Abstract
We describe herein a new conformationally constrained analog of PNA carrying an alternating α/β amino acid backbone consisting of (2′R,4′R)-nucleobase-subtituted proline and (1S,2S)-2-aminocyclopentanecarboxylic acid (acpcPNA). The acpcPNA has been synthesized and evaluated for DNA, RNA and self-pairing properties by thermal denaturation experiments. It can form antiparallel hybrids with complementary DNA with high affinity and sequence specificity. Unlike other PNA systems, the thermal stability of acpcPNA·DNA hybrid is largely independent of G+C contents, and is generally higher than that of acpcPNA·RNA hybrid with the same sequence. Thermodynamic parameters analysis suggest that the A·T base pairs in the acpcPNA·DNA hybrids are enthalpically stabilized over G·C pairs. The acpcPNA also shows a hitherto unreported behavior, namely the inability to form self-pairing hybrids. These unusual properties should make the new acpcPNA a potentially useful candidate for various applications including microarray probes and antigene agents.
Key words: peptide nucleic acid, nucleic acid, DNA recognition, RNA recognition, pre-organization, foldamer, α/β-peptide
Introduction
Almost 20 years ago, Nielsen et al. demonstrated an unusually high affinity as well as sequence specificity in recognition of DNA and RNA by a new class of DNA analog called peptide nucleic acid (PNA).1–3 In many aspects, PNA behaves just like a structural mimic of natural nucleic acids.4 However, PNA also displays many unique characteristics not found in DNA/RNA or analogs that derived from partial modification of the natural sugar-phosphate backbone.5 The electrostatically neutral backbone of PNA contributes to the enhanced stability of PNA·DNA hybrids, which is also relatively independent of salt concentrations In addition to binding to single stranded nucleic acid targets, PNA can bind, either by a triplex formation or strand invasion, to double stranded DNA7,8 as well as RNA targets.9 The unnatural backbone of PNA makes it stable towards degradation by nucleases and proteases.10 These properties make PNA a potential candidate for many applications ranging from nucleic acid detection probes11,12 to therapeutic agents via antisense or antigene applications.13,14
Until recently, the original PNA system derived from an achiral backbone of N-2-aminoethylglycine (now generally referred to as aegPNA) has been the most widely studied. Although a number of new PNA systems have been developed in the past decades, few of them offer comparable or better hybridization properties found in the original aegPNA.15–21 It has been argued that the binding properties should be improved if one could restrict the conformational flexibility of the PNA backbone, for example, by locking the PNA backbone into cyclic structures15–18 or by adding one or more substituents at a specific position of the aegPNA backbone.19–21 In addition to improving binding affinity and specificity, conformational restriction should offer a possibility to selectively recognize different types of nucleic acids. While aegPNA has been shown to bind marginally more strongly to RNA over DNA, the selectivity towards RNA has been improved by incorporating one or more cyclohexane rings into the PNA backbone.16,17 Kinetic selectivity in RNA recognition over DNA has been observed in some pyrrolidinyl PNA variants,22,23 although this behavior seems to be highly sequencedependent.24,25 In a much fewer instances, preference for DNA binding over RNA has also been reported.26
Recently, oligomers with α/β peptide backbone were recognized as a potential foldamer system due to their well defined helical conformation.27–29 We have recently developed a new class of pyrrolidinyl PNA consisting of an alternate sequence of nucleobase-modified proline and five-membered ring cyclic β-amino acids.30–33 These pyrrolidinyl PNA retained the basic property of nucleic acid mimics, i.e., the ability to bind to their complementary nucleic acid targets in a sequence-specific fashion. The specificity towards DNA target is in fact considerably better than the original aegPNA system.34 Applications of these pyrrolidinyl PNA as probes for DNA sequence determination have been successfully demonstrated Furthermore, our preliminary studies in a few representative sequences revealed additional unique features of these pyrrolidinyl PNA including the high directional specificity (parallel/antiparallel), the preference for binding to DNA over RNA, and the inability of two complementary PNA sequences to self-hybridize.33 Nevertheless, from the limited data available, it is difficult to conclude whether these properties are general or limited to certain sequences as observed in many other cases.24,25 Herein we describe a comprehensive study of DNA, RNA and self-pairing properties of the pyrrolidinyl PNA system with a backbone consisting of alternating nucleobase-modified proline with a (2′R,4′R)-configuration and (1S,2S)-2-aminocyclopentanecarboxylic acid (acpcPNA) (Fig. 1). The results clearly indicate that the high antiparallel selectivity, the lack of self-pairing, and the preference for binding to DNA over RNA are general properties of acpcPNA over a range of sequences with different G+C contents. They also further revealed an important new observation that the thermal stability of the acpcPNA·DNA hybrid is much less dependent on the G+C content than the corresponding acpcPNA·RNA hybrid.
Figure 1.
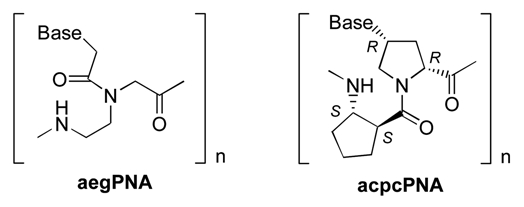
Structures of aegPNA and the pyrrolidinyl PNA carrying 2-aminocyclopen-tanecarboxylic acid spacer (acpcPNA); B = nucleobase (A, C, G or T).
Results and Discussion
Effect of stereochemistry on DNA binding.
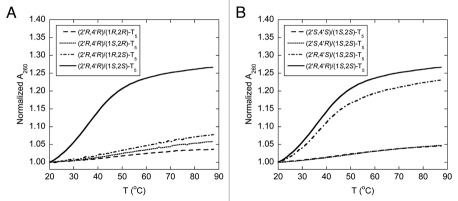
The backbone of the pyrrolidinyl PNA described in this study consists of an alternating nucleobase-modified proline and 2-aminocyclopentanecarboxylic acid, each of which carries two stereogenic centers. As a result, there are 16 possible stereoisomers of the pyrrolidinyl PNA. It is highly probable that the configuration of the monomers will exert significant influences on the ability of the PNA backbone to adopt the correct geometry required for binding with the nucleic acid targets. To systematically determine the effect of stereochemistry on the DNA binding properties, the spacer and the proline parts were optimized separately. Firstly, four diastereomers of pentathymidine (T5) pyrrolidinyl PNA with different stereochemistry at the 2-aminocyclopentanecarboxylic acid (acpc) part, namely (1S,2S)-, (1S,2R)-, (1R,2R)- and (1R,2S)-, but with the same (2′R,4′R)-configuration of the pyrrolidine part were synthesized. This particular configuration of the pyrrolidine part was chosen as the starting point because of its stereochemical similarity to natural nucleosides (assuming that the 2′ and 4′ positions in the pyrrolidine ring are equivalent to the 4′ and 1′ positions in natural nucleosides, respectively). Our previous results have also shown that conformationally restricted PNA derived from the (2′R,4′R)-pyrrolidine monomer can exhibit DNA and RNA binding properties.30,31 Melting experiments of the hybrid formed between each of the four T5 pyrrolidinyl PNA diastereomers and poly(dA) at 1:1 ratio of T and A nucleotides were carried out to evaluate the effect of the stereochemistry on the aminocyclopentanecarboxylic acid moiety to DNA binding. The thermal denaturation experiment (Fig. 2A) revealed that only the pyrrolidinyl PNA with a combination of (1S,2S)-acpc and (2′R,4′R)-pyrrolidine can form hybrids that are sufficiently stable to exhibit observable melting behavior above room temperature (Tm = 35.5°C). Accordingly, the (1S,2S)-acpc spacer was chosen for studying the effect of the pyrrolidine part. Three additional pyrrolidinyl PNAs with the same T5 sequence but different stereochemistry at the pyrrolidine ring, i.e., (2′S,4′S), (2′R,4′S) and (2′S,4′R), all of which carry the same (1S,2S)-acpc spacer, were synthesized. The UV melting results suggested that the acpcPNA with (2′R,4′S) configurations on the proline ring could also form a stable hybrid with poly(dA) (Tm = 36.5°C) (Fig. 2B). These results, together with our earlier reported UV titration data32 indicate that only the combination of (1S,2S)-acpc and (2′R,4′R)- or (2′R,4′S)-pyrrolidine could allow optimal binding with DNA. This paper will focus on the binding properties of the pyrrolidinyl PNA with (2′R,4′R)-proline and (1S,2S)-acpc backbone (Fig. 1), which will be simply referred as acpcPNA from this point onward. Synthesis and hybridization properties of the other pyrrolidinyl PNA diastereomer with (2′R,4′S)-pyrrolidine/(1S,2S)-acpc backbone that also exhibits DNA binding properties are the subject of a separated investigation.
