Abstract
Mammalian ribonucleases are emerging as cancer chemotherapeutic agents. Their cationicity engenders cell permeability, and their enzymatic activity destroys the biochemical information encoded by RNA. The pharmacologic potential of ribonucleases is, however, obviated by their high sensitivity to a cytosolic inhibitor protein (RI) and their small size, which limits their residence in serum. We reasoned that site-specific conjugation of a poly(ethylene glycol) (PEG) chain could both reduce sensitivity to RI and increase serum half-life. We found that appending a PEG moiety can enable bovine pancreatic ribonuclease (RNase A) to evade RI, depending on the site of conjugation and the length and branching of the chain. Although a pendant PEG moiety decreases antiproliferative activity in vitro, PEGylation discourages renal clearance in vivo and leads to nearly complete tumor growth inhibition in a mouse xenograft model. These data demonstrate that a pendant PEG moiety can be beneficial to the action of proteins that act within the cytosol, and that strategic site-specific PEGylation can endow a mammalian ribonuclease with potent antitumor activity.
Keywords: bioconjugation, biologic drug, cancer, cytotoxin, pharmacokinetics, poly(ethylene glycol), ribonuclease, ribonuclease inhibitor, site-directed mutagenesis, xenograft
Introduction
Homologs of bovine pancreatic ribonuclease (RNase A) exhibit innate antitumor activity both in vitro and in vivo.1–3 For reasons that are not yet clear, the toxicity of these enzymes is selective for cancer cells, stimulating their development as cancer chemotherapeutic agents. An amphibian homolog, ranpirnase, is currently in a Phase II clinical trial for the treatment of non-small cell lung cancer.
Unlike ranpirnase, mammalian pancreatic-type ribonucleases are highly susceptible to inhibition by the ubiquitous cytosolic ribonuclease inhibitor protein (RI) and are not cytotoxic.4–7 Variants of RNase A that have been engineered to evade RI are more potent cytotoxins in vitro than is ranpirnase.6 Because RNase A is less immunogenic than ranpirnase8 and RNase A variants can exhibit more selective cytotoxicity than ranpirnase,6 mammalian ribonucleases are more desirable chemotherapeutic agents than amphibian ones.
As small (12–14 kDa), parenterally administered proteins, ribonucleases are subject to rapid renal clearance.9 We reasoned that covalent modification of RNase A with poly(ethylene glycol) (PEG) could overcome this limitation and simultaneously reduce sensitivity to RI. A pendant PEG moiety increases hydrodynamic radius, which endows enhanced persistence in circulation and imparts resistance to proteolysis, improved solubility and reduced immunogenicity.10–13 A PEG moiety also provides steric bulk that could hinder the binding of RI.
The general utility of PEGylation is evidenced by the clinical efficacy of the many protein—PEG conjugates that are on the market or in late-stage clinical trials.13 Although these extant conjugates provide encouraging precedent for our work, a key difference exists. All extant PEGylated proteins act on an extracellular target. In contrast, a ribonuclease must gain access to intracellular RNA to exert its cytotoxic activity.
Early ribonuclease—PEG conjugates were made by randomly decorating the 11 amino groups of RNase A with linear PEG moieties.14–18 These RNase A-PEG conjugates exhibited enhanced persistence in circulation,16 as well as resistance to proteolytic degradation16,19 and a reduced affinity for anti-RNase A antibodies.15 In RNase A, the amino group of Lys41 has a low pKa of ∼8.8,20 and is thus especially reactive, but its modification reduces catalytic proficiency by 105-fold21 and obviates cytotoxicity.22,23 Indeed, extensive modification of the amino groups of RNase A with PEG led to the loss of 97% of its ribonucleolytic activity.15 Moreover, the necessary acylation of amino groups reduces net molecular charge, which is adverse to cellular binding and internalization, and hence cytotoxicity.24–26
Here, we report on the efficacy as a cancer chemotherapeutic agent of a mammalian ribonuclease that displays a site-specific PEG moiety. We find that these conjugates have somewhat less antiproliferative activity in vitro than expected from their ribonucleolytic activity in the presence of RI. These same conjugates, however, exert dramatic tumor growth inhibition at low dosing in xenograft mice bearing solid human tumors.
Results
Design of PEGylated variants of ribonuclease A.
We selected two distinct sites on RNase A for PEG conjugation. Residue 88 was chosen as a site within the interface of the RI·RNase A complex (Fig. 1), as single amino-acid substitutions there highly destabilize the RI·RNase A complex.6,27 Position 19 of RNase A was chosen as a site outside of the RI·RNase A interface because small-molecule appendages there do not lead to a detectable decrease in affinity for RI.28,29
Figure 1.
