Summary
The mechanisms by which ubiquitin ligases are regulated remain poorly understood. Here we describe a series of molecular events that coordinately regulate CHIP, a neuroprotective E3 implicated in protein quality control. Through their opposing activities, the initiator E2, Ube2w, and the specialized deubiquitinating enzyme (DUB), ataxin-3, participate in initiating, regulating and terminating the CHIP ubiquitination cycle. Monoubiquitination of CHIP by Ube2w stabilizes the interaction between CHIP and ataxin-3, which through its DUB activity limits the length of chains attached to CHIP substrates. Upon completion of substrate ubiquitination ataxin-3 deubiquitinates CHIP, effectively terminating the reaction. Our results suggest that functional pairing of E3s with ataxin-3 or similar DUBs represents an important point of regulation in ubiquitin-dependent protein quality control. In addition, the results shed light on disease pathogenesis in SCA3, a neurodegenerative disorder caused by polyglutamine expansion in ataxin-3.
Introduction
The efficient clearance of misfolded proteins is essential to ensure cellular homeostasis and counter the accumulation of toxic protein species in numerous human diseases. The chaperone and ubiquitin-proteasome systems (UPS) together maintain the balance between protein folding and protein degradation. A key component bridging these systems is the ubiquitin ligase (E3), C-terminus of Hsc70 interacting protein (CHIP), which interacts with chaperones to promote the degradation of misfolded proteins (Connell et al., 2001). CHIP binds chaperones through its tetratricopeptide repeat (TPR) domain and recruits ubiquitin-conjugating enzymes (E2s) through its U-box domain. Through these dual activities CHIP mediates the ubiquitination and degradation of chaperone client proteins, including many proteins implicated in neurodegenerative diseases (Dickey et al., 2007).
Much remains unknown about how the activity of CHIP and other E3s is regulated in cells. As with other E3s, CHIP activity is likely postranslationally regulated (Gao et al., 2004; Petroski and Deshaies, 2005). CHIP has been observed to be ubiquitinated in cells and in vitro (Imai et al., 2002; Jiang et al., 2001; Lees et al., 2003). Ubiquitination of E3s can modulate activity through proteolytic and non-proteolytic mechanisms (de Bie and Ciechanover, 2011), and ubiquitination of CHIP has been proposed to facilitate targeting of CHIP substrates to the proteasome for degradation (McDonough and Patterson, 2003). Consistent with this view, ubiquitination of CHIP does not promote its turnover (Jiang et al., 2001) and increases under conditions where CHIP levels and activity are increased (Sisoula et al., 2011). The mechanism by which ubiquitination of CHIP regulates its activity, however, remains unknown.
One potential regulatory role for ubiquitination of CHIP is the recruitment of CHIP cofactors (de Bie and Ciechanover, 2011; Fallon et al., 2006). E3 cofactors likely participate in restricting the type and length of ubiquitin chains produced. One such cofactor for CHIP is the Ubiquitin Interacting Motif (UIM)-containing proteasomal subunit S5a, which stimulates turnover of CHIP substrates by preventing the formation of forked linkage ubiquitin chains (Kim et al., 2009). CHIP also interacts with a second UIM-containing protein, ataxin-3 (Jana et al., 2005), but the function of this interaction is unknown. UIM proteins interact with other E3s including parkin and BRCA1 (Durcan et al., 2010; Fallon et al., 2006; Yan et al., 2007), and at least in the case of parkin this interaction is important for proper handling of E3 substrates (Fallon et al., 2006), suggesting that UIM proteins serve as key regulatory components in some E3 complexes.
The molecular events regulating ubiquitin chain formation and eventual termination also remain largely unknown. While ubiquitin chains added by E3’s to substrates must reach a critical length to ensure efficient targeting to the proteasome (Thrower et al., 2000), unchecked ubiquitin conjugation generating excessively long chains would be energetically costly and could deplete cellular ubiquitin stores (Hanna et al., 2006). The activity of the AAA ATPase, CDC48, provides one established mechanism by which excessive ubiquitin chain elongation is prevented (Richly et al., 2005), but it is reasonable to suspect that other mechanisms exist as well.
A potentially powerful, alternative mechanism to regulate ubiquitin chain production is the coordinated action of specific DUBs with E3s. Certain DUBs are known to regulate the levels of specific E3 substrates and E3s (Ventii and Wilkinson, 2008). To date, however, no functional CHIP/DUB pair has been described. CHIP interacts with the DUB ataxin-3 (Jana et al., 2005), and both CHIP and ataxin-3 are required for proper handling of CFTRΔF508 (Burnett and Pittman, 2005; Sha et al., 2009). Thus, ataxin-3 is a strong candidate to function with CHIP in promoting substrate turnover.
Ataxin-3 is an unusual DUB whose properties favor functional interactions with E3’s. Ataxin-3 contains an amino-terminal protease domain followed by three UIMs that bind longer ubiquitin chains. Ataxin-3 also trims longer chains, displaying little activity against chains of four or fewer ubiquitins (Burnett et al., 2003; Winborn et al., 2008). Like CHIP, ataxin-3 suppresses polyglutamine toxicity and does so in a manner linked to its ubiquitin-associated activities (Warrick et al., 2005; Williams and Paulson, 2008). Ataxin-3 also promotes the flux of substrates through degradation pathways (Kuhlbrodt et al., 2011; Zhong and Pittman, 2006), precisely the opposite action of other characterized DUBs that instead function to deubiquitinate substrates and rescue them from proteasomal delivery (Ventii and Wilkinson, 2008). Collectively these properties raise the possibility that ataxin-3 collaborates with CHIP, and perhaps other E3s (Durcan et al., 2010), to promote rather than inhibit clearance of ubiquitinated substrates.
In addition to its ubiquitin related functions, ataxin-3 also contains a polyglutamine domain that when expanded causes the neurodegenerative disease, Spinocerebellar Ataxia type 3 (SCA3). In the nine known polyglutamine disorders, polyglutamine expansion promotes aggregation and alters protein interactions normally engaged in by the disease proteins (Duvick et al.; Lam et al., 2006; Nedelsky et al., 2010). Therefore, identifying the protein complexes in which polyglutamine proteins normally function is critical to understanding disease pathogenesis. Polyglutamine expansion in ataxin-3 was recently shown to cause a decrease in parkin levels in a mouse model of SCA3, although the mechanism causing this decrease is unknown (Durcan et al., 2010). Defining the normal function of ataxin-3 may help us understand why neurodegeneration occurs in this polyglutamine disease.
Here we describe several molecular events that regulate CHIP activity, ranging from chain initiation to termination of the reaction. We show that the initiator E2, Ube2w, monoubiquitinates CHIP, which in turn facilitates the recruitment of ataxin-3. In cells, Ube2w promotes monoubiquitination of CHIP and enhances CHIP-mediated clearance of an established CHIP substrate. In CHIP ubiquitination assays, ataxin-3 functions to restrict the length of ubiquitin chains attached to CHIP substrates, terminating ubiquitin incorporation once chains reach a critical length. In response to substrate polyubiquitination, ataxin-3 deubiquitinates CHIP. Finally, we link this novel function of ataxin-3 to molecular events that may contribute to SCA3 disease pathogenesis: polyglutamine expansion increases the affinity of ataxin-3 for CHIP, which correlates with decreased levels of this neuroprotective E3 in a mouse model of SCA3. Together, Ube2w and ataxin-3 provide a mechanism by which an important quality control E3 is sequentially regulated at multiple steps of the ubiquitination reaction.