Figure 2.
Tm curves of the hybrids between poly(dA) and T5 pyrrolidinyl PNA with different stereochemistry on the aminocyclopentanecarboxylic acid part (A) and the proline part (B). Conditions: 10.0 µM PNA (by nucleotides), 10.0 µM DNA (by nucleotides), 10 mM sodium phosphate buffer pH 7.0, 0 mM NaCl, heating rate 1.0°C/min.
Hybridization studies of homothymine and homoadenine acpcPNA.
Thermal denaturation experiments of the hybrids formed between the homothymine acpcPNA (T5, T7, T9 and T10) and their complementary DNA (dA5, dA7, dA9 and dA10, respectively) were then carried out (Fig. 3). While the relatively short acpcPNA T5 binds to poly(dA) giving a hybrid with a Tm value of 35.5°C (Fig. 2A), it does not show any melting when hybridized with dA5. Evidently the hybrid formed between the short acpcPNA and DNA was too unstable. However, as observed with aegPNA, the thermal stability of the hybrid increased with the number of base pairs (Tm for 5-, 7-, 9- and 10-mer were <20, 55.5, 77.0 and >85°C, respectively). The Tm of the hybrid between the acpcPNA T10 and DNA was so high that the melting was not yet complete at 90°C. These Tm figures are considerably higher than the (PNA)2·DNA triplexes formed from homothymine aegPNA with comparable length (Tm for 6-, 8- and 10-mer were 31, 52 and 72°C, respectively).37 This dramatic increase in the melting temperature as the number of the base increases was further confirmed by variable temperature CD experiments (Sup. Figs. 34 and 35). In all cases the CD melting profiles were in good agreement with those obtained from UV melting studies. The melting process is completely reversible and shows very little hysteresis (Sup. Fig. 36), suggesting fast kinetics of the hybridization. The single transition observed in the melting curves, together with the data from CD and UV titration experiments (Sup. Figs. 37 and 39) suggested that 1:1 hybrids were formed between these homothymine acpcPNA and DNA. The formation of only acpcPNA·DNA duplexes with homopyrimidine sequences is in sharp contrast to the behavior of aegPNA and many other PNA systems whereby the formation of (PNA)2·DNA triplexes was usually observed.8,37,38 Steric bulkiness of the present acpcPNA backbone could be one important factor that make the binding of the third PNA strand to the PNA·DNA duplex initially formed difficult.
Figure 3.
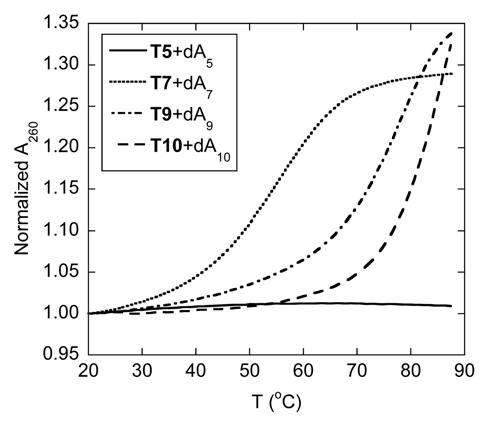
Tm curves of homothymine acpcPNA·DNA hybrids with different length. Conditions: 1.0 µM PNA, 1.0 µM DNA, 10 mM sodium phosphate buffer pH 7.0, 0 mM NaCl, heating rate 1.0°C/min.
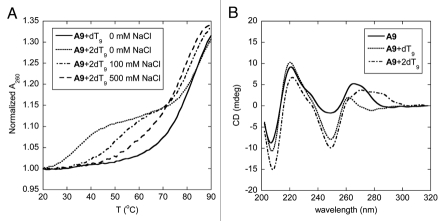
The melting behavior of the hybrid between homoadenine acpcPNA A9 and its complementary DNA is somewhat more complex than the corresponding homothymine sequence. When the DNA component (dT9) was present in excess, the melting proceeded in two steps (Fig. 4A). The first transition took place just above room temperature (∼34°C) whereas the other appeared at a much higher temperature (>80°C). Such double-transition melting curve is explained by the formation of a (DNA)2·PNA triplex. The triplex could form by Hoogsteen base pairing between the T base of the second strand of DNA to the originally formed Watson-Crick pA·dT pair. When the acpcPNA was present in excess, only the transition at the higher temperature remained, suggesting the presence of only the acpcPNA·DNA duplex in the solution (Fig. 4A). The triplex and duplex melting showed an interesting opposite salt-dependency. When the salt concentration was increased, the Tm of the triplex melting increased markedly. This could be explained by the screening effect of salt on the electrostatically repulsive interaction between the duplex and the third DNA strand. On the other hand, the Tm of the duplex melting slightly decreased at high salt concentrations, which is in accordance with the results observed in aegPNA·DNA hybrids, and a similar explanation based on counterion release upon duplex formation should be applicable here.6 At sufficiently high salt concentrations (∼500 mM or higher), the two transitions merged to give just one melting curve, indicating a simultaneous dissociation of the triplex to the three single strand components at the same temperature.
Figure 4.
Tm curves (A) and CD spectra (B) of homoadenine acpcPNA·DNA hybrids at different ratio. Conditions: (A) 1.0 µM acpcPNA, 10 mM sodium phosphate buffer pH 7.0, 0–500 mM NaCl, heating rate 1.0°C/min; (B) 2.0 µM acpcPNA, 10 mM sodium phosphate buffer pH 7.0, 0 mM NaCl, 25°C.
