Abstract
Atypical forms of bovine spongiform encephalopathy (BSE) may be caused by different prions from classical BSE (C-BSE). In this study, we examined the susceptibility of mice overexpressing mouse and hamster chimeric prion protein (PrP) to L-type atypical BSE (L-BSE). None of the transgenic mice showed susceptibility to L-BSE, except mice overexpressing hamster PrP. We also examined the transmission properties of L-BSE in hamsters. The incubation period of hamsters intracerebrally inoculated with L-BSE was 576.8 days, and that of the subsequent passage was decreased to 208 days. Although the lesion and glycoform profiles and relative proteinase K resistant core fragment of the abnormal isoform of PrP (PrPcore) of L-BSE were similar to that of C-BSE, the deposition of the abnormal isoform of PrP (PrPSc) and the molecular weight of PrPcore of L-BSE was different from than that of C-BSE. In hamster models, some prion strain characteristics of L-BSE were indistinguishable from those of C-BSE.
Key words: prion, atypical, L-BSE, PrPcore, hamster, transmission
Introduction
Bovine spongiform encephalopathy (BSE) is a fatal neurodegenerative disorder that is classified as a prion disease or a transmissible spongiform encephalopathy.1 BSE and the subsequent identification of variant Creutzfeldt-Jakob disease have raised important food safety issues. The incidence of BSE has decreased because of disease-control programs such as the feed ban;2 however, different phenotypes of BSE (atypical BSEs) have been identified in several countries.3 To date, these phenotypes are classified into two forms—L-type atypical BSE (L-BSE) or bovine amyloidotic spongiform encephalopathy (BASE) and H-type atypical BSE (H-BSE)—on the basis of the molecular weight of the proteinase K (PK)-resistant core fragment of the abnormal isoform of the prion protein, PrPSc (PrPcore).4,5
Several studies have indicated that prion strains causing L-BSE and H-BSE are different from those that cause classical BSE (C-BSE).6–11 The L-BSE prion was experimentally transmissible to cattle,8,10 bovinized prion protein (PrP)-overexpressing transgenic mice,6,11 and humanized PrP-transgenic mice9 with shorter incubation periods and more severe spongiform changes than the C-BSE prion. A transmission study with inbred mice has widely been used for prion strain classification; however, L-BSE could not be transmitted to wild-type mice,11,12 and subsequent passage in wild-type mice altered the characteristics of L-BSE prions12 and made comparison of strain characteristics difficult in L-BSE and C-BSE prions. Rodent-adapted L-BSE prions will help analyze L-BSE prion characteristics.
Conversion from cellular isoform of prion protein (PrPC) to PrPSc is a central event in prion pathogenesis. Amino acid differences between host PrPC and PrPSc of inoculums result in a species barrier in the interspecies transmission of prions. The mouse and hamster PrPcore regions differ with respect to eight amino acid substitutions.13 We examined the characteristics of C-BSE by using mice overexpressing mouse and hamster chimeric PrP.14 The mouse PrP131-188 sequence contributed to the susceptibility of mice with C-BSE prions. L-BSE prions may have a susceptible host range that is different from C-BSE prions, and thus an investigation of the susceptible host range using mice overexpressing mouse and hamster PrP may help reveal the host species susceptible to L-BSE prions. Here, we examined the species barrier to L-BSE prions using a chimeric PrP overexpression mouse model. However, none of the transgenic mice, except those overexpressing hamster PrP, were susceptible to L-BSE prions. We also confirmed this result by carrying out a transmission study in hamsters. Expectedly, the L-BSE prion but not the C-BSE prion was transmissible to hamsters. In this study, we also analyzed the disease phenotype and PrPcore characteristics of the L-BSE prion in Syrian hamsters.
Results
No susceptibility of L-BSE to transgenic mice overexpressing chimeric-PrP.
