Abstract
The Notch gene family encodes large transmembrane receptors that are components of an evolutionarily conserved intercellular signaling mechanism. To assess the role of the Notch4 gene, we generated Notch4-deficient mice by gene targeting. Embryos homozygous for this mutation developed normally, and homozygous mutant adults were viable and fertile. However, the Notch4 mutation displayed genetic interactions with a targeted mutation of the related Notch1 gene. Embryos homozygous for mutations of both the Notch4 and Notch1 genes often displayed a more severe phenotype than Notch1 homozygous mutant embryos. Both Notch1 mutant and Notch1/Notch4 double mutant embryos displayed severe defects in angiogenic vascular remodeling. Analysis of the expression patterns of genes encoding ligands for Notch family receptors indicated that only the Dll4 gene is expressed in a pattern consistent with that expected for a gene encoding a ligand for the Notch1 and Notch4 receptors in the early embryonic vasculature. These results reveal an essential role for the Notch signaling pathway in regulating embryonic vascular morphogenesis and remodeling, and indicate that whereas the Notch4 gene is not essential during embryonic development, the Notch4 and Notch1 genes have partially overlapping roles during embryogenesis in mice.
Keywords: Notch signaling pathway, angiogenesis, vasculogenesis, ephrins
The Notch signaling pathway is an evolutionarily conserved intercellular signaling mechanism, and mutations in its components disrupt cell fate specification and embryonic development in organisms as diverse as insects, nematodes, and mammals. This signaling pathway was first studied in Drosophila. The Notch gene of Drosophila encodes a large transmembrane receptor that, at the extracellular surface of a cell, interacts with membrane-bound ligands encoded by the Delta and Serrate genes. The signal induced by ligand binding is then transmitted at the intracellular surface in a process involving proteolytic processing of the receptor and nuclear translocation of the intracellular domain of the Notch protein (for recent reviews of the Notch signaling pathway, see Gridley 1997; Weinmaster 1997; Chan and Jan 1998; Greenwald 1998; Artavanis-Tsakonas et al. 1999).
Four Notch genes have been described in mice. The Notch4 gene was originally identified as a common viral integration site (originally termed the int3 locus) in mouse mammary tumor virus (MMTV)-induced mammary tumors (Gallahan and Callahan 1987; Robbins et al. 1992). In these tumors, proviral integration leads to the production of a truncated Notch4 transcript. This truncated transcript initiates within the 3′ MMTV long terminal repeat and includes the region encoding the intracellular domain of the Notch4 protein (Robbins et al. 1992; Gallahan and Callahan 1997), which is a constitutively active form of the Notch4 protein. Further work in transgenic mouse models and in tissue culture cells has demonstrated that unregulated expression of the cytoplasmic domain of the Notch4 protein inhibits mammary development (Smith et al. 1995; Uyttendaele et al. 1998). Cloning of the full-length Notch4 cDNA revealed that it encodes a protein of ∼200 kD with fewer epidermal growth factor (EGF)-like repeats and a shorter intracellular domain than the other mammalian Notch homologs (Uyttendaele et al. 1996; Gallahan and Callahan 1997; Shirayoshi et al. 1997). Somewhat surprisingly, expression analyses indicated that embryonic expression of the Notch4 gene was largely restricted to vascular endothelial cells (Uyttendaele et al. 1996; Shirayoshi et al. 1997).
Formation of the vascular system is one of the earliest and most important events during embryogenesis in mammals. During the early stages of vascular development in both the mammalian embryo and its extraembryonic membranes such as the yolk sac, endothelial cell precursors differentiate and coalesce into a network of homogeneously sized primitive blood vessels (the primary vascular plexus) in a process termed vasculogenesis (Risau and Flamme 1995). This primary vascular plexus is then remodeled by the process of angiogenesis, which involves the sprouting, branching, splitting, and differential growth of vessels in the primary plexus to form both the large and small vessels of the mature vascular system (Risau 1997; Gale and Yancopoulos 1999). A number of different intercellular signaling pathways have been implicated in the control of vasculogenesis and angiogenesis in mammals. These pathways include the vascular endothelial growth factor pathway, the transforming growth factor-β (TGF-β) pathway, the angiopoietin/Tie receptor pathway, and the ephrin/Eph receptor pathway (for reviews, see Folkman and D'Amore 1996; Hanahan 1997; Carmeliet and Collen 1999; Gale and Yancopoulos 1999; Neufeld et al. 1999).
A number of observations indicate that the Notch signaling pathway also plays a critical role in vascular development and homeostasis. In addition to the Notch4 gene, the Notch1 gene is expressed in endothelial cells in the embryonic vasculature (Franco del Amo et al. 1992; Reaume et al. 1992). Analysis of targeted mutations of genes encoding Notch ligands has shown that both Jag1 (Xue et al. 1999) and Dll1 (Hrabé de Angelis et al. 1997) homozygous mutant embryos die from vascular defects and hemorrhaging at approximately gestational day E10.5. Expression of the human JAG1 gene is induced in an in vitro angiogenesis model, and administration of Jag1 antisense oligonucleotides modulates in vitro angiogenesis (Zimrin et al. 1996). In adults, a role for the Notch pathway in vascular homeostasis has been suggested by the finding that the degenerative vascular disease CADASIL is caused by missense mutations in the Notch3 gene (Joutel et al. 1996).
In this study we examined whether the Notch4 gene plays an essential role during embryonic development in mice. Notch4-deficient mice were viable and fertile and exhibited no obvious mutant phenotype. However, the Notch4 null mutation genetically interacted with a mutation we had constructed previously in the Notch1 gene (Swiatek et al. 1994), indicating partial functional redundancy between the Notch1 and Notch4 genes during embryogenesis. Further characterization of the phenotypes of both Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos revealed extensive defects in angiogenic vascular remodeling that affected the embryo, the yolk sac, and the placenta. Characterization of the expression patterns of the genes encoding Notch ligands revealed that only the newly isolated Dll4 gene (Shutter et al. 2000) was expressed in a pattern consistent with that expected for a gene encoding a ligand for the Notch1 and Notch4 receptors in the early embryonic vasculature.
