Abstract
We report the cloning and characterization of a new member of the Delta family of Notch ligands, which we have named Dll4. Like other Delta genes, Dll4 is predicted to encode a membrane-bound ligand, characterized by an extracellular region containing several EGF-like domains and a DSL domain required for receptor binding. In situ analysis reveals a highly selective expression pattern of Dll4 within the vascular endothelium. The activity and expression of Dll4 and the known actions of other members of this family suggest a role for Dll4 in the control of endothelial cell biology.
Keywords: Notch signaling pathway, endothelium
Cell-to-cell communication is required for many biological processes such as differentiation, proliferation, and homeostasis. One system utilized by a wide range of eukaryotes is the Notch-signaling pathway. The Notch gene family consists of structurally conserved cell surface receptors that are activated by one or more membrane-bound ligands of the emerging Delta/Serrate/Lag-2 (DSL) gene family (Fleming 1998). Previous studies with invertebrates have shown that the Notch-signaling pathway allows a subset of cells within an equipotent precursor population to acquire a specific cell fate in a spatially and temporally restricted manner. In this process, known as lateral inhibition or specification, cells destined for one fate inhibit neighboring cells from adopting the same fate (Artavanis-Tsakonas et al. 1999). In Drosophila, for example, equipotent cells in the embryonic ectoderm give rise to both neural and epidermal cells. As cells assume a neural fate, expression of neurogenic genes results in increased expression of Delta on the cell surface, leading to an inhibition of these same genes in neighboring cells via activation of their Notch receptors. Notch-receptor activation results in the transactivation of the Enhancer of Split complex [E(spl)-C] which, in turn, suppresses the expression of downstream genes, including Delta, necessary for neurogenesis (Artavanis-Tsakonas et al. 1999). Accordingly, mutant flies lacking Notch have excessive numbers of neuronal cells, whereas activating mutations of Notch lead to an inhibition of neuron production (Struhl et al. 1993). Although Delta–Notch interactions in vertebrates are less well understood, the identification of several family members suggests that these genes play an important role in mammalian development.
Four mammalian Notch receptors have been identified, Notch1/TAN-1 (Ellisen et al. 1991; del Amo et al. 1993), Notch2 (Weinmaster et al. 1992), Notch3 (Lardelli et al. 1994), and Notch4/int-3 (Uyttendaele et al. 1996). Dysregulated Notch signaling has been shown to result in several developmental abnormalities and diseases. Chromosomal translocations resulting in the truncation of the human Notch1 homolog, TAN1, have been found in certain T-cell acute lymphoblastic leukemias (T-ALL) (Ellisen et al. 1991). A frequent site for insertional activation by the mouse mammary tumor virus (MMTV) in malignant mammary carcinomas is the Notch4 locus. The insertion of MMTV results in an aberrantly expressed intracellular form of Notch4 originally identified as Int-3 (Gallahan and Callahan 1987). Cerebral autosomal-dominant arteriopathy with subcortical infarcts and leucoencephalopathy (CADASIL) is a disorder that leads to ischaemic strokes and dementia in adults, and has been traced recently to missense mutations of the Notch3 receptor (Joutel et al. 1997).
Notch ligands are divided into two subclasses, the Delta family and the Serrate family. All Notch ligands share some structural features (Fleming 1998) including epidermal growth factor (EGF)-like repeats, a characteristic DSL domain necessary for Notch binding (Muskavitch 1994) and a transmembrane region. However, an extracellular cysteine-rich domain and insertions that interrupt some EGF-like repeats are common only to the Serrate family. It is these structural differences that categorize a Notch ligand as a Delta or Serrate family member. Mammalian ligands reported include two members of the Serrate family, Jagged1/JAG1 (Lindsell et al. 1995; Oda et al. 1997a) and Jagged2 (Shawber et al. 1996), and two members of the Delta family, murine Dll1 (Bettenhausen et al. 1995) and murine Dll3 (Dunwoodie et al. 1997).
