Abstract
Objective
To investigate TGFβ regulation of CCN3 expression in cells of the nucleus pulposus.
Methods
Real Time RT-PCR and Western blot analysis was used to measure CCN3 expression in the nucleus pulposus. Transfections were used to measure the effect of Smad3, MAPKs and AP1 on TGFβ mediated CCN3 promoter activity. Lentiviral knock down of Smad3 was performed to asses the role of Smad3 in CCN3 expression.
Results
CCN3 was expressed in embryonic and adult intervertebral discs. TGFβ decreased CCN3 expression and suppressed its promoter activity in nucleus pulposus cells. DN-Smad3, Smad3-siRNA or DN-AP1 had little effect on TGFβ suppression of CCN3 promoter activity. However, p38 and ERK inhibitors blocked suppression of CCN3 by TGFβ, suggesting involvement of these signaling pathways in the regulation. Interestingly, overexpression of Smad3, in absence of TGFβ increased CCN3 promoter activity. We validated the role of Smad3 in controlling CCN3 expression in Smad3 null mice and in nucleus pulposus cells transduced with lentiviral shSmad3. In terms of function, treatment with rCCN3 showed a dose dependent decrease in proliferation of nucleus pulposus cells. Moreover, CCN3 treated cells shows a decrease in aggrecan, versican, CCN2 as well as collagen type I expression.
Conclusion
The opposing effect of TGFβ on CCN2 and CCN3 expression and suppression of CCN2 by CCN3 in nucleus pulposus cells furthers the paradigm that these CCN proteins form an interacting triad, possibly important in maintaining extracellular matrix homeostasis and cell number.
Keywords: intervertebral disc, nucleus pulposus, CCN3, TGFβ, Smad3
INTRODUCTION
The intervertebral disc is a unique structure that permits rotation as well as flexion and extension of the human spine. It is comprised of a gel-like nucleus pulposus surrounded circumferentially by a ligamentous annulus fibrosus. Although cells comprise only around 1% of tissue volume, they are critically important in maintaining a healthy disc (1). Nucleus pulposus cells secrete and organize a complex extracellular matrix that mainly contains the proteoglycan aggrecan and a small proportion of fibrillar collagens. This macromolecular assembly provides a robust hydrodynamic system that accommodates applied biomechanical forces to the spine (2-4). During degenerative disc disease, loss of disc cells, limited proteoglycan synthesis and a shift towards synthesis of a fibrotic matrix decreases the water binding capacity of the disc and results in a failure of the tissue to resist compressive loads (5).
Growth factors, in particular TGFβ, are known to upregulate aggrecan synthesis in the disc and there is evidence that this molecule influences the activities of members of the CCN family of proteins (6,7). CCN proteins signal through heparan sulfate proteoglycans and integrins, interacting with, and regulating, the activity of many growth factors. They play important roles in processes such as cell differentiation, proliferation, extracellular matrix synthesis, and angiogenesis (8-12). The antiproliferative activities of CCN3 have been documented using embryonic fibroblasts, gliomal cells, vascular smooth muscle cells and osteogenic mesenchymal cells (13-16); this activity is most likely mediated through association with, and enhancement of, the activities of Notch-1 and connexin43 (15-17). In skeletal tissues, expression of a mutant, truncated CCN3 protein lacking the Von Willebrand factor C (VWC) domain shows enlarged axial and appendicular skeletal elements, increased bone mineralization, severe joint malformations and disrupted growth plate organization (18). In epiphyseal chondrocytes, CCN3 is upregulated early during differentiation and treatment with the recombinant CCN3 protein causes enhanced expression of collagen type X, suggesting a requirement for CCN3 during the chondrocyte maturation process (19). Other than a single report that indicates CCN3 expression during early development of the notochord, the anlagen of the nucleus pulposus (16), little is known about the regulation of CCN3 expression and function in the developing intervertebral disc.
The goal of this investigation was to examine the expression and regulation of CCN3 in cells of the nucleus pulposus and to determine if CCN3 expression is regulated by TGFβ. Results of this study show that TGFβ suppresses CCN3 gene expression in a Smad and AP1 independent fashion; importantly, Smad3 exerts an opposite effect on CCN3 expression. A second goal was to examine the effects of CCN3 on proliferation and on the expression of extracellular matrix components of nucleus pulposus tissue. These studies suggest that during the pathogenesis of degenerative disc disease, the interaction between TGFβ and members of the CCN family of proteins may provide a reparative function.
MATERIALS AND METHODS
Plasmids and reagents
Human CCN3 reporter plasmid (-1257/+317) was from Kurt Engeland, Universitat Leipzig. Plasmids were kindly provided by Drs. Bert Vogelstein, John Hopkins University [Smad2], Nancy Colburn, NIH [TAM67], Charles Vinson, NIH [DN-AP1; A-Fos], Silvio Gutkind, NIH [AP1 reporter], David Danielpour, Case Western Reserve University [pLVTHM-ShSmad3, pLVTHM], Jiahui Han, Scripps Research Institute, La Jolla [DN-p38 plasmids p38αAF, p38β2AF, p38γAF and p38δAF], Melanie Cobb, University of Texas Southwestern Medical Center, Dallas [DN-ERK1 (ERK1K71R) and DN-ERK2 (ERK2K52R)]. Expression plasmids for Smad3 (#11742), DN-ALK5 (#14834), CA-ALK5 (#14833) and for lentiviral packaging psPAX2 (#12260) and pMD2.G (#12259) were obtained from Addgene. As an internal transfection control, vector pGL4.10 (Promega) containing Renilla reniformis luciferase gene was used. The amount of transfected plasmid, the pre-transfection period after seeding, and the post-transfection period before harvesting, have been optimized for rat nucleus pulposus cells using pSV β-galactosidase plasmid (Promega) (20). Smad3 null embryonic fibroblasts were provided by Dr. Rik Derynck, University of California San Francisco. Anti-CCN3 rabbit polyclonal K19M antibody was used for CCN3 detection (21). Recombinant human TGFβ3 and CCN3 was purchased from R&D systems (Minneapolis, MN).
Isolation of nucleus pulposus cells and treatments of cells
Rat nucleus pulposus and annulus fibrosus cells were isolated using a method reported earlier by Risbud et al (20). Nucleus pulposus cells and MEFs were maintained in Dulbeccos Modified Eagles Medium (DMEM) and 10% fetal bovine serum (FBS) supplemented with antibiotics. In some experiments, cells were treated with rhTGFβ3 (10 ng/ml), rCCN3 (50 -500 ng/ml) and rCCN2 (100 ng/ml) all from R&D systems.