Results
Mice homozygous for a targeted mutation of the Notch4 gene are viable and fertile
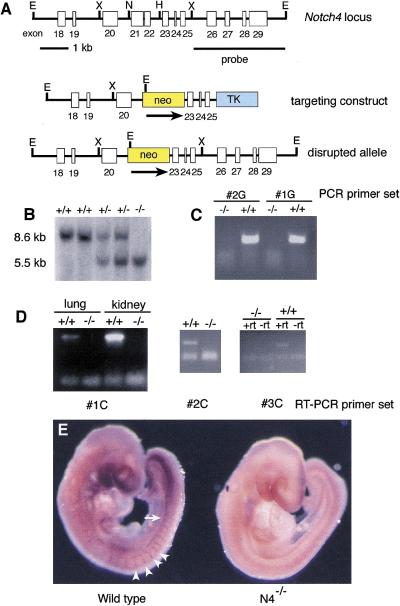
To analyze the role of the Notch4 gene, we created a deletion allele by gene targeting. A Notch4 targeting vector was constructed that deleted a 1.0-kb genomic fragment containing exons 21 and 22 encoding part of the extracellular domain of the protein (Fig. 1A). We refer to this allele as Notch4d1. Germ-line transmission of the Notch4d1 mutant allele was obtained for two targeted clones (Fig. 1B). Mice heterozygous for the Notch4d1 mutation appeared normal and were fertile.
Figure 1.
Targeted disruption of the Notch4 gene. (A) Targeting scheme. (Top) The genomic organization of a portion of the Notch4 gene. Exons are indicated by white boxes. (Middle) The structure of the targeting vector. The deleted exons 21 and 22 encode amino acids 1249–1434 in the extracellular domain of the Notch4 protein. (Bottom) The predicted structure of the Notch4 locus following homologous recombination of the targeting vector. (E) EcoRI; (H) HindIII; (N) NcoI; (X) XbaI. (B) DNA isolated from embryos of the intercross of Notch4+/− heterozygous mice was digested with EcoRI, blotted, and hybridized with the indicated probe. Sizes of hybridizing fragments are indicated. Genotypes of progeny are indicated at the top of the lane. (C) Genomic PCR analysis with primers located in region deleted in the Notch4d1 mutant allele. Genotypes are indicated at top. (D) RT–PCR analysis. RT–PCR primer sets are indicated at the bottom of each panel. Primer set #1C flanks the deleted region; primers sets #2C and #3C are located at the 3′ end of the Notch4 cDNA. Genotypes are indicated at the top of the lane. (+rt) Plus reverse transcriptase; (−rt) without reverse transcriptase. (E) Whole mount in situ hybridization of a wild-type and a Notch4−/− embryo with an antisense Notch4 riboprobe encoding the intracellular domain of the Notch4 protein. Notch4 expression in the wild-type embryo is observed in intersomitic blood vessels (arrowheads) and the dorsal aorta (arrow). No Notch4 expression is observed in the mutant embryo.
To examine whether mice homozygous for the Notch4d1 mutation were viable, heterozygous F1 animals were intercrossed and the genotypes of F2 progeny were determined 2–3 weeks after birth. Mice homozygous for the Notch4d1 mutation (designated Notch4−/−) were recovered at the expected frequency and by morphological and histological analyses did not differ from heterozygous and wild-type littermates. Genomic PCR analysis confirmed that the expected genomic fragment was deleted in the Notch4−/− mutant homozygotes (Fig. 1C). Mammary glands from the Notch4−/− mutant homozygotes were examined in particular detail, because the Notch4 gene was initially identified as the int3 gene, a common integration site for MMTV-induced mammary tumors (Robbins et al. 1992), and unregulated expression of the cytoplasmic domain of the Notch4 protein inhibits mammary development (Smith et al. 1995; Uyttendaele et al. 1998). However, morphological and histological analyses of mammary glands isolated from virgin, pregnant, and lactating females did not reveal any defects in the Notch4−/− homozygotes (data not shown).
We analyzed RNA isolated from Notch4−/− homozygous mutant adult animals for the presence of Notch4 transcripts by RT-PCR with several different primer sets (Fig. 1D), and analyzed Notch4−/− homozygous mutant embryos by whole-mount in situ hybridization with a Notch4 antisense riboprobe encoding the entire intracellular domain of the Notch4 protein (Fig. 1E). No Notch4 transcripts were detected in the homozygotes by either technique. Thus, unlike mouse embryos homozygous for targeted mutations in the Notch1 (Swiatek et al. 1994; Conlon et al. 1995) and Notch2 (Hamada et al. 1999; T. Gridley unpubl.) genes, embryos homozygous for a Notch4 null mutation develop normally. We next examined whether we could detect genetic interactions between the Notch4d1 mutation and a null mutation we had made previously in the Notch1 gene (the Notch1in32 mutation; Swiatek et al. 1994).
Notch1+/− Notch4−/− mutant mice are viable but show growth retardation
To examine possible genetic interactions between the Notch1 and Notch4 targeted mutations, we crossed mice heterozygous for the Notch1in32 mutation (designated Notch1+/−) to mice homozygous for the Notch4d1 mutation (Notch4−/−). Double heterozygous ( Notch1+/− Notch4+/−) animals were crossed with Notch4−/− mice to generate animals with the genotype Notch1+/− Notch4−/−. Mice with this genotype appeared smaller than their Notch1+/+ Notch4−/− littermates. Breeding studies indicated that Notch1+/− Notch4−/− adult mice were fertile. We therefore crossed Notch1+/− Notch4−/− mice with Notch4−/− mice and followed the growth rates of the progeny.
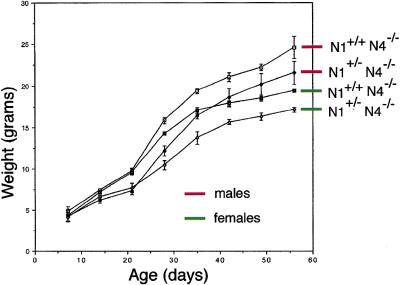
This analysis revealed postnatal growth retardation in the Notch1+/− Notch4−/− double mutant animals. At birth, no clear differences in the weight of the pups could be detected. By weaning age (around 3 weeks), however, the average weight of the Notch1+/− Notch4−/− animals was ∼80% that of their Notch1+/+ Notch4−/− littermates (Fig. 2). Although we have not determined the cause of the growth retardation in the Notch1+/− Notch4−/− mice, this analysis clearly revealed a genetic interaction between the Notch1 and Notch4 mutations.
