Abstract
The 17.7 kDa R2 module from Azotobacter vinelandii mannronan C5-epimerase AlgE6 has been isotopically labeled (13C,15N) and recombinantly expressed. Here we report the 1H, 13C, 15N resonance assignment of AlgE6R2.
Keywords: Alginate, Mannuronan C5-Epimerases, A and R-module
Biological context
In Azotobacter vinelandii a family of seven secreted and calcium-dependent, mannuronan C5–epimerases (AlgE1–7) has been identified (Ertesvåg et al. 1999). These enzymes catalyse the epimersation of β-d-mannuronic acid (M) to α-l-guluronic acid (G) in the polysaccharide alginate. The epimerases are composed of two different structural modules, designated A (~385 amino acids each, with 1 or 2 copies per enzyme) and R (~155 amino acids each, with one to seven copies per enzyme) and a C-terminal putative signal peptide (Ertesvåg et al. 1994, 1995). The A-modules alone are catalytically active, but their reaction rates are increased when covalently bound to at least one R-module (Ertesvåg and Valla 1999). The epimerases generate different monomer sequence distributions in their reaction products, and these patterns appear to a large extent to be controlled by the A-modules alone. The exact functional roles of the R-modules are not yet understood.
Previously the R-module from the smallest epimerase, AlgE4 (A–R), was found to fold as an all parallel β-roll protein similar to the repeats in toxin (RTX) proteins, but with a positively charged shallow grove on the front side with the ability to bind the polyanionic alginate (Aachmann et al. 2006, 2005). AlgE4 exhibit by a processive mode of action, while AlgE6 (A-R1-R2-R3) preferentially introduces GG-blocks in the alginate (Campa et al. 2004, Hartmann et al. 2002). Both the first and last R-module from AlgE6 has been studied by NMR (Buchinger et al. 2010, Aachmann and Skjåk-Bræk 2008) and the 3D structures are currently being determined. The sequence identity of AlgE6, R2 is 69% and 69% compared to R1 and R3, respectively. Some conserved chemical shift patterns seem to be shared in 15N HSQC fingerprint spectrum. Preliminary results from the structure determination confirm this observation, but it has also been observed additional structure elements, which are not found in R1 and R3. Therefore, it is interesting to study the structure of R2-module as well as R1 and R3 in detail in order to gain insight into its role in AlgE6 function. Altogether, access to the structures of the three AlgE6 R-modules and their affinities to different tailored made alginates will hopefully lead to new insights that may be used to deduce their functionality in all the seven epimerases. Here, we report the complete sequence-specific assignments of the AlgE6 R2 module.
Methods and experiments
The gene coding for the AlgE6R2-module (residues 534–693) has been codon optimized for Escherichia coli protein expression and synthesized de novo (GenScript, Piscataway, USA). The synthetic gene for AlgE6R2 was inserted into pTYB12 vector (IMPACT-CN system, New England Biolabs) using BsmI and PstI sites, hereafter purified via gel electrophoresis and ligated with T4 ligase at 289 K 2 h generating pFA12 plasmid. The pFA12 plasmid was confirmed by restriction mapping. This plasmid was transformed into the production host E. coli ER2566. For uniform labelling of AlgE6R2 (161 amino acids) the cells were grown in M9-medium supplemented with (15NH4)2SO4 (1 g/L), 13C6-d-glucose (2 g/L) (Sigma–Aldrich) with additional 0.2 mM CaCl2. The fusion protein was overexpressed by growing the cells at 310 K until an OD600 ~0.8 was reached and the expression was started by addition 1 mM IPTG (in toto) and subsequently incubated at 289 K for 16 h. The cells were harvested by centrifugation and resuspended in 20 mM HEPES pH 6.9, 500 mM NaCl, 5 mM CaCl2 and 0.1% Triton X-100 (Sigma–Aldrich). The cells were disrupted by sonication and centrifugated. The supernatant was applied onto chitin bead column (New England BioLabs). The column was washed with 20 mM HEPES pH 6.9, 500 mM NaCl and 5 mM CaCl2, whereafter followed by cleavage with 50 mM DTT at room temperature over ~16 h resulting in the release of the AlgE6R2 from the chitin bound intein tag. The eluted AlgE6R2 was dialysed against 10 mM HEPES, pH 6.9, 10 mM CaCl2 in order to remove a 1.6 kDa peptide that occurred as a by-product from the cleavage reaction.
The NMR spectra were recorded at 298 K on a Bruker Avance 600 spectrometer equipped with 5 mm Z-gradient TCI(H/C/N) cryogenic probe. 1H and 13C chemical shifts were referenced internal to the sodium salt of 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS), while 15N chemical shifts were referenced indirectly to DSS, based on the absolute frequency ratios (Zhang et al. 2003). Sequence-specific backbone and side-chain assignments of AlgE6R2 were accomplished using 15N HSQC, 13C HSQC, HNCA, HN(CO)CA, HNCO, HN(CA)CO, CBCANH, CBCA(CO)NH, HBHANH, HBHA(CO)NH, HCCH-TOCSY and HCCH-COSY spectra. The assignments of the aromatic side chains were obtained from 2D IP-COSY, TOCSY, NOESY and 13C HSQC experiments. The NMR data were recorded and processed with BRUKER XWinNMR version 3.5 or TopSpin 3.0 software and spectral analysis was performed using CARA version 1.4.1/1.5.1/1.8.4 (Keller 2004).
