Abstract
Nuclear entry of circadian oscillatory gene products is a key step for the generation of a 24-hr cycle of the biological clock. We have examined nuclear import of clock proteins of the mammalian period gene family and the effect of serum shock, which induces a synchronous clock in cultured cells. Previously, mCRY1 and mCRY2 have been found to complex with PER proteins leading to nuclear import. Here we report that nuclear translocation of mPER1 and mPER2 (1) involves physical interactions with mPER3, (2) is accelerated by serum treatment, and (3) still occurs in mCry1/mCry2 double-deficient cells lacking a functional biological clock. Moreover, nuclear localization of endogenous mPER1 was observed in cultured mCry1/mCry2 double-deficient cells as well as in the liver and the suprachiasmatic nuclei (SCN) of mCry1/mCry2 double-mutant mice. This indicates that nuclear translocation of at least mPER1 also can occur under physiological conditions (i.e., in the intact mouse) in the absence of any CRY protein. The mPER3 amino acid sequence predicts the presence of a cytoplasmic localization domain (CLD) and a nuclear localization signal (NLS). Deletion analysis suggests that the interplay of the CLD and NLS proposed to regulate nuclear entry of PER in Drosophila is conserved in mammals, but with the novel twist that mPER3 can act as the dimerizing partner.
Keywords: Circadian rhythm, mPER, nuclear entry, cytoplasmic localization domain, mCRY
Evidence accumulates that many organisms share a common oscillatory mechanism driving their circadian rhythms. This biological clock is comprised of a feed-back loop in which cyclically expressed clock gene products negatively regulate their own expression with an approximate 24-hr periodicity (Hastings 1997; Whitmore et al. 1998; Dunlap 1999). In the eukaryotic circadian model systems of Drosophila and Neurospora, nuclear entry of these oscillatory gene products preparatory to down-regulation of their own transcription constitutes one of the key steps in the control of the clock (e.g., Luo et al. 1998; for review, see Young 1998; Dunlap 1999). In Drosophila the product of the period (PER) gene oscillates in a circadian manner and appears to be essential for clock functioning. In mammals, three homologs have been identified, mPer1, mPer2, and mPer3 (Albrecht et al. 1997; Shearman et al. 1997; Sun et al. 1997; Tei et al. 1997; Takumi et al. 1998a,b; Zylka et al. 1998b). The transcript levels of all three genes oscillate in the suprachiasmatic nucleus (SCN) of the brain, the location of the master clock, as well as in peripheral tissues. Even in cultured cells, induction and subsequent rhythmic expression of mPer1 and mPer2 mRNA can be achieved by brief treatment with high levels of serum that either induce or synchronize individual cellular oscillators (Balsalobre et al. 1998).
Remarkably, available evidence suggests that the crucial regulatory step of nuclear entry of PER proteins is mediated differently in different organisms. In Drosophila, nuclear translocation of PER is triggered by the concealment of its cytoplasmic retention domain (cytoplasmic localization domain or CLD) by heterodimerization with TIMELESS (TIM) (Vosshall et al. 1994; Saez and Young 1996). The CLD region is preserved in mammalian PER proteins, but a functional role has not been established. In mammals, two homologs of plant cryptochromes (mCRY1 and mCRY2), members of the light-harvesting photolyase family (Hsu et al. 1996; Todo et al. 1996; van der Spek et al. 1996), have been reported recently to complex with mPER proteins and accomplish nuclear translocation, whereas heterodimerization of mPER members with each other also exerted some effect (Kume et al. 1999).
In the present study, we have explored the role of mPER proteins and the effect of serum shock on their nuclear relocation in COS7 cells. We report that nuclear entry can be accomplished by serum shock-induced heterodimerization of mPER1 and mPER2 with mPER3. In view of the role of mCRY1 and mCRY2 in translocation of mPER proteins into the nucleus (Kume et al. 1999), we also examined nuclear entry of mPER1 in cultured cells derived from mCry1/mCry2 double-deficient mice (van der Horst et al. 1999). We find that even in the absence of mCRY proteins, mPER1 translocates to the nucleus in a mPER3-dependent manner after serum shock in vitro. Also, we observed a clear nuclear localization of endogenous mPER1 in the SCN and liver of mCry1/mCry2 double-deficient mice. These findings establish a mCRY-independent route for nuclear translocation of mPER1 under physiological conditions.
Results
Serum shock stimulates heterodimerization of mPER proteins in COS7 cells
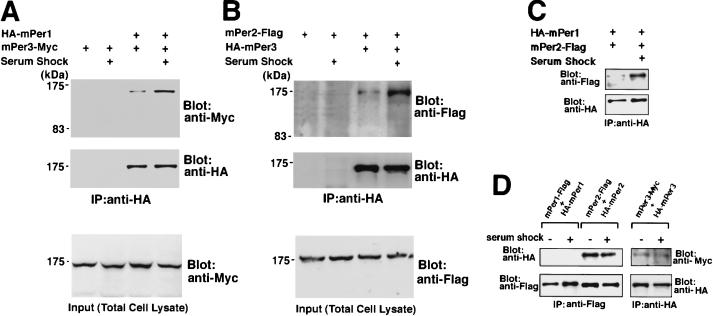
Epitope-tagged mPer genes were transfected in various combinations into COS7 cells, and the physical interactions between the encoded proteins were analyzed by immunoprecipitation. As brief exposure to high concentrations of horse serum (50%) has been shown to trigger mPer oscillation in cultured cells and induce clock functioning (Balsalobre et al. 1998); we also studied the effect of a serum shock on transiently expressed epitope-tagged mPER protein behavior. Following culture in serum-free medium for 12 hr but prior to serum shock, the amount of heterodimerized mPER proteins was very low (Fig. 1). A serum shock did not significantly change the amount of any mPER protein. However, 30 min after treatment, physical interactions of mPER1–mPER2, mPER1–mPER3, and mPER2–mPER3 had remarkably increased (Fig. 1A–C). In addition, we found strong homodimerization of mPER2 and weak homodimerization of mPER3, independent of the serum shock (Fig. 1D). Homodimerization of mPER1 was not detected under the conditions tested (Fig. 1D). Thus, we conclude that a serum shock significantly accelerates heterodimerization of mPER proteins in all combinations with no increase of homodimerization.