Figure 2.
Genetic interactions of Notch1 and Notch4 mutations. Growth curve of Notch1+/− Notch4−/− (♦,⋄) vs. Notch1+/+ Notch4−/− (▪, □) littermates. The weights of Notch1+/− Notch4−/− and Notch1+/+ Notch4−/− mutant animals are plotted against age. Males (red) and females (green) are plotted separately. Data presented are from four mice in each group. Error bars indicate the s.d..
Synergistic effects in Notch1−/− Notch4−/− double homozygous mutant embryos
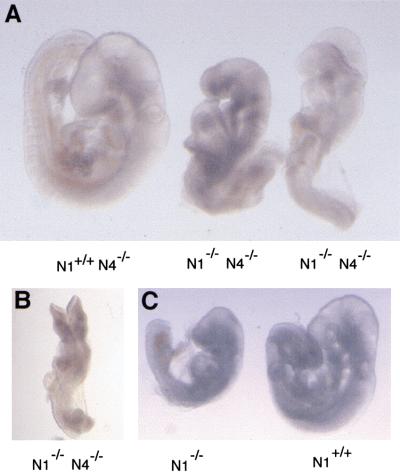
We next analyzed whether a genetic interaction between the Notch1 and Notch4 mutations was detectable in double homozygous mutant embryos. Because Notch1−/− embryos arrest in development at or shortly before E9.5 (Swiatek et al. 1994; Conlon et al. 1995), we isolated embryos at E9.5 from the intercross of Notch1+/− Notch4−/− mice (Fig. 3). Typically, Notch1−/− mutant embryos arrest in development after forming 16–20 somites, completing embryonic turning and completing closure of the anterior neural tube (Fig. 3C) (Swiatek et al. 1994; Conlon et al. 1995). Of Notch1−/− Notch4−/− double mutant embryos ∼50% were more severely affected than a typical Notch1−/− mutant embryo. These severely affected Notch1−/− Notch4−/− double mutant embryos had formed fewer somites, had not completed embryonic turning, and had open neural tubes (Fig. 3A,B). The remaining double mutant embryos appeared phenotypically like Notch1−/− mutant embryos. These data indicated that, despite the fact that the Notch4 gene is not essential for embryonic development, the Notch1 and Notch4 genes play partially redundant roles during embryogenesis.
Figure 3.
Synergistic effects in Notch1−/− Notch4−/− double mutant embryos. (A) Two Notch1−/− Notch4−/− double homozygous mutant embryos and littermate control isolated at E9.5. (B) Severely affected Notch1−/− Notch4−/− double homozygous mutant embryo. The double homozygous mutant embryos in both A and B have not completed embryonic turning and have open neural tubes. (C) A typical Notch1−/− embryo and control littermate at E9.5. The Notch1−/− mutant embryo has completed turning and has a closed neural tube.
Defects in angiogenic vascular remodeling in Notch1−/− and Notch1−/− Notch4−/− mutant embryos
Both the Notch1 (Franco del Amo et al. 1992; Reaume et al. 1992) and Notch4 (Uyttendaele et al. 1996; Shirayoshi et al. 1997; also see Fig. 1E) genes are expressed in vascular endothelial cells, and the Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos exhibit defects (e.g., pericardial edema) suggestive of a failure to maintain vascular homeostasis. We therefore examined vascular development in both the Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos. This analysis revealed substantial defects in angiogenic vascular remodeling in both the Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos.
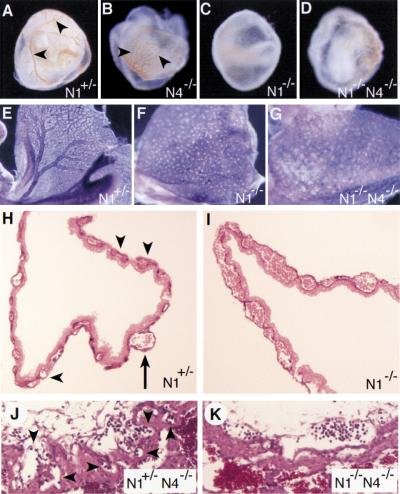
Vascular defects can first be observed in the yolk sacs of the mutant embryos. When viewed in whole mount at E9.5, yolk sacs from wild type (not shown), Notch1+/− (Fig. 4A), and Notch4−/− (Fig. 4B) embryos all exhibited large vitelline blood vessels. All Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos had pale yolk sacs lacking obvious blood vessels (Fig. 4C,D). We then visualized the vascular network of mutant embryos and littermate controls by staining with a monoclonal antibody to platelet endothelial cell adhesion molecule-1 (PECAM-1), a specific marker for vascular endothelial cells (Baldwin et al. 1994). In the yolk sacs of both Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos, the primary vascular plexus appeared to form normally, indicating that there were no apparent defects in vasculogenesis in the mutants. However, the mutant embryos failed to remodel the primary vascular plexus to form the large and small blood vessels of the mature yolk sac (Fig. 4E–G). Histological analysis of PECAM-1-stained yolk sacs demonstrated that both small capillaries and large vitelline collecting vessels differentiated in control yolk sacs (Fig. 4H), whereas both Notch1−/− (Fig. 4I) and Notch1−/− Notch4−/− (not shown) yolk sacs exhibited a disorganized, confluent vascular plexus.
Figure 4.