Results
CHIP ubiquitination correlates with cell stress
A fraction of the cellular pool of CHIP electrophoreses as a higher molecular weight species consistent with mono-ubiquitinated CHIP (Ub-CHIP) (Imai et al., 2002; Jiang et al., 2001; Lees et al., 2003). Similarly, we observe a higher molecular weight species consistent with Ub-CHIP in mouse brain and heart, two organs in which CHIP is especially abundant (data not shown). To determine whether this species represents Ub-CHIP, we expressed FLAG-tagged CHIP and HA-tagged ubiquitin in HEK293 cells, then immunoprecipitated CHIP under stringent conditions and probed for ubiquitinated CHIP. A HA-ubiquitin-positive band consistent with Ub-CHIP was detected, indicating that CHIP can be monoubiquitinated in cells (Fig 1a). To determine if ubiquitination of CHIP is important physiologically, we tested whether Ub-CHIP increased upon cell stress conditions in which CHIP function is known to be protective (Dai et al., 2003). Upon proteasome inhibition and heat shock, Ub-CHIP levels were observed to increase (Fig 1b and 1c) whereas polyubiquitinated CHIP was not detected (Fig S1a and b).
Figure 1. Ube2w monoubiquitinates CHIP.
(A) CHIP is mono-ubiquitinated in cells. FlagCHIP and HAUb were expressed in 293 cells and immunoprecipitated using anti-FLAG resin under stringent conditions. Cells expressing only HAUb were used as a negative control. (B) MG132 treatment increases the levels of Ub-CHIP. HEK293 cells were transfected with CHIP for 24 hours prior to addition of 30µM MG132 for either 1, 3, or 6 hours. Shown is a representative blot (n=3). (C) Thermal stress increases the levels of Ub-CHIP. HEK293 cells were transfected with CHIP for 24 hours prior to transfer of the cells to 41°C for either 1, 2, or 3 hours. Shown is a representative blot (n=3). (D) Ube2w rapidly and quantitatively monoubiquitinates CHIP. Ubiquitination reactions were performed for the indicated times with each E2 found to ubiquitinate CHIP (see figure S1). Assays were repeated three times with nearly identical results. (E) RNAi knockdown of Ube2w levels decreases ubiquitination of CHIP. HEK293 cells were transfected with FlagCHIP and shRNA vector targeting Ube2w or scrambled sequence (as negative control), then analyzed by western blot and quantified (n=3, *p =0.036). Error bars indicate Standard Error of the Mean (SEM). (F) Coexpression of Ube2w increases ubiquitination of CHIP in vivo. COS-7 cells were transfected with FlagCHIP and either Ube2w or Skp1 (as negative control) then analyzed by western blot and quantified by densitometry (n=3, **p = 0.0033). Error bars indicate SEM.
To identify which E2s can ubiquitinate CHIP, we tested all E2s containing a SPA motif, which is required for E2/CHIP interactions (Xu et al., 2008) (Fig 1d and Fig S1c and b). At a 1:1 molar ratio Ube2w rapidly and quantitatively ubiquitinated CHIP while no other E2 had comparable activity (Fig 1d, and Fig S1d and e). All tested E2s were functional proteins as they formed thioesters with similar efficiency (Fig S1f and g). Importantly, knockdown of Ube2w led to a decrease in Ub-CHIP (Fig 1e and FigS1h) while coexpression of Ube2w with CHIP increased Ub-CHIP levels in transfected cells (Fig 1f). Together, these results establish that a fraction of the CHIP pool is monoubiquitinated in vivo and that Ube2w can ubiquitinate CHIP in vitro and in vivo.
Ube2w reduces steady state levels of a CHIP substrate in cells
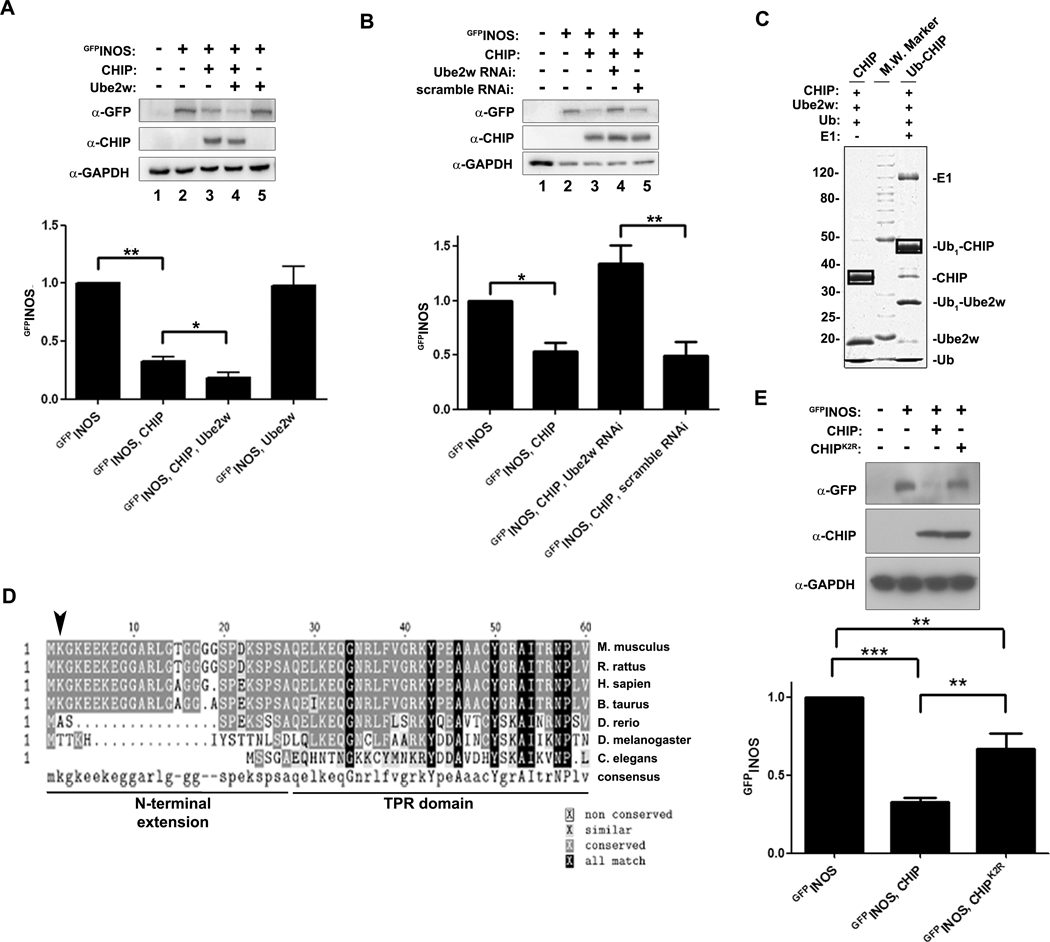
We next determined whether CHIP mono-ubiquitination has functional consequences for CHIP activity in cells. If Ube2w-mediated ubiquitination of CHIP facilitates clearance of substrates, then manipulating Ube2w levels should alter steady state levels of CHIP substrate. We tested this in transfected cells employing an established CHIP substrate, GFPINOS. As previously reported (Sha et al., 2009) CHIP overexpression decreased GFPINOS levels in transfected cells (Fig 2a, b, and e), and coexpression of Ube2w with CHIP decreased GFPINOS levels even further (Fig 2a). In contrast, knockdown of Ube2w stabilized GFPINOS levels even when CHIP was overexpressed (Fig 2b); this result was confirmed with a second shRNAi construct (data not shown). This effect of Ube2w required CHIP as overexpression of Ube2w alone did not decrease GFPINOS levels (Fig 2a). Thus, coexpression of Ube2w with CHIP facilitates clearance of a CHIP substrate.