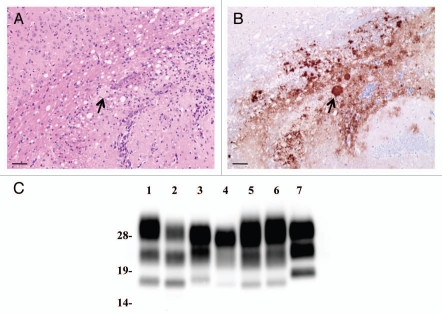
As shown in Table 1, L-BSE was not transmitted to MHM2, MH2M, wild-type (ICR) or tga20 mice. None of these mice showed clinical signs of L-BSE, and no PrPSc accumulation was observed. However, L-BSE was transmitted to 2 of 3 TgHaNSE mice incubated for 567 and 853 days. PrP plaque deposition was observed in the brain of L-BSE-affected TgHaNSE mice (Fig. 1). The molecular weight of PrPcore of L-BSE affected TgHaNSE was similar to that of L-BSE (Fig. 1). However, its glycoprofile was different from that of L-BSE cattle, but similar to that of C-BSE cattle (Fig. 1).
Table 1.
Transmissibility of L-BSE in mice
| Mice | Number of mice affected/inoculated | Incubation period (days) |
| Mouse | 0/5 | >700* |
| tga20 | 0/5 | >668* |
| MHM2 | 0/5 | >755* |
| MH2M | 0/5 | >853* |
| TgHaNSE | 2/3 | 567†, 853† |
The brain homogenate of L-BSE was intracerebrally inoculated into mice.
Mice showed no clinical signs, and tested negative for PrPSc in the brain.
PrPSc was present in the brain.
Figure 1.
Neuropathology and PrPcore characteristics of L-BSE-affected TgHaNSE mice. (A) hematoxylin and eosin staining of the corpus callosum of mice. (B) PrPSc deposition was detected in the semiserial sections. PrP-plaque is indicated by an arrow. PrP was detected by mAb SAF-84. Scale bars: 200 mm. (C) PrPSc in L-BSE affected TgHaNSE mice was detected by western blotting. Lane 1: C-BSE (cattle), lane 2: L-BSE (cattle), lane 3: C-BSE affected hamster, lane 4: L-BSE affected hamster, lanes 5 and 6: L-BSE affected TgHaNSE, lane 7: scrapie Obihiro-affected mouse. PrP was detected by mAb T2.
BSE transmission to hamsters.
C-BSE prions were not transmitted to hamsters. However, mouse-passaged C-BSE prions were transmissible to TgHaNSE mice and prions accumulated in the brains of TgHaNSE mice and were successfully transmitted to wild-type hamsters with an incubation period of 349.5 (6.6) days. A subsequent passage decreased the incubation period to 267 days. The incubation period of the third passage was 271 days (Table 2). In contrast, L-BSE was successfully transmitted to hamsters from the first passage. The attack rate with the primary passage and an incubation period of 576.8 days was 75%. The subsequent passage increased the attack rate (100%) and decreased the incubation period to 208 days.
Table 2.
Incubation period of hamsters inoculated with C-BSE and L-BSE
| Inoculum | Passage numbers | Number of hamsters diseased/inoculated | Incubation period mean (SD) (days) |
| C-BSE | 1st | 0/5 | >600* |
| Mouse passaged C-BSE† | 1st | 10/10 | 349.5 (6.6) |
| 2nd | 10/10 | 267.3 (19.2) | |
| 3rd | 10/10 | 271.6 (16.2) | |
| L-BSE | 1st | 3/4 | 576.8 (127.8) |
| 2nd | 4/4 | 208 (15.5) |
Hamsters showed no clinical signs and were negative for PrPSc in the brain.
After one passage of BSE from cattle in mice (incubation period, 408.6 (28.2) days) and subsequent transmission to TgHaNSE mice (incubation period, 153.1 (1.1) days).14 The brain of a diseased TgHaNSE mouse was inoculated in a hamster.
Neuropathology of L-BSE-affected hamsters.