The formation of a triplex structure between A9 and dT9 was also confirmed by UV titration and circular dichroism (CD) spectroscopy. The UV titration revealed two inflection points at acpcPNA:DNA ratio of 1:1 and 1:2 respectively (Sup. Fig. 38). The CD spectrum of single-stranded acpcPNA A9 (Fig. 4B) shows strong signals in the region of nucleobase absorption around 265 nm. This suggests that the single stranded homoadenine acpcPNA exists in a well-defined helical conformation, allowing effective stacking of the nucleobases. Addition of one equivalent of dT9 to this acpcPNA solution caused a substantial spectral change in both position and magnitude of the CD bands in the nucleobase absorption region. The positive CD bands at 265 nm in the original single stranded acpcPNA shifted to a shorter wavelength of 260 nm in the duplex. This is also accompanied by an increase in the intensity of the negative CD band at 250 nm. The CD spectrum of this A9·dT9 is remarkably similar to the CD spectrum of the T9·dA9 duplex reported earlier.32 Upon addition of the second equivalent of the DNA, a distinctive change was again observed in the nucleobase absorption region. The measured CD spectrum is clearly different from the sum of the CD spectra of each component, therefore it can be concluded that yet another species—most likely to be the triplex—is formed at 2:1 DNA:PNA ratio. In all cases, very little changes of the CD signal were observed in the peptide backbone region suggesting that there is little conformational change of the acpcPNA backbone upon hybrid formation.
Despite the extremely high stability of the homothymine and homoadenine acpcPNA hybrids with their complementary DNA, Tm data of mismatched acpcPNA·DNA hybrids (Table 2) revealed that the specificity is not compromised. It can be clearly seen that the base T in the acpcPNA T9 can recognize only the base A in DNA, and the base A in the acpcPNA A9 can recognize only the base T in DNA. Introduction of a mismatched base in the DNA strand caused a significant destabilization of the mismatched duplex as shown by the large decrease of Tm (ΔTm) ranging from −23.4 to −37.8°C. The Tm decreases per mismatch in these cases are considerably larger than those typically observed in aegPNA (∼10–15°C per mismatched base).4
Table 2.
Tm data for the hybrids between acpcPNA T9 and A9 with complementary and single mismatched DNA
| Entry | PNA | DNA | Tm (°C)a,b | ΔTm (°C)c | |
| 1 | T9 | dAAAAXAAAA | X = A | 76.8 | - |
| 2 | X = T | 47.6 | −29.2 | ||
| 3 | X = C | 47.7 | −29.1 | ||
| 4 | X = G | 39.0 | −37.8 | ||
| 5 | A9 | dTTTTYTTTT | Y = A | 54.6 | −25.3 |
| 6 | Y = T | 79.9 | - | ||
| 7 | Y = C | 56.5 | −23.4 | ||
| 8 | Y = G | 52.7 | −27.2 | ||
Conditions: 1.0 µM acpcPNA, 1.0 µM DNA, 10 mM sodium phosphate buffer pH 7.0, 0 mM NaCl, heating rate 1.0°C/min. Tm values are accurate to within ±0.5°C.
Data for T9 (entries 1–4) were taken from reference 33.
Tm (mismatched) − Tm (complementary).
Hybridization studies of mixed-base acpcPNA—directional preference and base pairing specificity.
We have previously shown that the mixed sequence PNA decamer M10a preferred hybridization with DNA in antiparallel orientation.33 To ascertain that the antiparallel directional preference is a general behavior of the present acpcPNA system, additional Tm experiments were performed with nine different 10mer mixed-base PNA sequences (Table 3). Although the stability of acpcPNA·DNA hybrids is not expected to be very much dependent to the ionic strength as in the case of DNA·DNA hybrids, the melting studies were performed in 10 mM sodium phosphate buffer both in the absence and presence of NaCl (100 mM) conditions. All antiparallel hybrids showed single-transition melting curves with Tm in the range of 57.0–65.5°C (at 0 mM NaCl) and 52.8–61.0°C (at 100 mM NaCl). The slightly lower Tm values at high salt conditions compared to the low salt conditions (∼4–5°C) are consistent with the behavior of aegPNA.6 On the other hand, no sigmoidal transtitions could be observed when parallel DNA were present under both low salt and high salt conditions in all cases. This indicates that the parallel hybrids would have Tm values of less than 20°C for all nine sequences tested. The Tm difference by more than 30°C suggests that the antiparallel PNA·DNA hybrids are much more stable than the corresponding parallel hybrids. This property is in sharp contrast to the original aegPNA which could form both antiparallel and parallel hybrids with DNA, although the antiparallel hybrid was slightly more stable (ΔTm antiparallel vs. parallel hybrids ∼13°C for a 10mer mixed base aegPNA with the same sequence as M10a).4 It has been observed that a modification of the aegPNA backbone with a short stretch of chiral monomer (“chiral box”) could greatly enhance the antiparallel preference by destabilization of the parallel hybrid.39 It appears that the presence of stereoregular backbone in these chiral PNAs allows binding to DNA in only one direction, thus enhancing the orientation selectivity relative to the achiral backbone of aegPNA.
Table 3.
Tm data for the binding between mixed base acpcPNA and DNA in antiparallel and parallel directionsa
| Entry | PNA | Tm (antiparallel DNA) (°C) | Tm (parallel DNA) (°C) | ||
| 0 mM NaCl | 100 mM NaCl | 0 mM NaCl | 100 mM NaCl | ||
| 1 | M10a | 57.0b | 53.3c | <20b | <20c |
| 2 | M10b | 60.4 | 54.5 | <20 | <20 |
| 3 | M10c | 59.6 | 54.2 | <20 | <20 |
| 4 | M10d | 57.2 | 52.8 | <20 | <20 |
| 5 | M10e | 60.6 | 56.1 | <20 | <20 |
| 6 | M10f | 65.2 | 60.8 | <20 | <20 |
| 7 | M10g | 65.1 | 59.1 | <20 | <20 |
| 8 | M10h | 65.5 | 61.0 | <20 | <20 |
| 9 | M10i | 65.3 | 59.8 | <20 | <20 |
The Tm of the complementary hybrid formed between the acpcPNA M10a and its complementary DNA was 53.3°C at 100 mM NaCl and 10 mM sodium phosphate. This figure is slightly higher than the corresponding aegPNA system with identical sequence (48.9°C) under comparable conditions.15 To determine the base pairing specificity, the acpcPNA M10a was hybridized, one by one, with twelve other DNA probes carrying a mismatched base between the positions 4 to 7 of the sequence. The results, which are summarized in Table 4, indicated that the acpcPNA·DNA hybridization strictly follows the Watson-Crick base pairing rules and is highly specific. As observed in the case of homothymine or homoadenine acpcPNA hybrids with DNA, the presence of one mismatched base in the DNA part caused a substantial decrease in Tm values (−22.3 to −29.5°C) of the mixed sequence acpcPNA·DNA hybrids. With few exceptions, the pyrimidine-pyrimidine mismatched pairs tend to be more stable than the purine-purine mismatched pairs. The potentially wobble G·T pairs40 showed a similar drop in Tm to other mismatched pairs. It is important to note that the Tm values of all single mismatched hybrids of acpcPNA are smaller than the corresponding mismatched hybrids of aegPNA. As a result, the differences in thermal stability between complementary and mismatched hybrids of the present acpcPNA system are significantly greater than those of the original aegPNA system (ΔTm= −12.9 to −16.9°C for aegPNA/DNA with the same sequences).15 The enhanced directional selectivity and discrimination of single base mismatched target of acpcPNA over aegPNA have also been independently verified by surface plasmon resonance (SPR) experiments in a different sequence.34 Thus it can be concluded that the more rigid acpcPNA backbone improves both the binding affinity and the specificity for single base mismatches discrimination towards DNA targets.