Figure 2. Ubiquitination of CHIP by Ube2w regulates CHIP function.
(A) Ube2w enhances CHIP’s ability to reduce cellular levels of a known CHIP substrate, INOS. Combinations of GFPINOS, CHIP and Ube2w were expressed in COS-7 cells by transfection. Coexpression of Ube2w enhances CHIP-mediated reduction of steady state levels of GFPINOS, whereas expression of Ube2w alone has no effect (n=3, *p < 0.05, **p < 0.005). Error bars indicate SEM. (B) RNAi knockdown of Ube2w increases levels of GFPINOS even when CHIP is overexpressed. HEK293 cells were transfected with combinations of GFPINOS, CHIP and vectors expressing shRNA against Ube2w or scrambled shRNA (n=3, *p < 0.05, **p < 0.005). Error bars indicate SEM. (C) In vitro ubiquitination of CHIP by Ube2w generates a single band corresponding to monoubiquitinated CHIP, which is absent in a control reaction lacking E1. Mass spectrometry identified K2 as the sole modified lysine on Ub-CHIP (see Fig S2). (D) The ubiquitinated lysine on CHIP, K2, is conserved in mammals but is absent in zebrafish and invertebrate species. The N-terminal sequences of CHIP orthologues were aligned using CLUSTAL-W (biology workbench); arrowhead indicates K2. (E) K2 is important for CHIP function in vivo. HEK293 cells were transfected to express GFPINOS alone or together with CHIP or CHIPK2R. Coexpressed CHIPK2R is less effective than wild type CHIP in decreasing steady state levels of GFPINOS (n=4, **p value <0.005, ***p value < 0.0005). Error bars indicate SEM.
In addition to ubiquitinating CHIP, Ube2w also can attach the initial ubiquitin to a substrate prior to chain extension (Christensen et al., 2007). To begin differentiating between these two roles of Ube2w, we sought to identify the lysine residue(s) in CHIP ubiquitinated by Ube2w in order to engineer mutant CHIP lacking this lysine or lysines. An N-terminal lysine, K2, proved to be the sole lysine on CHIP ubiquitinated by Ube2w, although upon removal of K2 other lysines on CHIP can be ubiquitinated in vitro (Fig 2c and d, Fig S2a and b, data not shown). In cells, mutant CHIP lacking K2 showed an impaired ability to reduce GFPINOS levels (Fig 2e). Additional mutation of two neighboring lysines, K4 and K7, that may compensate for K2 further impaired CHIP’s ability to reduce levels of GFPINOS (Fig S2c), suggesting that Ube2w imparts its function at least partly through modifying CHIP. The defect of CHIPK2,4,7R in reducing GFPINOS levels was not due to impaired E3 function because CHIPK2,4,7R retained full activity in ubiquitination assays (Fig S2d). These results support a model in which Ube2w accelerates the degradation of CHIP substrates by monoubiquitinating CHIP.
Ubiquitination of CHIP stabilizes its interaction with the DUB ataxin-3
CHIP is known to interact with two UIM proteins: the proteasomal subunit S5a, and ataxin-3 through its UIM-containing C-terminus (Connell et al., 2001; Jana et al., 2005). To determine whether CHIP interacts with full length ataxin-3 we performed co-IPs with CHIP and ataxin-3 in lysates from transfected cells. Indeed, CHIP coprecipitated with ataxin-3 (Fig 3a).
Figure 3. Ubiquitination of CHIP enhances its interaction with the DUB ataxin-3.
(A) CHIP and ataxin-3 interact in vivo. HEK293 cells co-transfected with FLAG-tagged ataxin-3 and untagged CHIP constructs were lysed 48 hours after transfection, and immunoprecipitated with anti-FLAG beads. (B) Mono-ubiquitination of CHIP stabilizes the interaction between CHIP and two unrelated UIM proteins, S5a and ataxin-3. Pull-downs of recombinant S5a or ataxin-3 were performed using empty beads, CHIP beads or Ub-CHIP beads, then analyzed by western blot. (C, D) Octet RED quantitative binding analyses of the interaction of ataxin-3 with CHIP (C) or Ub-CHIP (D). Biotinylated CHIP or Ub-CHIP was immobilized to streptavidin coated sensors and association and dissociation of ataxin-3 was monitored (KD =2.2 µM for CHIP, and KD =0.12 µM for Ub-CHIP).
We next sought to determine whether mono-ubiquitination of CHIP promoted interactions with UIM proteins. In pull-down assays, ubiquitination of CHIP enhanced its interaction with both S5a and ataxin-3 (Fig 3b). To confirm that ubiquitination of CHIP stabilizes its association with ataxin-3, we performed additional quantitative binding assays using the Octet RED biosensor platform. Though ataxin-3 does interact with unmodified CHIP, it binds with ~18 fold increase in affinity to Ub-CHIP (Fig 3c and d). CHIP’s interaction with ataxin-3 is of interest since the two proteins share substrates in cells (e.g. CFTRΔ508) and both are neuroprotective in vivo in a manner dependent on their ubiquitin-associated activities (Meacham et al., 2001; Warrick et al., 2005).
Because ubiquitination of ataxin-3 is known to stimulate its DUB activity (Nicastro et al.; Todi et al., 2010; Todi et al., 2009) we next determined whether ataxin-3 becomes ubiquitinated upon recruitment to CHIP. In a CHIP ubiquitination reaction, Ube2w monoubiquitinated ataxin-3 more rapidly and robustly than did a second E2, UbcH5c (Fig S3a). In some cases E2-mediated ubiquitination of UIM proteins can occur independent of E3s (Hoeller et al., 2007), thus we tested whether CHIP was necessary for Ube2w-dependent ubiquitination of ataxin-3. The presence of CHIP markedly enhanced Ube2w-mediated ubiquitination of ataxin-3 in a manner requiring full length CHIP (Fig S3b). Polyglutamine expansion did not alter ataxin-3 ubiquitination, as there was no difference in the ubiquitination of normal (Q22) versus expanded (Q80) ataxin-3 by Ube2w (Fig S3c).
In addition to stabilizing CHIP’s interaction with UIM proteins, monoubiquitination of CHIP potentially could stimulate CHIP activity directly, similar to the attachment of Nedd8 to the cullin-1 subunit of SCF (Furukawa et al., 2000; Morimoto et al., 2000; Podust et al., 2000; Read et al., 2000; Wu et al., 2000). However, we observed no difference in CHIP’s ability to ubiquitinate a model substrate, HSP90, when CHIP is ubiquitinated (Fig S3d).
