 has been identified as a member of an 11 gene ‘death-from-cancer’ signature that acts as a predictor of treatment resistance, tumour aggressiveness and metastatic probability in cancer patients (Glinsky, 2006; Liu et al, 2010).
Despite evidence about the function of USP22 in transcriptional regulation, cell-cycle progression and tumorigenesis, the mechanistic details of the way in which USP22 affects these processes remain unknown. We previously reported that USP22 participates in telomere maintenance by deubiquitinating non-histone telomere-associated proteins, such as telomeric-repeat-binding factor 1 (TRF1) (Atanassov et al, 2009). Given this observation, we sought to explore whether USP22 regulates other cellular processes, by altering the ubiquitination level of other chromatin-associated substrates. In a proteomic screen, we identified far upstream element (FUSE)-binding protein 1 (FBP1) as a protein with abundant ubiquitination upon USP22 ablation. Sequential binding of FBP family members—including FBP1, FBP2 and FBP3, as well as FBP-interacting repressor (FIR)—to FUSE sequences of target genes controls gene activation and repression (Chung et al, 2006; Liu et al, 2006). Proper timing of FBP1 binding to FUSE and its subsequent interaction with FIR ensures the accurate repression of target loci, such as c-Myc and p21 (Levens, 2008; Rabenhorst et al, 2009). Our results indicate that USP22 mediates deubiquitination of FBP1, which modulates its stable recruitment to target loci and affects the ability of FBP1 to regulate target gene expression.
Results And Discussion
USP22 regulates the ubiquitination levels of FBP1
His-tagged ubiquitin was overexpressed in USP22-depleted or control 293T cells. These cells were treated with the proteasomal inhibitor MG132 to stabilize the ubiquitinated proteins, and nuclear extracts were prepared in denaturing conditions. The ubiquitinated species obtained from both the USP22-depleted and control nuclear extracts were purified by Ni-NTA columns, and the eluted fractions were analysed by mass spectrometry for protein identification. The unique peptides purified from shUSP22 cells, but not shControl cells, provided candidates for proteins that have increased ubiquitination after USP22 loss (Fig 1A and Methods) and were considered potential USP22 substrates. Most of the proteins with abundant ubiquitination in USP22-depleted cells were transcriptional regulators (supplementary Table S1 online), including FBP1.
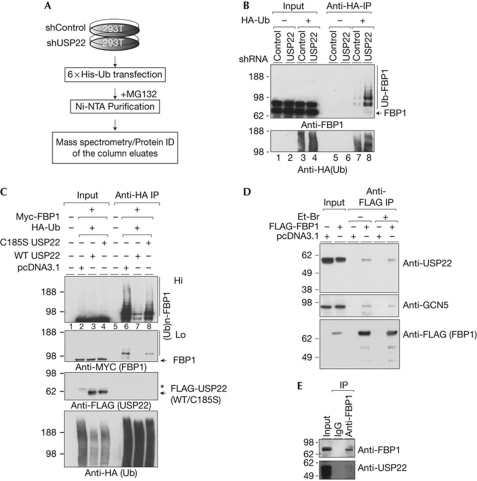
Figure 1.
USP22 targets FBP1 for deubiquitination. (A) Scheme for identification of substrates with abundant ubiquitination upon USP22 depletion. (B) Depletion of USP22 leads to increased FBP1 ubiquitination in 293T cells. shControl or shUSP22 293T cells were transfected with HA-Ub, ubiquitinated proteins were precipitated and blotted for FBP1. (C) Overexpression of WT but not catalytically dead (C185S) USP22 decreases FBP1 ubiqutination levels in vivo. The 293T cells were transfected with the indicated vectors, ubiquitinated proteins were purified with an anti-HA-resin, resolved by SDS–PAGE and blotted with the indicated antibodies. The asterisk indicates background bands. USP22 coprecipitates with FBP1. FLAG-FBP1 (D) or endogenous FBP1 (E) was purified, and purified fractions were analysed by immunoblot. Et-Br, ethidium bromide; FBP1, far upstream element (FUSE)-binding protein 1; HA, haemagglutinin; IgG, immunoglobulin G; IP, immunoprecipitation; Ni-NTA, nickel-nitriloacetic acid; shRNA, short-hairpin RNA; Ub, ubiquitin; USP22, ubiquitin-specific protease 22; WT, wild type.
