Abstract
Amyloid fibrils share a structural motif consisting of highly ordered β-sheets aligned perpendicular to the fibril axis.1, 2 At each fibril end, β-sheets provide a template for recruiting and converting monomers.3 Different amyloid fibrils often co-occur in the same individual, yet whether a protein aggregate aids or inhibits the assembly of a heterologous protein is unclear. In prion disease, diverse prion aggregate structures, known as strains, are thought to be the basis of disparate disease phenotypes in the same species expressing identical prion protein sequences.4–7 Here we explore the interactions reported to occur when two distinct prion strains occur together in the central nervous system.
Key words: prion, prions, strain, TSE, interaction, amyloid, LCP, neurodegeneration, aggregation
Diverse Misfolded Proteins in Disease
Protein misfolding and the accumulation of highly-ordered, β-sheet rich, insoluble aggregates are implicated in a diverse group of neurodegenerative diseases, including prion, Alzheimer, Parkinson and Huntington disease. In aged patients, often different aggregated proteins coexist. For example in Alzheimer disease (AD), amyloid-β plaques and tau neurofibrillary tangles often coexist. Whether the co-occurrence of multiple aggregated proteins indicates a failure in shared clearance pathways or a fibrillized protein nucleating a second protein is unclear.
In prion disease, two conformationally distinct subtypes of PrPSc, the misfolded prion protein, have been shown to co-exist in up to 50% of patients with sporadic Creutzfeldt-Jakob disease (sCJD).8–12 Each subtype was localized to a particular brain region, and the spatial distribution of the subtypes did not change due to the presence of the second subtype. Indeed, the two subtypes also differed in their biochemical characteristics, including the proteinase-K resistant core size,11 suggesting that the subtypes were structurally different.
In addition to sporadic human prion disease, the coexistence of prion strains commonly arises when prions cross species barriers13,14 and may be due to PrP sequence differences between the incoming PrPSc and the host cellular prion protein, PrPC.15 Sequence mismatches may shift the range of newly generated PrPSc conformations that can be accommodated by the initial seed template. The resulting mix of strains has implications for the treatment of infectious prion disease, as a drug may target only one strain and thus enable rarer strains to propagate.16 Strain mixtures can also have an impact on the pathogenesis, as interference between strains can alter prion replication.
Here we review the mechanisms of prion strain interference, interactions between conformationally distinct prion strains and new tools to track protein aggregates known as luminescent conjugated oligo- or polythiophenes (LCOs or LCPs).
Prion Strain Interference
Co-infection of two prion strains can result in interference where one prion strain extends the incubation period (ip) or completely blocks a second, superinfecting strain from causing disease.17,18 Prion interference has been described in mice and hamsters infected with a wide variety of strains and routes of inoculation suggesting that it is a common property of prions.18–22 Additionally, interference may be involved in the emergence of a dominant strain from a mixture that occurs following interspecies transmission.13,23–26 Interestingly, prion strain interference has been recapitulated in vitro using protein misfolding cyclic amplification (PMCA).27
Several parameters are known to govern prion strain interference. First, the blocking strain must be infectious in order to interfere with the superinfecting strain. Treatments that destroy prion infectivity eliminate the ability of the blocking strain to interfere.28 Second, the blocking-to-superinfecting strain ratio influences the interference, in that the higher the PrPSc quantity of the blocking strain, the more effectively the ip is prolonged.19,29 In some cases, the PrPSc from the superinfecting strain becomes undetectable.22,30 Third, increasing the interval between exposure to the two strains increases the interference ability of the blocking strain.17,18,30 Taken together, these studies suggest that replication of the blocking strain is required for strain interference to occur.30 Finally, recent reports have indicated that for prion interference to occur, the blocking and superinfecting strains must infect a common population of neurons.22,30 Overall, the relative onset of replication of interfering strains in a common population of neurons is the critical factor that determines which prion strain will emerge.