Yolk sac and placental defects in mutant embryos. (A–D) Morphology of embryos in their yolk sacs at E9.5. Large vitelline blood vessels (arrowheads) are observed in the Notch1+/− and Notch4−/− yolk sacs, but not in the Notch1−/− and Notch1−/− Notch4−/− mutants. (E–G) PECAM-1-stained yolk sacs. The yolk sacs of the Notch1−/− and Notch1−/− Notch4−/− mutant embryos are at the primitive vascular plexus stage and have not undergone vascular remodeling to form the large and small blood vessels of the mature yolk sac. (H,I) Histological sections of PECAM-1-stained yolk sacs. The Notch1+/− yolk sac (H) has differentiated both small capillaries (arrowheads) and large vitelline collecting vessels (arrow). The Notch1−/− yolk sac (I) exhibits a disorganized, confluent vascular plexus. (J,K) Histological sections of placentas at E9.5. In the Notch1+/− Notch4−/− control embryo (J), embryonic blood vessels containing nucleated erythrocytes (arrowheads) have invaded the labyrinthine layer of the placenta. In the Notch1−/− Notch4−/− mutant embryo (K), embryonic blood vessels are present at the edge of the placenta but have not invaded the labyrinthine layer.
Vascular defects were also observed in the labyrinthine layer of the placentas of the mutant embryos (Fig. 4J,K; data not shown). In both Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos at E9.5, the allantois of the mutant embryos fused with the chorionic plate, which is an initiating step in the formation of a functional chorioallantoic placenta. However, in the control embryos (e.g., the Notch1+/− Notch4−/− embryo shown in Fig. 4J, which is phenotypically wild type), blood vessels containing embryonic nucleated erythrocytes could be observed invading and interdigitating into the labyrinthine trophoblast layer of the developing placenta. In Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos, embryonic blood vessels containing nucleated erythrocytes remained at the periphery of the trophoblast layer and did not invade the labyrinthine layer (Fig. 4K). The failure of mutant endothelial cells to efficiently invade the labyrinthine region provides further evidence of defects in angiogenesis in the Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos.
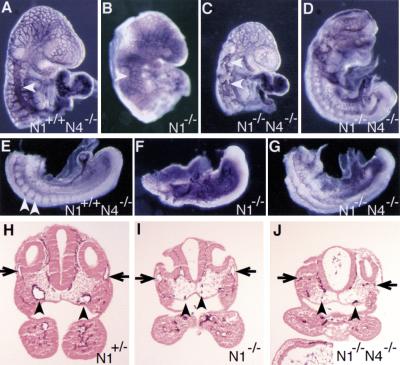
In addition to the vascular defects observed in the yolk sac and placenta, extensive defects in vascular morphogenesis were observed in the mutant embryos themselves (Fig. 5). A prominent feature in the Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos was malformation of the large blood vessels in the anterior of the embryo. The primary vessels for arterial flow to the head of the mammalian embryo are the dorsal aortae, and the primary vessels for venous return are the anterior cardinal veins. In the Notch4−/− (phenotypically wild type) PECAM-1-stained embryo shown in Figure 5A, the anterior cardinal vein (arrowhead) was easily visualized as a large vessel running in an anterior–posterior direction just beneath the surface ectoderm of the embryo. In the Notch1−/− embryo shown in Figure 5B, the anterior cardinal vein (arrowhead) had formed, but was smaller than in the control. Two Notch1−/− Notch4−/− double mutant embryos at E9.5 are shown in Figure 5, C and D. In the less severely affected double mutant embryo, discontinuous remnants of the anterior cardinal vein were observed (arrowheads in Fig. 5C). In the more severely affected Notch1−/− Notch4−/− double mutant embryo, the anterior cardinal vein appeared to be entirely missing (Fig. 5D). In the trunk of wild-type embryos at E9.5, intersomitic blood vessels were apparent along the boundaries between adjacent somites (Fig. 5E). On the dorsal side of the embryo, the intersomitic vessels branch to form a highly anastomosed capillary network. In both Notch1−/− mutant (Fig. 5F) and Notch1−/− Notch4−/− double mutant (Fig. 5G) embryos, intersomitic vessels were severely disorganized.
Figure 5.
Defects in vascular remodeling in Notch1−/− and Notch1−/− Notch4−/− mutant embryos. (A–G) PECAM-1-stained whole mount embryos. (A–D) Defective morphogenesis of the main trunk of the anterior cardinal vein (arrowhead) in Notch1−/− (B) and Notch1−/− Notch4−/− mutant embryos (C,D). The double mutant embryo in D is more severely affected than the embryo in C. (E–G) In the Notch1+/− Notch4−/− control embryo (E), intersomitic vessels (arrowheads) differentiate, whereas these vessels are not observed in the Notch1−/− (F) and Notch1−/− Notch4−/− (G) mutant embryos. (H–J) Histological sections of PECAM-1-stained embryos at the level of the otic vesicle. In the N1+/− control embryo (H), both the dorsal aortae (arrowheads) and the anterior cardinal veins (arrows) have open lumens and normal morphology. In the Notch1−/− Notch4−/− mutant embryo (J), endothelial cells have differentiated but both the dorsal aortae and the anterior cardinal veins have an abnormal, collapsed morphology. In the less-severely affected Notch1−/− mutant embryo (I), the anterior cardinal veins still have an open lumen but the dorsal aortae are collapsed.
Histological analyses of PECAM-1-stained embryos confirmed that dorsal aortae and anterior cardinal veins were frequently atretic in the mutant embryos (Fig. 5H–J). In the section of the Notch1+/− (phenotypically wild type) embryo shown in Figure 5H, both the dorsal aortae and the anterior cardinal veins had open lumens that were lined with a continuous layer of PECAM-1-positive endothelial cells. In the Notch1−/− embryo shown in Figure 5I, the anterior cardinal veins appeared relatively normal, but both dorsal aortae had a collapsed appearance. In the more severely affected Notch1−/− Notch4−/− double mutant embryo (Fig. 5J), endothelial cells were present at the correct positions of the anterior cardinal veins and dorsal aortae, but no vessel lumens were apparent.
