Abstract
We reported previously that heat or ethanol shock in Saccharomyces cerevisiae leads to nuclear retention of most poly(A)+ RNA but heat shock mRNAs (encoding Hsp70 proteins Ssa1p and Ssa4p) are efficiently exported in a process that is independent of the small GTPase Ran/Gsp1p, which is essential for most nucleocytoplasmic transport. To gain further insights into proteins essential or nonessential for export of heat shock mRNAs, in situ hybridization analyses to detect mRNA and pulse-labeling of proteins were used to examine several yeast mutant strains for their ability to export heat shock mRNAs following stress. Rip1p is a 42-kD protein associated with nuclear pore complexes and contains nucleoporin-like repeat sequences. It is dispensable for growth of yeast cells under normal conditions, but we report that it is essential for the export of heat shock mRNAs following stress. When SSA4 mRNA was induced from a GAL promoter in the absence of stress, it was efficiently exported in a strain lacking RIP1, indicating that Rip1p is required for export of heat shock mRNAs only following stress. Npl3p, a key mediator of export of poly(A)+ RNA, was not required for heat shock mRNA export, whereas Rss1p/Gle1p, a NES-containing factor essential for poly(A)+ RNA export, was also required for export of heat shock mRNAs after stress. High-level expression of the HIV-1 Rev protein, but not of Rev mutants, led to a partial block in export of heat shock mRNAs following stress. The data suggest a model wherein the requirement for Npl3p defines the mRNA export pathway, the requirement for Rip1p defines a pathway used for export of heat shock mRNAs after stress, and additional factors, including Rss1p/Gle1p and several nucleoporins (Rat7p/Nup159p, Rat2p/Nup120p, and Nup145p/Rat10p), are required in both pathways.
Keywords: RNA export, heat shock, RIP1, hnRNP, RSS1/GLE1, Rev
A distinguishing feature of eukaryotic cells is the nucleus, a distinct subcellular compartment separated from the cytoplasm by the double-membraned nuclear envelope. Embedded within the nuclear envelope are nuclear pore complexes (NPCs) that serve as the only known channels for transport between the nucleus and the cytoplasm (for review, see Davis 1995; Panté and Aebi 1996). Transport of macromolecules through NPCs is signal-mediated, saturable, and energy dependent. Considerable progress has been made in recent years in identifying (1) receptor molecules that recognize nuclear localization signals (NLSs) within karyophilic proteins and mediate interactions between these proteins and NPCs, (2) a small Ras-like GTPase (Ran in metazoan cells and Gsp1p/Gsp2p in Saccharomyces cerevisiae) and its accessory proteins, which play a central role in nuclear protein import, and (3) distinct components of NPCs required for nuclear protein import (for review, see Görlich and Mattaj 1996; Hicks and Raikhel 1996; Corbett and Silver 1997; Nigg 1997). The existence of permeabilized cell systems has allowed direct study of nuclear protein import in vitro and played an important role in elucidating the molecular details of this process (Adam et al. 1990; Moore and Blobel 1992; Schlenstedt et al. 1993).
In contrast, we know considerably less about the export of macromolecules from the nucleus. The same GTPase system (Ran/Gsp1p) is believed to be required for this process, as defects in RNA export occur rapidly when yeast or mammalian cells carrying temperature-sensitive alleles of Gsp1p/Ran, its GTPase activating protein (Rna1p), or its nucleotide exchange factor (Prp20p) are shifted to the nonpermissive temperature (Amberg et al. 1992, 1993; Forrester et al. 1992; Kadowaki et al. 1992; Shiokawa and Pogo 1974; Wong et al. 1997). Proteins that associate with nascent pre-mRNAs within the nucleus and package them into heterogeneous nuclear ribonucleoprotein (hnRNP) particles contain nuclear export signals (NESs) (Michael et al. 1995). For example, the yeast hnRNP protein Npl3p has been shown to be a key mediator of RNA export signals (Lee et al. 1996). In addition, several yeast nucleoporins (Nup82p, Nup84p, Nup85p, Nup120p, Nup133p, Nup145p, Nup159p, and Npl4p) have roles specifically in RNA export, as strains harboring mutant alleles of some or disruptions of others show temperature-dependent defects in mRNA export but not in nuclear protein import (Doye et al. 1994; Fabre et al. 1994; Wente and Blobel 1994; Aitchison et al. 1995; Gorsch et al. 1995; Heath et al. 1995; Hurwitz and Blobel 1995; Kraemer et al. 1995; Li et al. 1995; DeHoratius and Silver 1996; Goldstein et al. 1996; Murphy et al. 1996; Siniossoglou et al. 1996; Dockendorff et al. 1997).
Competition experiments indicate that export from the nucleus of distinct classes of RNA molecules requires class-specific factors (Jarmolowski et al. 1994). The existence of such factors creates the potential to regulate nuclear transport so that only certain types of macromolecules are transported under certain conditions or to enable the transport of RNA molecules that would otherwise be retained in the nucleus. For example, HIV-1 Rev protein facilitates the export of intron-containing mRNAs (Fischer et al. 1994). The export activity of Rev requires the presence of a leucine-rich NES in Rev (Malim et al. 1991) and appears to target Rev response element (RRE)-containing and intron-bearing HIV-1 mRNAs to an export pathway that shares components with those required for export of U snRNAs and 5S RNA, but not mRNA, tRNA, or rRNA (Fischer et al. 1995; Fritz et al. 1995).
Yeast Rip1p is a 42-kD polypeptide identified as a two-hybrid interactor with HIV-1 Rev (Stutz et al. 1995). Similar two-hybrid screens led to the identification of the related mouse and human proteins, Rab/hRIP (Bogerd et al. 1995; Fritz et al. 1995). Yeast Rip1p contains a unique carboxy-terminal domain of ∼80 amino acids. The amino-terminal 75% of Rip1p contains many XXFG repeats and is most closely related to the repeat domain of the yeast nucleoporin Rat7p/Nup159p (Gorsch et al. 1995; Kraemer et al. 1995). This sequence similarity suggests that Rip1p may also be a nucleoporin, and nuclear rim staining was seen using antibodies to Rip1p (Stutz et al. 1995). Mammalian hRIP/Rab was detected primarily in the nucleoplasm (Fritz et al. 1995). It is not yet clear whether yeast Rip1p and hRIP/Rab are homologs or whether Rip1p is a component of NPCs or an NPC-associated protein.
RNA export is regulated following heat shock and other forms of stress in S. cerevisiae (Saavedra et al. 1996). Following stress, poly(A)+ RNA accumulates within yeast nuclei, but stress also induces transcription of heat shock genes. Synthesis of inducible heat shock proteins requires that these heat shock mRNAs be exported to the cytoplasm. We showed previously that SSA4 mRNA, encoding stress-inducible Hsp70, is efficiently exported following heat or ethanol shock and that its export is unaffected by mutations in components of the Ran/Gsp1 system (Saavedra et al. 1996).
Here we report the identification of factors required for export of heat shock mRNAs following stress in yeast. Rip1p is required, but no defect in export of mRNA was seen under normal growth conditions (23°C–37°C) in cells disrupted at RIP1. In the absence of stress in cells lacking Rip1p, export of SSA4 mRNA expressed from a GAL promoter occurred efficiently, indicating that export of SSA4 mRNA requires Rip1p only following stress. High-level expression in yeast cells of HIV-1 Rev, but not of a mutant form of Rev lacking a functional NES, partially prevented the nuclear export of heat shock mRNAs. Export of heat shock mRNAs after stress was not dependent on functional Npl3p but did require Gle1p/Rss1p, an NES-containing protein essential for nuclear export of poly(A)+ RNA (Del Priore et al. 1996; Murphy and Wente 1996) and also shown by two-hybrid analyses to interact with Rip1p (Murphy and Wente 1996). The data suggest a model wherein the requirement for Npl3p defines the mRNA export pathway, the requirement for Rip1p defines a distinct pathway used for export of heat shock mRNAs after stress, and additional factors, including Rss1p/Gle1p and several nucleoporins (including Rat7p/Nup159p, Rat2p/Nup120p, Nup145p), are required in both pathways.