FBP1 regulates the expression levels of key oncogenes and cell-cycle controllers, such as c-Myc and p21 (Levens, 2008; Rabenhorst et al, 2009), and its expression level correlates with tumour growth and survival prognosis in cancer patients (Malz et al, 2009; Rabenhorst et al, 2009). Furthermore, FBP1 protein levels are tightly controlled by ubiquitination. This protein is ubiquitinated in a JTV-1 (also known as AIMP2 or p38)- and Parkin-dependent manner (Kim et al, 2003; Ko et al, 2006). Given that USP22 is involved in Myc transformation (Zhang et al, 2008) and its expression levels correlate with tumour aggressiveness and patient survival prognosis (Glinsky, 2006; Liu et al, 2010), we decided to further investigate how USP22 affects the ubiquitination levels of FBP1.
To confirm the results obtained in the proteomic screen, we overexpressed haemagglutinin-tagged ubiquitin (HA-Ub) in USP22-depleted and control 293T cells. Ubiquitinated proteins were then purified using an anti-HA affinity matrix, and the precipitated fractions were analysed by immunoblot for FBP1 (Fig 1B). Polyubiquitination of FBP1 (Ub-FBP1) was detected in both control and USP22-depleted cells, but the level of FBP1 ubiquitination was enriched in USP22-depleted cells (Fig 1B, compare lanes 7 and 8). The same results were obtained when FLAG-tagged FBP1 and HA-Ub were coexpressed in USP22-depleted cells (supplementary Fig S1A online). These findings indicate that USP22 deubiquitinates FBP1. To further confirm this possibility, we coexpressed Myc-tagged FBP1 and HA-Ub with wild-type or catalytically inactive USP22 in 293T cells. Ubiquitinated proteins were purified and analysed by immunoblot for FBP1 (Fig 1C). As before, ubiquitination of FBP1 was detected when ectopic FBP1 and ubiquitin were coexpressed in the cells (Fig 1C, lane 6, upper panels). Expression of wild-type–USP22 in these cells, however, led to a decrease in Ub-FBP1 levels, whereas expression of catalytically inactive USP22 had no effect on Ub-FBP1 levels (Fig 1C, lanes 7 and 8). Expression of wild-type but not catalytically inactive USP22 was also able to reduce the ubiquitination levels of endogenous FBP1 (supplementary Fig S1B online). These results further confirm that USP22 targets Ub-FBP1 for deubiquitination.
Next, we tested whether USP22 interacts with FBP1. FLAG-tagged FBP1 was immunoprecipitated from 293T nuclear extracts, and the eluted complexes were analysed by immunoblot for the presence of USP22. Endogenous USP22 was detected in the FBP1 immunoprecipitate fractions (Fig 1D, upper panel). This experiment was conducted in the presence of ethidium bromide and Dnase I treatment (supplementary Fig S1C online), to eliminate the possibility that the interaction between USP22 and FBP1 is mediated by DNA. USP22 was also detected in the endogenous FBP1 precipitate fractions (Fig 1E). It is worth mentioning that GCN5 was also found in FBP1-precipitated fractions (Fig 1D, middle panel), suggesting that USP22 interacts with and deubiquitinates FBP1 as part of the SAGA complex.