The Dll4 gene encodes the probable ligand for the Notch1 and Notch4 receptors in the early embryonic vasculature
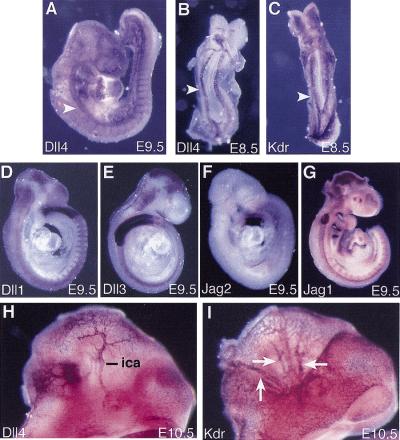
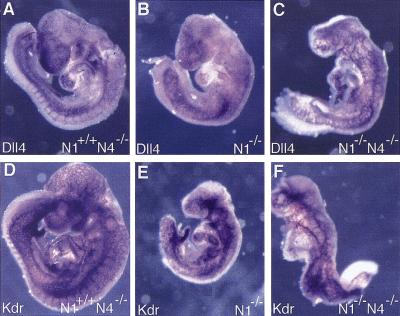
Previous work has shown that both Jag1 and Dll1 homozygous mutant embryos die from vascular defects and hemorrhaging (Xue et al. 1999; Hrabé de Angelis et al. 1997). However, in both these cases, embryonic lethality does not occur until E10.5, at least a day later than in Notch1−/− and Notch1−/− Notch4−/− mutant embryos. We therefore examined the RNA expression pattern of all the genes encoding Notch ligands at E8.5 and E9.5 (Fig. 6; data not shown). Only the Dll4 gene (Shutter et al. 2000) was expressed in a spatial and temporal pattern that suggested it could encode the ligand for the Notch1 and Notch4 receptors during early vascular development. At E8.5, Dll4 expression was observed in the dorsal aortae and the endocardium (Fig. 6B). Comparision of Dll4 expression with Kdr (formerly Flk1) expression (Fig. 6C), which is expressed in all endothelial cells and their precursors (Yamaguchi et al. 1993), indicated that Dll4 was expressed in only a subset of endothelial cells. Analysis of Dll4 expression at E9.5 and E10.5 (Fig. 6A,H) demonstrated that Dll4 was preferentially expressed in endothelial cells of the arterial system. In contrast, none of the other genes encoding Notch ligands exhibited obvious expression in the vascular system at E8.5 or E9.5 (Fig. 6D–G; data not shown), indicating that these ligands most likely are not involved in activation of the Notch1 and Notch4 receptors at these stages. We also examined Dll4 and Kdr expression in Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos. Dll4 expression was not down-regulated in the mutants, but the expression patterns of both Dll4 and Kdr revealed disruptions in vascular morphogenesis similar to those revealed by PECAM-1 staining of mutant embryos (Fig. 7).
Figure 6.
Expression pattern of the Dll4 gene suggests that it encodes the ligand for the Notch1 and Notch4 receptors in the embryonic vasculature. Whole-mount in situ hybridization with the indicated probes of embryos isolated at E8.5 (B,C), E9.5 (A,D–G), and E10.5 (H,I). Dll4 expression is observed in the dorsal aorta (arrowheads) and, at E9.5, the intersomitic vessels (A). The other genes encoding Notch ligands do not exhibit obvious expression in the vasculature at E9.5 (D–G). At E10.5, Dll4 expression is observed in the internal carotid artery (ica) (H), but is not observed in branches of the primary head vein (arrows in I), which express Kdr.
Figure 7.
Dll4 and Kdr expression in mutant embryos. Embryos of the indicated genotypes were hybridized with riboprobes for Dll4 (A–C) and Kdr (D–F). Note the more severe dysmorphogenesis of the Notch1−/− Notch4−/− double mutant embryos (C,F) compared with the Notch1−/− single mutant embryos (B,E). Dll4 expression was not downregulated in either the Notch1−/− or Notch1−/− Notch4−/− mutant embryos. However, the expression patterns of both Dll4 and Kdr reveal disruptions in vascular morphogenesis in the mutant embryos.
Discussion
The Notch4 gene is not essential, but the Notch4 mutation genetically interacts with a Notch1 mutation
To determine the biological role of the Notch4 gene in mice, we constructed and analyzed animals homozygous for a deletion that removed exons 21 and 22, which encode 186 amino acids of the extracellular domain of the Notch4 protein. The two closest remaining exons, exon 20 and exon 23, are out of frame with respect to each other. Therefore, a partially functional Notch4 protein that contains the intracellular (i.e., signal-transducing) domain of the Notch4 protein could not be produced by alternative splicing from exon 20 to exon 23. We also used RT-PCR and in situ hybridization analyses to assay for alternatively spliced Notch4 transcripts that would contain the region of the Notch4 cDNA encoding the intracellular domain of the Notch4 protein. We found no evidence of such transcripts in the Notch4−/− homozygous mutant mice. These results suggest that the Notch4d1 allele causes a null mutation in the Notch4 gene. Our results reveal that the Notch4 gene is dispensable for apparently normal embryonic growth in mice, and that Notch4−/− adults are viable and fertile. In contrast, both the Notch1 (Swiatek et al. 1994; Conlon et al. 1995) and Notch2 (Hamada et al. 1999; T. Gridley, unpubl.) genes are essential for embryonic development. It should be noted that all Notch ligands for which either spontaneous or targeted mutations have been isolated are also essential for proper embryonic development in mice. This includes mutations in the Jag1 (Xue et al. 1999), Jag2 (Sidow et al. 1997; Jiang et al. 1998), Dll1 (Hrabé de Angelis et al. 1997), and Dll3 (Kusumi et al. 1998) genes.
The Notch4 gene is primarily expressed in vascular endothelial cells during early embryogenesis in mice (Uyttendaele et al. 1996; Shirayoshi et al. 1997). Because the Notch1 gene is expressed in endothelial cells at the same stages as the Notch4 gene (Franco del Amo et al. 1992; Reaume et al. 1992), we generated Notch1/Notch4 double mutant mice and embryos to assess whether there were genetic interactions between these two mutations. These experiments revealed genetic interactions between the Notch1 and Notch4 mutations at two different stages. Adult Notch1+/− Notch4−/− double mutant mice were viable and fertile, but exhibited postnatal growth deficiency. Although Notch1+/− Notch4−/− double mutant mice were the same size as their Notch1+/+ Notch4−/− littermates at birth, their postnatal growth deficiency quickly became apparent. This was clearly seen at weaning age, when the weights of Notch1+/− Notch4−/− double mutant mice averaged ∼80% that of their littermates.