Table 4.
Comparison of Tm data to show specificity of binding between mixed base acpcPNA M10a and complementary/single base mismatched DNA
| Entry | DNA sequence (5′ → 3′) | Tm (°C)a | ΔTm (°C)b | Note |
| 1 | AGT GAT CTA C | 53.3c | - | complementary |
| 2 | AGT AAT CTA C | 26.4 | −26.9 | pC·dA mismatch |
| 3 | AGT TAT CTA C | 27.8 | −25.5 | pC·dT mismatch |
| 4 | AGT CAT CTA C | 31.0 | −22.3 | pC·dC mismatch |
| 5 | AGT GGT CTA C | 23.9c | −29.4 | pT·dG mismatch |
| 6 | AGT GTT CTA C | 29.4c | −23.9 | pT·dT mismatch |
| 7 | AGT GCT CTA C | 23.8c | −29.5 | pT·dC mismatch |
| 8 | AGT GAG CTA C | 26.5 | −26.8 | pA·dG mismatch |
| 9 | AGT GAA CTA C | 28.4 | −24.9 | pA·dA mismatch |
| 10 | AGT GAC CTA C | 28.8 | −24.5 | pA·dC mismatch |
| 11 | AGT GAT GTA C | 26.4 | −26.9 | pG·dG mismatch |
| 12 | AGT GAT ATA C | 24.2 | −29.1 | pG·dA mismatch |
| 13 | AGT GAT TTA C | 28.5 | −24.8 | pG·dT mismatch |
Conditions: 1.0 µM acpcPNA, 1.0 µM DNA, 10 mM sodium phosphate buffer pH 7.0, 100 mM NaCl, heating rate 1.0°C/min. Tm values are accurate to within ±0.5°C.
Tm (mismatched) − Tm (complementary).
Data taken from reference 36.
Hybridization studies of mixed-base PNA—comparison between DNA and RNA.
We have previously noted that the acpcPNA·DNA hybrids are more thermally stable than the corresponding acpcPNA·RNA hybrids in a few representative sequences.33 In the present study, a larger pool of acpcPNA and RNA sequences were included to obtain a clearer picture. Comparison between the results shown in Table 5 and Table 3 confirmed our previous observation that the acpcPNA·DNA hybrids are more stable than acpcPNA·RNA hybrids with identical sequences in all cases. The results also unexpectedly revealed an interesting relationship between Tm and G+C content, which can be observed more clearly in Figure 5. The Tm values of the antiparallel acpcPNA·RNA hybrids appears to show relationships to the %G+C content in the same way as normal DNA/RNA duplexes, i.e., duplexes with greater %G+C content possess higher Tm values. The nine antiparallel acpcPNA·RNA hybrids included in this study showed Tm values between 32.6 and 51.2°C (Tm difference = 18.6°C). On the other hand, the Tm values for the corresponding antiparallel acpcPNA·DNA hybrids span a much smaller temperature range between 52.8 and 61.0°C (Tm difference = 8.2°C) and do not show obvious correlations with %G+C content. It has been earlier noted that aegPNA·DNA duplexes derived from purine-rich aegPNA were much more stable than those derived from pyrimidine-rich aegPNA with identical G+C content.41 Such purine-stabilizing effect should be minimal here since all acpcPNA sequences used in this study contain a similar purine content of 40–60%. Furthermore, no correlation could be observed between thermal stability and purine content of the acpcPNA·DNA and acpcPNA·RNA hybrids used in this study and in other additional sequences.42 As shown in Figure 5, the difference in the stability between acpcPNA·DNA and acpcPNA·RNA hybrids is more pronounced at lower G+C contents than at higher ones. It is also interesting to note that the formation of parallel acpcPNA·RNA hybrids could be observed, especially when the G+C content is high. Nevertheless, the Tm values of these parallel acpcPNA·RNA hybrids are much lower than the corresponding antiparallel hybrids. Hence it can be concluded that (1) the acpcPNA binds more strongly to DNA over RNA, but the difference in thermal stability is dependent on the G+C content and (2) although the antiparallel binding mode is preferred in acpcPNA·RNA hybrids, the selectivity between the antiparallel/parallel binding modes is lower than the corresponding acpcPNA·DNA hybrids.
Table 5.
Tm data for the binding between mixed base acpcPNA and RNA in antiparallel and parallel directionsa
| Entry | PNA | %G+C | Tm (antiparallel RNA) (°C) | Tm (parallel RNA) (°C) |
| 1 | M10a | 40 | 42.3b | <20b |
| 2 | M10b | 70 | 48.0 | 32.9 |
| 3 | M10c | 20 | 32.6 | <20 |
| 4 | M10d | 40 | 40.8 | not determined |
| 5 | M10e | 40 | 40.9 | not determined |
| 6 | M10f | 70 | 49.3 | 30.2 |
| 7 | M10g | 70 | 51.2 | 28.5 |
| 8 | M10h | 20 | 39.7 | not determined |
| 9 | M10i | 20 | 37.7 | not determined |
Conditions: 1.0 µM acpcPNA, 1.0 µM DNA, 10 mM sodium phosphate buffer pH 7.0, 100 mM NaCl, heating rate 1.0°C/min. Tm values are accurate to within ±0.5°C.
Data taken from reference 36.
Figure 5.
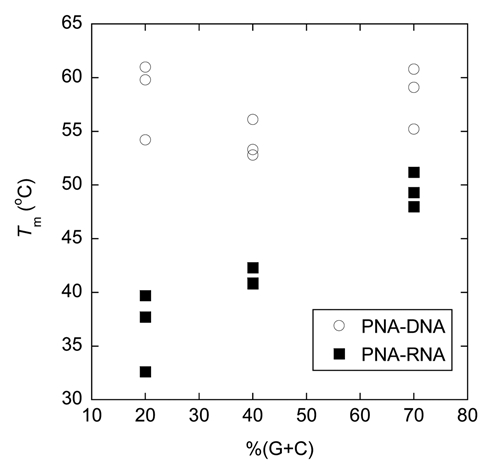
A plot showing relationship between G+C content and thermal stability of acpcPNA·DNA and acpcPNA·RNA hybrids. Conditions: 1.0 µM acpcPNA, 1.0 µM DNA, 10 mM sodium phosphate buffer pH 7.0, 100 mM NaCl, heating rate 1.0°C/min. Tm values are accurate to within ±0.5°C.