Immunohistological studies
Freshly isolated spines or whole embryos were immediately fixed in 4% paraformaldehyde in PBS and then embedded in paraffin. Transverse and coronal sections, 6-8 μm in thickness, were deparaffinized in xylene, rehydrated through graded ethanol and stained with alcian blue, eosin and hematoxylin. For localizing CCN3, sections were incubated with the anti-CCN3 antibody (K19M) in 2% bovine serum albumin in PBS at a dilution of 1:100 at 4 °C overnight. After thoroughly washing the sections, the bound primary antibody was incubated with Alexa fluor-488 conjugated anti-rabbit secondary antibody (Invitrogen), at a dilution of 1:200 for 45 min at room temperature. Sections were visualized using a fluorescence microscope (Olympus, Japan).
Real time RT-PCR analysis
Total RNA was extracted from nucleus pulposus cells using RNAeasy mini columns (Qiagen). Before elution from the column, RNA was treated with RNase free DNAse I (Qiagen). The purified, DNA-free RNA was converted to cDNA using Superscript III Reverse Transcriptase (Invitrogen). Template cDNA and gene specific primers were added (Rat CCN3 F: 5’tcattggaacctgtacctgccact 3’, R: 5’ tccctgggcacctgttacatttct 3’) to Fast SYBR Green master mix (Applied Biosystems) and mRNA expression was quantified using the StepOnePlus Real-Time PCR System (Applied Biosystems). β-actin and GAPDH were used to normalize the expression. Melting curves were analyzed to verify the specificity of the RT-PCR reaction and the absence of primer dimer formation. Each sample is analyzed in duplicate and included a template-free control. All the primers used were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Immunofluorescence microscopy
Cells were plated in flat bottom 96 well plates (4 × 103/ well) and treated with TGFβ for 6 h - 24 h. After incubation, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% triton-X 100 in PBS for 10 min, blocked with PBS containing 5% FBS, and incubated with antibodies against CCN3 (1:200), at 4° C overnight. As a negative control, cells were reacted with isotype IgG under similar conditions. After washing, the cells were incubated with Alexa fluor-488 conjugated anti-rabbit secondary antibody (Invitrogen), at a dilution of 1:200 for 45 min at room temperature. Cells were imaged using a laser scanning confocal microscope (Olympus Fluoview, Japan).
Western blotting
Cells were placed on ice immediately following treatment and washed with ice-cold HBSS. All the wash buffers and final re-suspension buffer included 1X protease inhibitor cocktail (Roche), NaF (5 mM) and Na3VO4 (200 μM). Total cell proteins were resolved on 8-12 % SDS-polyacrylamide gels and transferred by electroblotting to PVDF membranes (Bio-Rad, CA). The membranes were blocked with 5% non-fat dry milk in TBST (50 mM Tris, pH 7.6, 150 mM NaCl, 0.1% tween 20) and incubated overnight at 4 °C in 3% non-fat dry milk in TBST with the anti-CCN3 (1:600) or anti-β-tubulin antibody (1:2000, DSHB) or anti-GAPDH (1:2000, Cell Signaling). Immunolabeling was detected using the ECL reagent (Amersham Biosciences).
Transfections and dual luciferase assay
Cells were transferred to 24-well plates at a density of 4 × 104 cells/well one day before transfection. To measure the effect of TGFβ, cells were transfected with 500 ng of CCN3 reporter plasmids with 500 ng pRL-TK plasmid, in some wells cells were treated with the inhibitors SB203580 (10 μM), SB202190 (10 μM) and PD98059 (10 μM) (Calbiochem). To investigate the effect of Smad3 or AP1 on CCN3 promoter activity, cells were co-transfected with A-fos (100-300 ng), TAM67 (100-300ng), DN-Smad3 (100-300 ng), Smad3 (100-300 ng) or backbone vector with 400 ng CCN3 reporter and 300 ng pRL-TK plasmid in presence or absence of TGFβ (10 ng/ml). To investigate the effect of p38 and ERK signaling, cells were transfected with 100-300 ng of dominant negative p38α, β2, γ, or δ isoforms or 100-300 ng DN- ERK-1 or ERK-2. LipofectAMINE 2000 (Invitrogen) was used as the transfection reagent; for each transfection, plasmids were premixed with the transfection reagent. The next day, the cells were harvested and a Dual-Luciferase™ reporter assay system (Promega) was used for sequential measurements of firefly and Renilla luciferase activities. Quantification of luciferase activities and calculation of relative ratios were carried out using a luminometer (TD-20/20, Turner Designs, CA). At least three independent transfections were performed, and all analyses were carried out in triplicate.
Lentiviral particle production and viral transduction
HEK 293T cells were seeded in 10 cm plates (1.3 × 106 cells/plate) in DMEM with 10% heat-inactivated FBS two days before transfection. Cells were transfected with 2.5 μg of pLVTHM-LacZ, sh-Smad3 plasmids along with 1.875 μg psPAX2 and 0.625 μg pMD2.G. After 16 hours, transfection media was removed and replaced with DMEM with 5% heat-inactivated FBS and penicillin-streptomycin. Lentiviral particles were harvested at 48 and 60 hours post-transfection. Nucleus pulposus cells were plated in DMEM with 5% heat-inactivated FBS one day before transduction. Cells in 10 cm plates were transduced with 5 ml of conditioned media containing viral particles along with 6 μg/ml polybrene. After 24 hours, conditioned media was removed and replaced with DMEM with 5% heat-inactivated FBS with 5 μg/ml doxycycline. Cells were harvested for protein extraction 5 days after viral transduction.
MTT assay
Cell proliferation was measured by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. After treatment for 24 or 72 hours, MTT diluted in serum-free DMEM was added to the culture medium to a final concentration of 0.5 mg/ml. At the end of the incubation period (2 h at 37 °C), the medium was removed, and the precipitated formazan crystals were solubilized in dimethyl sufoxide. Product formation was measured by reading the absorbance at 560 nm using a microplate reader (Tecan, Spectra Flour Plus, NC).
Cell cycle analysis
Following treatment, single cell suspension was prepared from cell cultures and fixed in ice-cold 70% ethanol for 1 h. Cells were washed and resuspended in PBS with 5% FBS. Cells were incubated with 50 μM propidium iodide for 30 min at 37°C. Cell cycle was conducted using a Coulter Epics XL-MCL system using XL System II software.
Statistical analysis
All measurements were performed in triplicate, data is presented as mean + S.E. Differences between groups were analyzed by the student t test; *p< 0.05.