Figure 1.
Physical interaction of mPER proteins after serum shock. Heterodimerization detected by the immunoprecipitation in combinations mPER1–mPER3, (A), mPER2–mPER3, (B) and mPER1–mPER2. (C) Treatment with high concentrations of horse serum accelerates the heterodimerizations among mPERs in all combinations. No change was noted in the amount of mPER3-Myc (A, bottom) and mPER2-Flag (B, bottom) in total cell lysates showing that serum shock does not increase the amount of expressed proteins. (D) Homodimerization detected by immunoprecipitation. mPER2 and mPER3 form homodimers, but mPER1 does not homodimerize to a significant degree; serum shock has no effects on homodimerization.
Serum shock promotes nuclear entry of mPER1–mPER3 and mPER2–mPER3 heterodimers
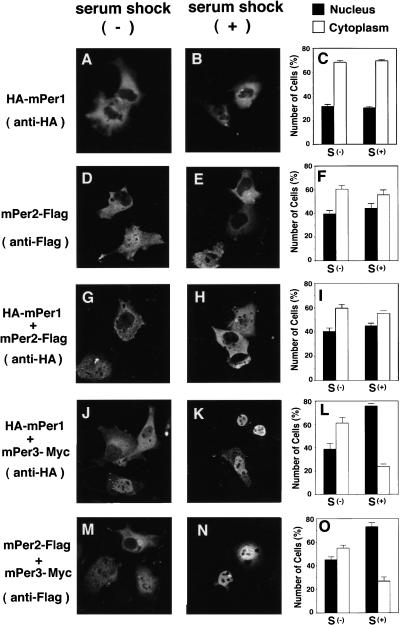
The fate of the heterodimerized mPERs in terms of subcellular localization was examined by immunofluorescence in COS7 cells transfected with Flag, Myc, or HA epitope-tagged mPer cDNAs. Figure 2 shows representative examples of immunostained single- and double-transfected cells with and without serum shock, as well as the ratio between cells with nuclear or cytoplasmic mPER staining. mPER1 or mPER2 alone showed predominantly cytoplasmic localization (Fig. 2A–F), whereas coexpression with mPer2 slightly enhanced the number of cells with nuclear mPER1 (Fig. 2G–I). The distribution patterns of mPER1 and mPER2 did not substantially alter after a serum shock. In cells transfected with mPer3–Myc, mPER3 was localized mostly in the cytoplasm (95%), and its subcellular localization was not altered after the serum shock (93%) (data not shown). When cells were cotransfected with combinations of mPer1/mPer3 or mPer2/mPer3 cDNA, a small increase in the number of cells with nuclear mPER1 and mPER2 was observed (Fig. 2J–O). Remarkably, after brief exposure to high serum, most cells show nuclear localization of the mPER1-HA and mPER2-Flag fusion proteins (75%–80%) (Fig. 2J–O). Thus, a serum shock promotes nuclear translocation of mPER1 and mPER2 proteins in COS7 cells only when mPER3 is available as a partner. On the basis of the immunoprecipitation and cellular localization data, we hypothesize that serum shock-induced heterodimerization of mPER3 with either mPER1 or mPER2 promotes nuclear entry of these clock proteins.
Figure 2.
Subcellular localization of heterodimerized mPER proteins. COS7 cells were transfected with HA-mPer1, (A–C), and mPer2-Flag (D,E,F); or cotransfected with HA-mPer1 and mPer2-Flag (G,H,I), HA-mPer1 and mPer3-His (J,K,L), and mPer2-Flag, and mPer3-His (M,N,O). Before (A,D,G,J,M) or 30 min after serum shock (B,E,H,K,N), cells were fixed and immunostained with anti-HA or anti-Flag M2 antibodies as indicated at left. Quantitative analyses are shown at right of each condition (C,F,I,L,O). Percentage of cells with predominant nuclear (N) and cytoplasmic (C) staining were determined as described. Three to five independent experiments were performed for each condition and one-hundred immunofluorescent cells from each cover glass were counted; means are shown +s.e.m.
The mPER3 protein has a functional CLD and NLS
In Drosophila, the translocation of PER into the nucleus is triggered by a rapid increase in heterodimerization between PER and TIM (Young 1998). Within PER and TIM, a nuclear localization signal (NLS) and CLD have been noted (Vosshall et al. 1994; Saez and Young 1996). Recently, sequences similar to the Drosophila CLD (amino acids 330–389 of mPER3) and NLS (residues 726–734) (Takumi et al. 1998b) were discerned in mPER3. In contrast, the deduced amino acid sequences of mPER1 and mPER2 do not show a typical single basic NLS, although mPER2 has sequences reminiscent of a bipartite basic NLS (Shearman et al. 1997). The absence of clearly distinguishable NLS sequences suggests that the strong serum-stimulated nuclear entry of mPER1 and mPER2 might be dependent on interactions with another protein(s). The data of Figure 2 suggest that mPER3 could fulfill this role.