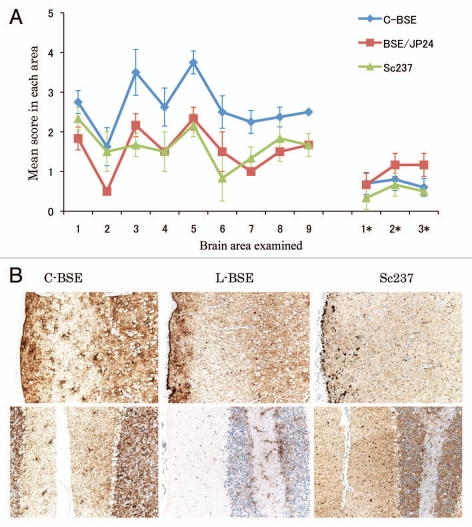
Though the L-BSE prion-affected hamster showed a lower vacuolation score of lesion profiling than C-BSE prion-affected hamsters, the targeted regions of the L-BSE prion were similar to those of the C-BSE prion (Fig. 2A). However, PrPSc distribution and patterns of L-BSE differed from those of C-BSE and scrapie (Fig. 2B). Most conspicuous pattern of PrPSc in C-BSE prion-affected hamsters was particulate and stellate deposits in the neuropil of the brain. In Sc237 prion-affected hamsters, plaque-like deposits were visible in the cerebral and cerebellar cortices. In contrast, the pattern of PrPSc deposition was characterized by the presence of sub-pial accumulation in the cerebral cortex and the absence of stellate and plaque forms in the brains of L-BSE prion-affected hamsters. In addition, PrPSc accumulation in the cerebellar cortex of L-BSE prion-affected hamsters was less common rather than that of C-BSE or Sc237.
Figure 2.
Neuropathological lesion profiling and PrPSc deposition in hamsters. (A) Vacuolar lesion profiles in Syrian hamster brains as observed for scrapie strains Sc237, C-BSE-affected hamsters (C-BSE) and L-BSE-affected hamsters (L-BSE). Gray matter scoring areas: 1, dorsal medulla; 2, cerebellar cortex; 3, superior colliculus; 4, hypothalamus; 5, medial thalamus; 6, hippocampus; 7, septum; 8, posterior cerebral cortex; 9, anterior cerebral cortex. White matter scoring areas: 1*, cerebellar white matter; 2*, midbrain white matter; 3*, cerebral peduncle. Mean (standard deviation) (n = 4). (B) PrPSc deposition in the brains of hamsters affected with C-BSE (second passage), L-BSE (second passage) and Sc237 (serial passage). MAb SAF84 was used for immunostaining. Upper, cerebral cortex; lower, cerebellum.
Molecular profile of PrPcore of L-BSE-affected hamsters.
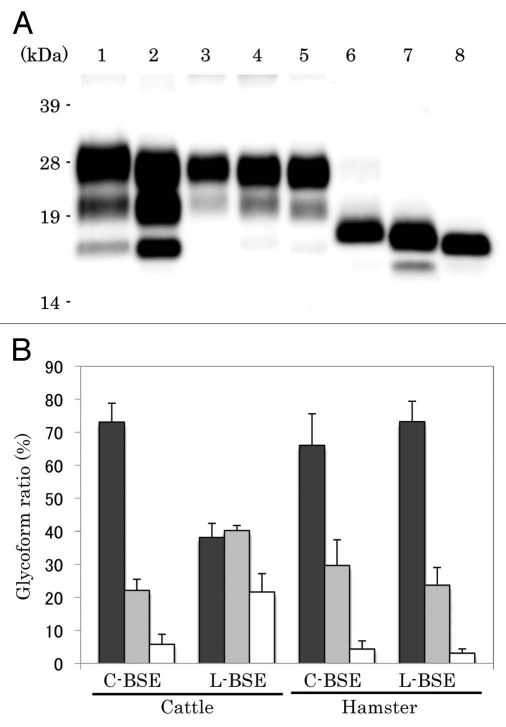
PrPcore was detected from both C-BSE- and L-BSE-affected hamsters (Fig. 3A). However, the PrPcore glycoform of L-BSE-affected hamsters changed relative to that of the original L-BSE cattle. The PrPcore glycoform of L-BSE-affected hamsters was similar to that of C-BSE- and scrapie-affected hamsters: dominant in the di-glycosylated form (Fig. 3A and lanes 4, 5; 3B). On the other hand, the molecular weight of PrPcore were conserved during hamster passage. After deglycosylation treatment of PrPcore, variations in molecular weight of PrPcore among Sc237-, C-BSE- and L-BSE-affected hamsters were evident (Fig. 3A and lanes 6–8). The molecular weight of PrPcore of L-BSE was less than that of C-BSE. C-BSE-affected hamsters harbored the additional truncated PrPcore band (Fig. 3A and lane 7),14 but this band was not present in L-BSE-affected hamsters (Fig. 3A and lane 8).