Ataxin-3 limits ubiquitin chain length on CHIP substrates
The interaction of ataxin-3 with CHIP suggests that ataxin-3 functions as a DUB when partnered with CHIP to modulate ubiquitination of CHIP substrates. We thus sought to determine whether ataxin-3 interacted with the CHIP substrate GFPINOS. In lysates from transfected cells, immunoprecipitation of ataxin-3 confirmed that ataxin-3 interacts with GFPINOS in cells (Fig 4a).
Figure 4. Ataxin-3 limits polyubiquitin chain length on CHIP substrates.
(A) Ataxin-3 interacts with GFPINOS. HEK293 cells were transfected with GFPINOS alone or with FLAG-tagged ataxin-3 or catalytically inactive C14Aataxin-3. Cells were lysed 48 hours after transfection and FLAG IP was performed. (B) Catalytically inactive ataxin-3 increases steady state levels of GFPINOS. COS-7 cells were transfected to express GFPINOS alone or together with ataxin-3 or C14Aataxin-3 (n=3, *p < 0.05). Error bars indicate SEM. (C) Ataxin-3 trims, but does not fully deubiquitinate, polyubiquitinated HSP90. Ubiquitination of HSP90 was carried out for 1 hour, the reaction stopped with 50mM EDTA, and ataxin-3 then added to the reaction for the indicated times. (D) When present during the ubiquitination reaction, ataxin-3 limits the length of polyubiquitin chains attached to HSP90 by CHIP. HSP90 ubiquitination reactions were performed in the presence or absence of ataxin-3 for the times indicated, with either UbcH5c as the E2 or a mutant form of UbcH5c (UbcH5cS22R) that is deficient in ubiquitin chain extension. (E) Both normal (Q22) and expanded (Q80) ataxin-3 limit polyubiquitin chain length in a manner that requires catalytic activity and ubiquitin chain binding via the UIMs; other tested DUBs had no effect on the length of chains added to substrate. Ubiquitination reactions were performed without DUB or with ataxin-3 (Q22 or Q80), C14Aataxin-3, UIM-mutated UIM1,2,3*ataxin-3, USP5, UCH-L1, or UCH-L3 for the indicated times. (F) Ataxin-3 trims polyubiquitin chains in cells. HEK293 cells were transfected with empty vector, FLAGataxin-3, or FLAGataxin-3C14A. Cells were then lysed and FLAG IP was performed. (G) Longer polyubiquitin chains compete with ataxin-3 binding to Ub-CHIP. Pull-downs of Ub-CHIP (1µm) were performed with ataxin-3 beads or empty beads as indicated, in the absence of ubiquitin or in the presence of 10µM mono-, di-, tri- or hexa-ubiquitin (all K63-linked).
We next determined whether coexpressing ataxin-3 affected turnover of CHIP substrates in vivo. To test this, we coexpressed GFPINOS with wild type ataxin-3 or catalytically inactive C14Aataxin-3. In contrast to previously described DUB/E3 pairs in which DUB activity stabilized substrate (Ventii and Wilkinson, 2008), catalytically inactive ataxin-3 stabilized GFPINOS levels whereas active ataxin-3 permitted normal clearance of GFPINOS (Fig 4b). Furthermore addition of Eeyarestatin 1, an inhibitor of ataxin-3 and VCP/p97 (Wang et al., 2008), stabilized GFPINOS and led to the accumulation of a species consistent with polyubiquitinated GFPINOS (Fig S4a). These results imply that the DUB activity of ataxin-3 promotes turnover of CHIP substrates.
In reconstitution assays employing free ubiquitin chains as substrate, ataxin-3 cleaves only longer ubiquitin chains and has very little activity toward chains shorter than 4–5 ubiquitins (Burnett et al., 2003; Winborn et al., 2008). Other DUBs tested by us behave differently: USP5 fully cleaves chains of any length whereas UCHL-1 and UCHL-3 show little activity towards free polyubiquitin chains (Fig S4b and data not shown). The chain length dependence to ataxin-3 DUB activity could serve to limit the length of ubiquitin chains added by CHIP to substrates. To address this possibility, we tested the ability of ataxin-3 to remove ubiquitin chains on two preubiquitinated CHIP model substrates, HSP90 (Fig 4c) and denatured luciferase (Fig S4c). Ataxin-3 effectively trimmed chains on preubiquitinated substrates but did not completely deubiquitinate them (Fig 4c and Fig S4c). When present during the ubiquitination reaction, ataxin-3 allowed ubiquitination of substrate yet prevented longer polyubiquitin chains from accumulating (Fig 4d, compare lanes 6 and 7). Because in this assay polyubiquitinated substrate cannot be distinguished from multiply mono-ubiquitinated substrate, we also tested the ability of ataxin-3 to cleave mono-ubiquitin from substrate ubiquitinated by UbcH5cS22R, a mutant form of UbcH5c that can initiate but not extend ubiquitin chains. Ataxin-3 was unable to remove ubiquitin from mono-ubiquitinated substrate, consistent with its primary role in cleaving longer ubiquitin chains (Fig 4d, compare lanes 13 and 14). When lysine-less ubiquitin (K0Ub) was used in reactions to determine whether multiple lysines on the test substrate (HSP90) are ubiquitinated by CHIP, we observed bands consistent with modification of multiple lysines on HSP90 (Fig S4d). Thus, polyubiquitinated HSP90 in our assay conditions likely contains multiple, shorter polyubiquitin chains rather than a single long chain.
We next investigated which domains of ataxin-3 are necessary for its chain trimming activity during ubiquitination reactions (Fig 4e). Ataxin-3’s catalytic activity and UIMs both proved essential for ataxin-3 to limit polyubiquitin chain length; in contrast, polyglutamine expansion had no effect (Fig 4e). Mutations in UIMs 1 and 2, but not UIM 3, impaired ataxin-3’s ability to limit ubiquitin chain length on CHIP substrates (Fig S4e). This chain trimming function may be unique to ataxin-3, as other tested DUBs (USP5, UCHL-1 and UCHL-3) were unable to trim ubiquitin chains from substrates (Fig 4e).
If ataxin-3 functions similarly in vivo to prevent excessive ubiquitination, then mice lacking ataxin-3 might be expected to have increased levels of polyubiquitinated proteins. Indeed, brain lysates from Atxn3 knockout mice show increased high molecular weight, ubiquitin-positive signal that electrophoreses at the top of the separating gel and stacking gel (Fig S4f). This result, independently reported in a second Atxn3 knockout mouse line (Schmitt et al., 2007), is consistent with an in vivo role for ataxin-3 in limiting the length of polyubiquitin chains added to substrates.
The above results indicate that ataxin-3 regulates the extent of substrate ubiquitination by CHIP. To determine if ataxin-3 behaves similarly in cells, we transfected cells with ataxin-3 or catalytically inactive ataxin-3, then performed immunoprecipitations on ataxin-3 and immunoblotted for ubiquitin. Catalytically inactive, but not catalytically active, ataxin-3 coprecipitated a strong ubiquitin signal in the stacking gel indicative of longer polyubiquitin chains (Fig 4f). These results support a model in which ataxin-3 effectively limits ubiquitin chain extension once chains reach a critical length.