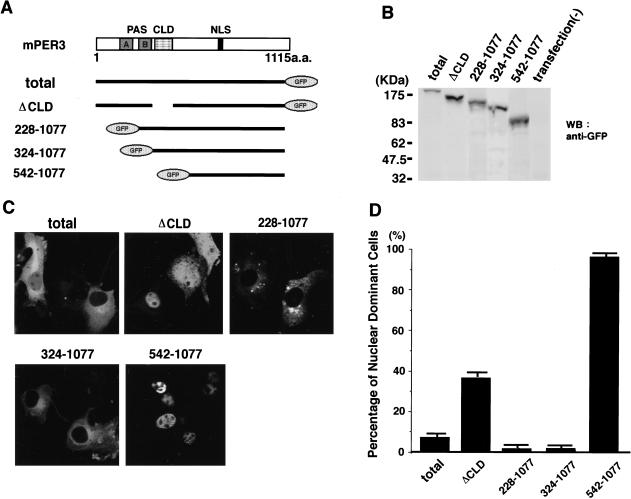
To examine this possibility and to determine whether the putative NLS and CLD of mPER3 are functional, we have generated the following panel of green fluorescent protein (GFP)–mPER3 fusion constructs as follows: (1) mPER3 (total), which contains the total sequence of mPER3 (1–1115 amino acids); (2) mPER3 (ΔCLD), which lacks amino acids 324–394 including the entire CLD region; (3) mPER3 (228–1077), which includes PAS-B, CLD, and NLS; (4) mPER3 (324–1077), which contains the CLD and NLS; (5) mPER3 (542–1077), which includes only the NLS (Fig. 3A). When expressed in COS7 cells, Western blot analysis using anti-GFP antibodies revealed single protein bands of the expected size (Fig. 3B). In the absence of a serum-shock mPER3 (total)–GFP fusion protein was expressed mainly in the cytoplasm and rarely located in the nucleus. The cytosolic-dominant distribution pattern was even more prominent for GFP–mPER3 fusion proteins [mPER3 (228–1077) and mPER3 (324–1077)] lacking one or more PAS domains but still containing the CLD and NLS (Fig. 3C). In contrast, a further size reduction of the amino-terminal half of mPER3, skipping the CLD, but retaining the NLS, [mPER3 (542–1077)] results in strong nuclear staining in >95% of the cells (Fig. 3C). Interestingly, a mPER3–GFP fusion protein lacking only the CLD portion [mPER3 (ΔCLD)] also showed clear nuclear staining in 40% of the cells (Fig. 3D). This result suggests that deletion of the CLD promotes entry of the mPER3–GFP protein into the nucleus. The weaker effect of mPER3 (ΔCLD) compared with mPER3 (542–1077) might result from interactions with other (endogenous) proteins via PAS-A and PAS-B domains. These data demonstrate that mPER3 has both a functional CLD and NLS and that, as is the case in Drosophila PER or TIM (Saez and Young 1996), the CLD is dominant to the NLS in dictating subcellular localization.
Figure 3.
CLD and NLS of mPER3 protein. Three truncated mutants of mPER3 fused with GFP were expressed in COS7 cells. (A) Schematic diagrams of the five constructs; (1) mPER3 (total), (2) mPER3 (ΔCLD), (3) mPER3 (228–1077) containing PAS-B, CLD, and NLS, (4) mPER3 (324–1077) containing CLD and NLS, and (5) mPER3 (542-1077) containing NLS and carboxy-terminal portion. (B) GFP fusion proteins analyzed by immunoblot confirmed the expression of expected GFP fusion proteins. (C) Example photos demonstrating the subcellular localization of each construct. GFP fusion proteins containing both the CLD and NLS [mPER3 (228–1077) and mPER3 (542–1077)] were localized only in the cytoplasm. Deletion of only the CLD region of mPER3 (ΔCLD) greatly increased the nuclear signals. (D) Summary of deletion study using truncated mutants of mPER3. Each bar indicates the percentage of cells expressing GFP-fusion proteins with predominant nuclear staining. All data were mean (+s.e.m.) in three independent experiments. In each experiment, 10 fields containing ∼200 cells altogether were examined.
mPER3 lacking the NLS fails to promote nuclear entry of mPER1 and mPER2
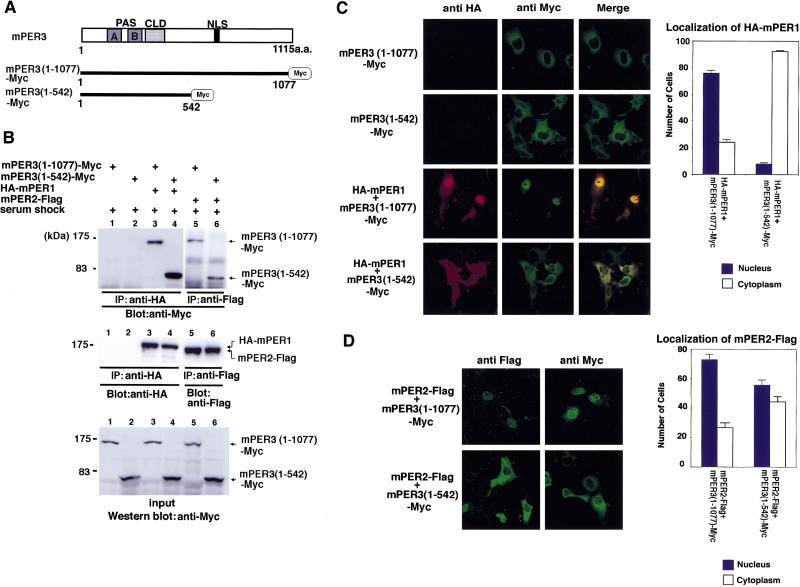
To further characterize the carboxy-terminal half of mPER3 containing the NLS, we made a truncated, Myc-tagged version [mPER3 (1–542); see Fig. 4A] and examined the effect of this deletion on serum shock-induced heterodimerization with, and nuclear localization of mPER1 and mPER2. Although heterodimerization with mPER1 after serum shock was unaffected by the deletion (Fig. 4B), truncated mPER3 almost completely abolished nuclear entry of mPER1/mPER3 heterodimers as evident from the reduced percentage of cells with nuclear staining (Fig. 4C). Similar results were obtained when Myc-tagged mPER3 (1–542) was replaced by a GFP-tagged version (data not shown). Analogous to the results for mPER1, truncated mPER3 (1–542) also retained the ability to heterodimerize with mPER2 after a serum shock (Fig. 4B). However, the effect of mPER3 truncation on mPER2 nuclear entry was weak compared with its effect on mPER1 (Fig. 4D). It is possible that this moderate effect on mPER2 nuclear localization is related to the putative bipartite basic NLS located in mPER2 (Shearman et al. 1997) that might retain some capacity for nuclear localization. In conclusion, these results demonstrate that the carboxy-terminal half of mPER3 has the capacity to strongly promote nuclear translocation of mPER1/mPER3 dimers and, to a lesser extent, mPER2/mPER3 dimers.