The mechanisms of strain interference are beginning to be deciphered in studies using the HY and DY strains of TME. Animal studies utilizing the sciatic nerve route of infection have determined that HY and DY prions are transported to the same populations of neurons in the lumbar spinal cord.31 Inoculation of the sciatic nerve with the DY prions prior to inoculation with HY completely blocks HY from infecting the spinal cord, while inoculation of HY prions into the opposite sciatic nerve leads to HY readily infecting the spinal cord.30 In these animals, the only observed difference is that when two strains are inoculated into the same sciatic nerve, neuronal deposits in the spinal cord consist of exclusively DY and lack any trace of HY PrPSc. Interestingly, HY and DY PrPSc each typically localize to the cell surface of neurons.32 Together, these in vivo data suggest that (1) DY and HY PrPSc are competing for a limiting host resource, (2) HY and DY prion interference likely occurs at the neuronal cell surface and (3) an immune response to the DY prions is not involved in the interference.
PMCA recapitulates strain interference between HY and DY prions in vitro similar to what is observed in vivo, indicating that the competing factors are present in this system.27 These in vitro studies suggest that prion strain interference is not due to DY PrPSc rapidly converting all of the available PrPC to PrPSc. After one round of PMCA, PrPSc consistently accumulates to higher levels in the HY compared to DY seeded reactions, consistent with the strain-specific in vivo rates of PrPSc accumulation.27 If DY PrPSc were converting all of the available PrPC to PrPSc in the PMCA reaction, then DY and HY PrPSc abundance would be predicted to be equal. A possible explanation is that DY PrPSc sequesters PrPC or another required co-factor (e.g., RNA, glycosaminoglycans), rendering it unavailable to HY PrPSc.33–36 This is consistent with the observation that a vast excess of DY PrPSc compared to HY PrPSc is required for interference.
LCPs Discriminate Distinct Prion Conformations In Vitro and In Vivo
Methods to visualize and track HY and DY or other prion strains in tissue sections have been limited. Small hydrophobic molecules such as thioflavin T (ThT) or Congo red have been used to detect protein aggregates having an extensive cross β-pleated sheet conformation. However, these conventional dyes are unable to distinguish heterogeneous populations of aggregated proteins because they are sterically rigid. A new class of amyloid ligands, the LCPs, consists of a polythiophene backbone that freely rotates and allows multiple rotational (torsion) angles between the thiophene rings.37,38 When bound to protein aggregates, the thiophene rings lock into a specific planarity and provide a direct correlation between the LCP geometry and the light emitted. Protein-bound LCPs with maximum emission at shorter wavelengths suggests that the polythiophene backbone is nonplanar and LCPs are loosely packed, whereas a shift toward longer wavelengths suggests that the backbone is planar and LCPs are tightly packed.39,40 Thus the LCPs provide structural insights regarding the morphology of bound protein deposits and can be used as a complementary technique to conventional staining protocols for characterizing protein aggregates.
In vitro, the LCPs bound to recombinant protein fibrils of the same protein can yield distinct spectral profiles. This was shown with recombinant mouse prion protein (mPrP) that had been converted into two chemically identical types of amyloid fibrils using varying conditions for fibrillation.41 The two fibril types could be distinguished by different PTAA emission spectra. The same procedure also distinguishes Aβ1–42 fibrils grown under different conditions in vitro.42