We also observed genetic interactions between the Notch4 and Notch1 mutations during embryonic development. Of Notch1−/− Notch4−/− double homozygous mutant embryos ∼50% had a more severe phenotype than Notch1−/− mutant embryos. These severely affected Notch1−/− Notch4−/− double mutant embryos formed fewer somites than Notch1−/− mutant embryos and did not complete embryonic turning or neural tube closure. Vascular defects were frequently more severe in the Notch1−/− Notch4−/− double mutant embryos than in Notch1−/− single mutant embryos (e.g., see Fig. 5). We hypothesize that the more severe phenotype of the Notch1−/− Notch4−/− double mutant embryos is due to a more severely compromised vascular system than in Notch1−/− mutant embryos, thereby leading to more severe growth retardation in the double mutant embryos. This interaction is not completely penetrant, as only ∼50% of the Notch1−/− Notch4−/− double homozygous mutant embryos exhibit the more severe phenotype. We do not know the cause of this incomplete penetrance. However, it may be due to the segregation of modifier loci in the mixed C57BL/6J × 129/SvImJ genetic background on which the mutant alleles are maintained.
Notch signaling is required for angiogenic vascular remodeling
Targeted mutations in components of a variety of signaling pathways (e.g., vascular endothelial growth factors, TGF-β1, angiopoietins, ephrins) have been shown to regulate vascular morphogenesis in mice (for review, see Folkman and D'Amore 1996; Hanahan 1997; Carmeliet and Collen 1999; Gale and Yancopoulos 1999; Neufeld et al. 1999). Our results demonstrate that another major intercellular signaling pathway, the Notch pathway, also regulates vascular morphogenesis and angiogenic vascular remodeling. We observed vascular defects in the placenta, yolk sac, and embryo proper of both Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos. In the yolk sac of the mutant embryos, the primary vascular plexus formed normally, indicating that there were no apparent defects in vasculogenesis in the mutants. However, both Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos failed to remodel the primary vascular plexus to form the large vitelline blood vessels, a process that occurs by angiogenesis. Defects in angiogenesis were also apparent in the placenta, where embryonic blood vessels failed to invade the placental labyrinth. In the embryo proper, defects in angiogenesis and vascular remodeling were apparent as malformations of major vessels such as the dorsal aortae, the anterior cardinal veins, and the intersomitic blood vessels.
Our expression analysis of the genes encoding ligands for Notch-family receptors indicated that only the newly cloned Dll4 gene (Shutter et al. 2000) was expressed in the embryonic vasculature early enough to encode the ligand responsible for activation of the Notch1 and Notch4 receptors. The preferential expression of Dll4 in endothelial cells of the arterial system is strongly reminiscent of the differential expression of ephrin ligands and their Eph receptors in the arterial and venous systems (Wang et al. 1998; Adams et al. 1999; Gerety et al. 1999; for review, see Gale and Yancopoulos 1999). During early vascular development, the ephrinB2 gene is expressed in arterial endothelium, whereas its receptor EphB4 is expressed in venous endothelium. Embryos homozygous for targeted mutations of ephrinB2 (Wang et al. 1998; Adams et al. 1999) and EphB4 (Gerety et al. 1999) exhibit defects in angiogenesis and vascular remodeling that are essentially identical to the vascular defects we observe in the Notch1−/− mutant and Notch1−/− Notch4−/− double mutant embryos. This suggests the intriguing possibility that the Notch pathway and the ephrin/Eph pathway functionally interact during the intercellular signaling events that must be involved in remodeling the embryonic vasculature. Further work, including genetic epistasis experiments with various Notch pathway and ephrin/Eph pathway mutants, will be required to distinguish whether these two contact-mediated intercellular signaling pathways interact, or whether they function independently, during vascular development and remodeling.
A role for the Notch pathway in vascular homeostasis in adult humans has been indicated by the finding that the degenerative vascular disease CADASIL is caused by missense mutations in the Notch3 gene (Joutel et al. 1996). Our studies reveal a previously unappreciated role for the Notch signaling pathway in regulating angiogenic vascular remodeling during embryonic development in mice. Taken together, these results demonstrate that the Notch signaling pathway is essential both for vascular development in embryos and maintenance of vascular homeostasis in adults.
Partial functional redundancy between the Notch1 and Notch4 genes
Notch genes form a multigene family in vertebrates, and our results provide the first evidence for functional redundancy in the Notch signaling pathway in mammals. Previous experiments in Caenorhabditis elegans had revealed functional redundancy in the Notch signaling pathway. Although only a single Notch gene is known in Drosophila, C. elegans contains two genes encoding Notch-family receptors, lin-12 and glp-1. Lambie and Kimble (1991) showed that the lin-12 and glp-1 genes were functionally redundant, as double homozygous mutant embryos exhibit a novel, and more severe, synergistic phenotype. The redundancy of glp-1 and lin-12 gene action was further supported by the finding that expression of glp-1 coding sequences under control of lin-12 regulatory sequences can rescue the lin-12 null phenotype (Fitzgerald et al. 1993), and by the ability of a particular neomorphic glp-1 allele to mimic dominant gain of function lin-12 mutations (Mango et al. 1991). These results suggested that the Glp-1 and Lin-12 proteins are biochemically interchangeable, and that their distinct roles during development are due largely to differences in gene expression. Our results extend these findings to mammals and demonstrate that although the Notch4 gene plays no essential role in laboratory mice, the Notch1 and Notch4 genes have partially overlapping roles during embryogenesis.
Materials and methods
Targeting vector construction
A Notch4 genomic clone (Gallahan and Callahan 1997) isolated from a P1 library was used for construction of the targeting vector. The sequence of the mouse Notch4 gene is available in GenBank (accession no. AF030001). For construction of the targeting vector, 3.3-kb EcoRI–NcoI fragment of the Notch4 gene was subcloned upstream of a PGK-neo expression cassette (Soriano et al. 1991), and a 1.1-kb HpaI–XbaI Notch4 fragment was subcloned downstream of the PGK-neo cassette. This resulted in the deletion of a 1.0-kb Notch4 genomic fragment containing exons 21 and 22 of the Notch4 gene. We refer to this allele as Notch4d1. The official nomenclature for the alleles used in these studies are Notch4d1: Notch4tm1Grid; Notch1in32 (Swiatek et al. 1994): Notch1tm1Grid. A HSV-tk cassette was introduced to allow negative selection against random integration of the targeting vector (Mansour et al. 1988).