Assignment and data composition
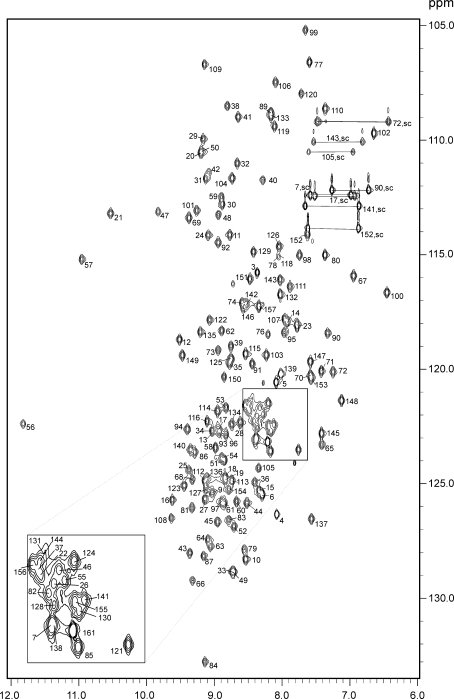
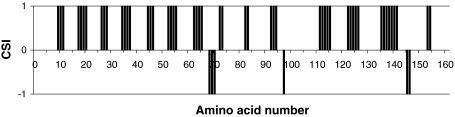
Here we report the resonance assignments of the R2-module of AlgE6R2. The 15N-HSQC spectrum of AlgE6R2, together with the assignments of the resonances, is shown in Fig. 1. The backbone and the side-chain assignments are essentially complete (HN,Hα, N, Cα, C’ > 97%; H and C side-chains > 94%). The amide groups (HN, N) of Asp1, Gln35, Ala75, and, Asp117 could not be found, although other nuclei of these residues were assigned. None of the exchangeable side-chain protons of Arg and Lys residues were assigned. Side-chain amide groups of all Asn and Gln were assigned. Most of the protons and the carbon atoms of aromatic side-chains were assigned. Results obtained for the chemical shift index (CSI) point toward mainly β-strands for the proteins secondary structure (See Fig. 2) that fit also well to an overall 3D β-roll structure common for the R-modules. The chemical shift data have been deposited in the BioMagResBank under the accession number 17249.
Fig. 1.
1H, 15N HSQC spectrum of the 13C, 15N-labelled AlgE6R2 subunit from Azotobacter vinelandii in 90:10 H2O:D2O at pH 6.9, 298 K. Residue numbers are indicated. Side-chain resonances of Asn and Gln residues are connected by lines. Other side-chain amine resonances are indicated with amino acid number and sc
Fig. 2.
The chemical shift index (CSI) for AlgE6R2 subunit from Azotobacter vinelandii
Acknowledgments
FLA acknowledges financial support from the Norwegian Research Council (NFR) via KMB project (182695/I40). EB acknowledges financial support from the Danish Agency for Science, Technology and Innovation.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aachmann FL, Skjåk-Bræk G. 1H, 15 N, 13C resonance assignment of the AlgE6R1 subunit from the Azotobacter vinelandii mannuronan C5-epimerase. Biomol NMR Assigm. 2008;2:123–125. doi: 10.1007/s12104-008-9101-0. [DOI] [PubMed] [Google Scholar]
- Aachmann FL, Svanem BIG, Valla S, et al. NMR assignment of the R-module from the Azotobacter vinelandii Mannuronan C5-epimerase AlgE4. J Biomol NMR. 2005;31:259. doi: 10.1007/s10858-004-7911-7. [DOI] [PubMed] [Google Scholar]
- Aachmann FL, Svanem BIG, Guntert P, et al. NMR structure of the R-module - A parallel beta-roll subunit from an Azotobacter vinelandii mannuronan C-5 epimerase. J Biol Chem. 2006;281:7350–7356. doi: 10.1074/jbc.M510069200. [DOI] [PubMed] [Google Scholar]
- Buchinger E, Skjak-Braek G, Valla S et al. (2010) NMR assignments of 1H, 13C and 15N resonances of the C-terminal subunit from Azotobacter vinelandii mannuronan C5-epimerase 6 (AlgE6R3). Biomol NMR Assign [DOI] [PMC free article] [PubMed]
- Campa C, Holtan S, Nilsen N, et al. Biochemical analysis of the processive mechanism for epimerization of alginate by mannuronan C-5 epimerase AlgE4. Biochem J. 2004;381:155–164. doi: 10.1042/BJ20031265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertesvåg H, Valla S. The A modules of the Azotobacter vinelandii mannuronan-C-5-epimerase AlgE1 are sufficient for both epimerization and binding of Ca2+ J Bacteriol. 1999;181:3033–3038. doi: 10.1128/jb.181.10.3033-3038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertesvåg H, Doseth B, Larsen B, et al. Cloning and expression of an Azotobacter vinelandii mannuronan C-5-epimerase gene. J Bacteriol. 1994;176:2846–2853. doi: 10.1128/jb.176.10.2846-2853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertesvåg H, Høidal HK, Hals IK, et al. A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol Microbiol. 1995;16:719–731. doi: 10.1111/j.1365-2958.1995.tb02433.x. [DOI] [PubMed] [Google Scholar]
- Ertesvåg H, Høidal HK, Schjerven H, et al. Mannuronan C-5-epimerases and their application for in vitro and in vivo design of new alginates useful in biotechnology. Metab Eng. 1999;1:262–269. doi: 10.1006/mben.1999.0130. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Duun AS, Markussen S, et al. Time-resolved 1H and 13C NMR spectroscopy for detailed analyses of the Azotobacter vinelandii mannuronan C-5 epimerase reaction. Biochim Biophys Acta. 2002;1570:104–112. doi: 10.1016/s0304-4165(02)00195-2. [DOI] [PubMed] [Google Scholar]
- Keller RLJ (2004) Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer Aided Resonance Assignment. CANTINA Verlag, Zürich
- Zhang HY, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR. 2003;25:173–195. doi: 10.1023/A:1022836027055. [DOI] [PubMed] [Google Scholar]