Figure 4.
Nuclear entry of a truncation mutant of mPER3 protein lacking the carboxy-terminal half containing the NLS. (A) Schematic diagrams of two mPER3-Myc constructs. (B) Immunoprecipitation showing that both mPER3 (1–1077)-Myc and mPER3 (1–542)-Myc associate with mPER1 and mPER2 protein. (C) Double-label immunofluorescence of cells coexpressing mPER3 (1–1077)-Myc and HA-mPER1, or mPER3 (1–542)-Myc and HA-mPER1. Although mPER1 coexpressed with mPER3 (1–1077) showed nuclear localization, truncated mPER3 (1–542)-Myc failed to allow nuclear entry of coexpressed mPER1 protein. Expression of only mPER3 (1–1077)-Myc or mPER3 (1–542)-Myc in COS7 cells revealed positive cytoplasmic staining to anti-Myc and no immunoreactivity against anti-HA antisera. Serum shock was performed in every case. Cell counts shown at right represent the mean of three independent experiments, and error bars indicate the s.e.m. One-hundred stained cells were observed and counted in each experiment. (D) Immunofluorescence of mPER3 (1–542)-Myc coexpressed with mPER2. mPER3 (1–542)-Myc also failed to promote nuclear entry of mPER2, but its effect on mPER2 was much weaker than that seen for mPER1. As above, serum shock was performed for all experiments. Results of cell counts represent the mean of three independent experiments, and error bars indicate the s.e.m. One-hundred stained cells were observed and counted in each experiment.
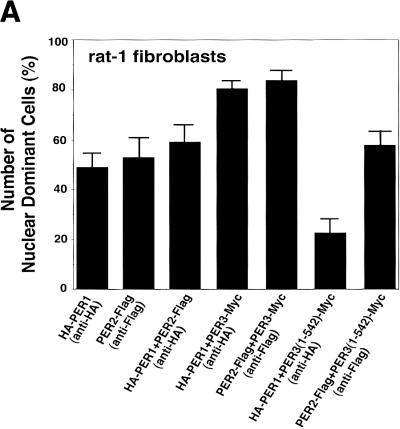
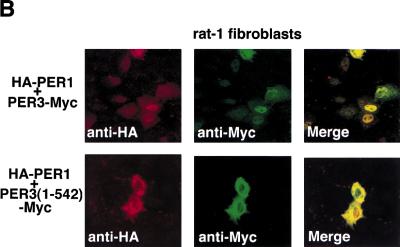
We also studied mPER3-dependent nuclear localization in rat-1 fibroblasts, known to produce a molecular rhythm following serum-shock (Balsalobre et al. 1998). Although in comparison to COS7 cells, rat-1 cells show higher levels of nuclear mPER1 and mPER2 after a single transfection with mPer1 and mPer2 cDNA, respectively, coexpression with mPer3 cDNA significantly increases nuclear localization of mPER1 and mPER2 (Fig. 5). Coexpression of mPER3 (1–542), lacking the NLS, again failed to stimulate nuclear localization of mPER1 and mPER2 (Fig. 5). Thus, mPER3-mediated mPER1 and mPER2 nuclear translocation appears not to be a COS7 cell-specific property, but rather might constitute a common phenomenon in various types of cells.
Figure 5.
Immunofluorescent study of subcellular localization of mPERs in rat-1 fibroblasts. (A) The graph shows the percentages of cells in which expressed mPER proteins show a predominantly nuclear distribution after serum shock. Coexpression with mPER3 significantly increases the ratio of mPER1 or mPER2 cells with a predominant nuclear staining, and coexpression with truncated mPER3 [mPER3 (1–542)] lacking the NLS significantly reduced the nuclear localization of mPER1. (B) Confocal images showing transfected cells with HA-mPER1/mPER3-Myc coexpression and HA-mPER1/mPER3 (1–542)-Myc coexpression. As shown in A, nuclear localization of expressed mPER1 was strongly enhanced by coexpression of mPER3 protein.
Nuclear entry of mPER1 in the absence of mCRY proteins
The above experiments strongly suggest that serum shock-induced nuclear translocation of mPER1 and mPER2 is promoted by heterodimerization with mPER3, but do not exclude the possibility that endogenous proteins in COS7 cells also mediate nuclear entry. In a recent study, Kume et al. (1999) demonstrated that mammalian CRY proteins are implicated in nuclear entry of mPER, but they also showed that mPER cellular localization is influenced by PER–PER heterodimerization. To further examine the role of mCRY and mPER3 proteins in mPER1 nuclear translocation, we investigated the subcellular localization of mPER1 in mCRY-deficient mouse embryonic fibroblasts (MEFs) derived from mCry1/mCry2 double-mutant mouse embryos (van der Horst et al. 1999).