Most other PNA systems reported to date bind more strongly to RNA than DNA, and many attempts have been made to further enhance the RNA binding with a view to antisense and other applications involving RNA targets.16,17,22,23 It is interesting to note that while the original aegPNA showed a preference for binding to RNA over DNA, substitution at the γ-position of the PNA backbone by a methyl group increased the stability of the DNA hybrids considerably more than the corresponding RNA hybrid, leading to a lower discrimination between DNA and RNA.19 The present acpcPNA system and its aza-analog described previously by our group,31 as well as a chimeric (aeg-pyrrolidine)PNA developed by Kumar,26 are among a few examples of PNA that show a preference for DNA binding.23 Although the structural basis of this preference is not available at present, it can be speculated that the preferential binding to DNA over RNA may be due to the more limited conformations space available in RNA duplexes compared to DNA duplexes as a result of the additional 2′-OH group present in RNA. Upon binding to RNA, it is likely that the acpcPNA could not adopt the same conformation as when it is bound to DNA. Works in this area, including molecular dynamics simulations43 and NMR spectroscopic studies are in progress.
Thermodynamic parameters of acpcPNA·DNA and acpcPNA·RNA hybridization.
The extremely high stability of homoadenine/homothymine acpcPNA·DNA hybrids compared to the mixed base acpcPNA·DNA of comparable length, as well as the sequence-independent stability of acpcPNA·DNA, but not acpcPNA·RNA, hybrids are quite unexpected. In an attempt to understand this unusual behavior, the melting curves of DNA and RNA hybrids of the acpcPNA M10a-M10c having different G+C contents between 20–70% were analyzed by nonlinear curve fitting44 and by van't Hoff analysis45 to extract the thermodynamic parameters (Table 6). The data suggested that despite the large enthalpy gain for G·C rich acpcPNA hybrids compared to A·T rich ones, the entropy terms are more negative (less favorable). This is true for both acpcPNA·DNA (entry 2) and acpcPNA·RNA hybrids (entry 5). In order to achieve the required geometry for the usual three hydrogen bonding pattern and/or maximum stacking of the G·C pairs, the structure of the base pairs must be distorted to some extent, resulting in a large entropy loss. On the other hand, the enthalpy term of the A·T rich acpcPNA·DNA hybrids (entry 3) were relatively large compared to the corresponding acpcPNA·RNA hybrid (entry 6), while the entropy terms were similar. Since Watson-Crick A·T pairs can form only two hydrogen bonds compared to three in G·C pairs, hydrogen bonding cannot be the main contributor to the enthalpy term. We propose that base stacking must be the main factor to stabilize the A·T pair relative to the G·C pairs in the acpcPNA·DNA hybrids. The stackinghydrogen bonding compensation46–48 might explain why the difference of the enthalpy change between G·C and A·T pairs is rather small. The same stabilization effects of A·T pair are absent in acpcPNA·RNA hybrids, perhaps due to the more limited conformation space available in acpcPNA·RNA hybrids, which may keep the spatial geometry from achieving maximum stacking. Additional structural studies, e.g., by X-ray or NMR are required to understand this behavior in details.
Table 6.
Thermodynamic parameters of hybrids of acpcPNA with DNA and RNAa
| Entry | PNA | Counterstrandb | %G+C | Tm (°C)c | ΔH° (kcal/mol)c | ΔS° (cal/mol·K)c | ΔG°298 (kcal/mol)c |
| 1 | M10a | DNA | 40 | 53.1 | −64.1 ± 1.0 (−64.1 ± 0.7) | −168 ± 3 (−167 ± 2) | −14.1 ± 0.1 (−14.1 ± 0.0) |
| 2 | M10b | DNA | 70 | 56.4 | −68.9 ± 1.9 (−69.0 ± 3) | −180 ± 6 (−180 ± 10) | −15.2 ± 0.3 (−15.2 ± 0.4) |
| 3 | M10c | DNA | 20 | 53.8 | −60.9 ± 1.0 (−61.7 ± 1.0) | −157 ± 3 (−160 ± 3) | −14.0 ± 0.0 (−14.1 ± 0.0) |
| 4 | M10a | RNA | 40 | 42.8 | −60.0 ± 0.2 (−60.1 ± 0.9) | −161 ± 0.5 (−161 ± 3) | −12.0 ± 0.1 (−12.0 ± 0.0) |
| 5 | M10b | RNA | 70 | 48.1 | −68.0 ± 2.6 (−67.9 ± 2.8) | −183 ± 8 (−183 ± 8) | −13.5 ± 0.3 (−13.5 ± 0.3) |
| 6 | M10c | RNA | 20 | 33.1 | −54.9 ± 3.1 (−54.3 ± 1.4) | −151 ± 10 (−148 ± 4) | −10.1 ± 0.3 (−10.0 ± 0.2) |
Conditions: 1.0 µM of each acpcPNA and DNA or RNA strand, 10 mM sodium phosphate buffer pH 7.0, 150 mM NaCl, heating rate 1.0°C/min.
Antiparallel complementary DNA or RNA.
Evidences for the absence of self-hybridization in acpcPNA.
In addition to the preference for binding to DNA over RNA, the acpcPNA system also displayed another rather unusual property, namely the inability of two complementary PNA strands to form self-pairing hybrids. This behavior has been previously demonstrated for acpcPNA with the sequence M10a and its antiparallel and parallel complementary acpcPNA sequences M10d and M10e.33 In this work, we extend the study to four other acpcPNA·acpcPNA pairs to demonstrate that the nonself pairing behavior is general over a range of sequences with different G+C contents (Table 7). Indeed, no melting could be observed in all of the antiparallel hybrids indicating that these hybrids were not stable above 20°C. On the other hand, some rather unstable parallel hybrids could be observed for sequences with ≥40% G+C content (entries 2 and 4). The low stability of both the antiparallel and parallel acpcPNA·acpcPNA hybrids is in sharp contrast to the situation of aegPNA, whereby the PNA·PNA hybrids are very stable. In fact, the stability of aegPNA·aegPNA antiparallel hybrids is generally much higher than the corresponding aegPNA·DNA and aegPNA·RNA hybrids.6 To the best of our knowledge this absence of self-pairing, despite the ability to cross-pair with both DNA and RNA, has not been reported in other PNA systems. Interestingly, our recent investigation revealed that the epimeric acpcPNA with the (2′R,4′S)-configuration on the proline ring could form an antiparallel self-pairing hybrid.36 Nevertheless, the self-pairing hybrid is considerably less stable than the corresponding acpcPNA·DNA hybrid. Although more studies are required to understand its structural basis, this unusual property is a highly desirable one. For example, the non-self-pairing acpcPNA may be useful for targeting dsDNA via a “double duplex invasion” mechanism whereby the two PNA strands designed to target two opposite DNA strands in the same region of the dsDNA target. The original aeg-PNA is not suitable for this purpose as the two complementary PNA strands binds to each other much more strongly than with their respective DNA targets.49,50 Current strategies to use aegPNA for double duplex invasion involve modification of the aegPNA by modifying the nucleobases49 and incorporation of multiple positive charges into the PNA backbone50 to provide a so-called pseudocomplementary PNA that binds more strongly to DNA than to itself. The present acpcPNA system does not require any modification to achieve this property. The absence of self-pairing in acpcPNA also means that there is a low propensity for secondary structure formation by intra- or interstrand self-pairing, which are often problematic in designing DNA and aegPNA sequences for various applications such as probes for DNA sequence determination. On the other hand, this property restricts the use of acpcPNA in applications that require self-organization of the PNA strand such as molecular beacons with the classical stem-loop design.51
Table 7.