LCPs have been additionally tested on brain sections of mice infected with different prion strains.41,43 We applied the anionic LCP polythiophene acetic acid (PTAA) to frozen brain sections and measured different spectral emission signatures depending on the strain. By calculating ratios of the intensity of the emitted light at certain wavelengths, prion aggregates associated with distinct prion strains were easily distinguished from each other, verifying the usefulness of spectral properties of LCPs for labeling specific strains. PTAA also binds to de novo prion aggregates, which do not stain with thioflavin T nor Congo red, indicating that LCPs could be used to identify a subset of protein deposits normally undetectable by conventional methods.44
Heterogenic protein aggregates are also found in other proteinopathies, such as AD or systemic amyloidoses. Upon application of several LCPs to transgenic mouse models having AD pathology, a striking heterogeneity in the characteristic plaques composed of the amyloid β (Aβ) peptide was identified. LCP staining of Aβ deposits in brain tissue sections revealed different sub-populations, observed as protein deposits with different spectral emissions.42 Furthermore, PTAA could be used to spectrally separate Aβ deposits associated with the Swedish and Arctic mutation as compared to the Swedish mutation in the amyloid precursor protein (APP), indicating that different point mutations of the amyloidogenic protein can give rise to various morphologies of protein deposits. The spectral profile from PTAA was also used for the structural sub-typing of systemic amyloidoses and similar to observations in earlier studies of prion deposits, some tissue samples exhibit mixtures of amyloid having separate PTAA spectra.45
LCPs in Studies of Prion Strain Mixtures
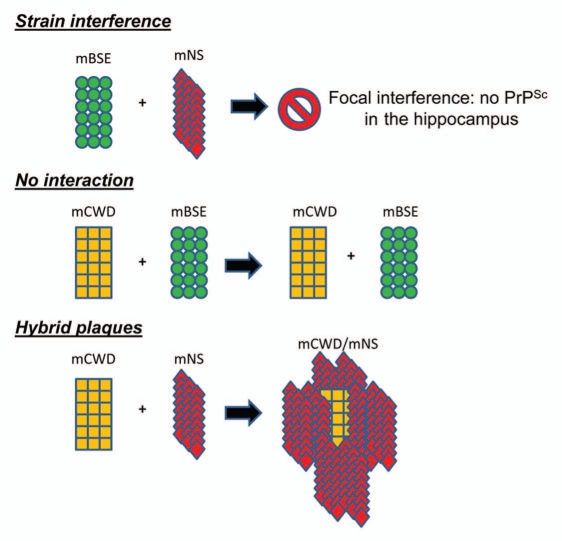
We recently characterized interactions that occur between prion strains in vivo using the LCPs.40 Three prion strains were used that had been previously characterized in mice histologically and biochemically: mouse-adapted chronic wasting disease (mCWD), natural sheep scrapie (mNS) and bovine spongiform encephalopathy (mBSE). Each individual strain and mixtures of two strains were intracerebrally inoculated into mice. The strain mixture consistently led to a delay in the incubation period as compared to that of the most rapid strain. Nevertheless, the lesion profile revealed that the affected regions essentially encompassed the combined sites of the two individual strains. The only exception was the mNS/mBSE strain mixture, in which interference occurred in a single region, the hippocampus, where mNS no longer accumulated.
To determine whether strains were colocalizing, frozen brain sections from mice infected with a mixture were stained with PTAA. The mCWD/mBSE mixture showed no evidence of interaction between aggregates, although both strains were present as seen by their characteristic plaque morphologies and the PTAA emission spectra.
PTAA bound to mCWD plaques emits light with a maximum intensity at ∼565 nm and appears yellow, whereas PTAA bound to mNS emits light with a maximum intensity at ∼595 and appears red. Surprisingly, the mCWD/mNS mixture led to the formation of hybrid plaques, wherein the plaque core was consistently composed of yellow mCWD, bordered by the red aggregates of mNS. The existence of the mCWD plaques may have facilitated mNS deposition by providing a scaffold, although hybrid plaques did not accelerate the mNS infection. We then investigated whether the hybrid plaques were composed of homotypic fibrils of one strain or heterotypic fibrils containing both strains. If the fibrils were homotypic, we would expect that on second passage, we would again observe the hybrid plaques containing each strain with its characteristic PTAA emission spectra. This was indeed the case as hybrid plaques were again observed. Thus, with three different strain combinations, interactions varied from strain inhibition (mNS/mBSE) no interaction (mBSE/mCWD), or hybrid plaque formation (mCWD/mNS) (Fig. 1).
Figure 1.
This schematic depicts interactions that have occurred in the brain among distinct prion strains as reported in reference 41.