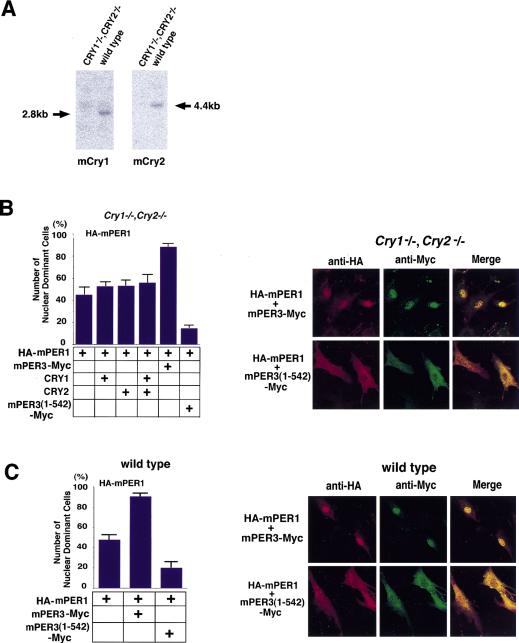
First, we confirmed by Northern blot analysis that intact mCry1 and mCry2 mRNAs were not detectable in these cells (Fig. 6A). Next, mCry1, hCry2, HA-tagged mPer1, and Myc-tagged mPer3 cDNAs were transfected in various combinations and subcellular localization was analyzed following a serum shock. Representative confocal micrographs (Fig. 6B, right) showed nuclear localization of exogenous mPER1 in the absence of endogenous mCRY in 45% of HA-mPer1 singly transfected cells (Fig. 6B, graph). Moreover, the percentage of mPER1 nuclear-stained cells was not significantly increased after coexpression with mCRY1, hCRY2, or a combination of the two. In contrast, coexpression with mPER3 dramatically promoted nuclear translocation of mPER1 (89% of nuclei are stained) (Fig. 6B, graph). Coexpression of mPER3 lacking the NLS [mPER3 (1–542)] with mPER1 not only failed to allow nuclear import, but instead promoted cytosolic localization of mPER1 (Fig. 6B, graph). As a control, we also analyzed mPER3-mediated import of mPER1 in wild-type MEFs containing endogenous mCRY proteins after serum shock. Comparable with the findings with mCry1/mCry2 double-deficient cells, 48% of the wild-type cells transfected with mPer1 show nuclear staining (Fig. 6C), whereas coexpression with mPER3 significantly promoted nuclear translocation of mPER1 (87%) and truncated mPER3 (1–542) prevented it (19.5%) (Fig. 6C). These results show that under the conditions tested, nuclear localization of mPER1 does not involve mCRY proteins and further support the hypothesis that mPER3 plays a prominent role in nuclear translocation of mPER1 after serum shock.
Figure 6.
Nuclear entry of mPER1/mPER3 in mCry1/mCry2 double knockout cells. The effect of mPER3 on nuclear entry of mPER1 was investigated in mCry1mCry2 double knockout cells. (A) Northern blot analysis of mCry1 (left) and mCry2 (right) MEFs from mCry1/mCry2 double knockout mouse and wild type. In the mCry1/mCry2 double knockout cells, no signals are seen at the appropriate molecular weight. (B) Analysis of nuclear translocation of mPER1 in mCry1/mCry2 double knockout cells after coexpression of mPER3, CRY1, CRY2, and/or mPER3 (1–542). The percentage of nuclear-positive cells is shown in the bar graph. About one-half of the cells (45%) show nuclear localization by single HA-mPER1 expression, perhaps due to partnering with endogenous proteins. Although transfection of CRY1/2 did not affect the nuclear entry of mPER1, cotransfection of mPER3 markedly increased nuclear entry of mPER1. Truncated mPER3 lacking the carboxy-terminal half including the NLS prevented the nuclear accumulation of mPER1. Confocal laser microscopic images are presented at right. (C) Control nuclear translocation experiments in MEFs originating from wild-type mice expressing endogenous mCry1 and mCry2. Note the very similar nuclear localization in all combinations in C, suggesting that mCRY proteins are not essential for nuclear translocation of mPER1. All results are the mean (+s.e.m.) of three independent experiments in B and C.
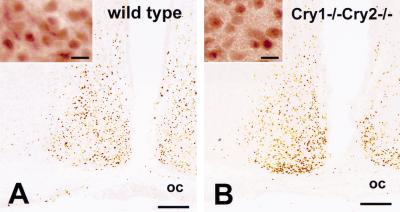
Nuclear localization of endogenous mPER1 protein in peripheral and central clock tissues of mCry1/mCry2 double-mutant mice
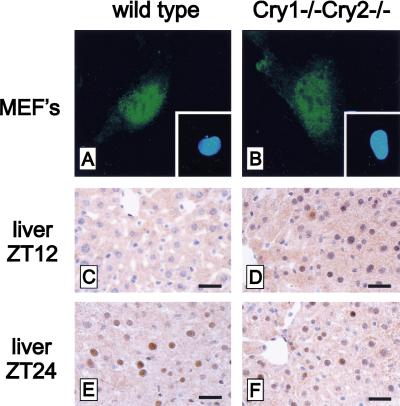
In the previous sections, in vitro evidence was obtained for nuclear localization of exogenously expressed mPER1 in mCry1/mCry2 double-deficient cells. To determine whether these results reflect the in vivo situation, we determined the expression and cellular localization of endogenous mPER1 protein in cells and tissues of wild-type and mCry1/mCry2 double-mutant mice using immunohistochemistry. In cultured embryonic fibroblasts, endogenous mPER1 was localized predominantly in the nucleus in both wild-type and mCry1/mCry2 double-mutant-derived cells (Fig. 7A,B). In the liver of wild-type mice, mPER1 could not be detected at ZT12, whereas a clear nuclear staining occurred at ZT24, which is indicative for the presence of a diurnal rhythm (Fig. 7C,E). In hepatocytes of mCry1/mCry2 double-deficient mice, endogenous mPER1 protein is constitutively expressed in the nucleus (Fig. 7D,F). Next, we examined the expression and localization of mPER1 protein in the SCN (the site of the master oscillator) of wild-type and mCry1/mCry2 double-mutant mice. In accordance with the literature (Hastings et al. 1999), we find in the SCN of wild-type mice high expression and nuclear localization of mPER1 at CT12–16 (see also Fig. 8A) and very low expression at CT2–6 (data not shown). In the SCN of mCry1/mCry2 double-mutant mice, mPER1 protein is expressed at a level comparable with that in wild-type mice at CT12–16 (Fig. 8B). In the absence of mCRY proteins, mPER1 protein can still be found in the nucleus (Fig. 8A,B, high magnification). These data clearly show that mCRY-independent nuclear translocation of mPER1 protein also occurs in the intact animal and are entirely consistent with the results of the cellular transfection studies.