Given ataxin-3’s strong binding preference for longer ubiquitin chains irrespective of linkage type (Burnett et al., 2003; Winborn et al., 2008), polyubiquitin chains conjugated to CHIP substrates might effectively compete ataxin-3 away from its binding to Ub-CHIP. To test this we performed pull-down assays to assess ataxin-3 binding to Ub-CHIP in the presence of mono-, di-, tri-, or hexa-K63 ubiquitin chains, a preferred substrate for ataxin-3 (Winborn et al., 2008). Of these, only hexa-ubiquitin efficiently prevented the interaction between ataxin-3 and Ub-CHIP (Fig 4g).
Together, these results indicate that ataxin-3 functions with CHIP to prevent the incorporation of additional ubiquitin to chains on substrates once they reach a certain length. Other tested DUBs in our hands do not do this. Moreover, in the presence of ubiquitin chains longer than three ubiquitins, such as those on polyubiquitinated CHIP substrates, ataxin-3 or other UIM proteins within the ubiquitinating complex likely would interact preferentially with polyubiquitin chains on the substrate rather than Ub-CHIP with its single conjugated ubiquitin.
Ataxin-3 deubiquitinates CHIP upon completion of substrate polyubiquitination
Remarkably, despite its established preference for cleaving longer ubiquitin chains ataxin-3 was able to remove monoubiquitin from Ub-CHIP in reactions in which further ubiquitin conjugation was inhibited (Fig 5a). In active ubiquitination assays containing ataxin-3, we observed over time that Ub-CHIP first appeared, then disappeared (Fig 5b). Intriguingly, deubiquitination of Ub-CHIP by ataxin-3 correlated temporally with the accumulation of ubiquitinated substrates and occurred whether ataxin-3 had a normal or expanded polyglutamine domain (Fig 5b).
Figure 5. Ataxin-3 deubiquitinates CHIP upon completion of substrate polyubiquitination.
(A) Ataxin-3 deubiquitinates Ub-CHIP in vitro. CHIP was monoubiquitinated by Ube2w, and the reaction was stopped with 50mM EDTA. Increasing concentrations of the indicated DUBs were added and loss of Ub-CHIP over time was assessed by western blot. Representative results are shown, with the graph depicting the mean results of two experiments. The dashed line represents extrapolated data for 10µM USP5. (B) Ataxin-3 deubiquitinates Ub-CHIP during ubiquitination reactions. Ubiquitination was carried out in the absence or presence of ataxin-3 as indicated. Ub-CHIP deubiquitination by ataxin-3 correlates temporally with the appearance of ubiquitinated substrate. Polyglutamine expansion has no discernible effect on ataxin-3’s ability to deubiquitinate Ub-CHIP. UbcH5c was employed as the sole E2 in these assays, so that the temporal nature of conjugation and deconjugation of ubiquitin to CHIP could be viewed more precisely. (C) The presence of ubiquitinated substrate enhances deubiquitination of Ub-CHIP byataxin-3. HSP90 ubiquitination reactions were performed in the absence or presence of ataxin-3 and/or UbcH5c for one hour, with increasing amounts of HSP90 (0 to 1µM.) (D) Ubiquitination of substrate markedly facilitates ataxin-3 deubiquitination of Ub-CHIP. CHIP, ataxin-3, and E1mix were incubated for one hour with additional components as indicated. Western blots were performed to assess the ubiquitination state of HSP90 and CHIP. (E) The UIMs of ataxin-3 are essential for deubiquitination of Ub-CHIP. Ubiquitination reactions were performed with ataxin-3, UIM-mutated ataxin-3 (UIM 1,2,3* in which all three UIMs are mutated) or no ataxin-3. (F) Ataxin-3 catalytic activity is required for deubiquitination of Ub-CHIP. Ubiquitination reactions were performed with ataxin-3 or catalytically inactive ataxin-3C14A.
To determine whether deubiquitination of Ub-CHIP by ataxin-3 requires the presence of ubiquitinated substrate, we performed CHIP ubiquitination reactions with increasing concentrations of HSP90 as substrate, in the presence or absence of ataxin-3 and the chain elongating E2, UbcH5c (Fig 5c), then assessed the ubiquitin state of CHIP at the end of the reaction. In the absence of ataxin-3, CHIP becomes monoubiquitinated and remains so throughout the reaction. When ataxin-3 is present, however, Ub-CHIP becomes deubiquitinated as the level of ubiquitinated substrate increases. In reactions containing ataxin-3 and Ube2w but not UbcH5c (thus preventing chain extension), ataxin-3 did not deubiquitinate Ub-CHIP (Fig 5c). These results suggest that polyubiquitin chain formation promotes ataxin-3 mediated cleavage of ubiquitin from Ub-CHIP. To exclude the possibility that this finding instead reflects a chaperoning effect of HSP90, we repeated the assay with a second substrate, HSP70, without an accompanying J-protein, which is required for efficient HSP70 chaperoning activity. With Hsp70 as substrate, we observed similar results (Fig S5a).
The above results do not exclude the possibility that ubiquitin chains per se rather than polyubiquitinated substrates facilitate deubiquitination of CHIP by ataxin-3. Accordingly, we assessed deubiquitination of Ub-CHIP when components were sequentially added to the reaction. The presence of UbcH5c, HSP90 or free K63-linked hexa-Ub chains alone did not stimulate Ub-CHIP deubiquitination by ataxin-3. Only the presence of both UbcH5c and HSP90, which generates polyubiquitinated HSP90, led to efficient deubiquitination of Ub-CHIP (Fig 5d). No other tested DUB behaved in a similar manner: USP5 deubiquitinated Ub-CHIP in an unregulated manner, while UCHL-1 and UCHL-3 did not deubiquitinate Ub-CHIP under any conditions (Fig 5d). We next tested whether the presence of UbcH5c, which is needed to polyubiquitinate HSP90, reduced the amount of ataxin-3 needed to deubiquitinate Ub-CHIP in active ubiquitination assays. As expected, the presence of this chain-elongating E2 markedly enhanced deubiquitination of CHIP by ataxin-3 (Fig 5Sb).
The fact that efficient deubiquitination of Ub-CHIP by ataxin-3 requires the presence of polyubiquitinated substrate suggests that the UIMs of ataxin-3 play an essential role in this process. Indeed, ataxin-3 mutated in all three UIMs was defective in deubiquitinating Ub-CHIP during the course of a ubiquitination reaction (Fig 5e) as was catalytically inactive ataxin-3 (Fig 5f). Collectively, these data show that ataxin-3 efficiently removes ubiquitin from Ub-CHIP in the presence of polyubiquitinated substrate. Because this activity of ataxin-3 requires both ubiquitinated substrate and the UIMs of ataxin-3, ataxin-3 is likely bound to polyubiquitinated substrate when it cleaves ubiquitin from Ub-CHIP.