Tm data for the binding between two complementary strands of acpcPNA in antiparallel and parallel directions
| Entry | PNA pairs | %G+C | Tm (°C)a | Note |
| 1 | M10a·M10d | 40 | <20 | antiparallel |
| 2 | M10a·M10e | 40 | 24.2 | parallel |
| 3 | M10b·M10f | 70 | <20 | antiparallel |
| 4 | M10b·M10g | 70 | 35.1 | parallel |
| 5 | M10c·M10h | 20 | <20 | antiparallel |
| 6 | M10c·M10i | 20 | <20 | parallel |
Conditions: 1.0 µM of each acpcPNA strand, 10 mM sodium phosphate buffer pH 7.0, 100 mM NaCl, heating rate 1.0°C/min. Tm values are accurate to within ±0.5°C.
In conclusion, we have developed new conformationally constrained pyrrolidinyl PNA systems based on an alternating nucleobase-subtituted proline and 2-aminocyclopentanecarboxylic acid backbone. The specific stereochemical requirements of both the proline and the 2-aminocyclopentanecarboxylic acid parts that allow effective binding to DNA have been identified. The PNA system with (2′R,4′R)-proline/(1S,2S)-2-aminocyclopentanecarboxylic acid backbone, which is named acpcPNA, binds to DNA only in antiparallel orientation with exceptionally high affinity and sequence specificity following the Watson-Crick base pairing rules. In addition, the stability of the acpcPNA·DNA hybrids is largely independent of the base sequence. This particular property should render the acpcPNA system highly useful as probes in multiplex SNP analysis and microarray applications whereby several DNA targets are to be analyzed simultaneously.35,49 The acpcPNA also binds to complementary RNA, albeit with lower hybrid stability and directional specificity compared to DNA. Consequently, the present acpcPNA system may be less useful for applications that require targeting RNA such as RNA detection or antisense applications. The difference in stability between acpcPNA·DNA and acpcPNA·RNA hybrids is more pronounced in A·T rich than G·C rich sequences, which could be explained by enthalpic stabilization of A·T pairs over G·C pairs in acpcPNA·DNA hybrids. Finally, the unique absence of self-pairing between two complementary strands of acpcPNA suggests that it should be a potential candidate for targeting double stranded DNA by double duplex invasion, which could be useful for antigene applications.
Materials and Methods
Materials and reagents.
Oligonucleotides were obtained from Biodesign (Pathumthani, Thailand) and were used as received. The concentration of oligonucleotides and acpcPNAs was determined from the absorbance at 260 nm using the reported molar extinction coefficients at 260 nm (ε260) for DNA monomers.53
PNA synthesis.
Synthesis and characterization data of acpcPNA monomers have either been previously described [(2′R,4′R) and (2′R,4′S) diastereomers]33,36 or can be found in the supporting information [(2′S,4′S) and (2′S,4′R) diastereomers]. All acpcPNAs were synthesized manually from the corresponding Fmoc-protected acpcPNA monomers and β-amino acids spacers by Fmoc-solid phase peptide synthesis on a Tentagel resin carrying the acid-labile Rink amide linker as described previously.33 L-Lysinamide was incorporated at the C-termini of the acpcPNAs to improve water solubility and the PNA was end-capped by acetylation. The acpcPNAs were purified by reverse-phase HPLC and characterized by MALDI-TOF mass spectrometry. The sequences and MALDI-TOF analysis data of the synthesized PNA are shown in Table 1.
Table 1.
Sequences and characterization data of acpcPNA used in this study
| Entry | PNA | PNA sequencea (N → C) | m/z (calcd for [M+H]+) | m/z (found) |
| 1 | T5 | TTT TT | 1848.9 | 1848.8 |
| 2 | T7 | TTT TTT T | 2513.2 | 2513.5 |
| 3 | T9 | TTT TTT TTT | 3177.5 | 3177.6 |
| 4 | T10 | TTT TTT TTT T | 3467.6 | 3468.8 |
| 5 | A9 | AAA AAA AAA | 3258.6 | 3258.7 |
| 6 | M10a | GTA GAT CAC T | 3556.7 | 3555.9 |
| 7 | M10b | GCT ACG TCG C | 3533.7 | 3533.9 |
| 8 | M10c | TAT GTA CTA T | 3546.7 | 3546.9 |
| 9 | M10d | AGT GAT CTA C | 3556.7 | 3556.0 |
| 10 | M10e | CAT CTA GTG A | 3556.7 | 3556.5 |
| 11 | M10f | GCG ACG TAG C | 3582.7 | 3582.6 |
| 12 | M10g | CGA TGC AGC G | 3582.7 | 3582.6 |
| 13 | M10h | ATA GTA CAT A | 3564.7 | 3564.6 |
| 14 | M10i | ATA CAT GAT A | 3564.7 | 3564.5 |
All sequences carried a C-terminal lysinamide and were N-acetylated.
MALDI-TOF, linear mode, CCA matrix.
UV melting experiments.
UV melting experiments were performed on a CARY 100 UV Spectrophotometer (Varian Inc., Australia) equipped with a Peltier temperature controller and a thermal analysis software. The sample for thermal denaturation measurement was prepared by mixing calculated amounts of stock oligonucleotide, PNA and buffer solutions together to give the final concentration of PNA = 1.0 µM and DNA = 1.0 µM, sodium phosphate buffer pH 7.0 (10 mM) and sodium chloride (0 or 100 mM). The final volumes were adjusted to 1.0 mL by addition of deionized water. The samples were transferred to a 10 mm quartz cell with Teflon stopper and equilibrated at the starting temperature for 10 min. The A260 was recorded in steps from 20 to 90°C in two heating and one cooling cycles (20-90-20-90) with a temperature increment of 1.0°C/min and a hold time of 10 min at the end of each cycle. The recorded temperature was corrected by a linear equation obtained from the relationship between the recorded temperature and actual temperature measured by a built-in temperature probe. Only the result taken from the last heating cycle was used. This was normalized by dividing the absorbance at each temperature by the initial absorbance. Thermodynamic parameters were obtained from melting curves by non-linear curve fitting44 and by van't Hoff analysis.45 The data obtained are average of at least three melting curves for each PNA samples at a concentration of PNA = DNA = 1.0 µM (ct = 2.0 µM) in sodium phosphate buffer, pH 7.0 and 150 mM NaCl. The ΔG° at 298 K and Tm were calculated from the ΔH° and ΔS° values.
CD experiments.
CD measurements were performed on a JASCO Model J-715 or J-815 Spectropolarimeter (JASCO, Japan). The samples were prepared by mixing the calculated amounts of stock oligonucleotide and PNA solutions in an appropriate buffer in a quartz cell (path length = 10 mm). The spectra were measured at 25°C from 320 to 200 nm and averaged 4 times then subtracted from a spectrum of the buffer under the same conditions.