RESULTS
Expression of CCN3 in the intervertebral disc
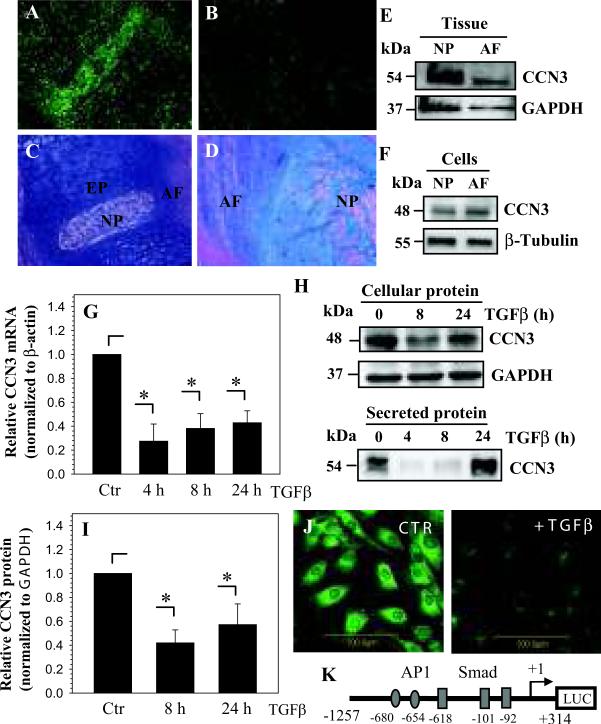
Saggital sections of neonatal (Fig. 1A) and skeletally mature rat discs (Fig. 1B) were stained with an antibody to CCN3 or counterstained with hemotoxylin, eosin and alcian blue (Fig. 1C, 1D). Supplemental Figure 1 shows negative control. Figures 1A and 1B indicate that CCN3 is expressed by cells of the nucleus pulposus, annulus fibrosus and cartilaginous endplate. In the embryonic nucleus pulposus, staining is localized to the cytosol as well as to the pericellular matrix. In contrast, at this early stage in the annulus fibrosus staining is primarily cellular. In skeletally mature animals, both discal tissues show cellular as well as pericellular staining. CCN3 expression in the embryonic discs is more prominent compared to discs of skeletally mature animals. Expression of CCN3 in native tissues and cultured cells was studied using Western blot analysis. Both nucleus pulposus and annulus fibrosus tissues express a 54 kDa band, representing the full length secreted CCN3 (Fig. 1E), while a prominent 48 kDa band, representing intracellular CCN3, is expressed in cultured nucleus pulposus and annulus fibrosus cells (Fig. 1F). Western blot analysis of conditioned media from nucleus pulposus cells, shows that full length CCN3 is secreted into the media (Fig. 1H).
Figure 1.
CCN3 expression in the intervertebral disc is suppressed by TGFβ. A-D. Saggital sections of embryonic (Mag. 20X) (A) and a skeletally mature rat discs (Mag. 10X) (B) were treated with an anti-CCN3 antibody or counterstained with H&E and alcian blue (C, D). CCN3 is expressed in the nucleus pulposus (NP) and annulus fibrosus (AF) of both the neonatal and skeletally mature rat. E-F. Western blot analysis of CCN3 expression in rat NP and AF tissue (E) and cultured NP and AF cells (F). The expression of a 54 kDa CCN3 band in tissue extracts is evident, while a 48 kDa band is present in cultured cells. G. Real-time RT-PCR analysis of CCN3 expression by NP cells treated with TGFβ shows decreased expression. H. Western blot analysis of CCN3 in NP cells and conditioned media. I. Densitometric analysis shows that TGFβ significantly decreases CCN3 expression. J. Immunofluorescence analysis of NP cells treated with TGFβ shows decreased CCN3 expression. K. Schematic of -1257/+317 bp CCN3 promoter construct with a few major transcription factor binding sites. Transcription start site is represented as + 1. Values shown are mean ± SE from three independent experiments, * p < 0.05.
TGFβ decreases CCN3 gene expression in nucleus pulposus cells
To explore the premise that TGFβ regulates CCN3 expression, nucleus pulposus cells were treated with TGFβ and expression of CCN3 analyzed using real-time RT-PCR. Figure 1G shows that treatment with TGFβ for 4, 8 and 24 hours significantly decreases CCN3 mRNA levels in nucleus pulposus cells. Western blot analysis indicates a similar decline in CCN3 protein expression with TGFβ treatment (Fig. 1H). TGFβ treatment also decreases levels of secreted CCN3 (Fig. 1H), though secreted levels return to baseline at 24 hours. Densitometry analysis of Western blots from three independent experiments was performed showing significant suppression of CCN3 protein levels by TGFβ (Fig. 1I). Likewise, immunofluorescence microscopy of TGFβ–treated nucleus pulposus cells shows that treatment causes a strong decrease in CCN3 expression (Fig. 1J).
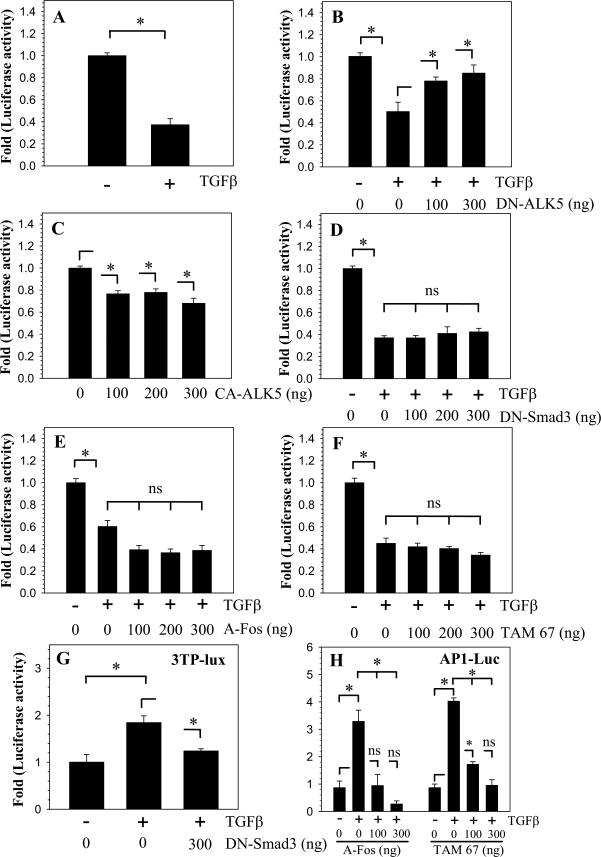
To investigate if TGFβ regulates CCN3 promoter activity, we transfected nucleus pulposus cells with a -1257/+317 bp human CCN3 promoter construct (Fig. 1K) and measured promoter activity following treatment of cells with TGFβ. CCN3 promoter activity significantly decreases with TGFβ treatment (Fig. 2A). To confirm the involvement of TGFβ signaling, cells were co-transfected with increasing doses of dominant negative ALK5 (DN-ALK5), the primary TGFβ type I receptor in nucleus pulposus cells. Figure 2B shows that co-expression of the DN-ALK5 blocks suppression of CCN3 promoter activity by TGFβ. CCN3 promoter activity also decreases when cells were co-transfected with constitutively active ALK5 (CA-ALK5) in the absence of TGFβ (Fig. 2C), suggesting a role for TGFβ in regulation of CCN3 expression in nucleus pulposus cells.