Results
Rip1p is essential for nuclear export of heat shock mRNAs following cellular stress
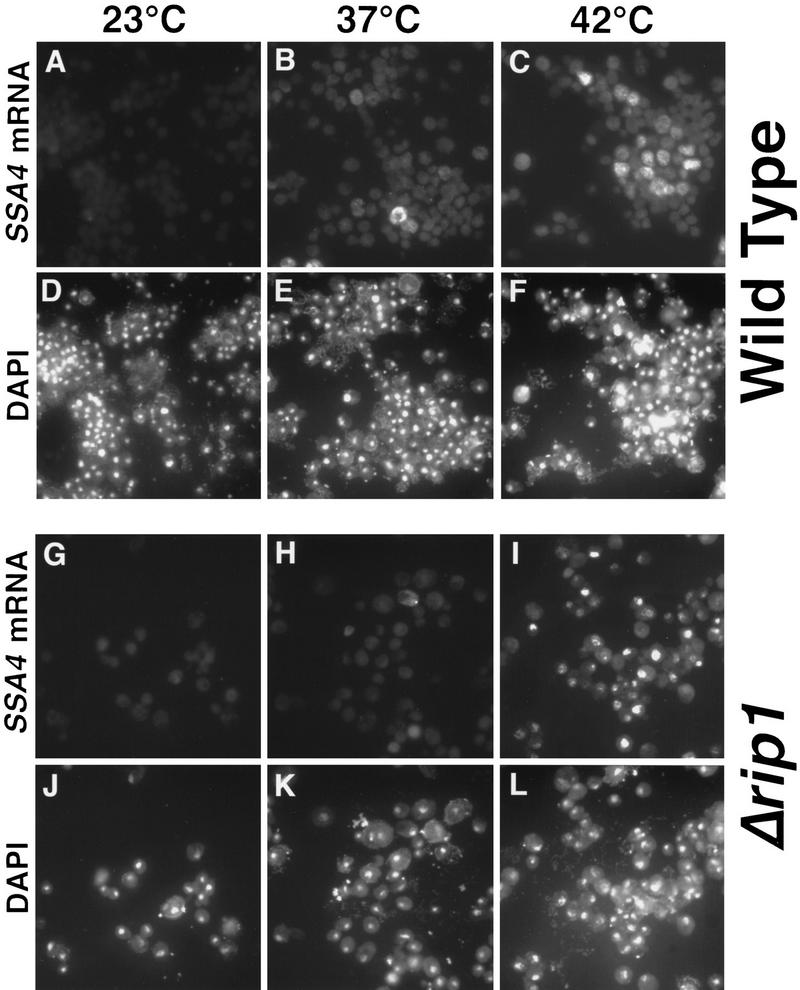
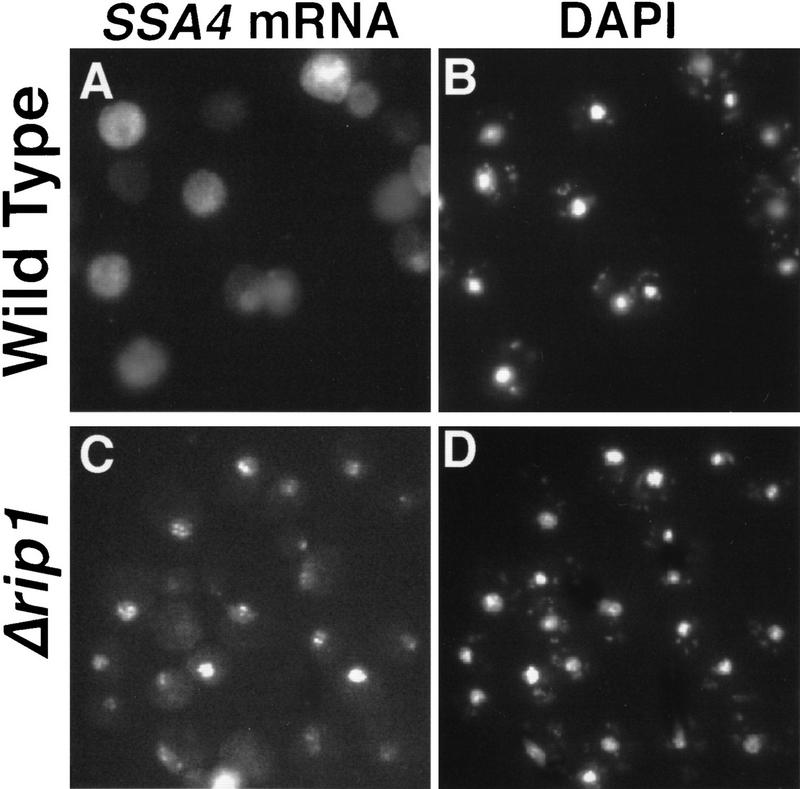
In a previous report, we showed that cellular stress (heat or ethanol shock) causes accumulation of poly(A)+ RNA within yeast nuclei but that heat shock mRNAs are transcribed and exported efficiently (Saavedra et al. 1996). During the course of those studies we learned that yeast cells lacking the small NPC-associated protein Rip1p are partially compromised for Rev-dependent export of RRE-containing RNAs but showed no defects in cellular growth or nuclear processes at either 23°C or 37°C (Stutz et al. 1995). We tested whether Rip1p might be involved in the selective export of heat shock mRNAs following stress by comparing the patterns of localization of poly(A)+ RNA and SSA4 mRNA (expressed from a high-copy plasmid and encoding stress-inducible Hsp70) in wild-type cells and in cells deleted for RIP1. Interestingly, SSA4 mRNA accumulated in nuclei of Δrip1 cells heat-shocked at 42°C for 1 hr (Fig. 1I). Very little SSA4 mRNA was visible when these cells where grown at 23°C (Fig. 1G) or shifted to 37°C (Fig 1H). Furthermore, in Δrip1 cells shifted to 42°C for 1 hr, the poly(A)+ RNA signal was exclusively nuclear (data not shown), whereas in wild-type cells shifted to 42°C, a faint signal for poly(A)+ RNA is detected in the cytoplasm following heat shock (Saavedra et al. 1996; data not shown) and most likely reflects the export of a variety of heat shock mRNAs produced following stress. We also examined the effect of ethanol shock on export of SSA4 mRNA at 23°C in Δrip1 cells (Fig. 2). Following ethanol shock, SSA4 mRNA accumulated within nuclei of Δrip1 cells, indicating that the requirement for Rip1p for export following stress does not merely reflect a high-temperature threshold for inactivation of the export function of NPCs. Together, these results provide evidence that Rip1p plays an essential role in the export of heat shock mRNA following stress.
Figure 1.
Rip1p is required for export of SSA4 mRNA following heat shock. Wild-type (FY23) or Δrip1 (CHY119) cells were maintained at 23°C (A,D,G,J) or shifted to 37°C (B,E,H,K) or 42°C (C,F,I,L) for 1 hr before processing for in situ hybridization to detect SSA4 mRNA (A–C, G–I). Cells were also stained with DAPI (D–F, J–L) to permit visualization of the DNA within nuclei. Shown are the same fields of cells examined by in situ hybridization or stained with DAPI. The SSA4 mRNA fluorescent signal was variable from cell to cell, most likely reflecting varying copy numbers of the SSA4 plasmid in different cells.
Figure 2.
Rip1p is required for export of SSA4 mRNA following ethanol shock. Wild-type (FY23) or Δrip1 cells (CHY119) were incubated at 23°C and shocked by addition of ethanol to 10% for 1 hr. Cells were processed for in situ hybridization and were also stained with DAPI to permit detection of DNA within nuclei. (A) Wild-type cells, in situ hybridization; (B) the same field of cells as in A stained with DAPI; (C) Δrip1 cells, in situ hybridization; (D) the same field of cells as in C stained with DAPI.
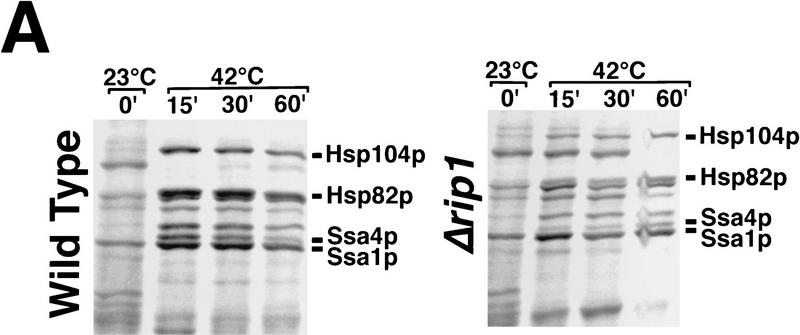
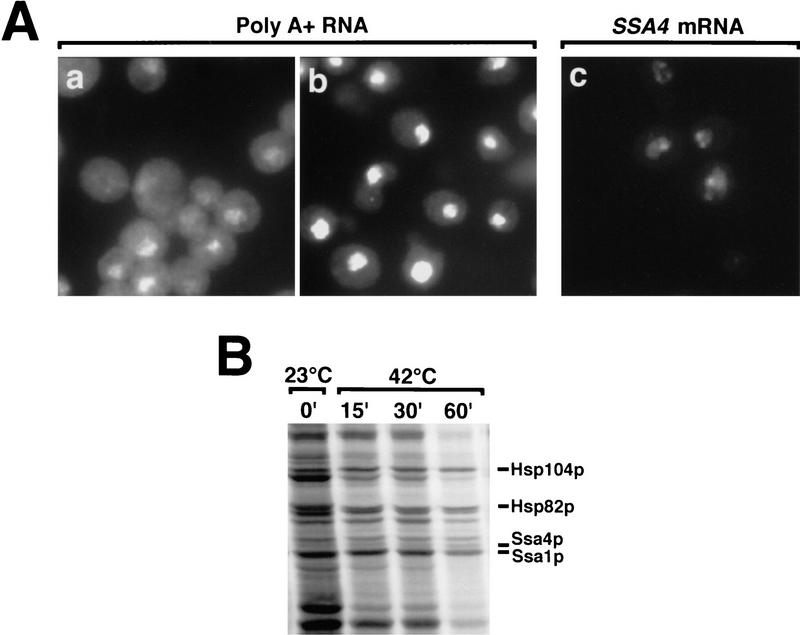
In our previous studies, we examined the trafficking of SSA4 mRNA and the closely related SSA1 mRNA following stress but did not examine expression of other heat shock genes. To determine whether Rip1p is required for the export of other classes of heat shock mRNA, we radiolabeled cells with [35S]methionine plus [35S]cysteine and compared the patterns of protein synthesis in wild-type and Δrip1 cells (Fig. 3A). Cells were labeled for 10-min periods prior to or at different times following heat shock. In wild-type cells, a range of heat shock proteins comprised the primary labeled products synthesized as early as 15 min after heat shock (Fig. 3A). Some of these reflect enhanced expression of proteins produced at lower levels under normal growth conditions (e.g., Ssa1p), whereas others represent heat shock proteins that are synthesized only or primarily following stress (e.g., Ssa4p, Hsp104p). Following heat shock, the background labeling, representing a broad spectrum of cellular proteins synthesized under normal growth conditions, virtually disappeared. In contrast, in Δrip1 cells a pattern of translation persisted after heat shock that resembles that seen in both wild-type and Δrip1 cells prior to stress but became less intense as cytoplasmic mRNAs present prior to heat shock turned over. Note particularly the dark band migrating just below Hsp104p in Δrip1 cells; this band was nearly undetectable in wild-type cells heat-shocked for 15 min but persisted during the heat shock period in Δrip1 cells.