Specificity of Aggregation Among Other Misfolded Proteins
Interactions between pre-formed aggregates have also been studied in yeast prions.46,47 Yeast that propagate two different protein aggregates frequently showed aggregate co-localization, even when the proteins did not seed the fibrillization of each other. Although the proteins co-localized, they did not seem to coassemble, indicating that the proteins more efficiently converted a homologous rather than a heterologous protein. This colocalization does not indicate any promotion of aggregate formation and is consistent with our observations of colocalization in the prion strains where there was no disease acceleration. Rajan and colleagues also studied the aggregation of unrelated proteins expressed in the same cell. Here they found that the aggregates were homogenous, indicating that aggregation of single proteins was exquisitely specific.48
Conclusion
Prion strain mixtures are common in certain sporadic and infectious prion diseases, as most recently described in chronic wasting disease (CWD) in mule deer.15 Yet detecting a second strain can be a challenge since one prion strain may mask a second strain in western blot or immunohistochemical assays. Although prion strains have been shown to interfere or aggregate with a second strain, other interactions may be possible, such as the formation of heterotypic fibrils. Staining with the conformationally sensitive LCPs presents a powerful technique for exploring the range of possible interactions between protein aggregates. Finally, the LCPs can provide insights into the structural arrangements of protein aggregates and can be used to track aggregates in vivo in real time.49
Acknowledgments
We thank Magdalini Polymenidou, Cyrus Bett and Shivanjali Joshi-Barr for their helpful comments on the manuscript. Research in our laboratories is supported by the National Institutes of Health Grants R21NS055116, R01NS069566 and U54AI065359 (CJS), R01NS052609 (JCB), as well as the Swedish Foundation for Strategic Research (KPRN), the European Union FP7 HEALTH (Project LUPAS) (KPRN), Institutional Grants for Younger Researchers from The Swedish Foundation for International Cooperation in Research and Higher Education (STINT) (CJS, KPRN), and the Knut and Alice Wallenberg Foundation (KPRN).
References
- 1.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 2.Eanes E, Glenner GG. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 3.Makarava N, Ostapchenko VG, Savtchenko R, Baskakov IV. Conformational switching within individual amyloid fibrils. J Biol Chem. 2009;284:14386–14395. doi: 10.1074/jbc.M900533200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguzzi A, Heikenwalder M, Polymenidou M. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol. 2007;8:552–561. doi: 10.1038/nrm2204. [DOI] [PubMed] [Google Scholar]
- 5.Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 6.Bessen RA, Marsh RF. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol. 1992;66:2096–2101. doi: 10.1128/jvi.66.4.2096-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce ME, McConnell I, Fraser H, Dickinson AG. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol. 1991;72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 8.Puoti G, Giaccone G, Rossi G, Canciani B, Bugiani O, Tagliavini F. Sporadic Creutzfeldt-Jakob disease: co-occurrence of different types of PrP(Sc) in the same brain. Neurology. 1999;53:2173–2176. doi: 10.1212/wnl.53.9.2173. [DOI] [PubMed] [Google Scholar]
- 9.Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol. 2005;4:805–814. doi: 10.1016/S1474-4422(05)70225-8. [DOI] [PubMed] [Google Scholar]
- 10.Uro-Coste E, Cassard H, Simon S, Lugan S, Bilheude JM, Perret-Liaudet A, et al. Beyond PrP res type 1/type 2 dichotomy in Creutzfeldt-Jakob disease. PLoS Pathog. 2008;4:1000029. [PubMed] [Google Scholar]