Figure 2.
TGFβ decreases CCN3 promoter activity in nucleus pulposus cells independent of Smad and AP1 signaling. A. Nucleus pulposus cells transfected with a CCN3 promoter construct and treated with TGFβ (10 ng/ml) show decrease in CCN3 promoter activity. B. Cells were transfected with DN-ALK5 in the presence of TGFβ. Increasing doses of DN-ALK5 blocked TGFβ-mediated suppression of CCN3 promoter activity. C. CCN3 promoter activity decreases in cells transfected with CA-ALK5 in absence of TGFβ. D-F. Cells were transfected with increasing doses of DN- Smad3 (D), A-Fos (E), or TAM 67 (F) in the presence of TGFβ. In all cases there are no significant changes in CCN3 promoter repression by TGFβ. Control experiments were performed to validate dominant negative plasmids: G. 3TP-lux increases with TGFβ treatment and decreases with addition of DN-Smad3. H. AP1 reporter activity increases with TGFβ treatment and decreases significantly with TAM67 and A-Fos expression. Values shown are mean ± SE from three independent experiments, * p < 0.05.
CCN3 suppression by TGFβ is independent of Smad and AP1 signaling
To elucidate the mechanism of CCN3 suppression by TGFβ we first investigated the involvement of Smad signaling pathway. Nucleus pulposus cells were co-transfected with DN-Smad3 (Fig. 2D) or siRNA against Smad3 (Supplemental Fig. 2) and then treated with TGFβ. Figure 2D shows that blocking Smad3 function has no effect on TGFβ suppression of CCN3 promoter activity. Next, we examined another important mediator of TGFβ action, the AP1 pathway, which we showed previously to be induced by TGFβ in the nucleus pulposus (22). Cells were co-transfected with dominant negative forms of the Fos (A-Fos) and Jun (TAM67) subunits of AP1, which inhibit transcriptional activity of endogenous AP1. Figures 2E and 2F indicates that co-expression of either of the dominant negative AP1 subunits has no effect on TGFβ dependent action on CCN3 promoter activity. Control experiments were performed using Smad responsive 3TP-Lux and AP1 responsive reporters to validate inhibitory function of DN-Smad3 (Fig. 2G), TAM67 and A-Fos plasmids (Fig. 2H).
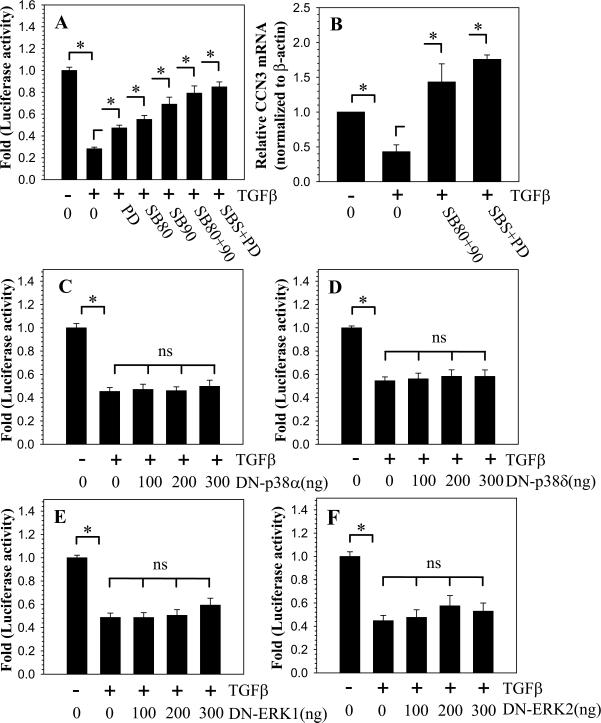
Role of MAPK signaling in TGFβ suppression of CCN3
We next investigated the involvement of the MAPK signaling pathway in TGFβ mediated suppression of CCN3 promoter activity. Cells were treated with p38 inhibitors, SB203580 or SB202190 or the ERK inhibitor, PD98059 or their combination, in the presence of TGFβ. Figure 3A shows that treatment with the individual p38 and ERK inhibitors partially counteracted the suppressive effects of TGFβ on CCN3 promoter activity. The combination of p38 and ERK inhibitors rescue CCN3 promoter activity almost to the basal level (Fig. 3A). As a control, cells were treated with p38 and ERK inhibitors without TGFβ, which did not cause significant change in CCN3 promoter activity (Supplemental Figure 3). Quantitative real-time PCR analysis on cells treated with the combination of MAPK inhibitors in the presence of TGFβ confirmed this regulation at the mRNA level (Fig. 3B). We then co-transfected nucleus pulposus cells with plasmids encoding DN-p38 isoforms α, β, γ or δ to examine which isoform is important for mediating TGFβ suppression of CCN3. Figure 3C-D shows that, individually, neither of the dominant negative isoforms of p38 blocked the suppressive effects of TGFβ on CCN3 promoter activity (DN-p38α and DN-p38γ results not shown). Similarly, co-transfections with DN-ERK1 and DN-ERK2 also showed little effect on CCN3 promoter activity (Fig. 3E,F).
Figure 3.
TGFβ-mediated suppression of CCN3 is partially dependent on p38 and ERK signaling. A. Cells were transfected with CCN3 promoter and treated with TGFβ in presence of inhibitors of p38 (SB203580; SB80 or SB202190; SB90) and ERK (PD98059; PD) signaling individually and in combination (used at 10 μM each). Suppression of CCN3 promoter activity is partially blocked by individual inhibitors and almost fully blocked when the inhibitors are combined. B. Real-time RT-PCR analysis of nucleus pulposus cells treated with TGFβ and similar combinations of MAPK inhibitors (SB80+90 and SBS+PD) show restoration of CCN3 expression. C-F. Cells were transfected with dominant negative isoforms of p38 α (C), δ (D), or dominant negative ERK-1 (E) or ERK-2 (F) in the presence of TGFβ. Suppression of individual MAPK isoform function did not alter CCN3 promoter activity. Values shown are mean ± SE from three independent experiments. * p < 0.05.