USP22 depletion does not affect FBP1 stability
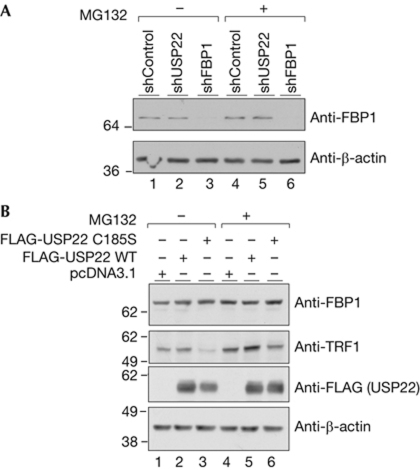
As USP22 affects FBP1 ubiquitination, we reasoned that its stability would be altered upon USP22 ablation. To test this, we monitored FBP1 protein levels in control and USP22-depleted cells (Fig 2A). Surprisingly, reduction of USP22 had no significant effect on FBP1 steady-state levels (Fig 2A, compare lanes 1 and 2), and inhibition of proteasomes by MG132 did not substantially change its cellular abundance (Fig 2A, lanes 4 and 5). This result was surprising, as USP22 depletion affects Ub-FBP1 levels. To further confirm the effect of USP22 on FBP1 steady-state levels, we overexpressed wild-type USP22 and catalytically inactive USP22 in USP22-depleted cells, and monitored FBP1 steady-state levels. Overexpression of neither of these USP22 affected FBP1 stability (Fig 2B, upper panel). However, as we reported previously (Atanassov et al, 2009), USP22 overexpression affects TRF1 protein stability, and this effect is abolished by inhibition of the proteasomes (Fig 2B, second panel, compare lanes 1 and 3, and 4 and 6).
Figure 2.
USP22 activity does not affect FBP1 steady-state level. (A) FBP1 steady-state levels were monitored by immunoblot in USP22-depleted or control cells before and after MG132 treatment. (B) FBP1 and TRF1 steady-state levels in USP22-depleted cells after transfection with the indicated vectors. Immunoblots were performed with the indicated antibodies. FBP1, far upstream element (FUSE)-binding protein 1; USP22, ubiquitin-specific protease 22; WT, wild type.
Finally, we used cycloheximide to inhibit protein synthesis in HeLa and MCF7 cells and monitored FBP1 stability over time. As a control for the cycloheximide treatment, we monitored the levels of p53, a protein that is known to have a short half-life (supplementary Fig S2A,C,D online middle panel, compare lanes 1–7 and 8–14). This experiment showed that depletion of USP22 (supplementary Fig S2B,E online) had no significant effect on FBP1 stability in these cells (supplementary Fig S2A,C,D online, upper panels, lanes 7–14 and Fig 2E).
USP22 modulates Lys 63-linked FBP1 ubiquitination
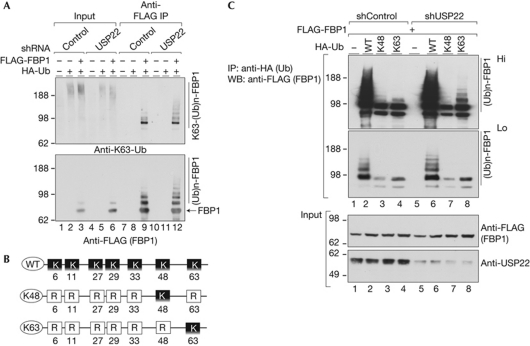
The minor effect of USP22 ablation on FBP1 protein stability, and the fact that Parkin is known to mediate Lys 63-linked polyubiquitination (Olzmann et al, 2007), prompted us to test whether FBP1 is Lys 63-polyubiquitinated. By using Lys 63-linkage-specific antibodies (Newton et al, 2008), we probed FBP1-purified from USP22-depleted or control cells after transfection with FBP1 and ubiquitin expression vectors. This experiment showed that at least a portion of FBP1 is Lys 63-polyubiquitinated. Most importantly, diminishing USP22 led to an enrichment of this ubiquitination (Fig 3A, upper panel, compare lanes 9 and 12). To further confirm these results, we transfected cells with constructs that express different ubiquitin mutants (Fig 3B) in combination with FLAG-FBP1. At 40 h after transfection of USP22-depleted or control cells with these vectors, cells were treated with MG132 for an additional 5 h and the Ub-FBP1 was precipitated (Fig 3C). As before, enhanced ubiquitination of FBP1 was detected upon USP22 reduction (Fig 3C, middle panel, compare lanes 2 and 6). Both Lys 48 and Lys 63-linked ubiquitinations were detected in control 293T cells (Fig 3C, upper panel, lanes 3 and 4). Interestingly, depletion of USP22 had no effect on Lys 48-linked ubiquitination levels (Fig 3C, upper panel, compare lanes 3 and 7), but did increase Lys 63-linked FBP1 ubiquitination (Fig 3C, upper panel, lanes 4 and 8). These results further confirm that USP22 modulates Lys 63-linked ubiquitination of FBP1.