Figure 3.
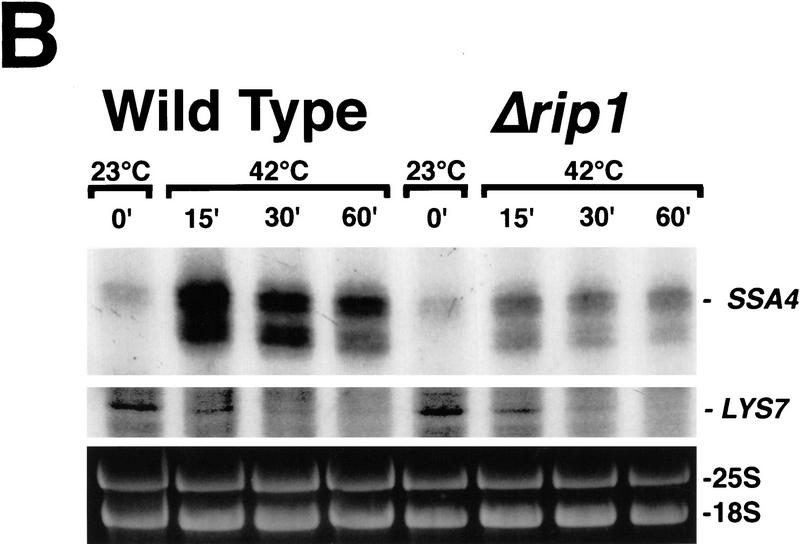
(A) Patterns of protein synthesis following heat shock. Wild-type cells and cells containing a disruption of RIP1 were grown at 23°C, processed as described in Materials and Methods, and radiolabeled for 10 min at 23°C or shifted to 42°C and labeled for the final 10 min of a 15-, 30-, or 60-min period of heat shock at 42°C. (B) Quantitation of specific mRNA levels by primer extension. SSA4 and LYS7 mRNA levels were examined in wild-type (lanes 1–4) and Δrip1 cells (lanes 5–8) immediately prior to heat shock (lanes 1,5) and following 15 (lanes 2,6), 30 (lanes 3,7), or 60 min (lanes 4,8) at 42°C. To ensure that all primer extension analyses were performed on RNA samples derived from approximately the same number of cells, rRNA levels in each sample were examined by electrophoresis of aliquots of each sample in a 1% agarose gel, which was stained with ethidium bromide and photographed.
Those bands that represent heat shock proteins produced constitutively continued to be translated during stress, reflecting their preferential translation. Heat shock proteins that reach high levels only following heat shock (e.g., Ssa4p and Hsp104p) were produced in Δrip1 cells at levels substantially lower than in wild-type cells and with different kinetics. Although the rates of synthesis of induced heat shock proteins in wild-type cells were declining within 60 min of a shift to 42°C, their levels were still increasing in Δrip1 cells. Although cells maintained at a very low density (e.g., OD600 < 0.10) contain essentially no SSA4 mRNA (Stutz et al. 1997), low levels of SSA4 mRNA are produced as cells reach higher densities, and it is probable that most of the Ssa4p synthesized following heat shock in Δrip1 cells derives from enhanced translation of SSA4 mRNA produced prior to heat shock. However, the increase in the rate of synthesis of Ssa4p 60 min after heat shock in Δrip1 cells may also reflect some export of SSA4 mRNA late in the heat shock period. The in situ hybridization results (Fig. 1) clearly show that most SSA4 mRNA is prevented from leaving the nucleus in Δrip1 cells. The data shown in Figure 3A indicate that the production of several other classes of heat shock proteins was dramatically reduced in Δrip1 cells, suggesting that Rip1p is important for the export of multiple classes of heat shock mRNA.
We also examined SSA4 mRNA levels using a primer extension assay (Fig. 3B). Analysis of rRNA levels in each sample (Fig. 3B) indicated that primer extension analyses were performed using approximately equal cell equivalents of RNA. Very low but detectable levels of SSA4 mRNA were present in both wild-type and Δrip1 cells grown at 23°C. SSA4 mRNA increased dramatically following the shift to 42°C, but to a lower extent in Δrip1 cells than in wild-type cells. This may reflect a lower stability for the SSA4 mRNA that accumulates in the nuclei of Δrip1 cells. As a control, we also analyzed LYS7 mRNA. This mRNA declined from similar levels and with essentially identical kinetics in wild-type and Δrip1 cells (Fig. 3B).
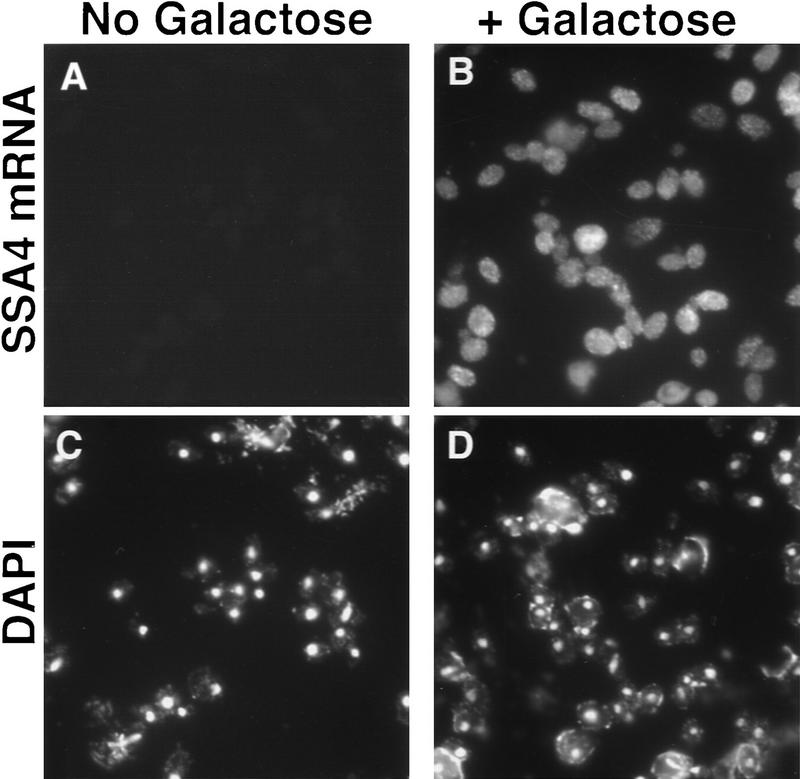
To determine whether export of SSA4 mRNA was dependent on Rip1p under all conditions or only following stress, we examined the localization of SSA4 mRNA expressed from the GAL1 promoter in Δrip1 cells. No SSA4 mRNA was detected when cells were grown on glucose (data not shown) or raffinose (Fig. 4A). When cells were incubated for 2 hr at 23°C in the presence of 2% galactose, SSA4 mRNA was present throughout the cell (Fig. 4B). This indicates that SSA4 mRNA can be exported from the nucleus by at least two pathways: at 23°C, Rip1p is not required for export of SSA4 mRNA, and this mRNA may be exported using the conventional poly(A)+ RNA export machinery; at 42°C, SSA4 mRNA export requires Rip1p (Fig. 1) and is also independent of the Ran/Gsp1p GTPase machinery (Saavedra et al. 1996).
Figure 4.
Export of SSA4 mRNA under nonstress conditions does not require Rip1p. Δrip1 cells, containing a CEN plasmid encoding Ssa4p under control of a GAL promoter, were grown on media containing 2% raffinose (A,C) or shifted to media containing 2% galactose for 30 min (B,D) before processing for in situ hybridization using an SSA4 probe (A,B). The same fields of cells shown in A and B were also stained with DAPI (C,D).
Because Rip1p appears to be a nucleoporin whose repeat region is not essential, we wondered whether disruption of RIP1 caused any abnormalities in NPC distribution that could be involved in transport defects. We compared wild-type cells and cells disrupted for RIP1 by indirect immunofluorescence using either an anti-Rat7p/Nup159p antibody or the RL1 monoclonal antibody that recognizes multiple repeat-containing yeast nucleoporins and observed a normal nuclear rim-staining pattern (C. Heath and C. Cole, unpubl.).
The essential yeast hnRNP protein Npl3p is not required for export of heat shock mRNAs
Npl3p is an essential mediator of the export of poly(A)+ RNA in yeast (Lee et al. 1996). To determine whether Npl3p is required for export of heat shock mRNAs, we examined the localization of poly(A)+ RNA and SSA4 mRNA in cells carrying the npl3-1 and npl3-17 alleles and expressing SSA4 from its own promoter from a high-copy plasmid. As reported previously, poly(A)+ RNA accumulated in the nuclei of npl3-1 cells shifted to 37°C for 1 hr (Fig. 5A, h), but the block to export was incomplete as some cytoplasmic signal remained. Following a shift to 42°C, poly(A)+ RNA was detected primarily in nuclei, indicating that less poly(A)+ RNA was exported in npl3-1 cells following heat shock than at the nonpermissive temperature of 37°C. Little or no SSA4 mRNA was detected at 23°C (Fig. 5A, a) or 37°C (Fig. 5A, b), whereas it was primarily cytoplasmic following heat shock at 42°C (Fig. 5A, c). This indicates that Npl3p is not required for efficient export of SSA4 mRNA following stress. Essentially identical results were obtained with the npl3-17 allele (data not shown), even though this is a tighter allele than npl3-1, and the defect in poly(A)+ RNA export at 37°C in this strain was more complete than in npl3-1 cells. We also examined the pattern of protein synthesis in heat-shocked npl3-17 cells (Fig. 5B) and found it to be indistinguishable from the wild-type pattern. We conclude that Npl3p is not required for the export of heat shock mRNAs following stress.
Figure 5.