Electroporation, selection, and screening of ES cells and mouse genotyping
CJ7 ES cells were electroporated with 25 μg of linearized targeting vector, selected, screened, and injected into blastocysts from C57BL/6J mice as described previously (Swiatek and Gridley 1993). For Southern blot analysis, a 2.4-kb XbaI–EcoRI fragment was used as probe. Germ-line transmission was obtained for two independently targeted ES cell clones. Animals were genotyped by PCR or by Southern blot analysis. PCR primers for the wild-type Notch4 allele were (5′-CCTATGACCAGTACTGCCGAG-3′) and (5′-GAGGAATCCCTAGCTCCACTC-3′), yielding an amplification product of 312 bp; the primers for the Notch4d1 allele were (5′-GAGGAGTGTCTCTTTGATGGC-3′) and (5′-AAGCGCATGCTCCAGACTGCC-3′), yielding an amplification product of ∼500 bp.
For confirmation that the expected fragment of the Notch4 gene was deleted in the Notch4d1 homozygous mutant mice, genomic DNA was isolated from Notch4d1 adult homozygotes and wild-type controls and analyzed with PCR primers from the deleted region. Two primer sets were used: #1G forward (5′-CTTTCCCTTCCCACAGGTTTG-3′) and #1G reverse (5′-TTGGTTCTATCTCTCCCAGCGAGG-3′), which amplify a 257-bp product; #2G forward (5′-AGGTTTGTGGTAGTGATGGGAGTG-3′) and #2G reverse (5′-ACTGAATAGATGGGACAGGGGG-3′), which amplify a 322-bp product.
RT–PCR analysis
Total RNA was isolated from kidney and lung tissues of adult homozygous Notch4d1 mutants and wild-type littermates. For the RT–PCR analysis, cDNA was synthesized with oligo(dT) primer, and Notch4 cDNAs were detected by PCR. Two RT–PCR primer sets were used. One primer set was chosen to flank the region deleted in the Notch4d1 mutant allele: #1C forward (5′-ACCCTGCTCCAATGGAGGAT-3′, encoding amino acids 1049–1055) and #1C reverse (5′-AGAACCTCCGATTCACACTCC-3′, encoding amino acids 1564–1570), generating an amplification product of 1.3 kb (Smith et al. 1995).
Additional primer sets were chosen to amplify the 3′ end of any potential alternatively spliced transcripts from the Notch4d1 mutant allele: #2C forward (5′-TGGAGCGGATAAAGATGCCC-3′, encoding amino acids 1752–1758) and #2C reverse (5′-AGCGTTAGCAGGTCCCAGTGAC-3′), encoding amino acids 1806–1813), generating an amplification product of 185 base pairs; #3C forward (5′-CCAAGAGATTCCCTTAAACTCGG-3′, encoding amino acids 1952–1958) and #3C reverse (5′-CCAGAGTTTAGGGATTCTCG-3′, located in the 3′ untranslated region), generating an amplification product of 260 bp. Because both primer #3C forward and #3C reverse are located in exon 29, the large 3′ terminal exon of the Notch4 gene (Gallahan and Callahan 1997), total RNA was treated with ribonuclease-free DNase prior to synthesis of cDNA with oligo(dT) primer.
Histology, in situ hybridization, and immunohistochemistry
Organs from adult mice were fixed by immersion in Fekete's acid–alcohol–formalin and were processed as described (Relyea et al. 1999). Embryos were dissected and DNA was prepared from the yolk sacs or tails for genotyping by PCR or by Southern blot analysis. Embryos for whole-mount in situ hybridization were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) overnight at 4°C. Embryos for immunohistochemistry were fixed in 4% paraformaldehyde in PBS and stained with a monoclonal anti-PECAM-1 antibody (clone MEC 13.3; Pharmingen).
Acknowledgments
We thank J. Barker, T. O'Brien, B. McCright, and E. Carver for comments on the manuscript. This work was supported by grants to T.G. from the NIH (NS36437) and the March of Dimes Birth Defects Foundation (FY99-290), and to J.K. from the NIH (HL62454). L.T.K. was supported by a NRSA postdoctoral fellowship from NHLBI and a Training Grant from NICHD to The Jackson Laboratory. This work was also supported by a grant (CA34196) from the National Cancer Institute to The Jackson Laboratory.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gridley@jax.org; FAX 207-288-6077.