Figure 3.
USP22 modulates Lys 63-linked FBP1 ubiquitination. (A) FLAG-FBP1 was purified from the indicated cells and blotted with the Lys 63-linkage-specific or FLAG (FBP1) antibody. (B) Schematic representation of the HA-ubiquitin expression vectors used in C. (C) Enhanced Lys 63-linked FBP1 polyubiquitination on USP22 depletion. Immunoblot of the HA-precipitated fractions after transfections with the indicated vectors. FBP1, far upstream element (FUSE)-binding protein 1; HA, haemagglutinin; IP, immunoprecipitation; shRNA, short-hairpin RNA; Ub, ubiquitin; USP22, ubiquitin-specific protease 22; WB, western blot; WT, wild type.
USP22 alters the expression of FBP1 target genes
After confirming that USP22 depletion leads to abundant K63-polyubiquitination of FBP1, we sought to understand whether this altered ubiquitination abrogates the abilities of FBP1 to regulate the expression of target genes. The transcriptional profile of FBP1-depleted cells has been reported previously (Chung et al, 2006), and we selected several genes that were shown to be affected upon FBP1 ablation for further analysis. For instance, the expression of matrix metalloproteinase 1 (MMP1) is enhanced upon FBP1 depletion (Chung et al, 2006). Myc was also previously described as a main target of FBP regulation (Liu et al, 2006) and, most recently, p21 was added to the list of FBP1-controlled genes (Rabenhorst et al, 2009). We prepared total RNA from control, USP22- and FBP1-depleted normal human fibroblasts or HeLa cells and performed quantitative reverse transcription–PCR (qRT–PCR) to measure the expression levels of these genes (Fig 4A,B). The results showed that those genes the expression of which was altered upon FBP1 reduction also showed significant changes upon USP22 ablation. Surprisingly, silencing of FBP1 or USP22 had no significant effect on global c-Myc levels in either cell line. Experiments with serum-starved HeLa cells and normal human fibroblasts, however, revealed an aberrant pulse of c-Myc expression upon serum addition in both USP22- and FBP1-depleted cells, compared with control cells (supplementary Fig S3A–C online). Thus, these results indicate that the abundant FBP1 ubiquitination observed after USP22 loss compromises the ability of FBP1 to regulate the expression of downstream target genes.
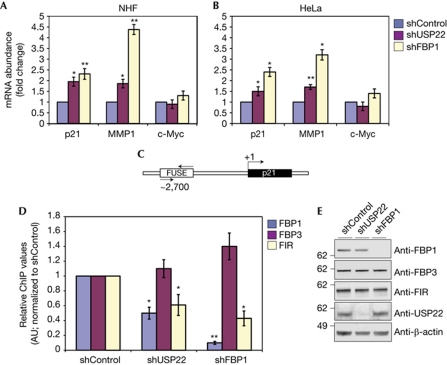
Figure 4.