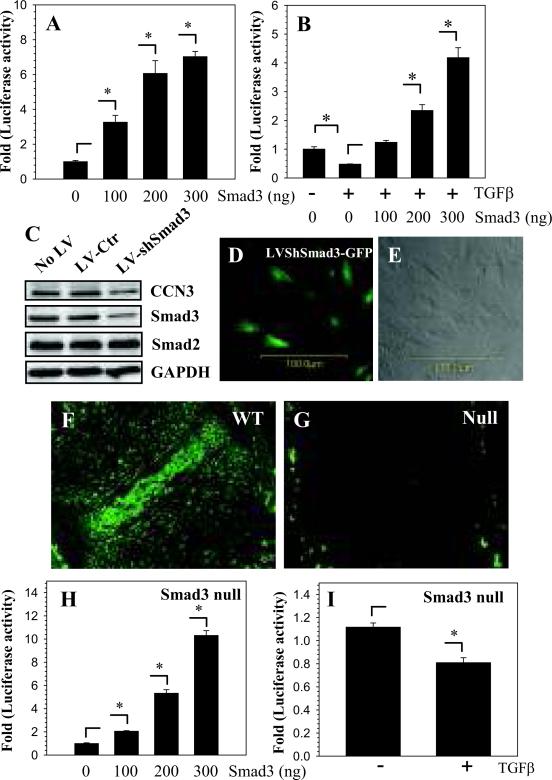
Smad 3 positively regulates CCN3 expression
To further determine if Smad3 exerts an effect independent of TGFβ signaling on CCN3 promoter activity, we overexpressed Smad3 in nucleus pulposus cells in the absence of exogenous TGFβ. Surprisingly, co-transfection with Smad3, results in an almost 7 fold increase in CCN3 promoter activity (Fig. 4A). Figure 4B shows that overexpression of Smad3 in the presence of TGFβ rescues suppression of CCN3 promoter activity by the growth factor. We suppressed Smad3 expression in nucleus pulposus cells by transduction with lentivirus expressing Smad3-shRNA. Figure 4C shows that there is significant decrease in Smad3 expression in nucleus pulposus cells and that shRNA mediated suppression is specific to Smad3. CCN3 expression is reduced in the sh-Smad3 transduced cells compared to non-tranduced cells or cells transduced with control shRNA (Fig. 4C). Figure 4D shows a robust GFP expression from virally transduced cells indicating high level of transgene expression. Moreover, comparison of GFP image to transmitted light image shows that most of the cells are transduced with the lentivirus (Fig. 4D-E). To confirm these results, sections of intervertebral disc from wild type (Fig. 4F) and Smad3 null mice (Fig. 4G) were stained with a CCN3 antibody. The Smad3 null mouse disc shows less CCN3 expression than the wild type disc (Fig. 4G). Similarly, when Smad3 is overexpressed in embryonic fibroblasts derived from Smad3 null mice, a significant increase in CCN3 promoter activity is seen (Fig. 4H). Lastly, treatment of Smad3 null mouse embryonic fibroblasts with TGFβ decreases CCN3 expression (Fig. 4I).
Figure 4.
Smad3 increases CCN3 promoter activity. A. Transfection of NP cells with Smad3 increases CCN3 promoter activity dose dependently. B. Smad3 overexpression counteracts suppression of CCN3 promoter activity by TGFβ. C. Western blot analysis of NP cells transduced with sh-Smad3 lentivirus. Smad3 suppression results in a concomitant decrease in CCN3 expression. Smad3 silencing has no effect on Smad2 expression. No Lentiviurs (No-LV), control lentivirus (LV-Ctr), shSmad3 lentivirus (LV-shSmad3). D. GFP reporter expression from cells transduced with sh-Smad3 lentivirus compared with transmitted light image of the cells (E) shows efficient viral transduction. Saggital sections of intervertebral disc from wild type (F) and Smad3 null mice (G) treated with an antibody against CCN3. Wild type (WT) disc shows a higher expression of CCN3 than discal tissues from Smad3 null mice (Mag. 20X). H. Smad3 null embryonic fibroblasts transfected with increasing doses of Smad3 show strong dose-dependent increase in CCN3 promoter activity. I. Smad3 null mouse embryonic fibroblasts treated with TGFβ show decrease in CCN3 promoter activity. Values shown are mean ± SE from three independent experiments, * p < 0.05.
CCN3 modulates proliferation of nucleus pulposus cells
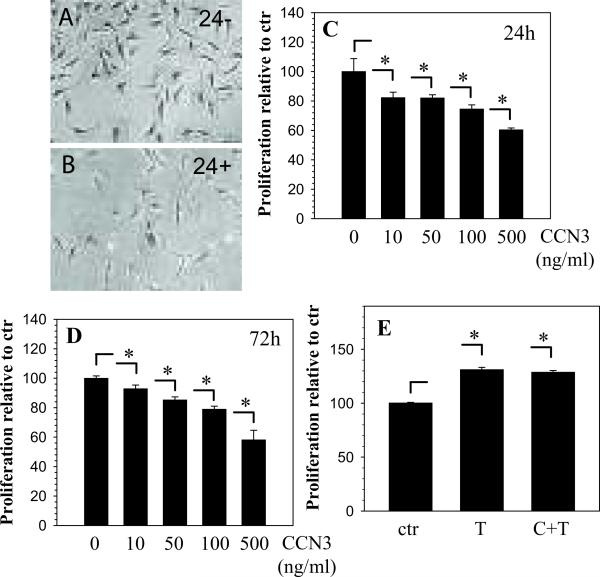
To delineate the effect of CCN3 on nucleus pulposus cell proliferation, we treated cells with an increasing dose of rCCN3 (10-500 ng/ml) for 24 or 72 hours. Figures 5A-B show phase contrast images of cells treated with 100 ng/ml of CCN3 for 24 hours (Fig. 5A) compared with control (Fig. 5B). There is no appreciable difference in cellular morphology between untreated and treated groups, although cell density appeared lower in treated group. MTT assay shows that cell proliferation decreases with increasing doses of CCN3 at 24 and 72 hours (Fig. 5C, D). To examine if the effect on proliferation was due to modulation of the cell cycle, we treated cells with 100 ng/ml of CCN3 and cell cycle analysis was performed using fluorescence-activated cell sorting. The results from four independent experiments show that CCN3 has no significant effect on the cell cycle status of nucleus pulposus cells (Control: G0/G1 81.34 ± 4.7%, S 5.37 ± 2%, G2/M 12.12 ± 5.91%, CCN3 treated: G0/G1 76.9 ± 8.2%, S 8.02 ± 3.5%, G2/M 14.83 ± 5.2%). To examine the effect CCN3 on TGFβ dependent cell proliferation, we treated the cells with TGFβ or TGFβ together with CCN3 for 72 hours. Figure 5E shows that TGFβ treatment results in a small increase in cell proliferation, but when combined with CCN3 there was no significant difference in cell proliferation.
Figure 5.