Ligand mutations can also result in the disruption of the Notch-signaling pathway, leading to developmental abnormalities. Mutations of human JAG1 have been attributed to the development of Alagille syndrome (AGS), an autosomal dominant disorder characterized by developmental abnormalities of the heart, skeleton, muscle, liver, and eyes (Li et al. 1997; Oda et al. 1997b). Mice homozygous for the pudgy (pu) mutation exhibit severe deformities of the ribs and vertebrae, and this mutation has been mapped to the Dll3 locus (Kusumi et al. 1998). Dll1-deficient mice generated by homologous recombination exhibit neonatal lethality (Hrabe de Angelis et al. 1997), and disruption of murine Jagged2 revealed an essential role for this gene in limb, craniofacial, and thymic development (Jiang et al. 1998).
The pleiotropic effects of dysregulated Notch signaling validate this pathway as an important regulator of development, and the existence of multiple ligands and receptors suggests a more specialized role for these genes in mammals than in lower eukaryotes. Here we report the molecular cloning and characterization of Dll4, the third mammalian member of the Delta family of Notch ligands. The ability to activate Notch receptors and the specific expression pattern of Dll4 implicate a role for this gene in the regulation of vascular biology.
Results and Discussion
cDNA cloning
An EST database from a murine, white adipose cDNA library was found to contain a clone with homology to known members of the DSL family of Notch ligands. This EST consisted of 450 bp of sequence encoding a DSL domain (Tax et al. 1994) followed by an EGF-like repeat. RACE of double-stranded DNA synthesized from white adipose tissue yielded a 3.4-kb cDNA. Analysis of the insert sequences from multiple clones revealed a 2058-bp ORF which was predicted to encode a 686 amino acid protein (Fig. 1). These clones also contained ∼300 bp of 5′ untranslated region (UTR) and 1000 bp of 3′ UTR, including a polyadenylation signal sequence, and a poly(A) tail (data not shown). The human DLL4 gene was also isolated and the ORF is ∼86% identical at the nucleotide level and 87% identical at the amino acid level to murine Dll4.
Figure 1.
Murine Dll4 nucleotide sequence with murine and human-deduced amino acid sequence shown. The DSL domain is framed, the eight EGF-like repeats are shaded, and the predicted transmembrane region is underlined. For the human amino acid sequence, only the residues differing from mouse are shown. Human and mouse amino acid sequences are ∼86% identical. The GenBank accession nos. for human and mouse are AF253468 and AF253469, respectively.
Dll4 is a novel Delta family member
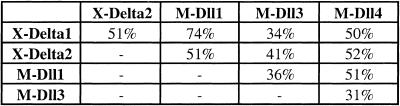
The predicted protein sequences for both human DLL4 and murine Dll4 exhibit all of the hallmarks of the Delta family of Notch ligands. The extracellular region contains 8 EGF-like repeats, as well as a DSL domain that is conserved among all Notch ligands and is necessary for receptor binding. The predicted protein also contains a transmembrane region, and a cytoplasmic tail lacking any catalytic motifs. The absence of an extracellular cysteine-rich domain and the lack of insertions between the EGF-like repeats clearly exclude Dll4 from the Jagged family of ligands. Sequence comparisons between Dll4 and the mu-rine Dll1 and Dll3, and Xenopus X-Delta1 (Chitnis et al. 1995) and X-Delta2 (Jen et al. 1997) proteins are shown in Figure 2. The Dll1 and X-Delta1 homologs are >70% identical at the amino acid level, whereas each of these genes is ∼50% identical to Dll4. Similarly, Dll4 is ∼50% identical to X-Delta2; suggesting Dll4 is not a mammalian homolog of the X-Delta2 gene, but rather a novel fourth member of the Delta gene family.
Figure 2.
Comparison of amino acid identity between vertebrate Delta genes reveals Dll4 is a novel family member. Murine Dll4 is ∼30%–50% identical to known Delta genes, whereas homologs (i.e., X-Delta1 and murine Dll1) are >70% identical. Dll3 remains the most divergent family member.
Dll4 is expressed in arterial endothelium
Northern blots containing mRNA from a variety of murine tissues were hybridized with a Dll4 probe. Expression of the 4.3-kb Dll4 message was highest in lung, followed by heart, kidney, skeletal muscle, and brain (Fig. 3). Transcripts were barely detectable in spleen and testis, but hybridization with GAPDH showed little detectable mRNA in these samples. Northern analysis of whole mouse embryos demonstrated increased expression of Dll4 from E7 through E17 of development. Analysis by RT–PCR revealed a low level of Dll4 expression in all tissues examined with higher levels in lung, brown and white adipose, and adrenal (data not shown).