Figure 3.
Western blotting analysis of PrPcore from C-BSE- and L-BSE-affected hamsters. (A) Lane 1: C-BSE-affected cattle (natural case); lane 2: L-BSE-affected cattle (natural case); lanes 3 and 6: Sc237-affected hamsters; lanes 4 and 7: C-BSE-affected hamsters; lanes 5 and 8: L-BSE-affected hamsters; lanes 6–8: PNGaseF treatment. (B) The relative amount (%) of di-, mono- and non-glycosylated PrPcore. The results are the mean (standard deviation) of three experiments. Bar diagram: di- (black), mono-(grey) and nonglycosylated form (white).
Relative PK resistance of PrPcore of L-BSE.
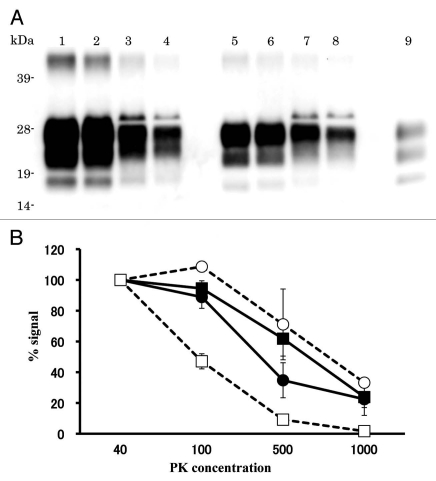
The relative PK resistance of PrPcore of L-BSE in cattle was weaker than that of C-BSE; this has also been observed in the case of TgBoPrP mice.11 We analyzed the relative PK resistance of PrPcore for C-BSE and L-BSE in hamsters. The signal intensity decreased in 1,000 µg/ml of the PK condition; however, PrPcore was still detected from both C-BSE and L-BSE samples (Fig. 4 and lanes 4 and 8). PK resistance of PrPcore for L-BSE in hamsters was remarkably different from original L-BSE (Fig. 4B).
Figure 4.
Relative PK resistance of PrPcore in prion-affected hamsters. (A) Western blot results. Lanes 1–4: hamster-adapted C-BSE; lanes 5–8: L-BSE-affected hamster. Lane 9: mouse scrapie prion. The samples were treated with 40 (lanes 1 and 5), 100 (lanes 2 and 6), 500 (lanes 3 and 7) and 1,000 (lanes 4 and 8) µg/ml of PK at 37°C for 1 h. PrPcore was detected with mAb 6H4. Molecular markers are shown on the left (kDa). (B) Relative amount (%) of PrPcore after different PK concentration were indicated. Black circle: C-BSE affected hamster, black square: L-BSE affected hamster, white circle: C-BSE affected cattle, white square: L-BSE affected cattle. Cattle results are obtained from previous study in reference 11.
Discussion
This study showed that the host range for L-BSE prions was different from that of C-BSE prions. The C-BSE prion was not transmitted to hamsters (Table 2). The mouse and hamster PrPcore subregions differ with respect to 8 amino acid substitutions (Table 3). Yokoyama et al.14 showed that the PrP131-188 subregion contributed to the susceptibility of mice to C-BSE. Once the C-BSE prion was passaged in wild-type mice, it could be transmitted to hamsters; further, its characteristics are similar to the previously reported hamster-adapted C-BSE prion.27,28 On the other hand, neither MH2M nor MHM2 mice were susceptible to L-BSE prions (Table 1). Three amino acid substitutions (V203I, M205I and V215T) at PrP189-231 or in the authentic hamster PrP sequence may be required for conversion of the PrPSc of the L-BSE prion (Table 3).
Table 3.