Antiproliferative effect of CCN3 on nucleus pulposus cells. A-B. Phase contrast images of untreated control cells (A) and cells treated with 100 ng/ml rCCN3 for 24 hours (B) stained with crystal violet after treatment. C-D. MTT assay shows the effect of increasing doses of rCCN3 on cell proliferation for 24 (C) and 72 hours (D). Cell proliferation decreased in a dose dependent manner with increasing doses of CCN3 at all time points. E. Nucleus pulposus cells were treated with TGFβ (T) and CCN3 in combination with TGFβ (C+T) for 72 hours. TGFβ increases cell proliferation while addition of CCN3 along with TGFβ shows no significant change. Values shown are mean ± SE from three independent experiments, * p < 0.05.
CCN3 modulates expression of critical extracellular matrix genes in the nucleus pulposus
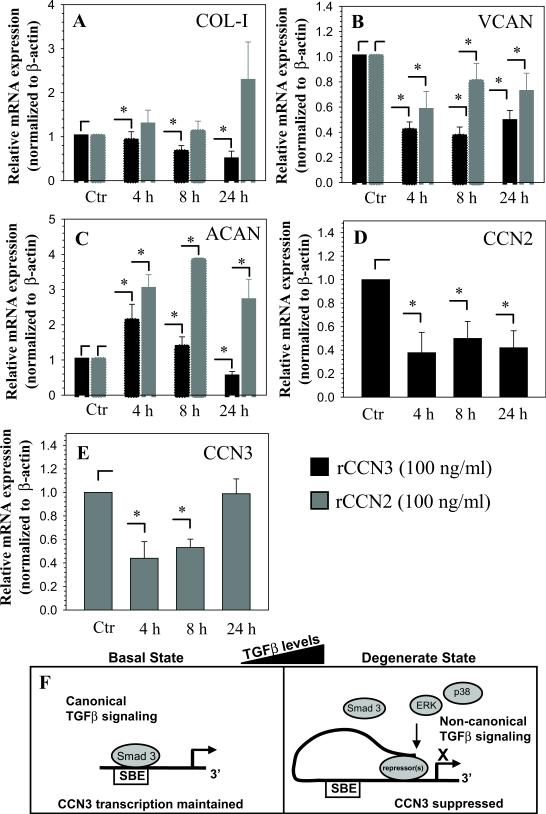
Finally, we examined the effect of CCN3 treatment on the expression of critical extracellular matrix genes in the nucleus pulposus cells. Figures 6A-C show that treatment with CCN3 decreases expression of versican, aggrecan and type I collagen after 24 hours. In addition, we treated cells with rCCN2 and measured expression of the same matrix genes (Fig. 6A-C). In contrast to CCN3, CCN2 strongly induces aggrecan mRNA expression at all time points. To explore the possibility that these CCN proteins could regulate each other as speculated in other cell types (25, 30), we treated the cells with either CCN2 or CCN3 and measured the expression of corresponding CCN. CCN3 treatment decreased CCN2 expression by nucleus pulposus cells (Fig. 6D), while CCN2 treatment decreased CCN3 expression transiently at 4 and 8 hours, but showed no effect at 24 hours (Fig. 6E).
Figure 6.
CCN3 decreases expression of extracellular matrix genes by NP cells. Real-time RT-PCR analysis of cells treated with 100 ng/ml of rCCN3 (black) and 100 ng/ml of rCCN2 (grey) show changes in expression of type I collagen (COL-I) (A), versican (VCAN) (B) aggrecan (ACAN) (C) and CCN2 (D). E. Cells treated with CCN2 show a decrease in CCN3 mRNA levels at 4 and 8 hours, returning to baseline at 24 hours. F. Proposed model for the regulation of CCN3 expression by Smad3 and TGFβ. In basal state when TGFβ levels are low, binding of Smad3 to Smad binding elements (SBE) maintains baseline CCN3 expression. When TGFβ levels are elevated, phosphorylation of the type I receptor (ALK5) activates the non-canonical MAPK signaling pathways as well as phosphorylating the canonical effector Smad3. Enhanced non-canonical ERK and p38 signaling and possible recruitment of as yet unknown repressor to the promoter may limit interaction of the Smads with the promoter resulting in overall suppression of CCN3 expression. Values shown are mean ± SE from three independent experiments, * p < 0.05
DISCUSSION
The experiments described in this investigation demonstrated for the first time that CCN3 was expressed in nucleus pulposus cells of the intervertebral disc and its basal expression was regulated by TGFβ. Growth factor suppression of CCN3 transcription was partially mediated through p38 and ERK signaling pathways, independent of Smad and AP1 signaling. When treated with CCN3, nucleus pulposus cells elicited a dose-dependent decrease in cell number and lowered expression of versican, aggrecan, type I collagen, and CCN2. These studies also revealed that TGFβ signaling induces CCN3 promoter activity through Smad3 while also enhancing CCN3 repression via MAPK signaling. We propose that the repressive activity of MAPK signaling and as yet unknown transcription factors exceeds the stimulatory response generated by the Smads. To our knowledge, this is the first instance in which Smad and TGFβ signaling is decoupled thereby promoting opposing effects on target gene transcription. We suggest that during disc degeneration, an increase in TGFβ activity and the corresponding changes in CCN3 and CCN2 expression, as we reported previously, could trigger a reparative response that may influence the remodeling response of the extracellular matrix and proliferative status of the cells (22).
The expression and localization of CCN3 in the intervertebral disc was age dependent. In the embryonic disc, there was robust expression in both annulus fibrosus and nucleus pulposus cells. On the other hand, in the adult disc, there was a lower level of CCN3 protein expression in both tissues. Immunofluorescence analysis of the mature disc indicated that staining was both cellular and pericellular, suggesting that the protein was being secreted and possibly trapped in the pericellular matrix. This notion was substantiated by Western blot analysis of adult discal tissues where a prominent 54 kDa band, corresponding to the full length secreted CCN3 protein was seen (23). From this perspective, CCN3, like CCN2, must be considered to be a matrix constituent of the disc. That intracellular levels of CCN3 are regulated by TGFβ was evident from studies of cultured nucleus pulposus cells. Treatment with TGFβ suppressed CCN3 mRNA as well as the protein expression. We confirmed that TGFβ suppressed CCN3 activity by using a dominant negative TGFβRI/ALK5 receptor construct. When cells were transfected with this construct in the presence of TGFβ, there was a dose dependant restoration of CCN3 promoter activity. Likewise, in the absence of the growth factor, transfection with a constitutively active ALK5 construct decreased CCN3 promoter activity. Results of these experiments suggest that TGFβ is concerned with the basal regulation of CCN3 in nucleus pulposus cells of the intervertebral disc.