Figure 3.
Murine Dll4 is expressed in most tissues. Northern analysis shows a transcript of ∼4.3 kb in several adult tissues, with lung expressing the highest levels. Expression is also seen in embryos, with increasing levels from embryonic day 9 through day 17. GAPDH hybridization provides for variations in loading and helps explain the lack of signal in spleen and testis.
In situ hybridization was performed on a variety of mouse tissues with an antisense Dll4 riboprobe, and a strikingly specific expression pattern was observed. In both embryonic and adult tissues, the predominant site of Dll4 expression was the vasculature (Fig. 4). Although it is difficult to rule out expression in the underlying smooth muscle, the expression of Dll4 in the choroid plexus (Fig. 4a), which lacks smooth muscle cells, suggests that the endothelium is the predominant site of Dll4 expression. In brain, Dll4 expression was observed in blood vessels (Fig. 4b), although some non-vascular expression was also seen in neural epithelium (not shown). Non-vascular expression was also observed in adrenal cortex, spleen, and lymph node (not shown). The precise cell types that express Dll4 in these tissues requires further investigation. Expression of Dll4 was observed in the glomeruli of the kidney (Fig. 4c) and the vasculature of the lung (Fig. 4d). Within the vasculature, Dll4 is relatively restricted to arteries, arterioles, and capillaries. This is illustrated strikingly in the umbilical cord (Fig. 4e), in which high expression is observed within the umbilical artery but not in the adjacent vein. The arterial expression of Dll4 is further illustrated by whole-mount in situ analysis of developing embryos (see the accompanying paper by Krebs et al.). In situ analysis of human DLL4 was also performed, and a similar expression pattern was seen (data not shown).
Figure 4.
Murine Dll4 is highly specific for arterial endothelial cells. In situ analysis was performed on murine tissues with an antisense murine Dll4 riboprobe. Dll4 expression in (a) choroid plexus; (b) brain; (c) kidney; (d) lung; (e) umbilical cord. The bright field and dark field views are displayed on the left and right, respectively, for each tissue section. Endothelial cells expressing Dll4 are indicated with arrows. See text for details.
Chromosomal localization
To determine whether the human or mouse gene mapped near any human disease syndromes or pre-existing mouse mutants, the chromosomal localization of both genes was performed. The human gene was mapped using the Stanford G3 human–hamster radiation hybrid panel, and the mouse gene was localized by FISH. The human DLL4 gene was located on 15q21.1, and the mouse Dll4 gene was mapped to chromosome 2E3, a region that shows conservation of synteny with human chromosome 15q. A search of human syndromes (OMIM 1999) and mouse mutants (MGD 1999) that map near these regions did not reveal any phenotypes apparently attributable to the Notch-signaling pathway.
Dll4 activates Notch1 and Notch4
In developing Xenopus embryos, activation of the Notch-signaling pathway results in the inhibition of neurogenesis (Coffman et al. 1993), which can be visualized by staining for the presence of N-tubulin. The ability of Dll4 to activate the Xenopus Notch receptor (X-Notch) was examined by injecting Dll4 RNA into a single blastomere of two-celled albino embryos, which were subsequently labeled for N-tubulin expression at stage 14. Consistent with X-Notch activation, Dll4 suppresses N-tubulin formation in approximately one-half of the embryos tested when compared with uninjected controls (Fig. 5a,b). To further verify Notch activation by Dll4, expression levels of ESR-1 (Wettstein et al. 1997) and ESR-7 (Deblandre et al. 1999) in neural ectoderm were quantified in animal cap experiments in the presence or absence of Dll4 (Fig. 5c, columns 1–6). The Xenopus ESR genes are related to Drosophila E(spl)-C, and are transcriptionally activated by the Notch-signaling pathway. Dll4 activated endogenous X-Notch, leading to increases in ESR-1 and ESR-7 levels. The coexpression of murine Notch1 with Dll4 resulted in higher ESR levels, similar to those resulting from activation of X-Notch by X-Delta1. This effect is most likely due to a higher affinity between the murine genes, as coexpression of murine Notch1 with X-Delta1 did not significantly increase ESR gene expression. The Notch4 receptor is highly expressed in the vascular endothelium of both embryos and adults (Uyttendaele et al. 1996). To investigate the potential of Dll4 as a Notch4 ligand, we performed animal cap experiments with murine Notch4 (Fig. 5c, columns 7–10). Similar to the Notch1 signaling seen previously, the activation of ESR gene expression upon Dll4/Notch4 coinjection was >3.5-fold higher than injection of Dll4 alone. Although the specificity of Dll4 for each of the Notch receptors has yet to be elucidated, Dll4 clearly functions as a ligand for Notch1 and Notch4.