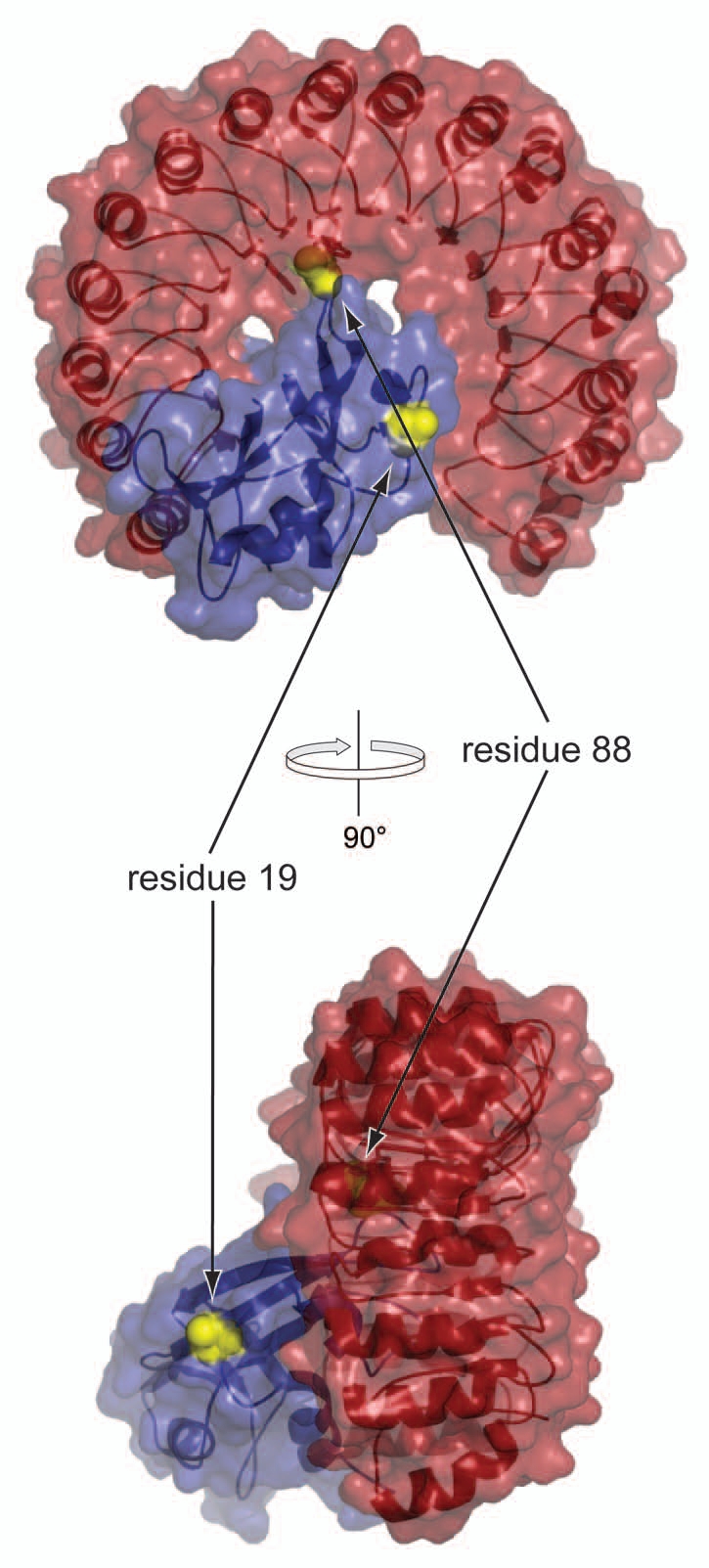
Three-dimensional structure of the complex between RNase A (blue) and RI (red). Virtual A19C and G88C substitutions (yellow spheres) were made in RNase A. Images were made with PyMol (Delano Scientific) and Protein Data Bank entry 1dfj.50
The size10 and branching19 of PEG chains can impact the biochemical properties of conjugates in vitro19 and their behavior in vivo.30 Hence, we chose 2 PEGs of different size: 2 kDa (Fig. 2A) and 20 kDa (Fig. 2B and C). With the 20-kDa mPEG, we used both a linear polymer (Fig. 2B) and one with two equivalent branches (Fig. 2C).
Figure 2.
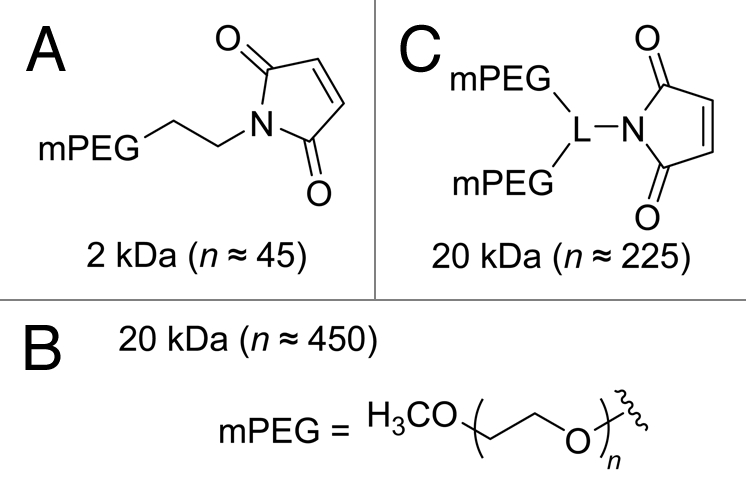
Chemical structures of the mPEG maleimides used for site-specific conjugation to Cys19 or Cys88 of RNase A variants. L = linker.
Physicochemical analyses of RNase A-PEG conjugates.
The observed molecular mass of each RNase A-PEG conjugate was consistent with its containing a single PEG moiety (Table S1). Discrepancies between expected and observed m/z values for the PEGylated variants of RNase A reflect the polydispersity of PEG moieties and the ensuing broad ion peaks.
The PEGylated variants of RNase A migrated more quickly during cation-exchange chromatography (Fig. S1) and more slowly during electrophoresis than expected from their molecular mass (Fig. S2 and Table S1). These observations are consistent with the extended structure and extensive hydration of the PEG moiety, as well as its shielding of the charge of the protein,31 which is highly basic.32 The effect of PEGylation was even more apparent during gel-filtration chromatography than during electrophoresis (Fig. S3 and Table S1).
Conformational stability.
Pancreatic-type ribonucleases must maintain their 3-dimensional structure in order to exert their cytotoxic enzymatic activity.33 Conformational stability was assessed by the value of Tm, which is the temperature at the midpoint of the thermal transition between the folded and unfolded states. Values of Tm for wild-type RNase A and its G88R variant are indistinguishable from those reported previously in reference 27. All PEGylated forms of the A19C and G88C variants exhibited values of Tm that were indistinguishable from that of wild-type RNase A (Fig. S4 and Table 1). Likewise, 20-kDa mPEG-D38R/R39D/N67R/G88C RNase A had a Tm value of 55°C (Table 1), which is not distinguishable from that of the unmodified variant.6 We conclude that appending a PEG moiety had a negligible effect on conformational stability, and that all conjugates retained their structure at physiological temperature.