- 11.Cali I, Castellani R, Alshekhlee A, Cohen Y, Blevins J, Yuan J, et al. Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt-Jakob disease: its effect on the phenotype and prion-type characteristics. Brain. 2009;132:2643–2658. doi: 10.1093/brain/awp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi A, Mizukoshi K, Iwasaki Y, Miyata H, Yoshida Y, Kitamoto T. Co-occurrence of types 1 and 2 PrP(res) in sporadic Creutzfeldt-Jakob disease MM1. Am J Pathol. 178:1309–1315. doi: 10.1016/j.ajpath.2010.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimberlin RH, Walker CA. Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J Gen Virol. 1978;39:487–496. doi: 10.1099/0022-1317-39-3-487. [DOI] [PubMed] [Google Scholar]
- 14.Mahal SP, Browning S, Li J, Suponitsky-Kroyter I, Weissmann C. Transfer of a prion strain to different hosts leads to emergence of strain variants. Proc Natl Acad Sci USA. 107:22653–22658. doi: 10.1073/pnas.1013014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angers RC, Kang HE, Napier D, Browning S, Seward T, Mathiason C, et al. Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science. 328:1154–1158. doi: 10.1126/science.1187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghaemmaghami S, Ahn M, Lessard P, Giles K, Legname G, DeArmond SJ, et al. Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 2009;5:1000673. doi: 10.1371/journal.ppat.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickinson AG, Fraser H, McConnell I, Outram GW, Sales DI, Taylor DM. Extraneural competition between different scrapie agents leading to loss of infectivity. Nature. 1975;253:556. doi: 10.1038/253556a0. [DOI] [PubMed] [Google Scholar]
- 18.Dickinson AG, Fraser H, Meikle VM, Outram GW. Competition between different scrapie agents in mice. Nat New Biol. 1972;237:244–245. doi: 10.1038/newbio237244a0. [DOI] [PubMed] [Google Scholar]
- 19.Bartz JC, Aiken JM, Bessen RA. Delay in onset of prion disease for the HY strain of transmissible mink encephalopathy as a result of prior peripheral inoculation with the replication-deficient DY strain. J Gen Virol. 2004;85:265–273. doi: 10.1099/vir.0.19394-0. [DOI] [PubMed] [Google Scholar]
- 20.Manuelidis L. Vaccination with an attenuated Creutzfeldt-Jakob disease strain prevents expression of a virulent agent. Proc Natl Acad Sci USA. 1998;95:2520–2525. doi: 10.1073/pnas.95.5.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manuelidis L, Lu ZY. Virus-like interference in the latency and prevention of Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA. 2003;100:5360–5365. doi: 10.1073/pnas.0931192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schutt CR, Bartz JC. Prion interference with multiple prion isolates. Prion. 2008;2:61–63. doi: 10.4161/pri.2.2.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattison IH. The relative susceptibility of sheep, goats and mice to two types of the goat scrapie agent. Res Vet Sci. 1966;7:207–212. [PubMed] [Google Scholar]
- 24.Bartz JC, Bessen RA, McKenzie D, Marsh RF, Aiken JM. Adaptation and selection of prion protein strain conformations following interspecies transmission of transmissible mink encephalopathy. J Virol. 2000;74:5542–5547. doi: 10.1128/jvi.74.12.5542-5547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science. 2009;327:869–872. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shikiya RA, Ayers JI, Schutt CR, Kincaid AE, Bartz JC. Co-infecting prion strains compete for a limiting cellular resource. J Virol. 2010;84:5706–5714. doi: 10.1128/JVI.00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimberlin RH, Walker CA. Competition between strains of scrapie depends on the blocking agent being infectious. Intervirology. 1985;23:74–81. doi: 10.1159/000149588. [DOI] [PubMed] [Google Scholar]
- 29.Dickinson AGaO GW. The scrapie replication-site hypothesis and its implications for pathogenesis. In: Prusiner SBaH WJ., editor. Slow transmissible diseases of the central nervous system. New York: Academic Press; 1979. pp. 13–31. [Google Scholar]