Figure 5.
Dll4 acts as a Notch ligand in Xenopus embryos. One blastomere of 2-cell albino embryos was injected with either (a) nLacZ RNA and 0.1–0.25 ng of Dll4 RNA, or (b) nLacZ RNA only. At stage 14, the embryos were fixed, stained in whole mount with X-gal (light blue reaction product), and then labeled by in situ hybridization for the expression of N-tubulin (dark blue). Embryos are presented anterior to the left with the injected side on the top. Note that N-tubulin expression is suppressed in the area of Dll4 injection (as visualized with lacZ). (c) Results of RNase protection assay to measure the levels of ESR-1 and ESR-7, targets of Notch signaling, in neural ectoderm isolated from embryos injected with the indicated RNAs. Ectoderm was neuralized by injection of Noggin. RNAs were extracted at stage 13 and the ability of Dll4 to activate Notch targets were examined. For quantification, specific band intensities in each lane were normalized to the amount of EF1-α RNA to control for sample recovery, and the gene expression results are represented relative to the uninjected controls. Note that Dll4 weakly activates ESR-1 and ESR-7 expression via endogenous X-Notch activation (column 3), whereas ESR gene activation by Dll4 is strongly potentiated by coinjection of murine Notch1 (column 4) to reach the levels induced by X-Delta1 (column 5). In a separate experiment (columns 7–10), the ability of Dll4 to signal through the murine Notch4 receptor was tested. The activation of ESR-1 and ESR-7 expression by Dll4 (column 9) is strongly potentiated by coinjection with murine Notch4 (column 10). In both experiments, activation of targeted genes was increased >3.5-fold upon coinjection of Dll4 with either murine Notch1 or Notch4.
The isolation and characterization of Dll4 further expands the DSL family of mammalian Notch ligands. The structural characteristics of the protein clearly establish Dll4 as a novel Delta gene, and the expression pattern implicates Notch signaling in the regulation of blood vessel biology. The specific expression of Dll4 in arterial endothelium is reminiscent of ephrin-B2; another membrane-bound ligand (Wang et al. 1998). During vascular development, ephrin-B2 is expressed in arterial endothelial cells, whereas its cognate receptor, Eph-B4, is expressed only in venous endothelium. Targeted disruption of the murine ephrin-B2 gene affects angiogenesis, but not vasculogenesis, in developing embryos. Because the Notch-signaling pathway is an important regulator of neurogenesis and myogenesis (Nye and Kopan 1995) a similar involvement in the regulation of vascularization could be envisioned. In fact, the study of receptor mutants and downstream genes is providing evidence that Notch signaling is pivotal in vascular development. The E(spl)-C genes are regulated by Notch activation, and a new E(spl)-C family member in zebrafish, gridlock, is required for development of the aorta (Zhong et al. 2000). CADASIL patients have missense mutations within the extracellular EGF domains of their Notch3 receptors. These mutations result in the addition or deletion of cysteine residues, possibly altering the formation of disulfide bonds necessary for normal Notch3 signaling. In the accompanying manuscript by Krebs et al. (2000), mice lacking the Notch1 and Notch4 receptors develop vascular defects, and expression analysis of known Notch ligands suggests that Dll4 is the only ligand that exhibits spatial and temporal expression consistent with the activation of Notch1 and Notch4 during vascular development. The identification of Dll4 reveals a candidate ligand for Notch receptors involved in blood vessel biology, although no vascular disorders map near the Dll4 locus in mice or humans. A more detailed examination of receptor-ligand specificities and the cell types involved should provide new insight into the role of the Notch-signaling pathway in the development and regulation of the vasculature.