Acknowledgments
Financial support to this work from the Thailand Research Fund (RTA5280002), the Thai government stimulus package 2 (TKK2555, SP2) under the Project for Establishment of Comprehensive Center for Innovative Food, Health Products and Agriculture,and the National Research University of CHE and the Ratchadaphiseksomphot Endowment Fund (AM1006A).
Supplementary Material
References
- 1.Nielsen PE. Peptide nucleic acid. A molecule with two identities. Acc Chem Res. 1999;32:624–630. [Google Scholar]
- 2.Ahlman E, Peyman A, Breipohl G, Will DW. PNA: Synthetic polyamide nucleic acids with unusual binding properties. Angew Chem Int Ed. 1998;37:2796–2823. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2796::AID-ANIE2796>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen PE, Haaima G. Peptide nucleic acid (PNA). A DNA mimic with a pseudopeptide backbone. Chem Soc Rev. 1997;26:73–78. [Google Scholar]
- 4.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 5.Leumann CJ. DNA analogues: From supramolecular principles to biological properties. Bioorg Med Chem. 2002;10:841–854. doi: 10.1016/s0968-0896(01)00348-0. [DOI] [PubMed] [Google Scholar]
- 6.Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen PE, Nordén B, Gräslund A. Ionic effects on the stability and conformation of peptide nucleic acid complexes. J Am Chem Soc. 1996;118:5544–5552. [Google Scholar]
- 7.Bentin T, Hansen GI, Nielsen PE. Structural diversity of target-specific homopyrimidine peptide nucleic acid-dsDNA complexes. Nucl Acids Res. 2006;34:5790–5799. doi: 10.1093/nar/gkl736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 9.Li M, Zengeya T, Rozners E. Short peptide nucleic acids bind strongly to homopurine tract of double helical RNA at pH 5.5. J Am Chem Soc. 2010;132:8676–8681. doi: 10.1021/ja101384k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchardt O, Sonnichsen SH, Nielsen PE. Stability of peptide nucleic acids in human serum and cellular extracts. Biochemical Pharmacology. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 11.Brandt O, Hoheisel JD. Peptide nucleic acids on microarrays and other biosensors. Trends Biotechnol. 2004;22:617–622. doi: 10.1016/j.tibtech.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Pellestor F, Paulasova P, Hamamah S. Peptide nucleic acids (PNAs) as diagnostic devices for genetic and cytogenetic analysis. Curr Pharm Des. 2008;14:2439–2444. doi: 10.2174/138161208785777405. [DOI] [PubMed] [Google Scholar]
- 13.Braasch DA, Corey DR. Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry. 2002;41:4503–4510. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- 14.Ray A, Nordén B. Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J. 2000;14:1041–1060. doi: 10.1096/fasebj.14.9.1041. [DOI] [PubMed] [Google Scholar]
- 15.Pokorski JK, Witschi MA, Purnell BL, Appella DH. (S,S)-trans-Cyclopentane-constrained peptide nucleic acids. A general backbone modification that improves binding affinity and sequence specificity. J Am Chem Soc. 2004;126:15067–15073. doi: 10.1021/ja046280q. [DOI] [PubMed] [Google Scholar]
- 16.Govindaraju T, Kumar VA, Ganesh KN. (SR/RS)-Cyclohexanyl PNAs: Conformationally preorganized PNA analogues with unprecedented preference for duplex formation with RNA. J Am Chem Soc. 2005;127:4144–4145. doi: 10.1021/ja044142v. [DOI] [PubMed] [Google Scholar]
- 17.Govindaraju T, Madhuri V, Kumar VA, Ganesh KN. Cyclohexanyl peptide nucleic acids (chPNAs) for preferential RNA binding: Effective tuning of dihedral angle β in PNAs for DNA/RNA discrimination. J Org Chem. 2006;71:14–21. doi: 10.1021/jo051227l. [DOI] [PubMed] [Google Scholar]
- 18.Kumar VA, Ganesh KN. Conformationally constrained PNA analogues: Structural evolution toward DNA/RNA binding selectivity. Acc Chem Res. 2005;38:404–412. doi: 10.1021/ar030277e. [DOI] [PubMed] [Google Scholar]
- 19.Dragulescu-Andrasi A, Rapireddy S, Frezza BM, Gayathri C, Gil RR, Ly DH. A simple γ-backbone modification preorganizes peptide nucleic acid into a helical structure. J Am Chem Soc. 2006;128:10258–10267. doi: 10.1021/ja0625576. [DOI] [PubMed] [Google Scholar]
- 20.Yeh JI, Shivachev B, Rapireddy S, Crawford MJ, Gil RR, Du SC, et al. Crystal structure of chiral γPNA with complementary DNA strand: Insights into the stability and specificity of recognition and conformational preorganization. J Am Chem Soc. 2010;132:10717–10727. doi: 10.1021/ja907225d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Englund EA, Appella DH. γ-Substituted peptide nucleic acids constructed from L-Lysine are a versatile scaffold for multifunctional display. Angew Chem Int Ed. 2007;46:1414–1418. doi: 10.1002/anie.200603483. [DOI] [PubMed] [Google Scholar]
- 22.Ngamwiriyawong P, Vilaivan T. Synthesis and nucleic acids binding properties of diastereomeric aminoethylprolyl peptide nucleic acids (aepPNA) Nucleosides Nucleotides and Nucleic Acids. 2011;30:97–112. doi: 10.1080/15257770.2010.547839. [DOI] [PubMed] [Google Scholar]
- 23.Tan THS, Hickman DT, Morral J, Beadham IG, Micklefield J. Nucleic acid binding properties of thyminyl and adeninyl pyrrolidine-amide oligonucleotide mimics (POM) Chem Comm. 2004:516–517. doi: 10.1039/b315768g. [DOI] [PubMed] [Google Scholar]
- 24.Tan THS, Worthington RJ, Pritchard RG, Morral J, Micklefield J. Homopolymeric pyrrolidine-amide oligonucleotide mimics: Fmoc-synthesis and DNA/RNA binding properties. Org Biomol Chem. 2007;5:239–248. doi: 10.1039/b613384n. [DOI] [PubMed] [Google Scholar]
- 25.Worthington RJ, O'Rourke AP, Morral J, Tan THS, Micklefield J. Mixed sequence pyrrolidine-amide oligonucleotide mimics: Boc(Z) synthesis and DNA/RNA binding properties. Org Biomol Chem. 2007;5:249–259. doi: 10.1039/b613386j. [DOI] [PubMed] [Google Scholar]
- 26.Lonkar PS, Ganesh KN, Kumar VA. Chimeric (aegpyrrolidine) PNAs: synthesis and stereodiscriminative duplex binding with DNA/RNA. Org Biomol Chem. 2004;2:2604–2611. doi: 10.1039/B407292H. [DOI] [PubMed] [Google Scholar]
- 27.Choi SH, Guzei IA, Spencer LC, Gellman SH. Crystallographic characterization of helical secondary structures in α/β-peptides with 1:1 residue alternation. J Am Chem Soc. 2008;130:6544–6550. doi: 10.1021/ja800355p. [DOI] [PubMed] [Google Scholar]