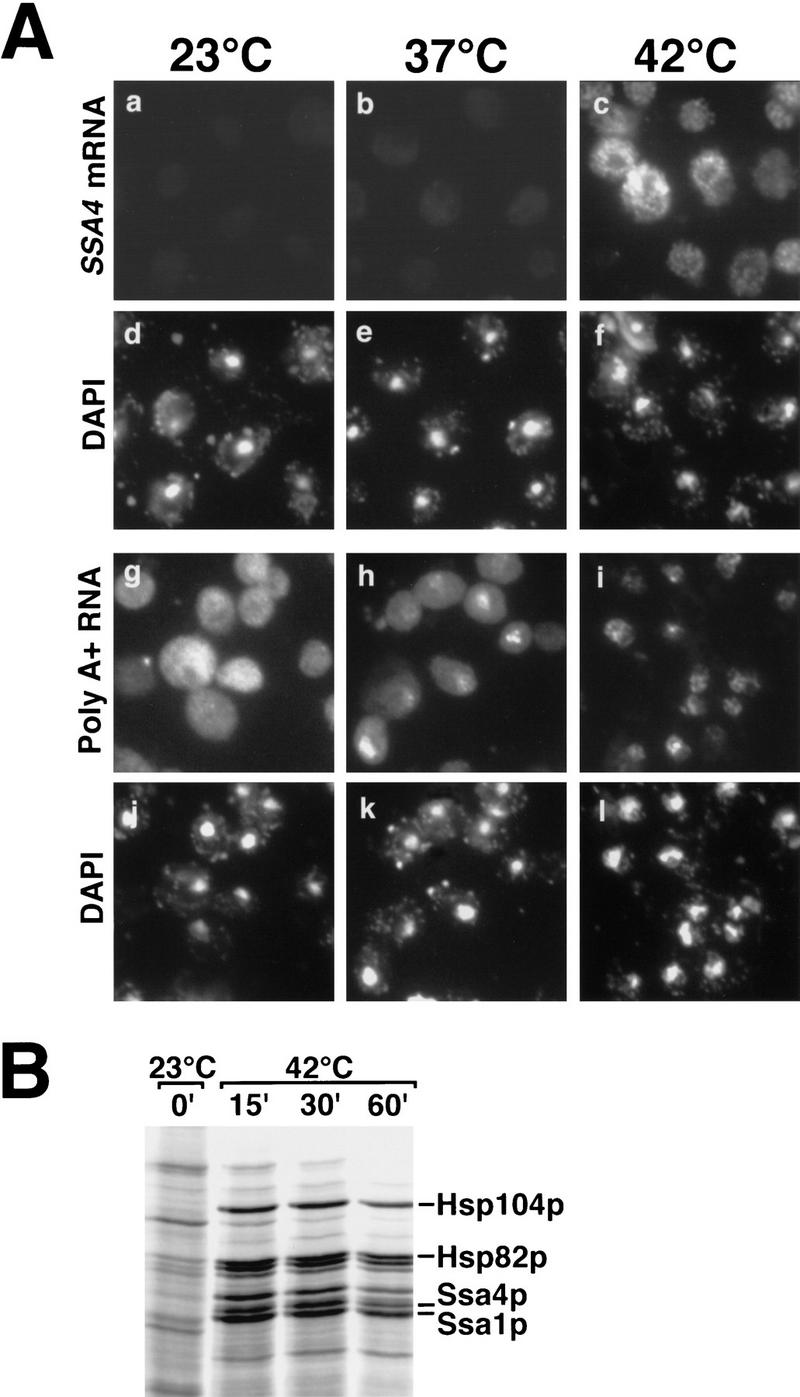
Npl3p is not required for export of heat shock mRNAs following heat shock. (A) In situ hybridization to detect SSA4 mRNA (a–c) and bulk poly(A)+ RNA (g–i) in npl3-1 cells incubated at 23°C or shifted to 37°C or 42°C for 1 hr. The same fields of cells analyzed by in situ hybridization were also stained with DAPI (d–f, j–l). (B) Pattern of protein synthesis in npl3-17 cells incubated at 23°C or shifted to 42°C for 15, 30, or 60 min.
High-level expression of HIV-1 Rev protein interferes with efficient export of SSA4 mRNA following stress in yeast
HIV-1 Rev possesses a nuclear export signal required for the export of Rev itself and RRE-containing mRNAs (Malim et al. 1991; Fischer et al. 1994, 1995). Microinjection into Xenopus oocyte nuclei of a high level of the leucine-rich Rev NES conjugated to bovine serum albumin (BSA) inhibited the nucleocytoplasmic export of 5S RNA and U small nuclear RNAs (snRNAs) but did not affect export of rRNA, tRNA, or poly(A)+ RNA (Fischer et al. 1995). Because export of SSA4 mRNA following stress is dependent on the presence of Rip1p (Fig. 1) and Rev functions to facilitate the export of RRE-containing mRNAs even in yeast (Stutz and Rosbash 1994), we examined the effects of high-level expression of HIV-1 Rev on poly(A)+ and SSA4 mRNA export in wild-type yeast cells. Plasmids encoding either wild-type Rev or the M10 export-defective Rev mutant, under control of the GAL10 promoter, were introduced into yeast cells carrying a high-copy SSA4 plasmid. The M10 Rev allele contains two missense mutations within the Rev activation domain/NES and does not facilitate the export of RRE-containing RNAs in either yeast (Stutz et al. 1995) or mammalian cells (Malim et al. 1991). Although Rev interacts efficiently with Rip1p and hRIP/Rab in two-hybrid assays (Bogerd et al. 1995; Fritz et al. 1995; Stutz et al. 1995), the ability of mutant Rev activation domains to function in vivo closely correlates with their ability to interact with hRIP/Rab/Rip1p in two-hybrid assays (Bogerd et al. 1995; Stutz et al. 1996).
We induced production of wild-type or M10 Rev by addition of 2% galactose for 4 hr. As assayed by Western blotting using an anti-Rev antiserum, both Rev and M10 were induced to comparable levels by this treatment (Fig. 6A). Following induction at 23°C, cells were shifted to 42°C for 30 min to induce transcription from the SSA4 promoter. Expression of wild-type Rev caused partial inhibition of the export of SSA4 mRNA (Fig. 6B, cf. panel G with panel C), whereas SSA4 mRNA export was normal in heat-shocked cells maintained in raffinose (Fig. 6B, panel C). Expression of the M10 mutant did not interfere with the export of SSA4 mRNA (Fig. 6B, panel O). This suggests that wild-type Rev is able to titrate an important mediator of SSA4 mRNA export following stress. We note that only a minority (∼30%) of cells expressing wild-type Rev show clear nuclear accumulation of SSA4 mRNA. Most likely, there is a correlation between Rev levels and the degree of accumulation of SSA4 mRNA in nuclei. Natural cell-to-cell variation in copy numbers for both the Rev and SSA4 plasmids likely results in substantial differences in actual Rev and SSA4 mRNA levels.
Figure 6.
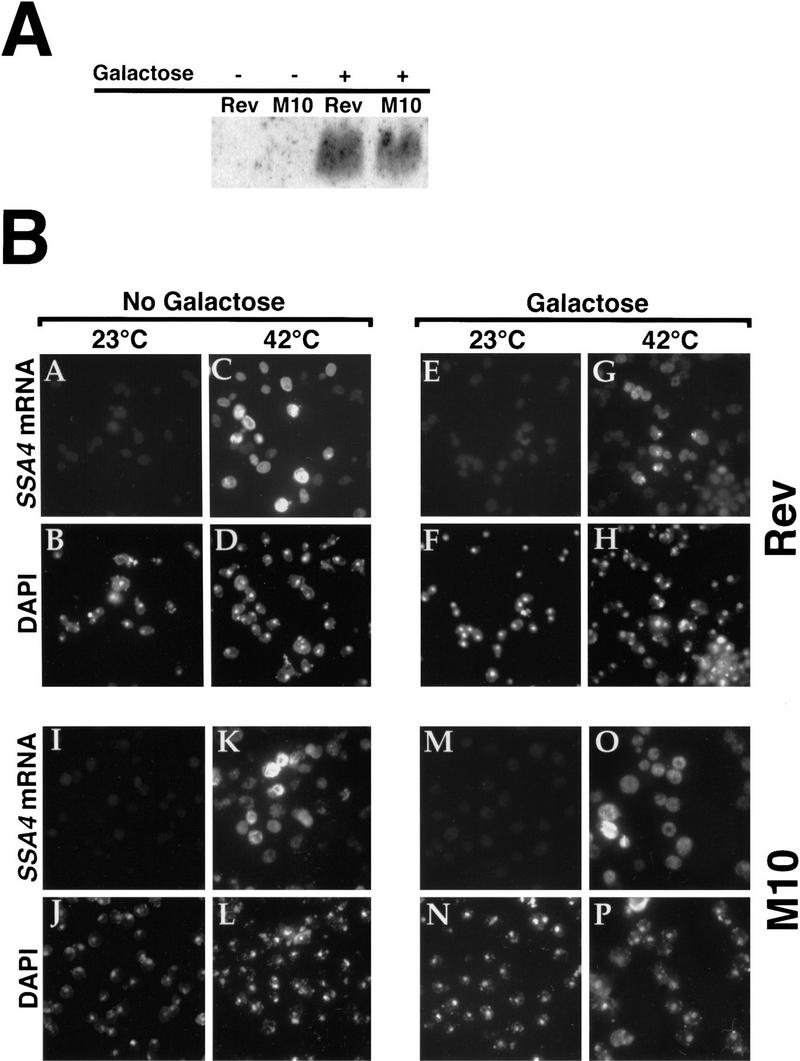
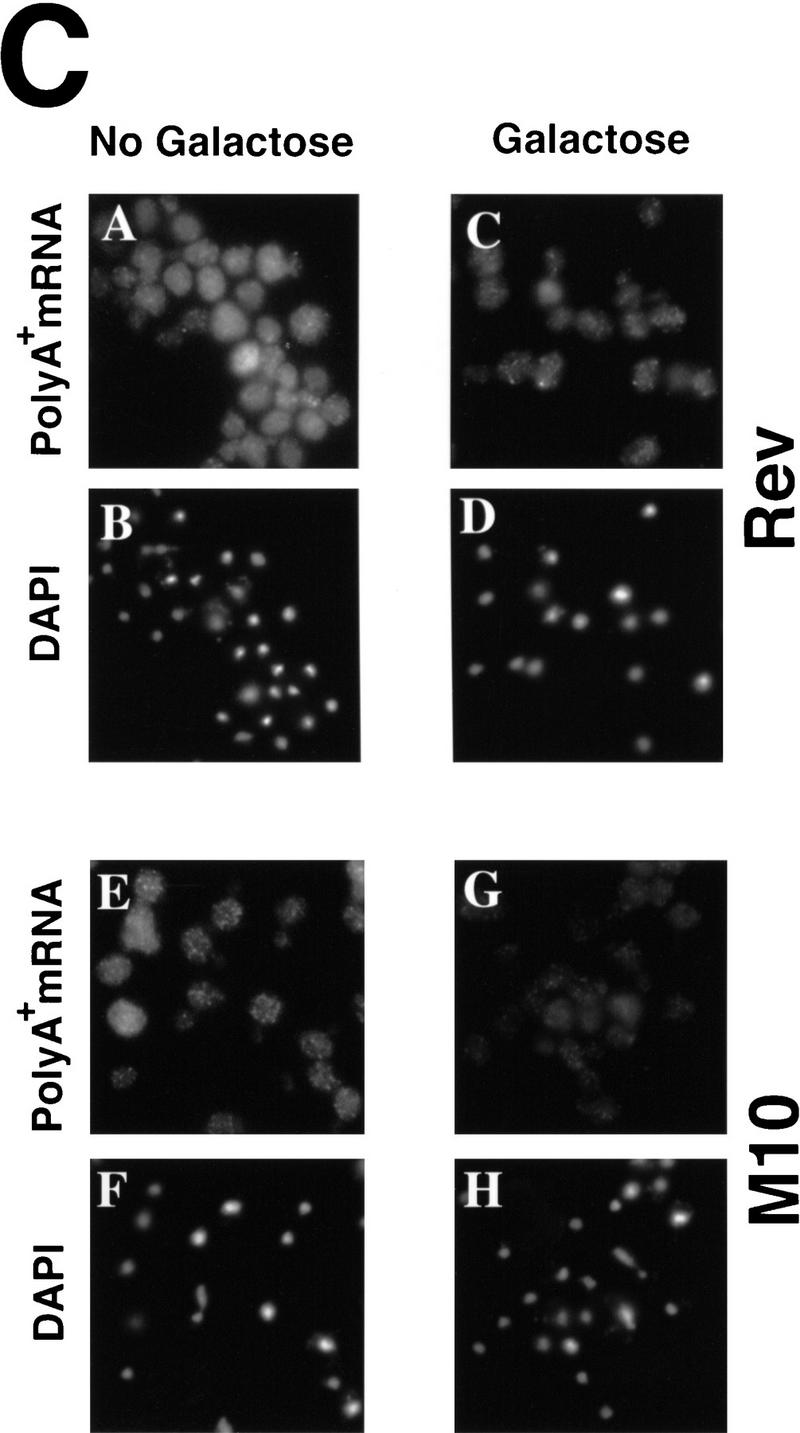
High-level expression of HIV-1 Rev interferes with export of heat shock mRNAs. Wild-type cells harboring plasmids encoding wild-type Rev or the M10 mutant of Rev, under control of a GAL promoter, were grown in media containing 2% raffinose. Expression of Rev or M10 was induced by shifting to 2% galactose for 4 hr. Subsequently, cells were shifted to 42°C for 30 min. (A) Western blot to examine levels of Rev and M10 protein. Extracts were prepared following 4 hr of incubation in media containing 2% galactose. (B) In situ hybridization to detect SSA4 mRNA (A,C,E,G,I,K,M,O). Cells were also stained with DAPI (B,D,F,H,J,L,N,P). (C) In situ hybridization to examine the distribution of poly(A)+ RNA in cells expressing high levels of Rev (A,C) or M10 (E,G). Cells were incubated overnight at 23°C in media containing 2% raffinose (A,E), shifted to media containing 2% galactose, and incubation was continued for 4 hr (C,G). Cells were then processed for in situ hybridization. The cells shown in A, C, E, and G were also stained with DAPI to show the location of the nucleus (B, D, F, and H, respectively).