Materials and methods
Murine Dll4 cDNA cloning
A murine adipose Marathon library (Clontech) was constructed and RACE was performed. For the 3′ RACE reaction, PCR was performed with a gene-specific primer (5′-TGCTGTGGGTAAGATTTGGCGAACA-3′) and the AP1 primer (5′-CCATCCTAATACGACTCACTATAGGGC-3′) using the Advantage PCR kit components (Clontech). A 5′ RACE was performed using a gene-specific primer (5′-GGTGAGTCCGCACAGGTCAAGGTAC-3′) and the AP1 primer. To generate the full-length Dll4 ORF, PCR was performed on murine adipose Marathon cDNA with the following oligos: (5′-AGCCACCATGACGCCTGCGTCCCG-3′) and (5′-TCTATTATACCTCTGTGGCAATCAC-3′).
Human Dll4 cDNA cloning
The murine Dll4 sequence was searched against the Genbank database (Wisconsin Package Version 9.1, Genetics Computer Group, Madison, WI) and a 409-bp sequence (accession no. T33811) from a human brain cDNA library was found that had 81.37% identity to the 3′ end of Dll4. Oligos were designed from this sequence (5′-AAGAAGGAGCTGGAAGTGGACTGTG-3′) and (5′-ATCAAACACACAGACTGGTACATGG-3′) and used to amplify a 245-bp product from a Marathon human brain cDNA library (Clontech). This fragment was used to screen a human fat cell 5′ Stretch Plus cDNA (gt10) library (Clontech) as per instructions. One phage clone was isolated using the Wizard Lambda Prep DNA Purification System (Promega). This clone contained ∼215 bp of 5′ UTR and 1980 bp of the DLL4 coding region, lacking the last 85 bp of the coding region. Oligos from the DLL4 phage clone sequence and from T33811 sequence (5′-ACCTGATTCCTGCCGCCCAGCT-3′) and (5′-GATGTCCCAGGTAGGCTCCTGC-3′), respectively, were used to amplify the full-length human DLL4 gene from a Marathon human lung cDNA library (Clontech).
Northern blot analysis
Nylon filters containing immobilized poly(A) RNA (2 μg/lane) from a variety of murine tissues (Clontech) were probed with a 275-bp fragment that spanned nucleotides 461–736 of the murine Dll4 sequence as per the manufacturer's instructions. Hybridization signals were visualized with a PhosphorImager (Molecular Dynamics).
In situ hybridization
A panel of normal embryonic (E10.5–E18.5) and adult mouse tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm. Prior to in situ hybridization, tissues were permeabilized with 0.2 m HCL, followed by digestion with Proteinase K, and acetylation with triethanolamine and acetic anhydride. Sections were hybridized overnight at 55°C with a 33P-labeled riboprobe corresponding to murine Dll4 ORF. Following RNase digestion, the slides were subjected to a high-stringency wash in 0.1× SSC at 55°C. Slides were dipped in Kodak NTB2 emulsion, exposed at 4°C for 2–3 weeks, developed, and counterstained. Sections were examined with darkfield and standard illumination to allow simultaneous evaluation of tissue morphology and hybridization signal.
Chromosomal localization
The human DLL4 gene was localized using the Stanford G3 RH panel (Research Genetics, Baltimore, MD). Oligonucleotides that amplified human, but not hamster DNA (5′-GCGCTACTGCTGCTGGTGGC-3′) and (5′-GGGTCCGGGCGAGACGACC-3′), were used to amplify RH panel DNA under the following PCR conditions: an initial denaturation at 94°C for 3 min; 35 cycles of 94°C at 30 sec, 68°C at 45 sec, and 72°C for 10 min. A score of 00000100000000000000000000000000000000000000000001000000101000010110000000000000020 was obtained with the StanfordG3 RH panel and was submitted to the appropriate server (http://www.shgc.stanford.edu/RH/rhserver_form2.html). These data place DLL4 within 15cR10000 of SHGC-5930 and SHGC-57568 (LOD 7.74 for each). The Genome Database (http://www.gdb.org) was used to determine the cytogenetic location of these markers and indicated that DLL4 maps to 15q21.1. For mouse chromosomal localization, DNA from a BAC clone (Genome Systems, St. Louis, MO) containing the murine Dll4 gene was labeled with digoxigenin dUTP by nick translation. Labeled probe was combined with sheared mouse genomic DNA and hybridized to metaphase chromosomes derived from a mouse fibroblast cell line in a solution containing 50% formamide, 10% dextran sulfate, and 2× SSC. Specific hybridization signals were detected by incubating the hybridized slides in fluoresceinated antidigoxigenin antibodies followed by counterstaining with DAPI, resulting in preliminary localization to chromosome 2. A second hybridization was then performed with a probe specific for the centromeric region of chromosome 2 as well as the Dll4 probe. Measurements of 10 specifically labeled chromosomes demonstrated that Dll4 was located at a position that is 58% of the distance from the heterochromatic–euchromatic boundary to the telomere of chromosome 2, an area that corresponds to band 2E3. A total of 80 metaphase cells were examined with 71 exhibiting specific labeling.