USP22 depletion alters the expression of FBP1 target genes. The expression levels of p21, MMP1 and c-Myc in normal human fibroblasts (A) and HeLa cells (B) after the indicated depletions. (C) Schematic representation of the p21 FUSE region. Arrows indicate the region amplified in D. (D) ChIP analyses using the indicated antibodies in control, USP22- or FBP1-depleted HeLa cells. In A, B and D, error bars indicate s.e.m., n=3. Asterisks indicate the statistically significant differences (*P<0.05, **P<0.01) relative to control samples, as determined by Student's two-tailed t-test. (E) Immunoblot analysis of the indicated proteins in nuclear extracts purified from control, USP22- or FBP1-depleted HeLa cells. AU, arbitrary units; ChIP, chromatin immunoprecipitation; FBP1, far upstream element (FUSE)-binding protein 1; FIR, FBP interacting repressor; MMP1, matrix metalloproteinase 1; mRNA, messenger RNA; NHF, normal human fibroblast; USP22, ubiquitin-specific protease 22.
To gain further insights about the mechanism by which ubiquitination of FBP1 compromises its ability to regulate target gene expression, we used chromatin immunoprecipitation (ChIP) to monitor FBP1 occupancy at its target loci. FBP1 binds to FUSE on target genes and loads the FIR to these regions, which in turn leads to reduced expression of these genes (Liu et al, 2006; Cukier et al, 2010). Given that depletion of USP22 leads to elevated expression of p21, we tested whether USP22 depletion alters FBP1 occupancy and compromises FIR loading at the previously described p21 FUSE (Rabenhorst et al, 2009; Fig 4C,D). We found that FBP1 occupancy of the p21 FUSE is diminished in HeLa cells upon USP22 loss. Furthermore, FIR occupancy at this region was also compromised when USP22 was depleted. Conversely, FBP3—which binds to the FUSE before FBP1—occupancy was not altered in either USP22- or FBP1-depleted cells. Importantly, loss of USP22 did not alter the levels of FBP1, FBP3 or FIR proteins (Fig 4E). These data indicate that the abundant ubiquitination of FBP1 upon USP22 ablation compromises its ability to bind to FUSE and to load the FIR on these sites, leading to aberrant expression of target genes such as p21.
Impaired proliferation of USP22-depleted cells
The expression levels of USP22 are elevated in several malignancies (Glinsky, 2006; Liu et al, 2010), and its function is required for proper G1/S phase transition during the cell cycle (Zhang et al, 2008). Interestingly, the effect of USP22 on cell-cycle progression is p53 independent (Zhang et al, 2008). As our results indicate that USP22 affects expression of p21—an important p53 target gene—through regulation of FBP1 ubiquitination, we asked whether depletion of USP22 or FBP1 affects cell proliferation and p21 expression equally in wild-type p53 (MCF7) and p53-null (H1299) cells. These experiments showed impaired proliferation in both cell lines after depletion of either USP22 or FBP1 (Fig 5A,B), and fluorescence-activated cell sorting (FACS) analysis showed an accumulation of cells in the G1 phase of the cell cycle (supplementary Fig S4 online). In both cases, FBP1 depletion had a more-severe effect on cell proliferation than USP22 depletion. This difference probably reflects the fact that USP22 depletion affects the ubiquitination status of only a portion of the cellular pool of FBP1. As expected, qRT–PCR analysis showed upregulation of p21 in both cell lines, despite their p53 status (Fig 5C,D). The upregulation of p21 upon USP22 or FBP1 depletion was more prominent in MCF7 cells, which is not surprising given that p53 activates p21 expression. Nevertheless, absence of p53 did not abolish the effect of USP22 or FBP1 depletion on p21 expression levels or on cell proliferation. Simultaneous depletion of USP22 and FBP1 did not further increase the p21 expression level in MCF7 cells (supplementary Fig S5 online). To further confirm that the growth retardation after USP22 and FBP1 ablation is caused by the p21 upregulation, we monitored cell proliferation in MCF7 cells, in which USP22 or FBP1 and p21 were simultaneously depleted. As shown in Fig 5E, p21 reduction rescues the growth retardation in USP22-depleted cells and restores the growth kinetic of FBP1-silenced cells to levels close to control cells. The incomplete growth restoration in shFBP1-treated cells probably reflects the efficiency of p21 silencing (Fig 5F), whereas this silencing efficiency is sufficient to rescue the proliferation of USP22-depleted cells.