We examined the mechanism by which TGFβ suppresses CCN3 expression by examining both Smad and non-Smad transduction pathways. With respect to MAPK and AP1, Lafont et al. have previously shown in adrenocortical cells that CCN3 suppression was dependent on AP1 signaling (24). To ascertain if a similar regulation was functional in the nucleus pulposus, we transfected cells with dominant negative Fos and Jun subunits of AP1. Surprisingly, neither of these proteins influenced growth factor dependent CCN3 promoter activity. In a parallel study, we found that treatment with p38 and ERK inhibitors partially blocked TGFβ mediated suppression of CCN3 promoter activity as well as mRNA expression. Thus, from a regulatory perspective, it would appear that in discal tissues, regulation of CCN3 expression by TGFβ is unique and more complex than was originally thought. Indeed, trying to delineate which MAPK isoforms are most likely to be required for suppression was not obvious. Transfection with individual dominant negative isoforms of p38 (p38α, p38β, p38γ andpδ38) or ERK (ERK1 and 2) showed no effect on CCN3 promoter activity. One interpretation of these results is that more than one isoform is required for CCN3 promoter suppression by TGFβ, or possibly, that there is some isoform redundancy. The importance of these and other non-canonical signaling pathways in mediating TGFβ suppression of CCN3 is under active investigation.
To examine the involvement of the Smad signaling pathway, the more conventional transducer of TGFβ signaling, we cotransfected cells with DN-Smad3 and Smad3 siRNA in the presence of TGFβ. The observation that neither strategy rescued CCN3 promoter activity suggested that Smad3 is not involved in the suppression of CCN3. This result was not surprising as earlier studies indicated that in both mesangial and adrenocortical cells, TGFβ suppression of CCN3 is independent of Smad3 (24-25). Interestingly, in the course of the current investigation, we observed that overexpression of Smad3 caused a dose-dependent activation of CCN3 promoter activity. This result was further confirmed using Smad3 null fibroblasts. When transfected with increasing doses of Smad3, there was a dose-dependent increase in CCN3 promoter activity. In terms of disc cells, we observed a decrease in CCN3 protein levels in both the nucleus pulposus and annulus fibrosus of Smad3 null mice compared to wild type littermates. We confirmed this Smad3 dependent regulation in nucleus pulposus cells by suppressing Smad3 levels using lentiviral transduction. Thus, it is evident that Smad3 is more concerned with maintenance of basal CCN3 expression in nucleus pulposus cells than its control when high levels of TGFβ are present. Since the net effect of TGFβ treatment is suppression of CCN3 expression, one obvious interpretation of this result is that stimulatory effect of Smad3 must be offset by inhibitory activities of TGFβ-induced repressor molecule(s). It is possible that this could be achieved by limiting the binding of Smad3 to its response element by activation of the non-canonical pathway. Based on these observations, we propose a model of CCN3 regulation by TGFβ in nucleus pulposus cells that involves both canonical (Smad) and non-canonical (MAPK) signaling pathways (Fig. 6F). Accordingly, TGFβ, signaling though ALK5, induces both activators and repressors of CCN3: the activating arm of this pathway is mediated by Smad3, while the repressive arm is through MAPK signaling. Activation of the latter arm and the generation of as yet unknown repressors could possibly override the stimulation generated by the Smads by limiting their access to the promoter.
In terms of a physiological role for CCN3, we observed that it influenced matrix gene expression as well as the proliferative response of nucleus pulposus cells. When treated with CCN3, there was a dose dependent decrease in cell number, similar to what has been observed in other cell types (13-17). FACS analysis indicated that the change in proliferation was not caused by an arrest of cell cycle progression in the G2/M phase, as was reported for glioma cells (26). As suggested by Sakamoto et al. and Fu et al., CCN3 may exert its anti-proliferative activity by its association with notch and notch ligands and the connexin43 signaling pathways (15-17). In concert with previous reports, we also found that TGFβ caused an increase in nucleus pulposus cell proliferation (27, 28). This effect was not blocked with addition of CCN3 indicating that TGFβ overrides the antiproliferative function of CCN3. Concerning its effects on matrix genes, CCN3 caused alterations in the expression of aggrecan, versican and type I collagen, matrix genes known to be expressed by cells of the nucleus pulposus. It also caused a similar decrease in the expression of CCN2, a CCN family member that we have shown to be up-regulated by TGFβ in the nucleus pulposus and to promote aggrecan synthesis (22, 29). Noteworthy is the opposite effect of the two CCN proteins on aggrecan expression- CCN3 decreases while CCN2 promotes aggrecan expression. The opposing effects of TGFβ on CCN2 and CCN3 expression, and the suppression of CCN2 by CCN3, lends considerable strength to the notion that these molecules may form a regulatory triad (25,30). Since, during disc degeneration, there is a trend towards increased TGFβ expression, the decrease in CCN3 expression and a concomitant increase in CCN2 may promote a reparative response, enhancing matrix synthesis and promoting changes in cell number - events common in all tissues undergoing fibrotic degeneration. Whether the effects of TGFβ on CCN2 and CCN3 expression vary according to degenerative stage is not known. However, it would not be unreasonable to assume that each of the components of the regulatory circuit provide novel pharmacological targets for the treatment of degenerative disc disease.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health AR050087 and AR055655. Cassie Tran is supported by NRSA training grant 1F31AG038125.