Embryos and injections
Embryos were obtained from Xenopus laevis adult frogs by hormone-induced egg-laying and in vitro fertilization by standard methods. Xenopus embryos were staged according to Neiuwkoop (Nieuwkoop and Faber 1967). Constructs cloned in pCS2+ vectors (Turner and Weintraub 1994) were transcribed into capped RNA using SP6 RNA polymerase. Templates for generating RNA encoding Dll4 were obtained by linearization of the CS2 construct by Asp718. The templates for generating the Notch1 and Notch4 RNAs (Kopan et al. 1996) were obtained by linearization by HpaI. For in situ hybridization analysis, synthetic n-lacZ RNA (20 pg), encoding a nuclear-localized form of β-galactosidase, along with 1–2-ng test RNAs (stated if otherwise) were injected into single blastomeres of albino embryos at the two-cell stage as described previously (Coffman et al. 1993). For animal caps, two-cell stage Xenopus embryos were injected in the animal region of each blastomere with 0.5–2 ng of the indicated capped synthetic RNAs. To generate neuralized ectoderm, 0.1–0.5 ng of RNA encoding Noggin (Lamb et al. 1993) was coinjected in each blastomere.
Whole-mount in situ hybridization
Whole-mount in situ hybridization of Xenopus embryos was performed according to Harland (1991) with modifications described by Knecht et al. (1995) using digoxigenin-labeled anti-sense RNA probe for Xenopus N-tubulin (Oschwald et al. 1991). Prior to in situ hybridization or immunostaining, injected embryos were stained for β-galactosidase activity with X-gal (Biosynth, Naperville, IL) to localize the tracer.
Animal caps experiments and RNase protection assays
Animal caps were dissected at stage 9 and cultured on agarose-coated Petri dishes in 0.5 × MMR containing gentamycin until sibling controls reached stage 13. RNA was isolated and analyzed by RNase protection assay using 32P-labeled antisense RNA probes as described previously (Kintner and Melton 1987; Kintner and Dodd 1991). The template used for the ESR-7 probe corresponds to the PvuII–HindIII fragment containing most of the ORF and the first half of the 3′ UTR, whereas that for ESR-1 has been described previously (Wettstein et al. 1997). RNA samples isolated from eight animal caps were analyzed simultaneously along with the EF1-α probe to control for sample recovery during the assay. Quantification was carried out on a PhosphorImager (Molecular Dynamics), and for each lane, specific band intensities were normalized to the amount of EF1-α RNA.