We also examined the distribution of poly(A)+ RNA in cells (23°C) induced to express wild-type Rev or the M10 mutant from a GAL promoter. Induction of Rev and M10 expression for 4 hr caused no detectable alteration in the distribution of poly(A)+ RNA (Fig. 6C), demonstrating that the partial inhibition of SSA4 mRNA export in cells expressing a high-level of Rev and shifted to 42°C is not likely to be attributable to any general inhibition of RNA export by Rev.
Rss1p/Gle1p is required for export of both poly(A)+ RNA under normal conditions and heat shock mRNAs following stress
Previously, we identified Rss1p as a high copy suppressor of the rat7-1 mutation (Del Priore et al. 1996). Murphy and Wente (1996) also identified this protein (which they called Gle1p) and demonstrated that Rss1p/Gle1p contains a NES essential for its function. Strong nuclear accumulation of poly(A)+ RNA was seen when cells were depleted for Rss1p/Gle1p (Del Priore et al. 1996) or in cells carrying temperature-sensitive alleles of RSS1/GLE1 shifted to the nonpermissive temperature (Murphy and Wente 1996). To understand further the role of Rss1p, we prepared a series of temperature-sensitive alleles of RSS1 (V. Del Priore and C.N. Cole, unpubl.).
We examined mutant cells carrying the rss1-37 allele for their ability to export SSA4 mRNA when heat-shocked at 42°C (Fig. 7). There was modest nuclear accumulation of poly(A)+ RNA in mutant cells grown at 23°C (Fig. 7A, a) and strong accumulation of poly(A)+ RNA in the nuclei of 100% of the cells when they were shifted to 37°C for 1 hr (panel b). In this strain, SSA4 mRNA accumulated in the nuclei of cells shifted to 42°C (panel c), and there was little or no increase in the rate of synthesis of those heat shock proteins whose increased production requires export of newly transcribed heat shock mRNAs (Fig. 7B, cf. Fig. 3). This indicates that Rss1p is required for efficient export of RNAs both through the conventional mRNA export pathway and through the distinct pathway used for export of heat shock mRNAs following stress. We also examined the effect of loss of functional Rss1p/Gle1p on NPC distribution by comparing wild-type cells and cells carrying the rss1-37 allele. No changes in NPC distribution were seen in cells carrying the rss1-37 mutant allele incubated at 23°C or shifted to 37°C or 42°C for 1 hr (data not shown).
Figure 7.
Rss1p/Gle1p is required for export of heat shock mRNAs following stress. (A) In situ hybridization in rss1-37 cells. (a) poly(A)+ RNA in cells incubated at 23°C; (b) poly(A)+ RNA in cells shifted to 37°C for 1 hr; (c) SSA4 mRNA in cells heat-shocked at 42°C for 1 hr. (B) Pattern of protein synthesis in rss1-37 cells incubated at 23°C or shifted to 42°C for 15, 30, or 60 min. Radiolabeling occurred during the last 10 min of incubation.
Discussion
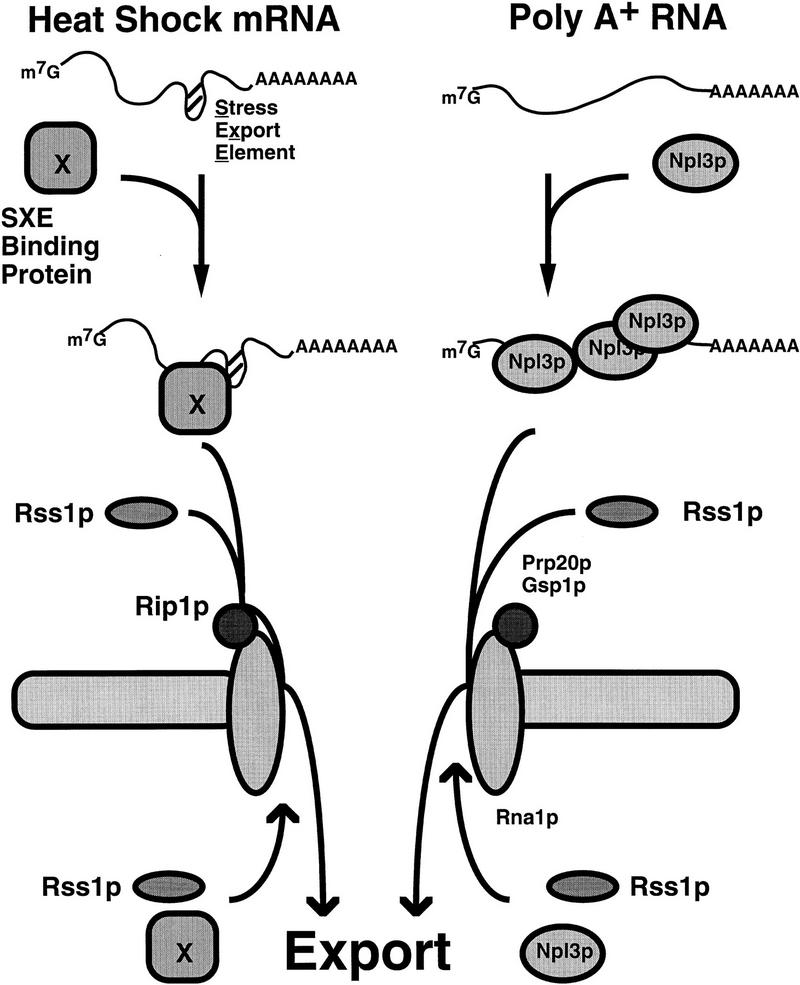
Cells respond to stress at multiple levels to enable them to adjust to rapid changes in their environment and to recover from stress when stress conditions have passed (for review, see Lindquist 1986; Jost and Lindquist 1988; Lindquist and Petersen 1990; Morimoto et al. 1994; Morimoto 1993; Panniers 1994; Sierra and Zapata 1994). The experiments presented in this paper indicate that stress-induced heat shock mRNAs are exported through a pathway that is independent of several but not all of the factors required for normal mRNA export. A model for these partially overlapping transport pathways is shown in Figure 8. In S. cerevisiae, export of heat shock mRNAs (Fig. 8, left), but not bulk poly(A)+ RNA (Fig 8, right), depends on the nucleoporin-like NPC-associated Rip1p (Fig. 1) but does not require Npl3p (Fig. 5), a key mediator of poly(A)+ RNA export (Lee et al. 1996) or components of the Ran/Gsp1p GTPase system (Saavedra et al. 1996). The NES-containing protein Rss1p/Gle1p is required for the export of both poly(A)+ RNA under nonstress conditions (Del Priore et al. 1996; Murphy and Wente 1996) and heat shock mRNAs following stress (Fig. 7). This factor may also have roles in the export of other classes of RNA. There are likely to be additional factors required for export of both bulk poly(A)+ RNA under normal conditions and SSA4 mRNA following stress, and these as yet unidentified factors may be required for export of other classes of RNA as well. In addition, there is likely to be at least one factor (X in Fig. 8) which binds specific elements in heat shock mRNAs and has a role analogous to that of Npl3p for export of poly(A)+ RNA under normal conditions. The same subset of nucleoporins required for export of poly(A)+ mRNA is also required for export of heat shock mRNAs following stress and may also be needed for export of other classes of RNA.