Polyglutamine expansion in ataxin-3 increases affinity for CHIP
A leading model of polyglutamine disease pathogenesis posits that polyglutamine expansion perturbs the balance of normal protein-protein interactions engaged in by the disease protein (Gu et al., 2009; Nedelsky et al., 2010; Zoghbi and Orr, 2009). To determine whether polyglutamine expansion alters ataxin-3’s affinity for CHIP, we performed quantitative binding assays between CHIP and normal (Q22) or expanded (Q80) ataxin-3. Polyglutamine-expanded ataxin-3 displayed a ~6 fold increase in binding affinity for CHIP compared to normal ataxin-3 (Fig 6a). Interestingly, unlike normal ataxin-3, polyglutamine-expanded ataxin-3 no longer bound Ub-CHIP with higher affinity than unmodified CHIP (Fig 3c and d, Fig 6a).
Figure 6. Polyglutamine expansion increases ataxin-3 affinity for CHIP and results in decreased CHIP levels in SCA3 mice.
(A) Quantitative binding analysis between CHIP or Ub-CHIP and normal or expanded ataxin-3 (Q22 or Q80). Biotinylated CHIP or Ub-CHIP was immobilized to streptavidin-coated sensors and association and dissociation of ataxin-3 (Q22 or Q80) was monitored (Kd =2.2 µM for Q22 to CHIP, Kd =0.39 µM for Q80 to CHIP, and Kd=0.75 µM for Q80 to Ub-CHIP). (B) CHIP levels are decreased in SCA3 Q84.2 transgenic mouse brain. CHIP levels were assessed in whole brain lysates from 4-month-old nontransgenic or Q84.2 hemizygous transgenic mice. CHIP levels are significantly reduced in Q84.2 mice (n=3, *p = 0.0372). Error bars indicate SEM. (C) Immunohistochemistry reveals decreased brain CHIP levels in Q84.2 mice. Brains from Q84.2 hemizygous mice or nontransgenic littermates were sectioned and immunostained with CHIP antibody. DAB immunohistochemistry was carried out simultaneously and identically on sections from nontransgenic and Q84 mice. (D) CHIP levels are not altered in Atxn3 knockout mice. Whole brain lysates from 2 month old wild-type or Atxn3 knockout mice were analyzed by SDS-PAGE and western blot. Error bars indicate SEM.
Ataxin-3 is known to regulate the flux of substrates through degradation pathways (Kuhlbrodt et al., 2011; Zhong and Pittman, 2006). Our results here suggest that it does so in part by modulating CHIP activity. The increased affinity of expanded ataxin-3 for CHIP could alter the dynamics of this functional interaction and inadvertently target CHIP for degradation. To address this possibility, we analyzed CHIP levels in brain lysates from wild-type and SCA3 transgenic mice that express human ataxin-3(Q84). SCA3 mice showed a significant decrease in CHIP levels (Fig 6b). This reduction was apparent throughout the brain by immunohistochemical staining for CHIP (Fig6c and data not shown). In contrast, CHIP levels remain unchanged in ataxin-3 knockout mice (Fig 6d), supporting a model in which CHIP/ataxin-3 interactions normally do not target CHIP for degradation but rather modulate CHIP activity within the ubiquitinating complex.
Discussion
The mechanisms by which the activity of CHIP and other E3s are regulated remain poorly understood. Our results lead us to propose a model of CHIP regulation that incorporates modulating functions for the initiator E2 Ube2w, and the specialized DUB ataxin-3: Ube2w, and perhaps other E2s, stimulate CHIP/ataxin-3 complex formation by adding mono-ubiquitin to CHIP, and ataxin-3 in turn regulates the extent and duration of substrate ubiquitination by CHIP (Fig 7). Through their opposing ubiquitinating and deubiquitinating activities, Ube2w and ataxin-3 participate sequentially in initiating, regulating and terminating the CHIP ubiquitination cycle, as outlined in figure 7. Our findings and recent reports (Durcan et al., 2010; Kim et al., 2009) lead us to favor a model in which mono-ubiquitination of CHIP promotes recruitment of ataxin-3 to the complex before chain formation on substrates. Alternatively, in some cases ataxin-3 may be recruited to CHIP after substrate ubiquitination is complete (as illustrated in Fig S6a). Similar mechanisms may regulate other E3s. Ataxin-3’s function as a DUB to modulate ubiquitin ligase activity may extend to other E3s with which ataxin-3 interacts, including parkin and E4b (Durcan et al., 2010; Kuhlbrodt et al., 2011; Matsumoto et al., 2004). Our results also provide new insight into SCA3 disease pathogenesis: polyglutamine-expanded ataxin-3 binds CHIP with higher affinity, and this change is associated with decreased levels of this neuroprotective E3 in a mouse model of SCA3.
Figure 7. Propsed model of the Ube2w/CHIP/ataxin-3 ubiquitination cycle.
1. CHIP recruits misfolded proteins via heat shock proteins and directly recruits Ube2w, which in turn monoubiquitinates both CHIP and chaperone-bound substrate. 2. Ubiquitination of CHIP facilitates its interaction with ataxin-3. Ube2w ubiquitinates ataxin-3, which may stimulate ataxin-3’s DUB activity. 3. A second E2 (e.g. UbcH5) is recruited to the complex where it extends the ubiquitin chain on misfolded substrate. 4. Once the ubiquitin chain is 4 ubiquitins or longer, ataxin-3 preferentially binds the polyubiquitinated substrate. 5. When bound to polyubiquitinated substrate, ataxin-3 trims the polyubiquitin chain and/or restricts further chain extension. Upon completion of substrate ubiquitination, ataxin-3 deubiquitinates CHIP, effectively terminating the ubiquitination cycle. 6. Upon completion of CHIP deubiquitination, ataxin-3 may escort the ubiquitinated substrate to its destination, leaving CHIP free to undergo another round of substrate ubiquitination.
Ataxin-3 regulates activity of CHIP complexes
Protein quality control ubiquitin ligases like CHIP are highly regulated by the accessory proteins with which they interact. A number of cofactors are known to influence CHIP activity. HSJ1a and Bag-1 promote proteasomal degradation of CHIP substrates (Luders et al., 2000; Westhoff et al., 2005), Bag-3 targets substrates for chaperone mediated autophagy (Arndt et al., 2010), and Bag-2 and Bag-5 stabilize CHIP substrates (Dai et al., 2005; Kalia et al., 2011). Here we show that ataxin-3 participates in targeting CHIP substrates for degradation and describe a novel molecular mechanism by which a DUB and an E3 collaborate in substrate ubiquitination.
Previously described DUBs in E3/DUB pairs promote complete deubiquitination of substrate or the E3 itself. In contrast, ataxin-3 plays a far different role with CHIP: it limits the length of ubiquitin chains on CHIP substrates and deubiquitinates Ub-CHIP once polyubiquitinated substrates have formed. In CHIP ubiquitinating complexes, ataxin-3 trims polyubiquitin chains on substrates but cannot remove chains completely or cleave monoubiquitin from substrates (Fig 4c, d and e). These properties are consistent with ataxin-3’s activity against free ubiquitin chains (Fig S4a (Winborn et al., 2008)): ataxin-3 does not readily cleave ubiquitin chains shorter than 5 ubiquitins.