This study was supported by NIH grants # R01AR050087, R01AR055655 (MVR and IMS) and T32 AR052273 and F31AG038125 (CMT)
REFERENCES
- 1.Roberts S, Evans H, Trivedi J, Menage J. Histology and pathology of the human intervertebral disc. J Bone Joint Surg Am. 2006;88(Suppl 2):10–4. doi: 10.2106/JBJS.F.00019. [DOI] [PubMed] [Google Scholar]
- 2.Feng H, Danfelter M, Stromqvist B, Heinegard D. Extracellular matrix in disc degeneration. J Bone Joint Surg Am. 2006;88(Suppl 2):25–9. doi: 10.2106/JBJS.E.01341. [DOI] [PubMed] [Google Scholar]
- 3.Setton LA, Chen J. Mechanobiology of the intervertebral disc and relevance to disc degeneration. J Bone Joint Surg Am. 2006;88(Suppl 2):52–7. doi: 10.2106/JBJS.F.00001. [DOI] [PubMed] [Google Scholar]
- 4.Ng L, Grodzinsky AJ, Patwari P, Sandy J, Plaas A, Ortiz C. Individual cartilage aggrecan macromolecules and their constituent glycosaminoglycans visualized via atomic force microscopy. J Struct Biol. 2003;143:242–57. doi: 10.1016/j.jsb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Urban JP, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5:120–30. doi: 10.1186/ar629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YJ, Kong MH, Song KY, Lee KH, Heo SH. The Relation Between Sox9, TGF-beta1, and Proteoglycan in Human Intervertebral Disc Cells. J Korean Neurosurg Soc. 2008;43:149–54. doi: 10.3340/jkns.2008.43.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009;41:771–83. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holbourn KP, Acharya KR, Perbal B. The CCN family of proteins: structure-function relationships. Trends Biochem Sci. 2008;33:461–73. doi: 10.1016/j.tibs.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holbourn KP, Perbal B, Ravi Acharya K. Proteins on the catwalk: modelling the structural domains of the CCN family of proteins. J Cell Commun Signal. 2009;3:25–41. doi: 10.1007/s12079-009-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubota S, Takigawa M. CCN family proteins and angiogenesis: from embryo to adulthood. Angiogenesis. 2007;10:1–11. doi: 10.1007/s10456-006-9058-5. [DOI] [PubMed] [Google Scholar]
- 11.Cicha I, Goppelt-Struebe M. Connective tissue growth factor: context-dependent functions and mechanisms of regulation. Biofactors. 2009;35:200–8. doi: 10.1002/biof.30. [DOI] [PubMed] [Google Scholar]
- 12.Perbal B. CCN3: Doctor Jekyll and Mister Hyde. J Cell Commun Signal. 2008;2:3–7. doi: 10.1007/s12079-008-0028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, et al. Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol Cell Biol. 1992;12:10–21. doi: 10.1128/mcb.12.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimoyama T, Hiraoka S, Takemoto M, Koshizaka M, Tokuyama H, Tokuyama T, et al. CCN3 inhibits neointimal hyperplasia through modulation of smooth muscle cell growth and migration. Arterioscler Thromb Vasc Biol. 2010;30:675–82. doi: 10.1161/ATVBAHA.110.203356. [DOI] [PubMed] [Google Scholar]
- 15.Fu CT, Bechberger JF, Ozog MA, Perbal B, Naus CC. CCN3 (NOV) interacts with connexin43 in C6 glioma cells: possible mechanism of connexin-mediated growth suppression. J Biol Chem. 2004;279:36943–50. doi: 10.1074/jbc.M403952200. [DOI] [PubMed] [Google Scholar]
- 16.Katsube K, Ichikawa S, Katsuki Y, Kihara T, Terai M, Lau LF, et al. CCN3 and bone marrow cells. J Cell Commun Signal. 2009;3:135–45. doi: 10.1007/s12079-009-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamoto K, Yamaguchi S, Ando R, Miyawaki A, Kabasawa Y, Takagi M, et al. The nephroblastoma overexpressed gene (NOV/ccn3) protein associates with Notch1 extracellular domain and inhibits myoblast differentiation via Notch signaling pathway. J Biol Chem. 2002;277:29399–405. doi: 10.1074/jbc.M203727200. [DOI] [PubMed] [Google Scholar]
- 18.Heath E, Tahri D, Andermarcher E, Schofield P, Fleming S, Boulter CA. Abnormal skeletal and cardiac development, cardiomyopathy, muscle atrophy and cataracts in mice with a targeted disruption of the Nov (Ccn3) gene. BMC Dev Biol. 2008;8:18. doi: 10.1186/1471-213X-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafont J, Jacques C, Le Dreau G, Calhabeu F, Thibout H, Dubois C, et al. New target genes for NOV/CCN3 in chondrocytes: TGF-beta2 and type X collagen. J Bone Miner Res. 2005;20:2213–23. doi: 10.1359/JBMR.050818. [DOI] [PubMed] [Google Scholar]
- 20.Risbud MV, Guttapalli A, Stokes DG, Hawkins D, Danielson KG, Schaer TP, et al. Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J Cell Biochem. 2006;98:152–9. doi: 10.1002/jcb.20765. [DOI] [PubMed] [Google Scholar]
- 21.Kyurkchiev S, Yeger H, Bleau AM, Perbal B. Potential cellular conformations of the CCN3(NOV) protein. Cell Commun Signal. 2004;2:9. doi: 10.1186/1478-811X-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran CM, Markova D, Smith HE, Susarla B, Ponnappan RK, Anderson DG, et al. Regulation of CCN2/CTGF expression in the nucleus pulposus of the intervertebral disc: Role of smad and AP1 signaling. Arthritis Rheum. 2010;62:1983–92. doi: 10.1002/art.27445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazar N, Manara C, Navarro S, Bleau AM, Llombart-Bosch A, Scotlandi K, et al. Domain-specific CCN3 antibodies as unique tools for structural and functional studies. J Cell Commun Signal. 2007;1:91–102. doi: 10.1007/s12079-007-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafont J, Laurent M, Thibout H, Lallemand F, Le Bouc Y, Atfi A, et al. The expression of novH in adrenocortical cells is down-regulated by TGFbeta 1 through c-Jun in a Smad-independent manner. J Biol Chem. 2002;277:41220–9. doi: 10.1074/jbc.M204405200. [DOI] [PubMed] [Google Scholar]
- 25.Riser BL, Najmabadi F, Perbal B, Peterson DR, Rambow JA, Riser ML, et al. CCN3 (NOV) is a negative regulator of CCN2 (CTGF) and a novel endogenous inhibitor of the fibrotic pathway in an in vitro model of renal disease. Am J Pathol. 2009;174:1725–34. doi: 10.2353/ajpath.2009.080241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bleau AM, Planque N, Lazar N, Zambelli D, Ori A, Quan T, et al. Antiproliferative activity of CCN3: involvement of the C-terminal module and post-translational regulation. J Cell Biochem. 2007;101:1475–91. doi: 10.1002/jcb.21262. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R, Ruan D, Zhang C. Effects of TGF-β1 and IGF-1 on proliferation of human nucleus pulposus cells in medium with difference serum concentrations. J Orthop Res. 2006;26(1):9. doi: 10.1186/1749-799X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakai T, Mochida J, Sakai D. Synergistic role of c-Myc and ERK1/2 in the mitogenic response to TGFβ-1 in cultured rat nucleus pulposus cells. Arthritis Res Ther. 2008;10:R140. doi: 10.1186/ar2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aoyama E, Hattori T, Hoshijima M, Araki D, Nishida T, Kubota S, et al. N-terminal domains of CCN family 2/connective tissue growth factor bind to aggrecan. Biochem J. 2009;420:413–20. doi: 10.1042/BJ20081991. [DOI] [PubMed] [Google Scholar]
- 30.Kawaki H, Kubota S, Suzuki A, Lazar N, Yamada T, Matsumura T, et al. Cooperative regulation of chondrocyte differentiation by CCN2 and CCN3 shown by a comprehensive analysis of the CCN family proteins in cartilage. J Bone Miner Res. 2008;23:1751–64. doi: 10.1359/jbmr.080615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.