Figure 8.
Multiple overlapping pathways for nucleocytoplasmic export of RNA in S. cerevisiae. The pathway for export of poly(A)+ RNA is shown on the right, and for heat shock mRNAs after stress on the left. X is a hypothetical RNA binding, NES-containing protein that recognizes specific elements in heat shock mRNAs. The subset of nucleoporins essential for both general export of poly(A)+ RNA under normal conditions and for export of heat shock mRNAs after stress is not shown.
Previous evidence for the existence of multiple RNA export pathways comes primarily from studies in Xenopus oocytes, where it was shown that radiolabeled RNAs microinjected into nuclei were exported efficiently and that their export could be competed by unlabeled RNAs of the same class but not by RNAs of other classes (Jarmolowski et al. 1994). This indicates that, at least in Xenopus, the limiting export factors are specific for single classes of RNAs. Other studies focused on the HIV-1 Rev protein, a small RNA-binding protein that shuttles in and out of nuclei (Meyer and Malim 1994) and is able to facilitate the export of intron-containing mRNAs that contain sequences to which Rev can bind (RRE) (Fischer et al. 1994). BSA coupled to the Rev NES was exported following microinjection into Xenopus oocyte nuclei and was able to inhibit the export of coinjected U snRNA and 5S RNA but not mRNA, rRNA or tRNA (Fischer et al. 1995). These and other studies suggest that Rev directs intron-containing HIV-1 mRNAs to an RNA export pathway normally used for export of nonpoly(A)+ RNAs (Fridell et al. 1996; Fritz and Green 1996), most likely 5S RNA and U snRNAs. The machinery involved in Rev-dependent export appears to be conserved among distantly related organisms, as Rev functions in S. cerevisiae in a manner that is also dependent on RREs within RNAs and the presence of a functional NES within Rev (Stutz and Rosbash 1994).
In the studies reported here, we used two assays to monitor RNA export. The first involves in situ hybridization to detect poly(A)+ (Amberg et al. 1992) or specific mRNAs (Saavedra et al. 1996). In situ assays are quite sensitive for detection of partial defects in RNA export, as modest accumulation of RNA in nuclei is readily seen, even when cytoplasmic RNA levels remain high. However, this assay is insensitive for determining whether a block in RNA export is complete or partial, as nuclear signals for poly(A)+ RNA differ only modestly when there is a complete versus incomplete block in RNA export. In addition, it is difficult to distinguish between low concentrations of cytoplasmic RNA and background fluorescence in in situ hybridization (Fay et al. 1997).
The second assay uses pulse-labeling of proteins. This assay complements the in situ hybridization assay, as it readily detects increased production of heat shock proteins, even when there is a partial block in export of heat shock mRNAs. However, it provides no information about the extent to which heat shock mRNAs are accumulating in nuclei under various conditions. Using both assays to examine RNA distribution and protein synthesis in various mutant strains and under various environmental conditions has allowed us to determine whether various gene products are required for export of heat shock mRNAs following stress.
The radiolabeling assay also demonstrated that export of mRNAs encoding other classes of heat shock proteins (e.g., Hsp104p and Hsp82p) occurred preferentially following stress and was dependent on the same factors (Rip1p, Rss1p/Gle1p) and nucleoporins (Rat7p/Nup159p, Rat2p/Nup120p and Nup145p; Saavedra et al. 1996) as export of SSA4 and SSA1 mRNAs. Thus, heat shock mRNAs as a class appear to share an RNA export mechanism. Stutz and Rosbash observed a partial reduction in Rev-dependent export in yeast cells lacking Rip1p (Stutz and Rosbash 1994; Stutz et al. 1995), suggesting that Rev mediates export of RRE-containing RNAs through both Rip1p-dependent and Rip1p-independent pathways. Coupled with the observation that the Rev NES can inhibit export of U snRNAs and 5S RNAs in Xenopus, these results suggest that a Rip1p-dependent RNA export pathway is used for export of both heat shock mRNAs following stress and for other classes of RNA (perhaps 5S RNA) but that a separate pathway, independent of Rip1p, is available, at least under normal growth conditions, for export of those classes of RNA that use the Rip1p-dependent pathway during normal growth.
Inhibition of export of heat shock mRNAs by high-level expression of HIV-1 Rev
The partial inhibition of SSA4 mRNA export when Rev was expressed from a strong inducible promoter (Fig. 6) further strengthens the hypothesis that heat shock mRNAs exit the nucleus via a pathway that is distinct from that normally used for poly(A)+ RNA export. What might be the mechanism by which Rev expression partially inhibits export of heat shock mRNAs? Because wild-type Rev, but not the M10 mutant, interacts with Rip1p in the two-hybrid system, high-level expression of Rev could titrate Rip1p, thereby reducing export of heat shock mRNAs. However, it now appears that the interaction of Rev and Rip1p may be indirect, as two-hybrid interactions between Rev and Rip1p were eliminated by loss-of-function mutations in Crm1p (M. Neville et al. 1997). Most likely, Rev titrates one or more cellular NES-binding proteins. We tested whether overexpression of Rss1p/Gle1p, which contains a Leu-rich NES similar to Rev’s, would affect RNA export but saw no effect on heat shock mRNA export induced following a shift to 42°C or on poly(A)+ RNA export at 23°C or 37°C (C. Heath and C. Cole, unpubl.). Although Rev expression had no effect on export of poly(A)+ RNA within the first 4 hr of induction (Fig. 6C), cells containing either Rev or the M10 mutant under control of the GAL10 promoter failed to form colonies on galactose plates, indicating eventual growth inhibition by both Rev and M10 (C. Hammell and C. Cole, unpubl.). In contrast, little or no growth inhibition was seen when Rss1p was expressed from a 2μ plasmid, or when cells containing a galactose-inducible RSS1 plasmid were grown on galactose (Del Priore et al. 1996; C. Hammell, C. Heath, and C. Cole, unpubl.). Possibly Rss1p/Gle1p levels never reach those attained by Rev or M10. Alternatively, Rev may titrate a critical cellular factor that does not interact with Rss1p/Gle1p. Another possibility is that Rev interacts more strongly with a factor that also interacts with Rss1p/Gle1p.
Interestingly, Rss1p/Gle1p is required for both bulk poly(A)+ RNA export under normal growth conditions (Del Priore et al. 1996; Murphy and Wente 1996) and export of heat shock mRNAs after stress (Fig. 7). Because Rss1p/Gle1p contains an NES similar to that of HIV-1 Rev (Murphy and Wente 1996), this protein is most likely a soluble export factor that shuttles between the nucleus and the cytoplasm. Because ts mutations of RSS1 affect heat shock export dramatically (Fig. 7), we suspect that Rss1p/Gle1p plays a similar role in export of all mRNAs, and perhaps in export of other classes of RNA as well.
Following the observation of Stutz et al. (this issue), we found that rss1-37 was synthetically lethal with disruption of RIP1 (C. Heath and C. Cole, unpubl.). This means that in the presence of ts alleles of RSS1, Rip1p becomes essential for growth even at the permissive temperature (23°C). This could reflect a nonessential role for Rip1p during normal mRNA export. However, there are other possible explanations. We do not yet know whether Rss1p is important for export of other classes of RNA, and it could well be involved in all RNA export. Assays to examine export of other classes of RNA in S. cerevisiae have not yet been developed. Some RNAs could exit the nucleus using both Rip1p-dependent and Rip1p-independent pathways. If cells lack Rip1p, export solely through the other pathway may be insufficient to allow growth in the presence of mutant alleles of RSS1. Rip1p may be a structural component of NPCs, most likely anchored within NPCs through its unique carboxyl terminus. NPCs lacking Rip1p may be structurally altered so that export efficiency is compromised, perhaps through important binding sites on certain nucleoporins becoming less accessible. Under these conditions, mutations of Rss1p, or other factors important for RNA export, might readily lead to insufficient export under permissive conditions, and thus to the observed synthetic lethality. The observation that the carboxy-terminal domain of Rip1p, lacking all nucleoporin repeats, suppresses the synthetic lethality between Δrip1 and temperature-sensitive mutations of RSS1 (Stutz et al. 1997) is consistent with this explanation. Murphy and Wente (1996) isolated GLE1/RSS1 because of its synthetic lethality with a disruption of NUP100, which encodes a nonessential nucleoporin. We tested Δnup100 cells for defects in export of heat shock mRNA after stress and saw no differences from wild-type; the same result was obtained with disruption of NUP2, another nonessential nucleoporin (C. Heath, C. Hammell, and C. Cole, unpubl.).