References
- Adams RH, Wilkinson GA, Weiss C, Diella F, Gale NW, Deutsch U, Risau W, Klein R. Roles of ephrinB ligands and EphB receptors in cardiovascular development: Demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes & Dev. 1999;13:295–306. doi: 10.1101/gad.13.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Baldwin HS, Shen HM, Yan HC, De Lisser HM, Chung A, Mickanin C, Trask T, Kirschbaum NE, Newman PJ, Albeda SM, Buck CA. Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31): Alternatively spliced, functionally distinct isoforms expressed during mammalian cardiovascular development. Development. 1994;120:2539–2553. doi: 10.1242/dev.120.9.2539. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Collen D. Role of vascular endothelial growth factor and vascular endothelial growth factor receptors in vascular development. Curr Top Microbiol Immunol. 1999;237:133–158. doi: 10.1007/978-3-642-59953-8_7. [DOI] [PubMed] [Google Scholar]
- Chan Y-M, Jan YN. Roles for proteolysis and trafficking in Notch maturation and signal transduction. Cell. 1998;94:423–426. doi: 10.1016/s0092-8674(00)81583-4. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K, Wilkinson HA, Greenwald I. glp-1 can substitute for lin-12 in specifying cell fate decisions in Caenorhabditis elegans. Development. 1993;119:1019–1027. doi: 10.1242/dev.119.4.1019. [DOI] [PubMed] [Google Scholar]
- Folkman J, D'Amore PA. Blood vessel formation: What is its molecular basis? Cell. 1996;87:1153–1155. doi: 10.1016/s0092-8674(00)81810-3. [DOI] [PubMed] [Google Scholar]
- Franco del Amo F, Smith DE, Swiatek PJ, Gendron-Maguire M, Greenspan RJ, McMahon AP, Gridley T. Expression of Motch, a mouse homolog of Drosophila Notch, suggests an important role in early postimplantation mouse development. Development. 1992;115:737–745. doi: 10.1242/dev.115.3.737. [DOI] [PubMed] [Google Scholar]
- Gale NW, Yancopoulos GD. Growth factors acting via endothelial cell-specific receptor tyrosine kinases: VEGFs, angiopoietins, and ephrins in vascular development. Genes & Dev. 1999;13:1055–1066. doi: 10.1101/gad.13.9.1055. [DOI] [PubMed] [Google Scholar]
- Gallahan D, Callahan R. Mammary tumorigenesis in feral mice: Identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987;61:66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4) Oncogene. 1997;14:1883–1890. doi: 10.1038/sj.onc.1201035. [DOI] [PubMed] [Google Scholar]
- Gerety SS, Wang HU, Chen ZF, Anderson DJ. Symmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol Cell. 1999;4:403–414. doi: 10.1016/s1097-2765(00)80342-1. [DOI] [PubMed] [Google Scholar]
- Greenwald I. LIN-12/Notch signaling: Lessons from worms and flies. Genes & Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling in vertebrate development and disease. Mol Cell Neurosci. 1997;9:103–108. doi: 10.1006/mcne.1997.0610. [DOI] [PubMed] [Google Scholar]
- Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman JR, Tsujimoto Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 1999;126:3415–3424. doi: 10.1242/dev.126.15.3415. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Signaling vascular morphogenesis and maintenance. Science. 1997;277:48–50. doi: 10.1126/science.277.5322.48. [DOI] [PubMed] [Google Scholar]
- Hrabé de Angelis M, McIntyre J, II, Gossler A. Maintenance of somite borders in mice requires the Delta homologue Dll1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T. Defects in limb, craniofacial and thymic development in Jagged2 mutant mice. Genes & Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cécillion M, Maréchal E, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- Kusumi K, Sun E, Kerrebrock AW, Bronson RT, Chi D-C, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES. The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat Genet. 1998;19:274–278. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- Lambie EJ, Kimble J. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development. 1991;112:231–240. doi: 10.1242/dev.112.1.231. [DOI] [PubMed] [Google Scholar]
- Mango SE, Maine EM, Kimble J. Carboxy-terminal truncation activates glp-1 protein to specify vulval fates in Caenorhabditis elegans. Nature. 1991;352:811–815. doi: 10.1038/352811a0. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: A general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 1999;13:9–22. [PubMed] [Google Scholar]
- Reaume AG, Conlon RA, Zirngibl R, Yamaguchi TP, Rossant J. Expression analysis of a Notch homologue in the mouse embryo. Dev Biol. 1992;154:377–387. doi: 10.1016/0012-1606(92)90076-s. [DOI] [PubMed] [Google Scholar]
- Relyea MJ, Miller J, Boggess D, Sundberg JP. Necropsy methods for laboratory mice. Biological characterization of a new mutation. In: Sundberg JP, Boggess D, editors. Systematic approach to evaluation of mouse mutations. Boca Raton, FL: CRC Press, Inc.; 1999. pp. 57–89. [Google Scholar]
- Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- Risau W, Flamme I. Vasculogenesis. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- Robbins J, Blondel BJ, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: A member of the notch gene family transforms mammary epithelial cells. J Virol. 1992;66:2594–2599. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayoshi Y, Yuasa Y, Suzuki T, Sugaya K, Kawase E, Ikemura T, Nakatsuji N. Proto-oncogene of int-3, a mouse Notch homologue, is expressed in endothelial cells during early embryogenesis. Genes & Cells. 1997;2:213–224. doi: 10.1046/j.1365-2443.1997.d01-310.x. [DOI] [PubMed] [Google Scholar]
- Shutter, J.R., S. Scully, G. Deblandre, C.R. Kintner, and K.L. Stark. 2000. Delta4, a novel Notch ligand expressed in arterial endothelium. Genes & Dev. (this issue). [PMC free article] [PubMed]
- Sidow A, Bulotsky MS, Kerrebrock AW, Bronson RT, Daly MJ, Reeve MP, Hawkins TL, Birren BW, Jaenisch R, Lander ES. Serrate2 is disrupted in the mouse limb-development mutant syndactylism. Nature. 1997;389:722–725. doi: 10.1038/39587. [DOI] [PubMed] [Google Scholar]
- Smith GH, Gallahan D, Diella F, Jhappan C, Merlino G, Callahan R. Constitutive expression of a truncated INT3 gene in mouse mammary epithelium impairs differentiation and functional development. Cell Growth Differ. 1995;6:563–577. [PubMed] [Google Scholar]
- Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- Swiatek P, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes & Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- Swiatek PJ, Lindsell CE, Franco del Amo F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes & Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- Uyttendaele H, Soriano JV, Montesano R, Kitajewski J. Notch4 and Wnt-1 proteins function to regulate branching morphogenesis of mammary epithelial cells in an opposing fashion. Dev Biol. 1998;196:204–217. doi: 10.1006/dbio.1998.8863. [DOI] [PubMed] [Google Scholar]
- Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- Weinmaster G. The ins and outs of Notch signaling. Mol Cell Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Dumont DJ, Conlon RA, Breitman ML, Rossant J. flk-1, an flt-related receptor tyrosine kinase is an early marker for endothelial cell precursors. Development. 1993;118:489–498. doi: 10.1242/dev.118.2.489. [DOI] [PubMed] [Google Scholar]
- Zimrin AB, Pepper MS, McMahon GA, Nguyen F, Montesano R, Maciag T. An antisense oligonucleotide to the notch ligand jagged enhances fibroblast growth factor-induced angiogenesis in vitro. J Biol Chem. 1996;271:32499–32502. doi: 10.1074/jbc.271.51.32499. [DOI] [PubMed] [Google Scholar]