- 28.Hayen A, Schmitt MA, Ngassa FN, Thomasson KA, Gellman SH. Two helical conformations from a single foldamer backbone: “Split personality” in short α/β-peptides. Angew Chem Int Ed. 2004;43:505–510. doi: 10.1002/anie.200352125. [DOI] [PubMed] [Google Scholar]
- 29.Gogoi K, Kumar VA. Chimeric (α-amino acid + nucleoside-β-amino acid)n peptide oligomers show sequence specific DNA/RNA recognition. Chem Commun. 2008:706–708. doi: 10.1039/b716835g. [DOI] [PubMed] [Google Scholar]
- 30.Vilaivan T, Suparpprom C, Harnyuttanakorn P, Lowe G. Synthesis and properties of novel pyrrolidinyl PNA carrying β-amino acid spacers. Tetrahedron Lett. 2001;42:5533–5536. [Google Scholar]
- 31.Vilaivan T, Lowe G. A novel pyrrolidinyl PNA showing high sequence specificity and preferential binding to DNA over RNA. J Am Chem Soc. 2002;124:9326–9327. doi: 10.1021/ja012543u. [DOI] [PubMed] [Google Scholar]
- 32.Suparpprom C, Srisuwannaket C, Sangvanich P, Vilaivan T. Synthesis and oligodeoxynucleotide binding properties of pyrrolidinyl peptide nucleic acids bearing prolyl-2-aminocyclopentanecarboxylic acid (ACPC) backbones. Tetrahedron Lett. 2005;46:2833–2837. [Google Scholar]
- 33.Vilaivan T, Srisuwannaket C. Hybridization of pyrrolidinyl peptide nucleic acids and DNA: Selectivity, base-pairing specificity and direction of binding. Org Lett. 2006;8:1897–1900. doi: 10.1021/ol060448q. [DOI] [PubMed] [Google Scholar]
- 34.Ananthanawat C, Vilaivan T, Hoven VP, Su X. Comparison of DNA, aminoethylglycyl PNA and pyrrolidinyl PNA as probes for detection of DNA hybridization using surface plasmon resonance technique. Biosens Bioelectron. 2010;25:1064–1069. doi: 10.1016/j.bios.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 35.Boontha B, Nakkuntod J, Hirankarn N, Chaumpluk P, Vilaivan T. Multiplex mass spectrometric genotyping of single nucleotide polymorphisms employing pyrrolidinyl peptide nucleic acid in combination with ion-exchange capture. Anal Chem. 2008;80:8178–8186. doi: 10.1021/ac801336q. [DOI] [PubMed] [Google Scholar]
- 36.Taechalertpaisarn J, Sriwarom P, Boonlua C, Yotapan N, Vilaivan C, Vilaivan T. DNA-, RNA- and self-pairing properties of a pyrrolidinyl peptide nucleic acid with a (2′R,4′S)-prolyl-(1S,2S)-2-aminocyclopentanecarboxylic acid backbone. Tetrahedron Lett. 2010;51:5822–5826. [Google Scholar]
- 37.Egholm M, Buchardt O, Nielsen PE, Berg RH. Peptide nucleic acids (PNA). Oligonucleotide analogues with an achiral peptide backbone. J Am Chem Soc. 1992;114:1895–1897. [Google Scholar]
- 38.Kim SK, Nielsen PE, Egholm M, Buchardt O, Berg RH, Nordén B. Right-handed triplex formed between peptide nucleic acid PNA-T8 and poly(dA) shown by linear and circular dichroism spectroscopy. J Am Chem Soc. 1993;115:6477–6481. [Google Scholar]
- 39.Sforza S, Corradini R, Ghirardi S, Dossena A, Marchelli R. DNA binding of a D-lysine-based chiral PNA: Direction control and mismatch recognition. Eur J Org Chem. 2000:2905–2913. [Google Scholar]
- 40.Igloi GL. Variability in the stability of DNA-peptide nucleic acid (PNA) single-base mismatched duplexes. Real-time hybridization during affinity electrophoresis in PNA-containing gels. Proc Natl Acad Sci USA. 1998;95:8562–8567. doi: 10.1073/pnas.95.15.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen A, Nielsen PE. Unique properties of purine/pyrimidine asymmetric PNA·DNA duplexes: Differential stabilization of PNA·DNA duplexes by purines in the PNA strand. Biophys J. 2006;90:1329–1337. doi: 10.1529/biophysj.105.073213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. In an extreme case, the Tm difference between two complementary acpcPNA·DNA hybrids with purine rich (AGG TAA CGA G) and pyrimidine rich (CTC GTT ACC T) sequences is only 8.4°C compared to a difference of 26.0°C for the corresponding aegPNA·DNA hybrids under identical conditions (data extracted from reference 41). [Google Scholar]
- 43.Siriwong K, Chuichay P, Saen-oon S, Suparpprom C, Vilaivan T, Hannongbua S. Insight into why pyrrolidinyl peptide nucleic acid binding to DNA is more stable than the DNA·DNA duplex. Biochem Biophys Res Commun. 2008;372:765–771. doi: 10.1016/j.bbrc.2008.05.102. [DOI] [PubMed] [Google Scholar]
- 44.Kawakami J, Tanaka Y, Kishimoto K. Accurate curve fitting procedure for UV melting analysis of highly thermostable RNA hairpins. Nucleic Acids Symp Series. 2009;53:227–228. doi: 10.1093/nass/nrp114. [DOI] [PubMed] [Google Scholar]
- 45.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 46.Guckian KM, Schweitzer BA, Ren RXF, Sheils CJ, Tahmassebi DC, Kool ET. Factors contributing to aromatic stacking in water: Evaluation in the context of DNA. J Am Chem Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bommarito S, Peyret N, SantaLucia J., Jr Thermodynamic parameters for DNA sequences with dangling ends. Nucl Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugimoto N, Kierzek R, Turner DH. Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry. 1987;26:4554–4558. doi: 10.1021/bi00388a058. [DOI] [PubMed] [Google Scholar]
- 49.Demidov VV, Protozanova E, Izvolsky KI, Price C, Nielsen PE, Frank-Kamenetskii MD. Kinetics and mechanism of the DNA double helix invasion by pseudocomplementary peptide nucleic acids. Proc Natl Acad Sci USA. 2002;99:5953–5958. doi: 10.1073/pnas.092127999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishizuka T, Yoshida J, Yamamoto Y, Sumaoka J, Tedeschi T, Corradini R, et al. Chiral introduction of positive charges to PNA for double-duplex invasion to versatile sequences. Nucleic Acids Res. 2008;36:1464–1471. doi: 10.1093/nar/gkm1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 52.Ahlborn C, Siegmund K, Richert C. Isostable DNA. J Am Chem Soc. 2007;129:15218–15232. doi: 10.1021/ja074209p. [DOI] [PubMed] [Google Scholar]
- 53.Egholm M, Nielsen PE, Buchardt O, Berg RH. Recognition of guanine and adenine in DNA by cytosine and thymine containing peptide nucleic acids (PNA) J Am Chem Soc. 1992;114:9677–9678. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.