Table 1.
Biochemical parameters of wild-type RNase A and its variants
| Ribonuclease A | Tma (°C) | kcat/KMb (107 M−1s−1) | Kdc (nM) | IC50d (µM) |
| Wild-type | 64 | 6.7 ± 0.3 | 44 × 10−6 | >25 |
| G88R | 63 | 7.4 ± 0.2 | 1.2 ± 0.2 | 3.6 ± 0.4 |
| D38R/R39D/N67R/G88R | 56 | 3.8 ± 0.6 | (1.4 ± 0.1) × 103 | 0.17 ± 0.01 |
| 2-kDa mPEG-A19C | 61 | 5.0 ± 0.4 | <1.2 | >25 |
| 20-kDa mPEG-A19C | 61 | 7.5 ± 0.2 | <1.2 | >25 |
| 20-kDa mPEG2-A19C | 61 | 6.9 ± 0.3 | <1.2 | >25 |
| 2-kDa mPEG-G88C | 64 | 4.0 ± 0.3 | 8.0 ± 1.3 | 10.4 ± 0.7 |
| 20-kDa mPEG-G88C | 64 | 6.8 ± 0.6 | 8.9 ± 0.1 | >25 |
| 20-kDa mPEG2-G88C | 64 | 6.1 ± 0.4 | 37 ± 2 | >25 |
| 20-kDa mPEG-D38R/R39D/N67R/G88C | 55 | 2.9 ± 0.1 | (3.3 ± 0.2) × 103 | 1.9 ± 0.2 |
Values of Tm (±2°C) were determined in PBS by UV spectroscopy. G88R RNase A, in reference 27; D38R/R39D/N67R/G88R RNase A.6
Values of kcat/KM (±SE) for wild-type RNase A and its variants are for catalysis of 6-FAM - dArU(dA)2 - 6-TAMRA cleavage in 0.10 M MES - NaOH buffer (pH 6.0) containing NaCl (0.10 M). D38R/R39D/N67R/G88R RNase A.6
Values of Kd (±SE) are for the complex with human RI in PBS. Wild-type RNase A, reference 49; G88R RNase A;48 D38R/R39D/N67R/G88R RNase A.6
Values of IC50 (±SE ) are for incorporation of [methyl-3H]thymidine into the DNA of K-562 cells (Fig. 3).
Ribonucleolytic activity.
Ribonucleolytic activity is essential to the antiproliferative activity of pancreatic-type ribonucleases.22 Conjugation of the A19C and G88C variants with linear or branched PEG moieties had little effect on their ability to catalyze the cleavage of a small substrate, 6-FAM-dArUdAdA-6-TAMRA (Table 1). Only 20-kDa mPEG-D38R/R39D/N67R/G88C RNase A showed a noticable (i.e., 2.3-fold) decrease in catalytic activity compared with the wild-type enzyme, but this decrease is in accord with that experienced by the analogous (unmodified) D38R/R39D/N67R/G88R variant.6 Values of kcat/KM for wild-type RNase A and its G88R variant were in agreement with those reported previously in reference 27.
Similarly, conjugation caused only a modest (i.e., < 2-fold) decrease in the ability of a ribonuclease to catalyze the cleavage of a large substrate, poly(C) (Table 2). This decrease was the result of a larger KM value, which is consistent with some occlusion of the larger substrate from the enzymic active site by the PEG moiety.
Table 2.
Steady-state kinetic parameters for catalysis of poly(C) cleavage by wild-type RNase A and its PEGylated variantsa
| Ribonuclease | kcat (s−1) | KM (µM) | kcat/KM (106 M−1s−1) |
| Wild-type RNase A | 368 ± 11 | 33 ± 3 | 11.2 ± 0.6 |
| 2-kDa mPEG-G88C RNase A | 422 ± 12 | 49 ± 6 | 8.8 ± 0.8 |
| 20-kDa mPEG-G88C RNase A | 428 ± 21 | 54 ± 4 | 8.0 ± 0.2 |
| 20-kDa mPEG2-G88C RNase A | 402 ± 16 | 55 ± 2 | 7.4 ± 0.6 |
Values (±SE) were obtained from assays in 0.10 M MES-NaOH buffer (pH 6.0) containing NaCl (0.10 M), poly(C) (0.010–1.5 mM), and enzyme (2 nM).
Ribonuclease inhibitor binding.
The site of PEG attachment affected the ability of RNase A to evade RI (Table 1). The high affinity of the A19C RNase A conjugates for RI was beyond the resolution of the assay (Kd < 1.2 nM). In contrast, the G88C conjugates were much more evasive of the inhibitor. For example, the 2-kDa mPEG-G88C RNase A conjugate was almost 7-fold more evasive than the G88R variant. Attaching a linear 20-kDa PEG chain reduced the affinity for RI slightly more, and attaching a branched 20-kDa PEG chain reduced the affinity still further. This 20-kDa mPEG2 moiety at residue 88 was 31-fold more effective at precluding the binding of RI than was the side chain of an arginine residue. Attaching a PEG group at position 88 in the context of other amino-acid substitutions (D38R/R39D/N67R) that were known to disrupt the RI·RNase A complex6 resulted in the most RI-evasive monomeric RNase A reported to date (Kd = 3.3 µM).
Cell proliferation.