Figure 5.
Impaired cell proliferation on USP22 and FBP1 depletion. Impaired proliferation in MCF7 (A) and H1299 (B) cells after depletion of USP22 or FBP1. USP22 or FBP1 depletion leads to p21 upregulation in MCF7 (C) and H1299 (D) cells. Error bars in A–D indicate s.e.m., n=3. Asterisks in C and D indicate the statistically significant differences (P<0.05) between control samples and denoted depletions, on the basis of Student's two-tailed t-test. (E) Ablation of p21 rescues the growth retardation of USP22- or FBP1-depleted MCF7 cells. The data points from two independent experiments were plotted. (F) Efficiency of the USP22, FBP1 and p21 silencing in the cells used in E. FBP1, far upstream element (FUSE)-binding protein 1; mRNA, messenger RNA; shRNA, short-hairpin RNA; USP22, ubiquitin-specific protease 22.
On the basis of the results presented here, we propose a model in which USP22 function is required for FBP1 Lys 63-linked deubiquitination. Deubiquitination of FBP1 is needed for its proper binding to target loci, and ablation of USP22 abrogates this binding. Lower FBP1 occupancy then compromises the FUSE/FBP/FIR system and, as a result, leads to misregulation of target genes such as p21 and MMP1. A growing body of evidence indicates that overexpression of both FBP1 and USP22 occur in several types of tumour (Glinsky, 2006; Malz et al, 2009; Rabenhorst et al, 2009; Liu et al, 2010). Our data reveal a potential mechanism by which USP22 overexpression and FBP1 deubiquitination might affect tumour progression. Active repression of cell-cycle inhibitors such as p21 potentiates fast proliferation of tumour cells (Gartel & Radhakrishnan, 2005; Wilson et al, 2008); further, by deubiquitinating FBP1, USP22 contributes to this repression. We do not know how Lys 63-linked polyubiquitination, which is not associated with protein stability, affects FBP1 recruitment to its target loci. Perhaps this type of ubiquitination compromises the DNA-binding ability of FBP1 or perhaps K63 ubiquitination promotes FBP1 interaction with other proteins and/or complexes, which in turn prevent its loading at FUSE.
Our results also highlight the role of USP22 in deubiquitination of non-histone substrates. USP22 probably has additional substrates, including other transcriptional regulators, as indicated by our proteomic experiments. Future studies will confirm such substrates and will determine whether changes in their ubiquitination status influence cell-cycle regulation, transcriptional control, tumour development or cancer progression.
Methods
Antibodies. FBP1 (H-42 and Z37), FBP3 (I-20), β-actin (Santa Cruz), Myc-tag, p21 and GCN5 (Cell Signaling), haemagglutinin (Roche), FLAG (M2; Sigma), USP22 (Zhao et al, 2008), ubiquitin Lys 63-specific (clone Apu3; Millipore) and FIR (Proteintech group) antibodies were used.
Preparation of lysates. Nuclear extracts from cells transfected with the indicated expression vectors or blank cells were prepared in immunoprecipitation lysis buffer (10 mM Tris–HCl, pH7.9, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP40, 1 mg/ml sodium deoxycholate and 10% glycerol) containing protease inhibitors, 25 mM N-ethyl maleimide and 10 mM 2-iodoacetamide. To inhibit the proteasomal activity, cells were treated with MG132 (10 μM final concentration) for 5 h before collecting.