Several important questions remain regarding differential export of mRNA following stress. One concerns the mechanism by which export of most poly(A)+ RNAs (Saavedra et al. 1996) and import of some nuclear proteins (Liu et al. 1996; Saavedra et al. 1996) are blocked. In addition, we do not know whether the pathway used for export of heat shock mRNAs following stress contains any components that must be activated for the pathway to function. Although we have reported that the Gsp1p/Ran system is not required for export of heat shock mRNAs after stress (Saavedra et al. 1996), recent observations in our laboratory indicate that this system remains functional under these conditions, so its inactivation does not appear to be part of the mechanism for altering nucleocytoplasmic transport following stress (C. Saavedra, C. Hammell, and C. Cole, unpubl.). Another question concerns the nature of the factors that recognize heat shock mRNAs and presumably play a critical role, analogous to that of Npl3p for poly(A)+ RNA, in mediating the export of heat shock mRNAs. Finally, it will be necessary to determine the identity of the factors that play the same role for export of heat shock mRNA that the Ran/Gsp1p system performs for export of most classes of RNA.
Materials and methods
Strains, plasmids, and growth conditions
Yeast strains are listed in Table 1. Except where noted, cells were grown to early exponential phase at 23°C in YPD-rich medium or in synthetic complete medium lacking leucine (SC − Leu) or uracil (SC − Ura) (Rose et al. 1989). For induction of the heat shock response, cells in the exponential phase of growth (5 × 106 to 2 × 107 cells/ml) were transferred to water baths at the appropriate temperature. In some experiments, the stress response was also induced by the addition of ethanol to 10% (vol/vol). Strains FY23 and FY86, derived from S288C, were used as wild types (Winston et al. 1995). Yeast transformations were performed by electroporation using a Gene-Pulser (Bio-Rad Laboratories, Melville, NY); cells were allowed to recover in rich medium containing 1 m sorbitol at 23°C for at least 1 hr before plating. E. coli strain DH5α was used for most cloning procedures. Plasmids used in these studies are listed in Table 2. A strain containing an integrated copy of the npl3-17 allele was generously provided by Margaret Lee and Pamela Silver (Dana Farber Cancer Institute, Boston, MA).
Table 1.
Yeast strains used in this study
| Strain
|
Genotype
|
Source
|
|---|---|---|
| FY23 | MATa trp1Δ63 leu2Δ1 ura3-52 | Winston et al. (1995) |
| FY86 | MATα his3Δ200 leu2Δ1 ura3-52 | Winston et al. (1995) |
| ACY1 | MATα/MATa his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 ura3-52/ura3-52 trp1Δ63/TRP1 | Anita Corbett (Dana Farber Cancer Institute, Boston, MA) |
| CHY119 | MATα rip1::HIS3 his3Δ200 ura3-52 leu2Δ1 | this study |
| PSY361 | MATα npl3-1 ura3-52 leu2-3,112 trp1-Δ901 ade2-101 his3 Δ200 his4-519 lys2-801 | Pamela Silver (Dana Farber Cancer Institute, Boston, MA) |
| PSY1026 | MATα np13-17 ade2-101 can1-100 his3Δ200 leu2Δ1 lys1-1 ura3-52 ade8 | Margaret Lee (Dana Farber Cancer Institute, Boston, MA) |
Table 2.
Plasmids used in this study
| Plasmid
|
Markers
|
Comments
|
Source
|
|---|---|---|---|
| YIplac211 | URA3 AmpR | integration vector | |
| YCplac33 | ARS/CEN URA3 AmpR | low-copy episomal vector | |
| YCplac111 | ARS/CEN LEU2 AmpR | low-copy episomal vector | |
| pVDP21 | RSS1 ARS/CEN LEU2 AmpR | Del Priore et al. (1996) | |
| pVDP29 | rss1-37 ARS/CEN LEU2 Amp | temperature-sensitive allele of RSS1 | Cole laboratory |
| pEC702 | 2μ LEU2 AmpRSSA4 | SSA4 gene in Yep351 | E. Craig (University of Wisconsin, Madison) |
| (GAL1-SSA4)2 | 2μ LEU2-d AmpR TetR | Gal-inducible SSA4 | E. Craig |
| pDAD-1 | 2μ URA3 AmpR TetR | GAL1 promoter vector, polylinker PHO5 terminator | G. Fink (Whitehead Institute, Cambridge, MA) |
| pCS120 | HIV-1 Rev open reading frame in pDAD-1 | Gal-inducible Rev expression vector | this study |
| pCS121 | HIV-1 M10 mutant open reading frame in pDAD-1 | Gal-inducible M10 expression vector | this study |
Construction of a strain disrupted for RIP1
A PCR-based gene deletion approach (Baudin et al. 1993) was used to disrupt the RIP1 gene. We generated a HIS3 cassette by PCR amplification that was flanked at its ends by 45 nucleotides identical to sequences just upstream and downstream of the RIP1 open reading frame (ORF). The upstream oligonucleotide sequence was 5′-GTAATGTCAGCTTTCGGTAACCCATTCACGTCGGGTGCTAAGCCTGGCCTCCTCTAGTACACTC-3′ and the downstream oligonucleotide sequence was 5′-CACTTACTATGCAACCAATGCAGGTGGTGGAGGTATATCAGGGTCGCGCGCCTCGTTCAGAATG-3′. For both oligonucleotides, the last 19 nucleotides are homologous to the HIS3 selectable marker. We used 20 ng of the HIS3-containing plasmid pBM2815 (obtained from P. Silver, Dana Farber Cancer Institute, Boston, MA) to generate the deletion construct in the PCR reaction. The 50 μl PCR reaction consisted of 1× PCR buffer (10 mm Tris-HCl, 15 mm MgCl2, 500 mm KCl at pH 8.3), 0.8 mm dNTP (0.2 mm each dATP, dGTP, dCTP, and dTTP), 1 μm of the upstream and downstream oligonucleotides, and 2 units of Taq DNA polymerase (Boehringer Mannheim Biochemicals, Indianapolis, IN). The reaction was subjected to a 30-cycle amplification consisting of 1.5 min at 94°C, 2.0 min at 50°C, and 2.0 min at 72°C. The PCR product was transformed into a wild-type haploid strain FY86, containing the deletion allele his3Δ200. The flanking upstream and downstream sequences target the deletion construct to the RIP1 such that its ORF is replaced with HIS3 by homologous recombination. Recombinants were selected on SC − His plates, and recombination was verified by PCR.
Construction of plasmids for inducible expression of HIV-1 Rev and M10 mutant Rev
Plasmids containing the ORFs of Rev and M10 mutant Rev, fused to glutathione S-transferase, were obtained from Michael Malim (University of Pennsylvania, Philadelphia). The ORFs of wild-type and M10 mutant Rev were amplified by PCR, and the resulting 350-bp DNA fragments were subcloned into pDAD-1 between the HindIII and EcoRI sites in its polylinker region, thereby placing Rev and M10 Rev under control of the yeast GAL10 promoter.
Construction of the RSS1 temperature-sensitive alleles
The RSS1 temperature-sensitive alleles were made using PCR mutagenesis (Cadwell and Joyce 1992). The RSS1 gene between base pair 246 and 1966 was PCR amplified using the following primers: 5′-GGTATATAATTGATAACAAGAGAATCGTCGGGCCGTTGCTACAGGATATTC-3′ and 5′-GTGTATTAGCATTTTTAATGTGCATATATAAGTTCAGAATTTTC-3′. The following changes were made to the standard PCR conditions: 7 mm MgCl2, 0.5 mm MnCl2, 5 units of Taq polymerase, 0.2 mm dGTP, 0.2 mm dATP, 1 mm dCTP, and 1 mm dTTP. These changes led to enhanced misincorporation of nucleotides. pVDP21 was digested with XhoI(318) and BspEI(1909), and the gapped vector was gel-isolated. A mixture of the vector and the PCR fragment was transformed into VDPY111, and the transformants were plated on SC − Leu medium. Colonies that came up on the SC − Leu plates were replica plated onto 5-FOA containing medium to select for loss of the plasmid-borne wild-type copy of RSS1. The colonies that grew on 5-FOA were replica plated to SC − Leu plates and incubated at 23°C and 37°C. Mutants that grew at 23°C but not at 37°C were selected for further studies.
In situ hybridization assays
In situ hybridization assays to detect poly(A)+ RNA or SSA4 RNA were performed as described previously (Amberg et al. 1992; Gorsch et al. 1995; Saavedra et al. 1996).
In vivo protein labeling
Cells were grown at 23°C in SC − Met and containing 0.85 grams/liter of yeast nitrogen base without amino acids and ammonium sulfate and 3.3 grams/liter of yeast nitrogen base without amino acids. Cells were grown to an OD600 of 0.5, collected by centrifugation, washed, and resuspended in one tenth the original volume in medium lacking sulfate. After incubation for 10 min at 23°C, 1-ml aliquots were either maintained at 23°C or shifted to 42°C for 15, 30, or 60 min. Cultures were radiolabeled by addition of 100 μCi/ml of Trans-35S-label (ICN Pharmaceuticals) for the final 10 min of the incubation period. Incorporation of radiolabel was stopped by the addition of 9 ml of ice-cold sodium azide (20 mm). Samples were then washed in sodium azide and resuspended in 0.2 ml of buffer A (Bram and Kornberg 1985). Cells were lysed with 0.75 grams of 0.45-mm glass beads. Supernatant was removed and centrifuged at 12,000g for 20 min. Total soluble protein was measured using a BioRad protein assay kit. Five micrograms of total soluble protein was boiled for 3 min in 5 × loading buffer (Bram and Kornberg 1985) and fractionated on a 7.5% SDS–polyacrylamide gel (Laemmli 1970). Gels were then dried and exposed to a Phosphor screen (Molecular Dynamics) and imaged using ImageQuant software (Molecular Dynamics).