Comparison of the PrP amino acid sequences in mouse and hamster
In order to clarify the characteristics of this prion, an L-BSE prion-adapted rodent model will be a useful tool. On the basis of the result of the transmission study in transgenic mice, we inoculated L-BSE prions into hamsters. L-BSE prion-affected hamsters showed PrPSc distribution and patterns that differed from those of C-BSE prion-affected hamsters (Fig. 1B). The targeted lesions of L-BSE were similar to those of C-BSE; however, the degree of spongiform change differed (Fig. 2A).
Glycoform of PrPcore has been used to classify prion strains and/or compare prion characteristics.29 In cattle, the molecular weight of PrPcore of L-BSE was less than that of C-BSE, and there were glycoprofile differences in their PrPcore (Fig. 3). However, the PrPcore of L-BSE in hamsters was different. Although the molecular weight of PrPcore of L-BSE was similar to that of L-BSE-affected cattle, the glycoprofile and relative PK resistance of L-BSE in hamsters differed from those of the original L-BSE (Fig. 3). A similar PrPcore glycoprofile within different prion strains was also observed in converted PrPSc in vitro.30 These results suggest that the glycoform of PrPcore in hamsters may be influenced by host-species characteristics rather than prion-strain characteristics. It has been recently reported that hamsters were susceptible to L-BSE and that, in this experimental model, L-BSE differed from classical BSE by its lower apparent molecular mass, whereas glycoforms proportions were similar.31 Our results in hamsters and in hamster transgenic mice confirmed these observations.
The biological characteristics of L-BSE and C-BSE prions differed in cattle and TgBoPrP mice.8,11 In cattle and bovinized mice, L-BSE showed severe spongiform changes, unlike C-BSE. On the other hand, in hamsters, the pathology of L-BSE was less severe than that of C-BSE. L-BSE was transmissible to hamsters (576.8 days), but C-BSE was not (>600 days). Further, the incubation period of C-BSE was approximately 270 days at the third passage, whereas that of L-BSE was shorter (208 days), even in the second passage (Table 3). The incubation period was shorter in L-BSE-affected animals than in C-BSE-affected animals; this observation was consistent among several animal species, including hamsters, cattle,8,10 bovinized PrP-overexpressing mice,6,11 humanized PrP-overexpressing mice,9,32 and primates.7
Interestingly, L-BSE-affected TgHaNSE mice and hamsters showed a different neuropathology: the former had PrP plaque (Fig. 1), but the latter did not (Fig. 2). The same neuropathological difference was observed between cattle and TgBoPrP mice.11 Therefore, we think that the formation of the PrP plaque was not only because of the prion strain and host PrP gene but also a factor that has not yet been identified.
Capobianco et al. reported that serial passage of the BASE prion in wild-type mice showed that the accumulated prion and transmissibility were identical to the C-BSE prion. In wild-type mice, the BASE prion strain was converted to the C-BSE-like prion strain during passages.12 Further, BASE-inoculated ovine PrP-expressing mice showed indistinguishable phenotypic traits with C-BSE.33 This study showed that some characteristics (lesion profile, glycoform) of L-BSE in hamsters resembled those of C-BSE, whereas other characteristics such as PrPSc deposition and molecular weight of PrPcore differed between C-BSE and L-BSE. This partial similarity in hamster-passaged C-BSE and L-BSE may be linked to the results of the previous transmission study, which showed that the subsequent passage of BASE altered its characteristics.12,27
In summary, the biochemical nature of L-BSE is modified to some extent after passage in hamsters. This study revealed the unstable phenotypic properties of L-BSE prions in interspecies transmission. The conformational moiety of PrPSc, which is linked to the N-terminal cleavage site of PrPcore, differed from the moieties linked to the relative PK resistance and glycoprofile of PrPcore.
Materials and Methods
The study protocol was approved by the Animal Ethics Committee and Animal Care and Use Committee of the National Institute of Animal Health, Japan.
Animals and prions.