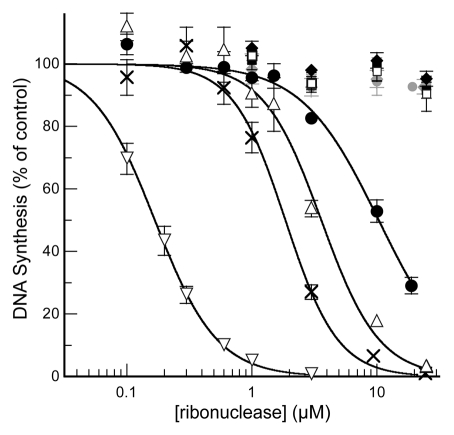
PEGylation affected the antiproliferative activity of RNase A in vitro, as revealed by assaying the inhibition of cellular DNA synthesis (Fig. 3 and Table 1). The 2-kDa mPEG-G88C RNase A and 20-kDa mPEG-D38R/R39D/N67R/G88C RNase A conjugates, which had IC50 values of 10.4 and 1.9 µM, respectively, retained much of the antiproliferative activity of the analogous G88R and D38R/R39D/N67R/G88R variants, which lack a PEG moiety. The IC50 values for these two variants were in agreement with those determined previously in reference 6. The remaining PEG conjugates had a lesser effect on the proliferation of K-562 cells. These data are consistent with the 2-kDa mPEG chain not being too detrimental to cytosolic uptake (vide infra), and the 20-kDa mPEG-D38R/R39D/N67R/G88C variant being especially evasive of RI (Table 1).
Figure 3.
Effect of RNase A with site-specific PEGylation on DNA synthesis by K-562 cells in vitro. Data points are the mean (±SE) of at least three separate experiments performed in triplicate. Values of IC50 are listed in Table 1. Wild-type RNase A (□), G88R RNase A (▵) and D38R/R39D/N67R/G88R RNase A (▿), 2-kDa mPEG-A19C RNase A (gray ●), 20-kDa mPEG-A19C RNase A (gray ▪), 20-kDa mPEG2-A19C RNase A (gray ◆), 2-kDa mPEG-G88C RNase A (black ●), 20-kDa mPEG-G88C RNase A (black ▪), 20-kDa mPEG2-G88C RNase A (black ◆) and 20-kDa mPEG-D38R/R39D/N67R/G88C RNase A (x).
DU145 cells were even less sensitive in vitro to PEGylated ribonucleases than were K-562 cells. 2-kDa mPEG-G88C RNase A, 20-kDa mPEG-G88C RNase A and 20-kDa mPEG2-G88C RNase A each had an IC50 value of >25 µM (data not shown). In contrast, D38R/D39R/N67R/G88R RNase A had an IC50 value of 0.2 µM, which is comparable to a previous report in reference 6.
Tumor growth inhibition.
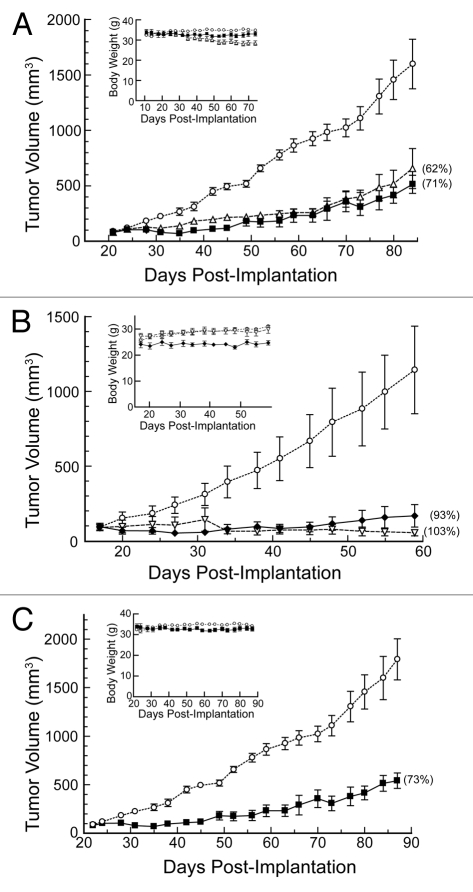
2-kDa mPEG-G88C RNase A was the simplest PEGylated variant to demonstrate antiproliferative activity in vitro (Fig. 3), and hence was used in initial assays of antitumoral activity in vivo. This variant (15 mg/kg; qd × 5; i.p.) inhibited tumor growth significantly (TGI = 71%; Fig. 4A). The inhibition was greater than that from a non-PEGylated variant that is much more RI-evasive (D38R/R39D/N67R/G88R RNase A, TGI = 62%). The 2-kDa mPEG-G88C variant also caused less loss in body weight than did the non-PEGylated variants (−2% vs. −16 and −6%, respectively).
Figure 4.
Effect of RNase A with site-specific PEGylation on the tumor volume and body weight (insets) of Balb c(-/-) mice in xenograft models bearing human DU145 prostate tumors. Data points are the mean (±SE) for n mice; % TGI values are in parentheses. Vehicle control (○; n = 7). (A) 2-kDa mPEG-G88C RNase A (▪, 11.2 mg/kg; i.p., qdx 5, n = 3) and D38R/R39D/N67R/G88R RNase A (▵, 15 mg/kg; i.p., qdx 5, n = 7). (B) 20-kDa mPEG2-G88C RNase A (◆, 75 mg/kg; i.p., 1x wk; n = 5) and docetaxel (▿, 8 mg/kg; i.p., 1x wk, n = 7). One mouse treated with docetaxel died on day 42 and another on day 68. (C) 2-kDa mPEG-G88C RNase A (▪, 15 mg/kg; i.p., 2x wk, n = 3).