Acknowledgments
We thank Bethany Sutton for DNA sequencing, Tom Gridley, Melissa Graham, Laarni Ulias, Gerry Weinmaster, and Carol Hicks for helpful discussions, and the support of the Amgen Genome Project and the Dll4 Project Team. J.K. was supported in part by NIH grant no. HL62454.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kstark@amgen.com; FAX (805) 480-1368.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch Signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet JL, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Coffman CR, Skoglund P, Harris WA, Kintner CR. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell. 1993;73:659–671. doi: 10.1016/0092-8674(93)90247-n. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Wettstein DA, Koyano-Nakagawa N, Kintner C. A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development. 1999;126:4715–4728. doi: 10.1242/dev.126.21.4715. [DOI] [PubMed] [Google Scholar]
- del Amo FF, Gendron-Maguire M, Swiatek PJ, Jenkins NA, Copeland NG, Gridley T. Cloning, analysis, and chromosomal localization of Notch-1, a mouse homolog of Drosophila Notch. Genomics. 1993;15:259–264. doi: 10.1006/geno.1993.1055. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Henrique D, Harrison SM, Beddington RS. Mouse Dll3: A novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124:3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Fleming RJ. Structural conservation of Notch Receptors and ligands. Semin Cell Dev Biol. 1998;9:599–607. doi: 10.1006/scdb.1998.0260. [DOI] [PubMed] [Google Scholar]
- Gallahan D, Callahan R. Mammary tumorigenesis in feral mice: Identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987;61:66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: An improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M, McIntyre J, II, Gossler A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- Jen WC, Wettstein D, Turner D, Chitnis A, Kintner C. The Notch ligand, X-Delta-2, mediates segmentation of the paraxial mesoderm in Xenopus embryos. Development. 1997;124:1169–1178. doi: 10.1242/dev.124.6.1169. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes & Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssiere C, Cruaud C, Maciazek J, Weissenbach J, Bousser MG, et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet. 1997;350:1511–1515. doi: 10.1016/S0140-6736(97)08083-5. [DOI] [PubMed] [Google Scholar]
- Kintner CR, Melton DA. Expression of Xenopus N-CAM RNA in ectoderm is an early response to neural induction. Development. 1987;99:311–325. doi: 10.1242/dev.99.3.311. [DOI] [PubMed] [Google Scholar]
- Kintner CR, Dodd J. Hensen's node induces neural tissue in Xenopus ectoderm. Implications for the action of the organizer in neural induction. Development. 1991;113:1495–1505. doi: 10.1242/dev.113.4.1495. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Good PJ, Dawid IB, Harland RM. Dorsal-ventral patterning and differentiation of noggin-induced neural tissue in the absence of mesoderm. Development. 1995;121:1927–1935. doi: 10.1242/dev.121.6.1927. [DOI] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: Importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi DC, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES. The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat Genet. 1998;19:274–278. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Knecht AK, Smith WC, Stachel SE, Economides AN, Stahl N, Yancopolous GD, Harland RM. Neural induction by the secreted polypeptide noggin. Science. 1993;262:713–718. doi: 10.1126/science.8235591. [DOI] [PubMed] [Google Scholar]
- Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46:123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: A mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Database (MGD), Mouse Genome Informatics. The Jackson Laboratory, Bar Harbor, Maine. World Wide Web (URL: http://www.informatics.jax.org/). October, 1999.
- Muskavitch MA. Delta-notch signaling and Drosophila cell fate choice. Dev Biol. 1994;166:415–430. doi: 10.1006/dbio.1994.1326. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop, P.D. and J. Faber. 1967. Normal table of Xenopus laevis. North Holland, Amsterdam.
- Nye JS, Kopan R. Vertebrate ligands for Notch. Curr Biol. 1995;5:966–969. doi: 10.1016/s0960-9822(95)00189-8. [DOI] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Meltzer PS, Chandrasekharappa SC. Identification and cloning of the human homolog (JAG1) of the rat Jagged1 gene from the Alagille syndrome critical region at 20p12. Genomics. 1997a;43:376–379. doi: 10.1006/geno.1997.4820. [DOI] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997b;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Online Mendelian Inheritance in Man, OMIM (TM). Center for Medical Genetics, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD). World Wide Web URL: http://www.ncbi.nlm.nih.gov/omim/.1999.
- Oschwald R, Richter K, Grunz H. Localization of a nervous system-specific class II beta-tubulin gene in Xenopus laevis embryos by whole-mount in situ hybridization. Int J Int Biol. 1991;35:399–405. [PubMed] [Google Scholar]
- Shawber C, Boulter J, Lindsell CE, Weinmaster G. Jagged2: A serrate-like gene expressed during rat embryogenesis. Dev Biol. 1996;180:370–376. doi: 10.1006/dbio.1996.0310. [DOI] [PubMed] [Google Scholar]
- Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- Tax FE, Yeargers JJ, Thomas JH. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature. 1994;368:150–154. doi: 10.1038/368150a0. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes & Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- Wang HU, Chen Z-F, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Roberts VJ, Lemke G. Notch2: A second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Zhong TP, Rosenberg M, Mohideen M-APK, Weinstein B, Fishman M. gridlock, an HLH gene required for assembly of the aorta in Zebrafish. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]