Why might it be advantageous for ataxin-3 to limit ubiquitin chain length on CHIP substrates? An attractive possibility is that ataxin-3 restricts chain length to the minimal size needed for proteasomal targeting, which is four ubiquitins (Thrower et al., 2000)]. Alternatively, ataxin-3 could function to regulate the rate of proteasomal degradation of CHIP substrates through multiple rounds of ubiquitin conjugation. Consistent with this, ataxin-3 is known to interact with VCP (Zhong and Pittman, 2006) and VCP promotes secondary rounds of ubiquitination (Richly et al., 2005). Moreover, ataxin-3 acts synergistically with VCP in regulating ubiquitin-mediated proteolysis and aging in a manner dependent upon ataxin-3’s catalytic activity (Kuhlbrodt et al., 2011), suggesting that regulation of ubiquitin chain formation by ataxin-3 is critical for specific biological pathways.
Ataxin-3 is known to selectively cleave longer Ub chains (Burnett et al., 2003; Winborn et al., 2008). Remarkably, we found that ataxin-3 also efficiently removes monoubiquitin from CHIP but only when polyubiquitinated substrate is present (Fig 5). In our in vitro assays, the ubiquitination status of CHIP represents a balance between the ubiquitin conjugating activity of Ube2w and the deubiquitinating activity of ataxin-3. By binding ataxin-3 through its UIMs, polyubiquitinated substrate may tether ataxin-3 to the Ub-CHIP complex and thereby facilitate deubiquitination of the neighboring Ub-CHIP polypeptide. Collectively, our results with ataxin-3 suggest a novel mechanism by which a specialized DUB can monitor ubiquitin chain formation, prevent excessive ubiquitination of substrates, and facilitate termination of the ubiquitination reaction.
Similar to ataxin-3, other UIM proteins may also be targeted to their substrates by ubiquitin chain formation. For example, HSJ1a has two UIMs and a J-domain that stimulates ATP hydrolysis by HSP70 (Westhoff et al., 2005). Through its ubiquitin binding properties and J-domain, HSJ1a stimulates release of ubiquitinated cargo from HSP70 to allow degradation by the proteasome (Westhoff et al., 2005). It will be important to test whether other UIM-containing DUBs (USP25, 28, and 37) behave similarly to ataxin-3.
Posttranslational modifications as important modulators of E3 activity
Ube2w conjugates ubiquitin to a single lysine residue of CHIP. This lysine, K2, resides in an N-terminal extension of CHIP that is conserved in mammals and is dispensable for basal CHIP activity in vitro. This region is also a site for phosphorylation (Dephoure et al., 2008) and ubiquitination by a second E2, UbcH5, at a different lysine (Wang et al., 2005). The latter finding may explain why we observe only a modest reduction in Ub-CHIP upon Ube2w knockdown (Fig 1e); other E2s may also modify CHIP. Because mutating K2 impairs CHIP activity in vivo (fig 2e), we propose that ubiquitination at or near K2 enhances CHIP function in mammalian cells. Our results indicate that this is a highly dynamic, transient posttranslational modification (Fig 1 and Fig 5). Thus, we would anticipate that only a small percentage of CHIP would be ubiquitinated in vivo, which is indeed the case (Fig. 1).
Posttranslational modifications of E3s are known to critically regulate activity. Modification of the cullin subunit of SCF complexes with the ubiquitin-like molecule, Nedd8, is essential for viability in most model organisms and in human cells (Osaka et al., 2000; Read et al., 2000), while deneddlyation by the COP9 signalosome inhibits SCF activity. The HECT type E3, ITCH, undergoes a phosphorylation-dependent conformational change that stimulates its activity (Gallagher et al., 2006). Here we observed that deubiquitination of CHIP by ataxin-3 occurs in response to the completion of substrate ubiquitination. It will be interesting to determine whether completion of substrate ubiquitination is also coupled to the removal of posttranslational modifications on other E3s (e.g. deneddlyation of SCF and dephosphorylation of ITCH).
Implications for disease
Polyglutamine diseases comprise nine neurodegenerative disorders that are associated with disease protein misfolding and aggregation (Williams and Paulson, 2008). A prevailing theme in the field is that perturbations in native functions of the individual disease proteins contribute to polyglutamine toxicity (Lim et al., 2008; Nedelsky et al., 2010). Our results reveal an increase in binding affinity between CHIP and polyglutamine-expanded ataxin-3 without an accompanying defect in ataxin-3 enzymatic activity (Fig 4e, 5b and 6a). Increased affinity of polyglutamine-expanded ataxin-3 for CHIP correlates with a decrease in CHIP levels in a mouse model of SCA3 (Fig 6b and c). Recent studies of Parkin/ataxin-3 interactions suggest that this aberrant property of expanded ataxin-3 extends to other quality control E3s. In the same SCA3 mouse model, levels of Parkin, a quality control E3 that is partially redundant with CHIP (Morishima et al., 2008) and contains a ubiquitin interacting domain, are similarly reduced in brain (Durcan et al., 2010).
Finally this study sheds light on how nonpathogenic ataxin-3 may play a cytoprotective role in vivo. In Drosophila, wildtype ataxin-3 suppresses degeneration caused by expanded polyglutamine proteins in a manner linked to its ubiquitin associated properties (Warrick et al., 2005). We propose that ataxin-3 mediates this protective effect through regulating ubiquitin chain formation by quality control E3 ligases such as CHIP. CHIP likewise protects against neurodegeneration in models of numerous neurodegenerative diseases including the polyglutamine disorders (Williams and Paulson, 2008). Further defining the linked mechanisms by which these two neuroprotective proteins act may suggest therapeutic targets to enhance protein quality control in age-related neurodegenerative diseases.
Experimental Procedures
Additional details of the methods can be found in supplemental data.
Ubiquitination assays
Ubiquitination was typically performed for 1 hr at 37°C in 10 µl mixtures containing buffer A (50 mM Tris pH7.5, 50 mM KCl, 0.2 mM DTT), Ubmix (2.5 mM ATP, 5 mM MgCl2, 50 nM Ube1, and 100 µM ubiquitin), 1 µM indicated E2, 1 µM CHIP, 1 µM HSP90/luciferase prepared as previously described (Murata et al., 2005). 1 µM ataxin-3, ataxin-3 mutants, or S5a were used where indicated. Reactions were stopped by addition of SDS-Laemmli buffer and boiling, followed by separation of proteins by SDS-PAGE and visualization by Western blotting with appropriate antibodies.
Transfection experiments
Cells were transfected using Lipofectamine-LTX (Invitrogen). For western blotting, cells were lysed in 95°C Laemmli buffer, boiled for 4 min, sonicated, and loaded on SDS–PAGE gels. Blots were imaged using the VersaDoc 5000 MP (Bio-Rad). For semi-quantification, images were collected below-saturation levels, and quantified with Quantity One (Bio-Rad). Background was subtracted equally among lanes. Students t-test or one way anova was used for statistical analyses using Graphpad Prism software.
IP under stringent conditions
Cos7 cells were transfected with FLAG-CHIP and HA-Ub using Lipofectamine/PLUS (Invitrogen) as recommended by the manufacturer. 48 hrs after transfection, cells were collected in PBS + PI (SigmaFast; Sigma- Aldrich), lysed in RIPA+PI, denatured in final 1% SDS (30 min, RT), and renatured in final 4.5% Triton-X 100 (30 min, RT). Lysates were then incubated with anti-FLAG antibody-conjugated beads (2 hrs at 4 degrees), rinsed 4X with RIPA + PI, and eluted by boiling in Laemmli buffer.