20-kDa mPEG2-G88C RNase A was likewise assayed for antitumoral activity in vivo. A once-weekly dose of 20-kDa mPEG2-G88C RNase A was comparable in efficacy to docetaxel (8 mg/kg; 1x wk; i.p.; TGI = 103%), which is an antimitotic agent in common use (Fig. 4B). Notably, the weekly molar dose of 20-kDa mPEG2-G88C RNase A (33.7 kg/mol) was 4.4-fold less than that of docetaxel (808 g/mol). The 20-kDa mPEG2-G88C RNase A was well-tolerated, as indicated by a 2% gain in body weight over the duration of the study.
Finally, the 2-kDa mPEG-G88C RNase A was tested at a lower dose (15 mg/kg; 2x wk; i.p.). Even at this low dose, the PEG conjugate inhibited tumor growth significantly (Fig. 4C; TGI = 73%). Again, only minimal toxicity was seen, with body weight decreasing by 2% over the course of the study. Overall, the data indicate that the both 2-kDa mPEG-G88C RNase A and 20-kDa mPEG2-G88C RNase A are effective and safe anti-cancer agents in vivo.
Pharmacokinetics.
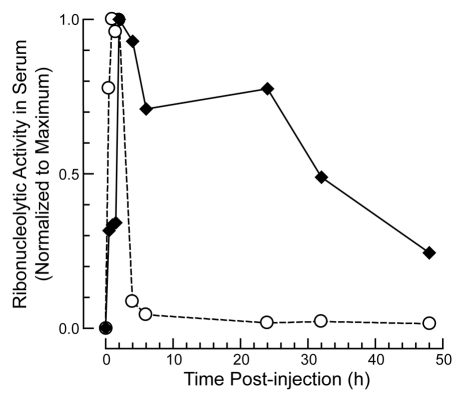
Wild-type RNase A clears in 5 and 3½ min from the serum of rats and mice, respectively.35 This rapid rate is consistent with that of other small proteins.36,37 Previously, a 5-kDa PEGylated RNase A had been shown to exhibit a 40- to 50-fold increase in circulation time in rats.16 A similarly large enhancement was observed in mice injected with 20-kDa mPEG2-G88C RNase A compared with that of G88R RNase A (Fig. 5). Peak serum levels were the same for both G88R RNase A and 20-kDa mPEG2-G88C RNase A, but the half-life increased from 0.4 to 62 h. Likewise the area under the curve, which is indicative of total exposure levels, also increased over 30-fold. These data confirm that PEG conjugation has enhanced the pharmacokinetic parameters of RNase A and might facilitate its anti-tumor activity.
Figure 5.
Effect of site-specific PEGylation of RNase A on its persistence in the circulation of mice. 20-kDa mPEG-G88C RNase A (◆; 15 mg/kg) G88R RNase A (○; 15 mg/kg).
Discussion
Mammalian ribonucleases are an emerging class of cancer chemotherapeutic agents,1–3 but limitations exist. Previously, we showed that variants of RNase A, engineered to evade RI, are potent cytotoxins.6,27,34,38 Nonetheless, the relatively small size of ribonucleases allows for their rapid clearance from circulation via glomerular filtration.37 Here, we set out to address simultaneously both the sub-optimal pharmacokinetic properties of RNase A and its sensitivity to RI by attaching PEG to a specific cysteine residue installed by site-directed mutagenesis. We found that this site-specific conjugation preserved both conformational stability and ribonucleolytic activity (Tables 1 and 2), which are essential to antiproliferative activity.
PEGylation can mediate RI evasion.
The influence of PEGylation on RI-binding had never been examined previously. We find that the site of PEGylation, and the length and branching order of the pendant PEG moiety influence the affinity for RI (Table 1). Our data are consistent with the known reduced affinity of PEGylated RNase A for anti-RNase A antibodies15 and lower susceptibility to proteolytic degradation.19 Moreover, the lower affinity of RI for 20-kDa mPEG-D38R/R39D/N67R/G88C RNase A than 20-kDa mPEG-G88C RNase A demonstrates that other amino-acid substitutions can augment RI-evasion gained through PEGylation.
PEGylation affects antiproliferative activity in vitro.
Based on its high conformational stability, catalytic activity and ability to evade RI (Table 1), 20-kDa mPEG2-G88C RNase A should be a potent inhibitor on cancer-cell proliferation.6,23,39 Yet, this conjugate demonstrated only weak antiproliferative activity in vitro (Fig. 3 and Table 1). PEGylation is known to reduce affinity for a target receptor, and a loss of in vitro activity is a common occurrence.12 We attribute the lower in vitro activity, at least in part, to the inefficient cellular association and internalization that results from the shielding of its cationicity by the PEG moiety. This hypothesis is supported by the reduced affinity of PEGylated variants for a cation-exchange resin (Fig. S1), which mimics the anionic glycans that play a critical role in the cellular internalization of ribonucleases.26,40–42
PEGylation enhances antitumoral activity.
Despite the attenuated bioactivity of mPEG-G88C RNase A in vitro, these conjugates are more effective than non-PEGylated variants at inhibiting tumor growth in a xenograft model (Fig. 4). Because PEGylation discourages renal clearance (Fig. 5), we conclude that the ensuing increase in persistence more than compensates for any decreased antiproliferative activity.
A biological implication.
Our data have an implication about natural human ribonucleases. Gly88 of RNase A is homologous to Asn88 of human pancreatic ribonuclease. Like Gly88 in RNase A,27 Asn88 in human pancreatic ribonuclease is a key RI-contact site.43 Asn88 is known to be N-glycosylated in humans.44 Given the resemblance of glycans to PEG, we speculate that a natural N-glycosylated ribonuclease could evade RI and provide humans with endogenous antitumor activity.
Materials and Methods
Materials.
The preparation of proteins and sources of other materials are described in the Supplemental Materials.
Assays of conformational stability.