- 30.Bartz JC, Kramer ML, Sheehan MH, Hutter JA, Ayers JI, Bessen RA, et al. Prion interference is due to a reduction in strain-specific PrPSc levels. J Virol. 2007;81:689–697. doi: 10.1128/JVI.01751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayers JI, Kincaid AE, Bartz JC. Prion strain targeting independent of strain-specific neuronal tropism. J Virol. 2009;83:81–87. doi: 10.1128/JVI.01745-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayers JI, Schutt CR, Shikiya RA, Aguzzi A, Kincaid AE, Bartz JC. The strain-encoded relationship between PrPSc replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Pathog. 2011;7:e1001317. doi: 10.1371/journal.ppat.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caughey B, Raymond GJ. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J Virol. 1993;67:643–650. doi: 10.1128/jvi.67.2.643-650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USA. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geoghegan JC, Valdes PA, Orem NR, Deleault NR, Williamson RA, Harris BT, et al. Selective incorporation of polyanionic molecules into hamster prions. J Biol Chem. 2007;282:36341–36353. doi: 10.1074/jbc.M704447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JI, Surewicz K, Gambetti P, Surewicz WK. The role of glycophosphatidylinositol anchor in the amplification of the scrapie isoform of prion protein in vitro. FEBS Lett. 2009;583:3671–3675. doi: 10.1016/j.febslet.2009.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson KP, Rydberg J, Baltzer L, Inganas O. Self-assembly of synthetic peptides control conformation and optical properties of a zwitterionic polythiophene derivative. Proc Natl Acad Sci USA. 2003;100:10170–10174. doi: 10.1073/pnas.1834422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsson KPR, Hammarstrom P, Ahlgren F, Herland A, Schnell EA, Lindgren M, et al. Conjugated polyelectrolytes-conformation-sensitive optical probes for staining and characterization of amyloid deposits. Chembiochem. 2006;7:1096–1104. doi: 10.1002/cbic.200500550. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson KPR, Andersson MR, Inganas O. Conformational transitions of a free amino-acid-functionalized polythiophene induced by different buffer systems. J Phys Condens Matter. 2002;14:10011–10020. [Google Scholar]
- 40.Nilsson KP, Joshi-Barr S, Winson O, Sigurdson CJ. Prion strain interactions are highly selective. J Neurosci. 2010;30:12094–12102. doi: 10.1523/JNEUROSCI.2417-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sigurdson CJ, Nilsson KP, Hornemann S, Manco G, Polymenidou M, Schwarz P, et al. Prion strain discrimination using luminescent conjugated polymers. Nat Methods. 2007;4:1023–1030. doi: 10.1038/nmeth1131. [DOI] [PubMed] [Google Scholar]
- 42.Nilsson KP, Aslund A, Berg I, Nystrom S, Konradsson P, Herland A, et al. Imaging distinct conformational states of amyloid-beta fibrils in Alzheimer's disease using novel luminescent probes. ACS Chem Biol. 2007;2:553–560. doi: 10.1021/cb700116u. [DOI] [PubMed] [Google Scholar]
- 43.Sigurdson CJ, Nilsson KP, Hornemann S, Manco G, Fernandez-Borges N, Schwarz P, et al. A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest. 2010;120:2590–2599. doi: 10.1172/JCI42051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sigurdson CJ, Nilsson KP, Hornemann S, Heikenwalder M, Manco G, Schwarz P, et al. De novo generation of a transmissible spongiform encephalopathy by mouse transgenesis. Proc Natl Acad Sci USA. 2009;106:304–309. doi: 10.1073/pnas.0810680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nilsson KP, Ikenberg K, Aslund A, Fransson S, Konradsson P, Rocken C, et al. Structural typing of systemic amyloidoses by luminescent-conjugated polymer spectroscopy. Am J Pathol. 2010;176:563–574. doi: 10.2353/ajpath.2010.080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derkatch IL, Liebman SW. Prion-prion interactions. Prion. 2007;1:161–169. doi: 10.4161/pri.1.3.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajan RS, Illing ME, Bence NF, Kopito RR. Specificity in intracellular protein aggregation and inclusion body formation. Proc Natl Acad Sci USA. 2001;98:13060–13065. doi: 10.1073/pnas.181479798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aslund A, Sigurdson CJ, Klingstedt T, Grathwohl S, Bolmont T, Dickstein DL, et al. Novel pentameric thiophene derivatives for in vitro and in vivo optical imaging of a plethora of protein aggregates in cerebral amyloidoses. ACS Chem Biol. 2009;4:673–684. doi: 10.1021/cb900112v. [DOI] [PMC free article] [PubMed] [Google Scholar]