Immunoprecipitation. Between 3 and 5 mg of nuclear extracts were used in each immunoprecipitation reaction. FLAG-precipitated complexes were eluted with 100 μg/ml 3 × FLAG peptide (Sigma) in TBS. For the mass spectrometric analysis, ubiquitin-conjugated species were precipitated as follows: 6 × His-Ub-transfected cell nuclei were lysed in 6 M guanidine-HCl (pH 8.0), 0.1 M phosphate buffer (pH 8.0) containing protease inhibitors, N-ethyl maleimide and 2-iodoacetamide (10 mM final concentration). The lysates were briefly sonicated to shear DNA and reduce viscosity, and were subjected to ultracentrifugation for 1 h at 100,000 g at 4°C. Precleared supernatants were subjected to His-ubiquitin purification by Ni-NTA beads (Qiagen, catalogue number 30210) overnight on a rocking platform at 4°C. After washes, the ubiquitinated species were eluted with 250 mM imidazole, resolved on NuPAGE gel and subjected to mass spectrometric analysis.
ChIP. ChIP experiments were conducted according to the Upstate protocol enclosed to the ChIP kit (catalogue number 17-259). Between 15 and 20 × 106 HeLa cells were used per ChIP reaction.
Details about the expression vectors, mass spectrometry, qRT–PCR, immunoprecipitation and ChIP procedure are presented in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank C. Hirsch and M. Napierala for critical reading of the manuscript, and C. Hirsch, E. Koutelou and M. Wilson-Pham for their contributions to this work. We also thank D. Levens (National Institutes of Health) for sharing materials and for many helpful discussions about FBP1 functions and regulation and B. Wang (UTMDACC) for the HA-Ub plasmids. This work was supported in large part by a grant from National Institute of General Medical Sciences to S.Y.R.D., GM067718.
Footnotes
The authors declare that they have no conflict of interest.
References
- Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY (2009) Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell 35: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Liu J, Dundr M, Nie Z, Sanford S, Levens D (2006) FBPs are calibrated molecular tools to adjust gene expression. Mol Cell Biol 26: 6584–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukier CD, Hollingworth D, Martin SR, Kelly G, Diaz-Moreno I, Ramos A (2010) Molecular basis of FIR-mediated c-myc transcriptional control. Nat Struct Mol Biol 17: 1058–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Radhakrishnan SK (2005) Lost in transcription: p21 epression, mechanisms, and consequences. Cancer Res 65: 3980–3985 [DOI] [PubMed] [Google Scholar]
- Glinsky GV (2006) Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle 5: 1208–1216 [DOI] [PubMed] [Google Scholar]
- Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H (2007) Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449: 1068–1072 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Park BJ, Kang YS, Kim HJ, Park JH, Kang JW, Lee SW, Han JM, Lee HW, Kim S (2003) Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat Genet 34: 330–336 [DOI] [PubMed] [Google Scholar]
- Ko HS, Kim SW, Sriram SR, Dawson VL, Dawson TM (2006) Identification of far upstream element-binding protein-1 as an authentic Parkin substrate. J Biol Chem 281: 16193–16196 [DOI] [PubMed] [Google Scholar]
- Levens D (2008) How the c-myc promoter works and why it sometimes does not. J Natl Cancer Inst Monogr 39: 41–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Kouzine F, Nie Z, Chung HJ, Elisha-Feil Z, Weber A, Zhao K, Levens D (2006) The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J 25: 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Yang YM, Xu H, Dong XS (2010) Increased expression of ubiquitin-specific protease 22 can promote cancer progression and predict therapy failure in human colorectal cancer. J Gastroenterol Hepatol 25: 1800–1805 [DOI] [PubMed] [Google Scholar]
- Malz M et al. (2009) Overexpression of far upstream element binding proteins: a mechanism regulating proliferation and migration in liver cancer cells. Hepatology 50: 1130–1139 [DOI] [PubMed] [Google Scholar]
- Newton K et al. (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134: 668–678 [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS (2007) Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol 178: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenhorst U et al. (2009) Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatology 50: 1121–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Wilson AJ et al. (2008) HDAC4 promotes growth of colon cancer cells via repression of p21. Mol Biol Cell 19: 4062–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB (2008) The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell 29: 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y et al. (2008) A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29: 92–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.