We purchased 3-week-old weanling female Syrian hamsters (SLC). We used transgenic mice that expressed mouse and hamster chimeric PrP (MHM2 and MH2M, respectively),15 tga20 mice overexpressing mouse PrP,16 and TgHaNSE mice overexpressing hamster PrP in their neurons.17 The PrP amino acid sequence of these Tg mice is shown in Table 3. PrP sequences are referred from previous reports: mouse,18 hamster19 and cattle.20 In MHM2 mice, amino acid substitutions are present at positions L109M and V112M. In addition to these amino acid substitutions, three other substitutions (I139M, Y155N and S170N) are present in MH2M mice. All mice were maintained by crossing with PrP-deficient mice21 in a PrP-null background. The expression level of PrP in these transgenic mice was approximately 4–10 times greater than that in wild-type mice.15–17 Brain samples of natural C-BSE22 and natural Japanese L-BSE (BSE/JP24),23 were used in this study.
Transmission experiments.
Animals were inoculated with 20 µl of 10% brain homogenate (w/v) of L-BSE. Clinically affected animals were sacrificed and used in the experiments. The hamster-adapted C-BSE prion (described below) was also used.
Hamster-adapted C-BSE prions.
For the control, we generated hamster-adapted BSE prions. C-BSE was not transmitted to hamsters; however, C-BSE was passaged in mice once, and this prion was transmissible to TgHaNSE with an incubation period of 153.1 days. The brains of diseased TgHaNSE mice were used as the origin of hamster BSE.
Histopathology and immunohistochemistry.
Half brains were fixed in 10% neutral buffered formalin and were then subjected to hematoxylin and eosin staining and immunohistochemical analysis for the detection of PrPSc, as reported previously in reference 11. The lesion profiles in the brain was determined in nine areas of gray matter and three areas of white matter as described previously in reference 24. A PrP signal was detected with the anti-PrP monoclonal antibody (mAb) SAF-84 (SPI-bio).
Western blot analysis.
Brain tissues were homogenized in a buffer containing 100 mM NaCl and 50 mM Tris-HCl (pH 7.6). The homogenate was mixed with an equal volume of detergent buffer containing 4% Zwittergent 3–14, 1% sarkosyl, 100 mM NaCl and 50 mM Tris-HCl (pH 7.6) and then incubated with 0.25 mg collagenase. The homogenate was then incubated with 40 µg/ml PK at 37°C for 30 min. PK digestion was terminated with 2 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (Pefabloc; Roche Diagnostics). The sample was mixed with 2-butanol:methanol (5:1) and then centrifuged at 20,000x g for 10 min. The extracted PrPcore was subjected to western blot analysis, according to a previously described method in reference 25. The PrP signal was detected with anti-PrP mAb 6H4 (Prionics) or mAb T2.26
Band profiles of PrPcore.
For band analysis, the relative quantities of the 3 PrPcore bands were measured using the Fluorochem software (Alpha-Innotech), as reported previously in reference 11.
Peptide N-glycosidase F digestion.
The PrPcore sample was deglycosylated with peptide N-glycosidase F (PNGase F; New England Biolabs), according to the manufacturer's instructions.
Relative PK resistance of PrPcore.
For comparing the relative PK resistance of PrPcore, the sample was subjected to PK digestion at various concentrations (40–1,000 µg/ml), as reported previously in reference 11.
Acknowledgments
We thank the laboratory staff at the Prion Disease Research Center for providing technical support and the animal care staff at the National Institute of Animal Health for maintaining the experimental animals. This study was supported by grants from the BSE control project of the Ministry of Agriculture, Forestry and Fisheries of Japan, and by grants from the Ministry of Health, Labour and Welfare of Japan and in part by a grant from the Bio-oriented Technology Research Advancement Institution (Tokyo, Japan).