Binding experiments
GSTCHIP (unmodified or monoubiquitinated) was immobilized on glutathione sepharose beads and incubated with ataxin-3 or S5a. Beads were then washed and eluted with Laemmli buffer, run on SDS-PAGE, and visualized by Western blotting.
Octet RED platform binding experiments were performed by immobilizing either Biotinylated CHIP or Ub-CHIP and monitoring the association and dissociation of ataxin-3 (Q22 or Q80).
Analysis of human and mouse tissue
For immunohistochemistry 40 µm free-floating brain sections were incubated overnight with α-CHIP antibody (1:500) and then processed with the Vector Elite kit (vector Laboratories) with a biotinylated anti-rabbit IgG. The peroxidase activity was developed using the Vector DAB peroxidase substrate kit.
Supplementary Material
Acknowledgements
We thank Drs. Rachel Klevit, Wade Harper, Saurav Misra, Bob Cohen, and Tony Eissa for reagents. This work was supported by 1F32NS064596 to KMS, 1K99NS064097 to SVT, R01NS038712 and R01AG034228 to HLP, RO1NS059690 to JEG and P50AG025688 to JP.
Literature Cited
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Furst DO, Saftig P, Saint R, Fleischmann BK, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- Burnett B, Li F, Pittman RN. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- Burnett BG, Pittman RN. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc Natl Acad Sci U S A. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- Dai Q, Qian SB, Li HH, McDonough H, Borchers C, Huang D, Takayama S, Younger JM, Ren HY, Cyr DM, et al. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J Biol Chem. 2005;280:38673–38681. doi: 10.1074/jbc.M507986200. [DOI] [PubMed] [Google Scholar]
- Dai Q, Zhang C, Wu Y, McDonough H, Whaley RA, Godfrey V, Li HH, Madamanchi N, Xu W, Neckers L, et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bie P, Ciechanover A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey CA, Patterson C, Dickson D, Petrucelli L. Brain CHIP: removing the culprits in neurodegenerative disease. Trends Mol Med. 2007;13:32–38. doi: 10.1016/j.molmed.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Durcan TM, Kontogiannea M, Thorarinsdottir T, Fallon L, Williams AJ, Djarmati A, Fantaneanu T, Paulson HL, Fon EA. The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick L, Barnes J, Ebner B, Agrawal S, Andresen M, Lim J, Giesler GJ, Zoghbi HY, Orr HT. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 67:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Zhang Y, McCarville J, Ohta T, Xiong Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol Cell Biol. 2000;20:8185–8197. doi: 10.1128/mcb.20.21.8185-8197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci U S A. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, Karin M. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- Gu XF, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. Serines 13 and 16 Are Critical Determinants of Full-Length Human Mutant Huntingtin Induced Disease Pathogenesis in HD Mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Jana NR, Dikshit P, Goswami A, Kotliarova S, Murata S, Tanaka K, Nukina N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- Kalia LV, Kalia SK, Chau H, Lozano AM, Hyman BT, McLean PJ. Ubiquitinylation of alpha-synuclein by carboxyl terminus Hsp70-interacting protein (CHIP) is regulated by Bcl-2-associated athanogene 5 (BAG5) PLoS One. 2011;6:e14695. doi: 10.1371/journal.pone.0014695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Uchiki T, Gygi SP, Goldberg AL. S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J. 2009;28:1867–1877. doi: 10.1038/emboj.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K, Janiesch PC, Kevei E, Segref A, Barikbin R, Hoppe T. The Machado-Joseph disease deubiquitylase ATX-3 couples longevity and proteostasis. Nat Cell Biol. 2011;13:273–281. doi: 10.1038/ncb2200. [DOI] [PubMed] [Google Scholar]
- Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun ED, Duvick LA, Orr HT, Botas J, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- Lees MJ, Peet DJ, Whitelaw ML. Defining the role for XAP2 in stabilization of the dioxin receptor. J Biol Chem. 2003;278:35878–35888. doi: 10.1074/jbc.M302430200. [DOI] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem. 2000;275:4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Yada M, Hatakeyama S, Ishimoto H, Tanimura T, Tsuji S, Kakizuka A, Kitagawa M, Nakayama KI. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 2004;23:659–669. doi: 10.1038/sj.emboj.7600081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Nishida T, Honda R, Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1) Biochem Biophys Res Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- Morishima Y, Wang AM, Yu Z, Pratt WB, Osawa Y, Lieberman AP. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum Mol Genet. 2008;17:3942–3952. doi: 10.1093/hmg/ddn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Minami M, Minami Y. Purification and assay of the chaperone-dependent ubiquitin ligase of the carboxyl terminus of Hsc70-interacting protein. Methods Enzymol. 2005;398:271–279. doi: 10.1016/S0076-6879(05)98022-1. [DOI] [PubMed] [Google Scholar]
- Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, Taylor JP. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67:936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro G, Todi SV, Karaca E, Bonvin AM, Paulson HL, Pastore A. Understanding the role of the Josephin domain in the PolyUb binding and cleavage properties of ataxin-3. PLoS One. 5:e12430. doi: 10.1371/journal.pone.0012430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Saeki M, Katayama S, Aida N, Toh EA, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. Embo J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci U S A. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Schmitt I, Linden M, Khazneh H, Evert BO, Breuer P, Klockgether T, Wuellner U. Inactivation of the mouse Atxn3 (ataxin-3) gene increases protein ubiquitination. Biochem Biophys Res Commun. 2007;362:734–739. doi: 10.1016/j.bbrc.2007.08.062. [DOI] [PubMed] [Google Scholar]
- Sha Y, Pandit L, Zeng S, Eissa NT. A critical role for CHIP in the aggresome pathway. Mol Cell Biol. 2009;29:116–128. doi: 10.1128/MCB.00829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisoula C, Trachana V, Patterson C, Gonos ES. CHIP-dependent p53 regulation occurs specifically during cellular senescence. Free Radic Biol Med. 2011;50:157–165. doi: 10.1016/j.freeradbiomed.2010.10.701. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todi SV, Scaglione KM, Blount JR, Basrur V, Conlon KP, Pastore A, Elenitoba-Johnson K, Paulson HL. Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117. J Biol Chem. 2010 doi: 10.1074/jbc.M110.181610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todi SV, Winborn BJ, Scaglione KM, Blount JR, Travis SM, Paulson HL. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. Embo J. 2009;28:372–382. doi: 10.1038/emboj.2008.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventii KH, Wilkinson KD. Protein partners of deubiquitinating enzymes. Biochem J. 2008;414:161–175. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Xu W, McGrath SC, Patterson C, Neckers L, Cotter RJ. Direct identification of ubiquitination sites on ubiquitin-conjugated CHIP using MALDI mass spectrometry. J Proteome Res. 2005;4:1554–1560. doi: 10.1021/pr050104e. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y. Inhibition of p97-dependent protein degradation by Eeyarestatin I. J Biol Chem. 2008;283:7445–7454. doi: 10.1074/jbc.M708347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, Bonini NM. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Westhoff B, Chapple JP, van der Spuy J, Hohfeld J, Cheetham ME. HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr Biol. 2005;15:1058–1064. doi: 10.1016/j.cub.2005.04.058. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Chen A, Pan ZQ. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J Biol Chem. 2000;275:32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J Biol Chem. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.