The conformational stability of PEGylated RNase A variants was determined by measuring their Tm value as described previously in reference 34.
Assays of catalytic activity.
Ribonucleolytic activities of the PEGylated RNase A variants were determined by assaying the cleavage of 2 RNA substrates, one small and one large. The small substrate, 6-FAM-dArUdAdA-6-TAMRA (20 nM), exhibits a ∼180-fold increase in fluorescence (λex = 493 nm; λem = 515 nm) upon cleavage.45 Assays of its cleavage were performed in 2.0 mL of 0.10 M MES-NaOH buffer (pH 6.0) containing NaCl (0.10 M). The MES used to prepare the assay buffer was purified by anion-exchange chromatography to remove trace amounts of oligomeric vinylsulfonic acid, which is a byproduct of commercial buffer synthesis and a potent inhibitor of RNase A.46 Values of kcat/KM were obtained as described previously in reference 45.
The large substrate, poly(cytidylic acid) (poly(C); ε = 6,200 M−1cm−1 per nucleotide at 268 nm), is hyperchromic, allowing its cleavage to be monitored by an increase in UV absorption (Δε = 2,380 M−1cm−1 at 250 nm).47 Assays of its cleavage were performed in 0.10 M MES-NaOH buffer (pH 6.0) containing NaCl (0.10 M),poly(C) (0.010–1.5 mM) and enzyme (2 nM). According to its supplier, the poly(C) had ≥250 nucleotides, with an average size of 300–800 nucleotides. Values of kcat and KM were calculated by fitting initial velocity data to the Michaelis-Menten equation.
Assays of ribonuclease inhibitor binding.
The affinity of PEGylated RNase A variants for human RI in PBS was determined by using the fluorescence-based competition assay described previously in reference 29 and 48.
Assays of cell proliferation.
Pancreatic-type ribonucleases can inhibit cell proliferation by evoking apoptosis.39 This activity was assessed by measuring the effect of ribonucleases on the incorporation of [methyl-3H]thymidine into the nascent DNA of two human cancer cell lines: K-562 (myelogenous leukemia) and DU145 (prostate cancer), as described previously in reference 27 and 34. Values of IC50 were calculated by fitting the curves using nonlinear regression to a sigmoidal dose—response curve.
Assays of tumor growth inhibition.
Variants and PEG conjugates of RNase A were tested for their ability to suppress the growth of human tumors implanted into the flanks of male athymic (nu/nu) mice. The DU145 tumor cell line was selected for its ability to proliferate in mice, low rate of spontaneous regression and clinical relevancy. The DU145 xenographs were treated intraperitoneally (i.p.) with both the RNase A variants and 2- and 20-kDa PEG conjugates. Assays of tumor growth inhibition (TGI) were performed as described previously in reference 34.
Pharmacokinetic studies.
The pharmacokinetic (PK) profile of the 20-kDa PEG conjugate and the parental RNase A variant was determined essentially as described previously in reference 35, that is, by measuring ribonucleolytic activity in serum. Normal male CD-1 mice (two or three) were injected once intravenously (i.v.) with a ribonuclease (15 mg/kg). Blood was collected at various time intervals after dosing. The blood was allowed to clot at 2–8°C. Within 30 min, samples were subjected to centrifugation at ∼1,500 g for 5–10 min to collect serum, which was stored frozen at −80°C. For analysis, frozen samples were thawed on ice. Ribonucleolytic activity within samples was assayed in a 96-well plate using the fluorogenic substrate 6-FAM-dArUdAdA-6-TAMRA. Pre-diluted serum (10 µL of a 1:10,000 dilution) was added to 160 µL of 0.10 M Tris-HCl buffer (pH 7.0) containing NaCl (0.10 M) and acetylated BSA (0.10 mg/mL). The assay was initiated by the addition of substrate (30 µL of a 1.33 µM stock solution). Fluorescence (λex = 490 nm; λem = 525 nm) was measured with a Tecan Safire plate reader.
Acknowledgments
We are grateful to Drs. S.M. Fuchs, J. Kalia, L.D. Lavis, J.E. Lee and B.D. Smith for contributive discussions. We also acknowledge L.D. Lavis for the synthesis of trifluoroacetate salt of 2-maleimidoethylamine(5). This work was supported by grant R01 CA073808 (NIH). T.J.R. was supported by NIH Biotechnology Training Grant T32 GM08349 and a William R. and Dorothy E. Sullivan Wisconsin Distinguished Graduate Fellowship. Instrumentation was purchased with grants S10 RR013790 (NIH) and BIR-9512577 (NSF), and by a grant from the W.M. Keck Foundation.