Abbreviations
- BASE
bovine amyloidotic spongiform encephalopathy
- BSE
bovine spongiform encephalopathy
- C-BSE
classical BSE
- H-BSE
H-type of atypical BSE
- L-BSE
L-type of atypical BSE
- PK
proteinase K
- PrP
prion protein
- PrPC
cellular isoform of prion protein
- PrPcore
PK resistant core fragment of PrPSc
- PrPSc
abnormal isoform of prion protein
References
- 1.Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 2.Ducrot C, Arnold M, de Koeijer A, Heim D, Calavas D. Review on the epidemiology and dynamics of BSE epidemics. Vet Res. 2008;39:15. doi: 10.1051/vetres:2007053. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs JG, Langeveld JP, Biacabe AG, Acutis PL, Polak MP, Gavier-Widen D, et al. Molecular discrimination of atypical bovine spongiform encephalopathy strains from a geographical region spanning a wide area in Europe. J Clin Microbiol. 2007;45:1821–1829. doi: 10.1128/JCM.00160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biacabe AG, Laplanche JL, Ryder S, Baron T. Distinct molecular phenotypes in bovine prion diseases. EMBO Rep. 2004;5:110–115. doi: 10.1038/sj.embor.7400054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, et al. Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA. 2004;101:3065–3070. doi: 10.1073/pnas.0305777101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buschmann A, Gretzschel A, Biacabe AG, Schiebel K, Corona C, Hoffmann C, et al. Atypical BSE in Germany-proof of transmissibility and biochemical characterization. Vet Microbiol. 2006;117:103–116. doi: 10.1016/j.vetmic.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Comoy EE, Casalone C, Lescoutra-Etchegaray N, Zanusso G, Freire S, Marce D, et al. Atypical BSE (BASE) transmitted from asymptomatic aging cattle to a primate. PLoS One. 2008;3:3017. doi: 10.1371/journal.pone.0003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda S, Iwamaru Y, Imamura M, Masujin K, Shimizu Y, Matsuura Y, et al. Intraspecies transmission of L-type-like Bovine Spongiform Encephalopathy detected in Japan. Microbiol Immunol. 2009;53:704–707. doi: 10.1111/j.1348-0421.2009.00169.x. [DOI] [PubMed] [Google Scholar]
- 9.Kong Q, Zheng M, Casalone C, Qing L, Huang S, Chakraborty B, et al. Evaluation of the human transmission risk of an atypical bovine spongiform encephalopathy prion strain. J Virol. 2008;82:3697–3701. doi: 10.1128/JVI.02561-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lombardi G, Casalone C, A DA, Gelmetti D, Torcoli G, Barbieri I, et al. Intraspecies transmission of BASE induces clinical dullness and amyotrophic changes. PLoS Pathog. 2008;4:1000075. doi: 10.1371/journal.ppat.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masujin K, Shu Y, Yamakawa Y, Hagiwara K, Sata T, Matsuura Y, et al. Biological and biochemical characterization of L-type-like bovine spongiform encephalopathy (BSE) detected in Japanese black beef cattle. Prion. 2008;2:123–128. doi: 10.4161/pri.2.3.7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capobianco R, Casalone C, Suardi S, Mangieri M, Miccolo C, Limido L, et al. Conversion of the BASE prion strain into the BSE strain: the origin of BSE? PLoS Pathog. 2007;3:31. doi: 10.1371/journal.ppat.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama T, Itohara S, Yuasa N. Detection of species specific epitopes of mouse and hamster prion proteins (PrPs) by anti-peptide antibodies. Arch Virol. 1996;141:763–769. doi: 10.1007/BF01718334. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama T, Masujin K, Iwamaru Y, Imamura M, Mohri S. Alteration of the biological and biochemical characteristics of bovine spongiform encephalopathy prions during interspecies transmission in transgenic mice models. J Gen Virol. 2009;90:261–268. doi: 10.1099/vir.0.004754-0. [DOI] [PubMed] [Google Scholar]