Western analyses
Cells were grown to a total OD600 of 0.75, pelleted by centrifugation at 2500 rpm for 2.5 min, and washed once with water. Two hundred micrograms of acid-washed glass beads and 200 μl of hot sample buffer (62.5 mm TrisCl at pH 6.8, 2% SDS, 10% glycerol, 8 m urea, 0.72 m 2-mercaptoethanol, 0.05% bromophenol blue) were added to the cell pellets. After brief vortexing, cells were transferred to a boiling water bath for 3 min. Cells were then lysed as follows: 10 sec vortexing and 50 sec incubation in the boiling water bath, repeated five times over 5 min. The supernatant was then transferred to a clean Eppendorf tube, and equal volumes of cell lysate were loaded onto a 10% polyacrylamide–SDS gel. Electrophoresis was carried out at 200 V for ∼50 min. Proteins in the gel were transferred to a PVDF membrane by electroblotting overnight at 100 mA in a cold room. The membrane was washed briefly with 1× phosphate-buffered saline (PBS), 0.1% Tween 20 solution (solution A), and then blocked for 1 hr with 1× PBS, 0.1% Tween 20, and 5% nonfat milk solution (solution B). The membrane was then incubated with anti-Rev antiserum (kindly provided by Dr. Michael Malim, Howard Hughes Medical Institute, University of Pennsylvania, Philadelphia) at a 1:5000 dilution in solution B for 2 hr followed by four washes of 10 min each with solution A. The membrane was then incubated with anti-rabbit antibody coupled to horseradish peroxidase (Amersham, Inc., Arlington Heights, IL), diluted 1:3000 in solution B for 1 hr. Washes were performed as described above. Signal was developed with the ECL kit (Amersham, Inc.) according to the manufacturer’s directions.
Analysis of SSA4 mRNA levels by primer extension
Both wild-type and Δrip1 cells were grown to an OD600 of 0.5. RNA extractions were performed as described by Pikielny and Rosbash (1985). The total RNA isolated from each sample was dissolved in 20 μl of distilled water. Identical aliquots from each sample were subjected to electrophoresis in a 1% agarose gel and stained with ethidium bromide to permit visualization of rRNAs. A primer with the sequence GTTGTACCTAAATCAATACC was used to specifically measure SSA4 mRNA levels. LYS7 mRNA levels were measured with primer AGGGGCTACTGAGCTTTCCA. Primer extension assays were performed using 6 μl of each isolated RNA sample. Both primers were added to each RNA sample and primer extension performed as described (Pikielny and Rosbash 1985). Extension products were analyzed on a 12% denaturing polyacrylamide gel. The relative levels of extension products were quantified using a Molecular Dynamics PhosphorImager using ImageQuant software.
Acknowledgments
We thank Elizabeth Craig, Margaret Lee, and Pamela Silver for plasmids and strains; Michael Malim for antibodies and plasmids; Veronica Del Priore for the rss1-37 temperature-sensitive mutant strain; Francoise Stutz and Michael Rosbash for discussions and sharing results of their experiments prior to publication; and members of our laboratory for discussions and advice. We thank Pamela Silver and Hildur Colot for critical review of the manuscript. This work was supported by a grant from the National Institute of General Medical Sciences, National Institutes of Health to C.N.C. (GM33998) and by a core grant to the Norris Cotton Cancer Center, Dartmouth Hitchcock Medical Center, from the National Cancer Institute (CA-16038).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL charles.cole@dartmouth.edu; FAX (603) 650-1128.
References
- Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Nup120p: A yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol. 1995;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DA, Goldstein AL, Cole CN. Isolation and characterization of RAT1, an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Fleischmann M, Stagljar I, Cole CN, Aebi M. Nuclear PRP20 protein is required for mRNA export. EMBO J. 1993;12:233–241. doi: 10.1002/j.1460-2075.1993.tb05649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Madore S, Cullen BR. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- Bram RJ, Kornberg RD. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci. 1985;82:43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods & Applic. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- DeHoratius C, Silver P A. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol Biol Cell. 1996;7:1835–1855. doi: 10.1091/mbc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Priore V, Snay CA, Bahr A, Cole CN. Identification of RSS1, a high-copy extragenic suppressor of the rat7-1 temperature-sensitive allele of the Saccharomyces cerevisiae RAT7/NUP159 nucleoporin. Mol Biol Cell. 1996;7:1601–1621. doi: 10.1091/mbc.7.10.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff TC, Heath CV, Goldstein AL, Snay CA, Cole CN. C-terminal truncations of the yeast nucleoporin Nup145p produce conditional mRNA export defects and alterations to nuclear structure. [Published erratum in Mol. Cell. Biol. 17: 2347–2350] Mol Cell Biol. 1997;17:906–920. doi: 10.1128/mcb.17.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EC. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- Fay FS, Taneja KL, Shenoy S, Lifshitz L, Singer RH. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A) Exp Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- Fischer U, Meyer S, Teufel M, Heckel C, Lührmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Forrester W, Stutz F, Rosbash M, Wickens M. Defects in mRNA 3′-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: Possible coupling of mRNA processing and chromatin structure. Genes & Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Bogerd HP, Cullen BR. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz CC, Green MR. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- Fritz CC, Zapp ML, Green MR. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, Snay CA, Heath CV, Cole CN. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol Biol Cell. 1996;7:917–934. doi: 10.1091/mbc.7.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Mattaj IW. Protein kinesis—Nucleocytoplasmic transport. Science. 1996;271:1512–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:9399–9355. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutations of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GR, Raikhel NV. Protein import into the nucleus: An integrated view. Annu Rev Cell Biol. 1996;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- Hurwitz ME, Blobel G. NUP82 is an essential yeast nucleoporin required for poly(A)+ RNA export. J Cell Biol. 1995;130:1275–1281. doi: 10.1083/jcb.130.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost HJ, Lindquist S. Translation of unspliced transcripts after heat shock. Science. 1988;242:1544–1548. doi: 10.1126/science.3201243. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Zhao Y, Tartakoff AM. A conditional yeast mutant deficient in mRNA transport from nucleus to cytoplasm. Proc Natl Acad Sci. 1992;89:2312–2316. doi: 10.1073/pnas.89.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee M, Henry M, Silver PA. A protein that shuttles between nucleus and cytoplasm, is an important mediator of mRNA export. Genes & Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Li O, Heath CV, Amberg DC, Dockendorff TC, Copeland CS, Snyder M, Cole CN. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol Biol Cell. 1995;6:401–417. doi: 10.1091/mbc.6.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Petersen R. Selective translation and degradation of heat-shock messenger RNAs in Drosophila. Enzyme. 1990;44:147–166. doi: 10.1159/000468754. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liang S, Tartakoff AM. Heat shock disassembles the nucleolus and inhibits nuclear protein import and poly(A)+ RNA export. EMBO J. 1996;15:6750–6757. [PMC free article] [PubMed] [Google Scholar]
- Malim MH, McCarn DF, Tiley LS, Cullen BR. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BW, Malim MH. The HIV-1 Rev trans-activator shuttles between the nucleus and cytoplasm. Genes & Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell. 1992;69:939–950. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Cells in stress: Transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- Morimoto R, Tissières A, Georgopoulos C. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1994. [Google Scholar]
- Murphy R, Wente SR. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville, M., L. Lee, F. Stutz, L.I. Davis, and M. Rosbash. 1997. Evidence that the importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export in S. cerevisiae. Curr. Biol. (in press). [DOI] [PubMed]
- Nigg EA. Nucleocytoplasmic transport: Signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Panniers R. Translational control during heat shock. Biochimie. 1994;76:737–747. doi: 10.1016/0300-9084(94)90078-7. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Molec Biol. 1996;31:153–199. doi: 10.3109/10409239609106583. [DOI] [PubMed] [Google Scholar]
- Pikielny CW, Rosbash M. mRNA splicing efficiency in yeast and the contribution of non-conserved sequences. Cell. 1985;41:119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saavedra C, Tung K-S, Amberg DC, Hopper AK, Cole CN. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes & Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Hurt E, Doye V, Silver PA. Reconstitution of nuclear protein transport with semi-intact yeast cells. J Cell Biol. 1993;123:785–798. doi: 10.1083/jcb.123.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiokawa K, Pogo KO. The role of cytoplasmic membranes in controlling the transport of nuclear messenger RNA and initiation of protein synthesis. Proc Natl Acad Sci. 1974;71:2658–2662. doi: 10.1073/pnas.71.7.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra JM, Zapata JM. Translational regulation of the heat shock response. Mol Biol Rep. 1994;19:211–220. doi: 10.1007/BF00986963. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Stutz F, Rosbash M. A functional interaction between Rev and yeast pre-mRNA is related to splicing complex formation. EMBO J. 1994;17:4096–4104. doi: 10.1002/j.1460-2075.1994.tb06727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- Stutz S, Izaurralde E, Mattaj I, Rosbash M. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz, F., J. Kantor, D. Zhang, T. McCarthy, M. Neville, and M. Rosbash. 1997. The yeast nucleoporin Rip1p contributes to multiple export pathways with no essential role for its FG-repeat region.Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Wente SR, Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Wong DH, Corbett AH, Kent HK, Stewart M, Silver PA. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]