Figure 7.
Nuclear localization of endogenous mPER1 in MEF's and liver of wild-type and mCry1/mCry2 double-mutant mice. Representative examples of immunofluorescent staining of mPER1 in wild-type (A) and mCry1/mCry2 double-mutant (B) MEFs (Insets) Hoechst stained nuclei. Immunohistochemical detection of mPER1 in the liver of a wild-type mouse at ZT12 (C), a mCry1/mCry2 double-mutant mouse at ZT12 (D), a wild-type mouse at ZT24 (E), and a mCry1/mCry2 double-mutant mouse at ZT24 (F). Animals were kept under LD conditions (12:12 hr). Bars in C–F 25 μm.
Figure 8.
Immunohistochemistry for endogenous mPER1 in the SCN of wild-type and mCry1/mCry2 double knockout mice. mPER1-immunoreactivity is found in neurons of the suprachiasmatic nucleus in both wild-type (A) and mCry1/mCry2 double-deficient (B) mice at ZT14. Note the absence of significant differences in immunoreactivity between wild-type and mCry1/mCry2 double knockout mice. Each high-magnification photograph shows that endogenous mPER1 is present in the nucleus of SCN cells in both wild-type (A) and mCry1/mCry2-deficient (B) mice. Bars, 100 μm for low-power micrographs, and 10 μm for high-power micrographs.
Discussion
Nuclear translocation of oscillator gene products is an essential step for the generation of a circadian negative feedback loop. Particularly, this nuclear entry step is considered to play a key role for determining the ∼24 hr period. In Drosophila, massive nuclear entry of PER occurs at about ZT18 (Young 1998) mediated by complexation with the TIM protein. In mammals, the circadian oscillator involves the products of three oscillator genes (mPer1, mPer2, and mPer3) (Albrecht et al. 1997; Shearman et al. 1997; Shigeyoshi et al. 1997; Sun et al. 1997; Tei et al. 1997; Takumi et al. 1998a,b; Zylka et al. 1998b). In the present study we show that in cultured COS7 cells constitutively expressing mPer cDNAs, mPER1 and mPER2 are predominantly localized in the cytoplasm. Similar results have been reported for NIH-3T3 cells (Kume et al. 1999). However, when COS7 cells are exposed to high levels of serum, a condition known to initiate rhythmic expression of rPer1 and rPer2 in cultured rat-1 cells (Balsalobre et al. 1998), mPER3 is shown to promote nuclear translocation, particularly of mPER1, and to a lesser extent of mPER2. This process appears to be coupled to a strong increase in heterodimerization of mPER proteins in all possible combinations (mPER1/mPER2, mPER1/mPER3, mPER2/mPER3). Because Drosophila PER can interact with each other through the PAS domains (Huang et al. 1995), interactions between mPER proteins may also involve these evolutionary highly conserved domains. The serum shock did not randomly facilitate mPER–mPER interactions, because mPER homodimers were detected at comparable levels before and after serum treatment. These data suggest that association with mPER3 forms a critical step in the nuclear translocation of mPER1 and mPER2. Accordingly, the rhythmic expression of rPer1 and rPer2 after serum shock observed by Balsalobre et al. (1998), starting with a brief, rapid induction followed by suppression of these clock genes, can be explained by rPER3 complex formation and nuclear translocation of rPER1 and rPER2. Recently, mPER1 and mPER3 were shown to coprecipitate from mouse SCN homogenates (Field et al., 2000), indicating that physical interactions between these proteins also occur under physiological conditions. Treatment of rat-1 cells with high concentrations of serum activate MAP kinases and CREB (Yagita and Okamura 2000). It is tentative to speculate that activation of cell signaling pathways may ultimately lead to phosphorylation of mPER proteins, which in turn may facilitate the interaction and subsequent nuclear entry of mPER proteins. It is noteworthy that in Drosophila, phosphorylation of dPER is essential for association with dTIM and nuclear translocation of the dPER/dTIM complex (Price et al. 1998).
mPER3 contains sequences similar to the CLD and NLS of Drosophila PER (Takumi et al. 1998b; Zylka et al. 1998b). In mPER1 and mPER2, the CLD is conserved, but these proteins do not possess a typical single basic NLS, although mPER2 may have a bipartite NLS (Shearman et al. 1997). Here we show both structural and functional conservation of the CLD and confirmed the dominance of the CLD over NLS activity in cellular localization of mPER3, as seen in Drosophila PER and TIM (Saez and Young 1996). According to the fly model for CLD function, PER and TIM associate to mask the CLD of PER, allowing TIM to escort PER into the nucleus (Vosshall et al. 1994; Saez and Young 1996). By analogy, we propose that in mammals, heterodimerization of mPER1/mPER3 or mPER2/mPER3 masks the CLD of each partner, allowing the NLS of mPER3 to direct the heterodimers to the nucleus.