- 15.Scott M, Groth D, Foster D, Torchia M, Yang SL, DeArmond SJ, et al. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell. 1993;73:979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 16.Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, et al. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 17.Race RE, Priola SA, Bessen RA, Ernst D, Dockter J, Rall GF, et al. Neuron-specific expression of a hamster prion protein minigene in transgenic mice induces susceptibility to hamster scrapie agent. Neuron. 1995;15:1183–1191. doi: 10.1016/0896-6273(95)90105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westaway D, Goodman PA, Mirenda CA, McKinley MP, Carlson GA, Prusiner SB. Distinct prion proteins in short and long scrapie incubation period mice. Cell. 1987;51:651–662. doi: 10.1016/0092-8674(87)90134-6. [DOI] [PubMed] [Google Scholar]
- 19.Oesch B, Westaway D, Walchli M, McKinley MP, Kent SB, Aebersold R, et al. A cellular gene encodes scrapie PrP 27-30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 20.Goldmann W, Hunter N, Martin T, Dawson M, Hope J. Different forms of the bovine PrP gene have five or six copies of a short, G-C-rich element within the protein-coding exon. J Gen Virol. 1991;72:201–204. doi: 10.1099/0022-1317-72-1-201. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama T, Kimura KM, Ushiki Y, Yamada S, Morooka A, Nakashiba T, et al. In vivo conversion of cellular prion protein to pathogenic isoforms, as monitored by conformation-specific antibodies. J Biol Chem. 2001;276:11265–11271. doi: 10.1074/jbc.M008734200. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi HK, Yokoyama T, Takata M, Iwamaru Y, Imamura M, Ushiki YK, et al. The N-terminal cleavage site of PrPSc from BSE differs from that of PrPSc from scrapie. Biochem Biophys Res Commun. 2005;328:1024–1027. doi: 10.1016/j.bbrc.2005.01.065. [DOI] [PubMed] [Google Scholar]
- 23.Hagiwara K, Yamakawa Y, Sato Y, Nakamura Y, Tobiume M, Shinagawa M, et al. Accumulation of mono-glycosylated form-rich, plaque-forming PrPSc in the second atypical bovine spongiform encephalopathy case in Japan. Jpn J Infect Dis. 2007;60:305–308. [PubMed] [Google Scholar]
- 24.Fraser H, Dickinson AG. Distribution of experimentally induced scrapie lesions in the brain. Nature. 1967;216:1310–1311. doi: 10.1038/2161310a0. [DOI] [PubMed] [Google Scholar]
- 25.Masujin K, Matthews D, Wells GA, Mohri S, Yokoyama T. Prions in the peripheral nerves of bovine spongiform encephalopathy-affected cattle. J Gen Virol. 2007;88:1850–1858. doi: 10.1099/vir.0.82779-0. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu Y, Kaku-Ushiki Y, Iwamaru Y, Muramoto T, Kitamoto T, Yokoyama T, et al. A novel anti-prion protein monoclonal antibody and its single-chain fragment variable derivative with ability to inhibit abnormal prion protein accumulation in cultured cells. Microbiol Immunol. 2010;54:112–121. doi: 10.1111/j.1348-0421.2009.00190.x. [DOI] [PubMed] [Google Scholar]
- 27.Thomzig A, Cardone F, Kruger D, Pocchiari M, Brown P, Beekes M. Pathological prion protein in muscles of hamsters and mice infected with rodent-adapted BSE or vCJD. J Gen Virol. 2006;87:251–254. doi: 10.1099/vir.0.81277-0. [DOI] [PubMed] [Google Scholar]
- 28.Thomzig A, Spassov S, Friedrich M, Naumann D, Beekes M. Discriminating scrapie and bovine spongiform encephalopathy isolates by infrared spectroscopy of pathological prion protein. J Biol Chem. 2004;279:33847–33854. doi: 10.1074/jbc.M403730200. [DOI] [PubMed] [Google Scholar]
- 29.Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 30.Kim JI, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, et al. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem. 2010;285:14083–14087. doi: 10.1074/jbc.C110.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicot S, Baron T. Strain-specific barriers against bovine prions in hamsters. J Virol. 2011;85:1906–1908. doi: 10.1128/JVI.01872-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beringue V, Herzog L, Reine F, Le Dur A, Casalone C, Vilotte JL, et al. Transmission of atypical bovine prions to mice transgenic for human prion protein. Emerg Infect Dis. 2008;14:1898–1901. doi: 10.3201/eid1412.080941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beringue V, Andreoletti O, Le Dur A, Essalmani R, Vilotte JL, Lacroux C, et al. A bovine prion acquires an epidemic bovine spongiform encephalopathy strain-like phenotype on interspecies transmission. J Neurosci. 2007;27:6965–6971. doi: 10.1523/JNEUROSCI.0693-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]