Abbreviations
- 6-FAM
6-carboxyfluorescein
- MES
2-(N-morpholino)ethanesulfonic acid
- MALDI-TOF
matrix-assisted laser desorption ionization-time-of-flight
- mPEG
monomethoxypoly(ethylene glycol)
- PBS
phosphate-buffered saline
- RI
ribonuclease inhibitor
- RNase A
bovine pancreatic ribonuclease
- PAGE
poly(acrylamide) gel electrophoresis
- SDS
sodium dodecyl sulfate
- 6-TAMRA
6-carboxytetramethylrhodamine
- Tris
tris(hydroxymethyl)aminomethane
- UV
ultraviolet
Supplementary Material
References
- 1.Arnold U, Ulbrich-Hofmann R. Natural and engineered ribonucleases as potential cancer therapeutics. Biotechnol Lett. 2006;28:1615–1622. doi: 10.1007/s10529-006-9145-0. [DOI] [PubMed] [Google Scholar]
- 2.Lee JE, Raines RT. Ribonucleases as novel chemotherapeutics: The ranpirnase example. BioDrugs. 2008;22:53–58. doi: 10.2165/00063030-200822010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang EF, Ng TB. Ribonucleases of different origins with a wide spectrum of medicinal applications. Biochim Biophys Acta. 2011;1815:65–74. doi: 10.1016/j.bbcan.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Murthy BS, Sirdeshmukh R. Sensitivity of monomeric and dimeric forms of bovine seminal ribonuclease to human placental ribonuclease inhibitor. Biochem J. 1992;281:343–348. doi: 10.1042/bj2810343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boix E, Wu Y, Vasandani VM, Saxena SK, Ardelt W, Ladner J, et al. Role of the N terminus in RNase A homologues: Differences in catalytic activity, ribonuclease inhibitor interaction and cytotoxicity. J Mol Biol. 1996;257:992–1007. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- 6.Rutkoski TJ, Kurten EL, Mitchell JC, Raines RT. Disruption of shape-complementarity markers to create cytotoxic variants of ribonuclease A. J Mol Biol. 2005;354:41–54. doi: 10.1016/j.jmb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Rutkoski TJ, Raines RT. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr Pharm Biotechnol. 2008;9:185–189. doi: 10.2174/138920108784567344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matoušek J, Souček J, Slavík T, Tománek M, Lee JE, Raines RT. Comprehensive comparison of the cytotoxic activities of onconase and bovine seminal ribonuclease. Comp Biochem Physiol C Toxicol Pharmacol. 2003;136:343–356. doi: 10.1016/j.cca.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Gad SC. Handbook of Pharmaceutical Biotechnology. Hoboken NJ: John Wiley and Sons, Inc.; 2007. [Google Scholar]
- 10.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 11.Greenwald RB, Choe YH, McGuire J, Conover CD. Effective drug delivery by PEGylated drug conjugates. Adv Drug Deliv Rev. 2003;55:217–250. doi: 10.1016/S0169-409X(02)00180-1. [DOI] [PubMed] [Google Scholar]
- 12.Fishburn CS. The pharmacology of PEGylation: Balancing PD with PK to generate novel therapeutics. J Pharm Sci. 2008;97:4167–4183. doi: 10.1002/jps.21278. [DOI] [PubMed] [Google Scholar]
- 13.Pasut G, Sergi M, Veronese FM. Anti-cancer PEGenzymes: 30 years old, but still a current approach. Adv Drug Deliv Rev. 2008;60:69–78. doi: 10.1016/j.addr.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 14.Veronese FM, Largajolli R, Boccu E, Benassi CA, Schiavon O. Surface modification of proteins. Activation of monomethoxy-polyethylene glycols by phenylchloroformates and modification of ribonuclease and superoxide dismutase. Appl Biochem Biotechnol. 1985;11:141–152. doi: 10.1007/BF02798546. [DOI] [PubMed] [Google Scholar]
- 15.Caliceti P, Schiavon O, Veronese FM, Chaiken IM. Effects of monomethoxypoly(ethylene glycol) modification of ribonuclease on antibody recognition, substrate accessibility and conformational stability. J Mol Recognit. 1990;3:89–93. doi: 10.1002/jmr.300030206. [DOI] [PubMed] [Google Scholar]
- 16.Schiavon O, Caliceti P, Sartore L, Veronese FM. Surface modification of enzymes for therapeutic use: Monomethoxypoly(ethylene glycol) derivatization of ribonuclease. Farmaco. 1991;46:967–978. [PubMed] [Google Scholar]
- 17.Matoušek J, Poucková P, Souček J, Škvor J. PEG chains increase aspermatogenic and antitumor activity of RNase A and BS-RNase enzymes. J Control Release. 2002;82:29–37. doi: 10.1016/S0168-3659(02)00082-2. [DOI] [PubMed] [Google Scholar]
- 18.Matoušek J, Poucková P, Hloušková D, Zadinová M, Souček J, Škvor J. Effect of hyaluronidase and PEG chain conjugation on the biologic and antitumor activity of RNase A. J Control Release. 2004;94:401–410. doi: 10.1016/j.jconrel.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 19.Monfardini C, Schiavon O, Caliceti P, Morpurgo M, Harris JM, Veronese FM. A branched monomethoxypoly(ethylene glycol) for protein modification. Bioconjug Chem. 1995;6:62–69. doi: 10.1021/bc00031a006. [DOI] [PubMed] [Google Scholar]
- 20.Murdock AL, Grist KL, Hirs CHW. On the dinitrophenylation of bovine pancreatic ribonuclease A. Kinetics of the reaction in water and 8 M urea. Arch Biochem Biophys. 1966;114:375–390. doi: 10.1016/0003-9861(66)90350-X. [DOI] [Google Scholar]
- 21.Messmore JM, Fuchs DN, Raines RT. Ribonuclease A. Revealing structure-function relationships with semisynthesis. J Am Chem Soc. 1995;117:8057–8060. doi: 10.1021/ja00136a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JS, Souček J, Matoušek J, Raines RT. Catalytic activity of bovine seminal ribonuclease is essential for its immunosuppressive and other biological activities. Biochem J. 1995;308:547–550. doi: 10.1042/bj3080547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bretscher LE, Abel RL, Raines RT. A ribonuclease A variant with low catalytic activity but high cytotoxicity. J Biol Chem. 2000;275:9893–9896. doi: 10.1074/jbc.275.14.9893. [DOI] [PubMed] [Google Scholar]