The role of mammalian TIM in the nuclear transportation step of mPER proteins is unknown. In Drosophila, the subcellular localization of the TIM protein is regulated in a time-dependent fashion, but in mammalian SCN cells, mTIM is predominantly in the nucleus and does not exhibit circadian rhythmicity (Hastings et al. 1999). Analogously, COS7 cells used in the present study constitutively express significant levels of endogenous mTim mRNA, even in the absence of a serum shock (data not shown). mPER3-dependent nuclear translocation seems not to correlate with the level of mTim expression, because mTim mRNA levels are high in COS7 and wild-type embryonic fibroblasts, but low in rat-1 cells and mCry1/mCry2 double knockout embryonic fibroblasts (data not shown).
According to the basic model for the molecular mechanism of the circadian pacemaker, mPER proteins—when in the nucleus—repress CLOCK/BMAL-mediated transcriptional activation of various clock genes (for review, see Reppert 1998; Dunlap 1999). Although mPER proteins inhibit transcription from mPer1- or vasopressin-promoter driven luciferase reporter constructs, repression is rarely complete (e.g., Sangoram et al. 1998; Zylka et al. 1998a; data not shown), indicating that additional factors are required to completely suppress CLOCK/BMAL function. Recently, mCRY1 and mCRY2, members of the light-harvesting cryptochrome/photolyase protein family (Hsu et al. 1996; Todo et al. 1996; van der Spek et al. 1996), were shown to be indispensable components of the molecular oscillator as mice with inactivated mCry1 and mCry2 genes completely lacking a biological clock (Okamura et al. 1999; van der Horst et al. 1999; Vitaterna et al. 1999). Subsequently, Kume et al. (1999) found that mCRY proteins strongly inhibit mPer1 and vasopressin promoter-driven luciferase expression in NIH-3T3 cells and that they act as dimerization partners for translocation of mPER1, mPER2, and mPER3 into the nucleus. However, heterodimerization of mPER by itself was reported to affect cellular localization also. Our findings extend the significance of mPER–mPER interactions for nuclear translocation: mPER3 promotes the nuclear entry of mPER1 and mPER2. The pronounced nuclear localization of exogenous mPER1 in the mCRY-deficient MEFs after serum shock indicates that mPER3 can also accomplish nuclear entry of mPER1 without the help of mCRY proteins. Remarkably, under our experimental conditions, overexpression of mCRY1 or hCRY2 or both in mCry1/mCry2 double-mutant cells did not increase the mPER1-nuclear entry. In contrast, mPER3 transfection enhances mPER1 nuclear import in mCRY-deficient cells, whereas NLS-deleted mPER3 transfection markedly decreased the nuclear localization. Thus, it appears that there may be more than one route for mPER protein import into the nucleus. Moreover, immunocytochemical analysis using anti-mPER1-specific antisera revealed that endogenous mPER1 also localized in the nucleus in cultured mCry1/mCry2 double-mutant cells, indicating that mCRY-independent nuclear translocation of exogenous mPER1 is not merely an artifact in cellular transfection studies. Importantly, we also demonstrated mCRY-independent nuclear localization of endogenous mPER1 in SCN and liver of mCry1/mCry2 double-mutant mice and thus is likely to be of physiological importance.
The absence of an effect of mCRY proteins on mPER subcellular localization when coexpressed in the mCry1/mCry2 double-deficient cells may be explained in various ways. First, we have used a serum shock, known to stimulate rhythmic expression of clock genes in cultured cells, which may involve additional or different factors for nuclear translocation of clock gene products. Second, different cell lines and different conditions were used. The COS7 cells used here are exceptional because they hardly express any endogenous mPer genes, virtually no mCry2, and moderate levels of mCry1 transcripts (K. Yagita and H. Okamura, unpubl.). In contrast, most other frequently used cell lines including rat-1 and NIH-3T3 fibroblasts highly express all endogenous mPer genes and NIH-3T3 cells and also highly express both mCry genes (K. Yagita and H. Okamura, unpubl.). As discussed above, it is conceivable that these differences in endogenous mPER and mCRY protein levels have an effect on the exogenous proteins. The nuclear localization of mPER1 in mCRY-deficient cells and tissues shows that the mCRY proteins are not indispensable for nuclear translocation of mPER1. Although mCRY proteins are very strong transcription suppressors of mPer1 in NIH-3T3 cells (Griffin et al. 1999; Kume et al. 1999) and in HepG2 cells (Yamaguchi et al., 2000), their role in nuclear migration of other clock proteins needs further exploration.
In conclusion, we have shown that there may be more than one route for mPER proteins to reach the nucleus. In addition to mCRY-mediated nuclear migration of mPERs, our data are consistent with a model in which nuclear entry is driven by masking of the CLD of mPER proteins through heterodimer formation of mPER3 with mPER1 or mPER2. In this scenario, the roles of the CLD and NLS in nuclear entry of clock components as described in Drosophila would be conserved in mammals, but the part played by TIM as the dimerizing partner for mPER1 and mPER2 might be taken over by mPER3.
Materials and methods
Plasmids
For construction of mPer1-Flag in a mammalian expression vector, the primers used were as follows, 5′-AAGCTTCCAGACATGAGTGGTCCCCTAGAAG-3′, 5′-GAATTCGCTGGTGCTGTTTTCTTCTGCAGGTAAAGC-3′. For construction of mPer2-Flag, nucleotides 1–3740 (codon 1–1207) were digested from a full-length cDNA of mPer2 by use of NotI and ClaI, and this fragment was subcloned into the pcDNA3–Flag vector. Nucleotides 1–3465 (codon 1–1077) of mPer3 were isolated from a full-length cDNA of mPer3 by use of NotI and BamHI and were subcloned into pcDNA3.1–Myc/His vector (Invitrogen). HA-mPer1, HA-mPer2, and HA-mPer3 constructions were gifts from Dr. T. Kishimoto (Sumitomo Electric Industries). Total coding regions of mPer1, mPer2, and mPer3 were cloned by PCR by use of an HA epitope sequence fused in 5′ primers, and subcloned into pTargeT vector (Promega).