- 24.Futami J, Maeda T, Kitazoe M, Nukui E, Tada H, Seno M, et al. Preparation of potent cytotoxic ribonucleases by cationization: Enhanced cellular uptake and decreased interaction with ribonuclease inhibitor by chemical modification of carboxyl groups. Biochemistry. 2001;40:7518–7524. doi: 10.1021/bi010248g. [DOI] [PubMed] [Google Scholar]
- 25.Chapman AP. PEGylated antibodies and antibody fragments for improved therapy: A review. Adv Drug Deliv Rev. 2002;54:531–545. doi: 10.1016/S0169-409X(02)00026-1. [DOI] [PubMed] [Google Scholar]
- 26.Johnson RJ, Chao TY, Lavis LD, Raines RT. Cytotoxic ribonucleases: The dichotomy of coulombic forces. Biochemistry. 2007;46:10308–10316. doi: 10.1021/bi700857u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leland PA, Schultz LW, Kim BM, Raines RT. Ribonuclease A variants with potent cytotoxic activity. Proc Natl Acad Sci USA. 1998;95:10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kothandaraman S, Hebert MC, Raines RT, Nibert ML. No role for pepstatin-A-sensitive acidic proteinases in reovirus infections of L or MDCK cells. Virology. 1998;251:264–272. doi: 10.1006/viro.1998.9434. [DOI] [PubMed] [Google Scholar]
- 29.Abel RL, Haigis MC, Park C, Raines RT. Fluorescence assay for the binding of ribonuclease A to the ribonuclease inhibitor protein. Anal Biochem. 2002;306:100–107. doi: 10.1006/abio.2002.5678. [DOI] [PubMed] [Google Scholar]
- 30.Yamaoka T, Tabata Y, Ikada Y. Distribution and tissue uptake of poly(ethylene glycol) with different molecular weights after intravenous administration to mice. J Pharm Sci. 1994;83:601–606. doi: 10.1002/jps.2600830432. [DOI] [PubMed] [Google Scholar]
- 31.Kurfürst MM. Detection and molecular weight determination of polyethylene glycol-modified hirudin by staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1992;200:244–248. doi: 10.1016/0003-2697(92)90460-O. [DOI] [PubMed] [Google Scholar]
- 32.Chao TY, Lavis LD, Raines RT. Cellular uptake of ribonuclease A relies on anionic glycans. Biochemistry. 2010;49:10666–10673. doi: 10.1021/bi1013485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klink TA, Raines RT. Conformational stability is a determinant of ribonuclease A cytotoxicity. J Biol Chem. 2000;275:17463–17467. doi: 10.1074/jbc.M001132200. [DOI] [PubMed] [Google Scholar]
- 34.Rutkoski TJ, Kink JA, Strong LE, Schilling CI, Raines RT. Antitumor activity of ribonuclease multimers created by site-specific covalent tethering. Bioconjug Chem. 2010;21:1691–1702. doi: 10.1021/bc100292x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarnowski GS, Kassel RL, Mountain IM, Blackburn P, Wilson G, Wang D. Comparison of antitumor activities of pancreatic ribonuclease and its cross-linked dimer. Cancer Res. 1976;36:4074–4078. [PubMed] [Google Scholar]
- 36.Venkatachalam MA, Rennke HG. The structural and molecular basis of glomerular filtration. Circ Res. 1978;43:337–347. doi: 10.1161/01.res.43.3.337. [DOI] [PubMed] [Google Scholar]
- 37.Maack T, Johnson V, Kau ST, Figueiredo J, Sigulem D. Renal filtration, transport and metabolism of low-molecular-weight proteins: A review. Kidney Int. 1979;16:251–270. doi: 10.1038/ki.1979.128. [DOI] [PubMed] [Google Scholar]
- 38.Haigis MC, Kurten EL, Abel RL, Raines RT. KFERQ sequence in ribonuclease A-mediated cytotoxicity. J Biol Chem. 2002;277:11576–11581. doi: 10.1074/jbc.M112227200. [DOI] [PubMed] [Google Scholar]
- 39.Haigis MC, Kurten EL, Raines RT. Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res. 2003;31:1024–1032. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soncin F, Strydom DJ, Shapiro R. Interaction of heparin with human angiogenin. J Biol Chem. 1997;272:9818–9824. doi: 10.1074/jbc.272.15.9818. [DOI] [PubMed] [Google Scholar]
- 41.Fuchs SM, Raines RT. Internalization of cationic peptides: The road less (or more?) traveled. Cell Mol Life Sci. 2006;63:1819–1822. doi: 10.1007/s00018-006-6170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turcotte RF, Lavis LD, Raines RT. Onconase cytotoxicity relies on the distribution of its positive charge. FEBS J. 2009;276:3846–3857. doi: 10.1111/j.1742-4658.2009.07098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson RJ, McCoy JG, Bingman CA, Phillips GN, Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007;368:434–449. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribó M, Beintema JJ, Osset M, Fernández E, Bravo J, de Llorens R, et al. Heterogeneity in the glycosylation pattern of human pancreatic ribonuclease. Biol Chem Hoppe Seyler. 1994;375:357–363. [PubMed] [Google Scholar]
- 45.Kelemen BR, Klink TA, Behlke MA, Eubanks SR, Leland PA, Raines RT. Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 1999;27:3696–3701. doi: 10.1093/nar/27.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith BD, Soellner MB, Raines RT. Potent inhibition of ribonuclease A by oligo(vinylsulfonic acid) J Biol Chem. 2003;278:20934–20938. doi: 10.1074/jbc.M301852200. [DOI] [PubMed] [Google Scholar]
- 47.delCardayré SB, Raines RT. Structural determinants of enzymatic processivity. Biochemistry. 1994;33:6031–6037. doi: 10.1021/bi00186a001. [DOI] [PubMed] [Google Scholar]
- 48.Lavis LD, Rutkoski TJ, Raines RT. Tuning the pKa of fluorescein to optimize binding assays. Anal Chem. 2007;79:6775–6782. doi: 10.1021/ac070907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee FS, Shapiro R, Vallee BL. Tight-binding inhibition of angiogenin and ribonuclease A by placental ribonuclease inhibitor. Biochemistry. 1989;28:225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]
- 50.Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.