For the construction of mPER3 (1–542)–Myc, total mPer3 cDNA cloned into pGEM-T Easy (Promega) was digested by NotI and KpnI, and cloned into pcDNA3.1–Myc/His vector.
The total coding region of mCry1 was obtained by RT–PCR and cloned into pcDNA3 vector (Invitrogen). The plasmid containing the partial human Cry2 was generously provided by Takahiro Nagase (KAZUSA DNA Research Institute). As the KIAA0658 gene lacks the 5′ end of the human Cry2-coding region, the further upstream 5′ fragment was obtained by PCR according to an EST clone (accession no. AL040215). The total coding region of human Cry2 was cloned into pcDNA3 vector.
Cell culture, transfection, and serum shock
COS7 cells were grown in DMEM supplemented with 10% FBS (GIBCO BRL) and penicillin–streptomycin (GIBCO BRL). MEF's were established from day 13.5 mCry1/mCry2 double-knockout and wild-type mouse embryos. Cells were grown in the DMEM containing 10% FCS at 37°C under 5% CO2. Cells were transfected with Lipofectamine-Plus (GIBCO BRL). Two and one-half hours after transfection, the medium was replaced with 5 ml of DMEM with 10% FCS. After 12 hr, medium was replaced with serum-free DMEM and incubated for 12 hr. Thirty minutes before harvest, cells were treated with DMEM containing 50% horse serum.
Immunoprecipitation and immunoblot
Cells were plated into 60-mm plates at 2 × 105 cells 24 hr prior to transfection. Cells in each plate were transfected with 2 μg (total) of DNA.
Immunoprecipitation was performed 24 hr after transfection by use of whole-cell lysates harvested with 0.2 ml of lysis buffer [50 mm Tris-HCl at pH 7.5, 150 mm NaCl, 1% NP 40, 50 mm NaF, 100 μm sodium vanadate, and complete mini-protease inhibitors (Boehringer)]. Total cell lysate was kept on ice for 1 hr and centrifuged at top speed for 10 min at 4°C, 10 μl of total-cell extract was used for each Western blot analysis. The supernatant was transferred to a fresh microtube and anti-Flag M2 (Kodak) or anti-HA antibodies (Boehringer) were added and incubated 4 hr at 4°C with mild agitation. After adding 30 μl of Protein-G Agarose equilibrated with lysis buffer, beads were collected (15,000 rpm, 20 sec) and washed twice with wash buffer (20 mm Tris-HCl at pH 7.5, 500 mm NaCl, 0.1% NP-40, 0.05% sodium deoxycholate). After removing the supernatant completely, 8 μl of 3 × SDS sample buffer was added and samples were boiled for 5 min. Immunoprecipitated samples were separated on 7% SDS–polyacrylamide gel, and transferred to polyvinylidene difluoride (PVDF) membranes. Immunoblotting was performed with anti-Flag M2 (Kodak, 500× dilution), anti-Myc (9E10; Santa Cruz, 5000× dilution), or anti-HA (Boehringer, 2000× dilution) antisera. Chemiluminescence was performed by use of Renaissance Western blot reagent plus (NEN, cat. no. NEL105).
GFP fusion proteins
The total coding region of mPer3 was cloned into pEGFP-N2 vector (Clontech). For construction of deletion mutants of mPER3 fused with GFP proteins, total mPer3 cDNA was digested by BamHI or DraI/BamHI or KpnI/BamHI. BamHI fragment (amino acids 228–1077), DraI–BamHI fragment (amino acids 324–1077), and KpnI–BamHI fragment (amino acids 542–1077) were cloned into pEGFP-C vectors (Clontech). For the clone mPER3 (ΔCLD), the total coding region of mPer3 was digested with DraI and NcoI, and then the PCR fragment encoding amino acids 395–476 was ligated in. This construct was cloned into the pEGFP-N2 vector.
COS7 cells were plated onto 60-mm plates at 2 × 105 cells 24 hr prior to the transfection. Cells transfected with indicated constructs were cultured for 24 hr after the transfection. Cells were harvested with 0.2 ml of lysis buffer. Total-cell lysates were prepared as described above. One hundred microliters of 3× SDS sample buffer was added and samples were boiled for 5 min. Ten microliters of samples were separated on 7% SDS–polyacrylamide gel, and transferred to a PVDF membrane. Immunoblotting was performed as described with anti-GFP (Clontech, 200× dilution), and detected by chemiluminescence.
Northern blot analysis
MEFs derived from both mCry1/2 double knock out and wild type were harvested in the TRIzol regent (GIBCO BRL). Ten micrograms of extracted total RNA was electrophoresed in a 1.2% agarose gel containing 2% formaldehyde. RNAs were transferred to Biodyne Nylon Membrane (Pall BioSupport, NY) and hybridized with probes. For probes, 730–1479 bp of mCry1 and 397–810 bp of mCry2 were used for templates. Probes were incubated with membranes at 42°C overnight, washed twice in 0.2× SSC/0.1% SDS at 60°C for 30 min, exposed to Imaging Plates, and analyzed by BAS 5000 (Fuji Film, Tokyo, Japan).
Antisera against mPER1
Polyclonal rabbit antibodies against mPER1 (code no. RY354) were raised using a synthetic peptide (GSSGNESNGPESRGASQRC) corresponding to amino acids 51–68 of the deduced amino acid sequence of mPer1 cDNA (Tei et al. 1997) and containing a carboxy-terminal cysteine residue for conjugation to keyhole limpet hemocyanin for immunization. Immunizations were performed using Freund's complete adjuvant for primary injection and Freund's incomplete adjuvant for boosts (given at 2-week intervals). Rabbits were bled from the marginal vein at 10 days after the fourth immunization. Peptide synthesis and subsequent immunization protocols were conducted at Yanaihara Institute (Fujinomiya, Japan). This antiserum was checked by immunoblots of nuclear extracts of mouse brain and the mPER1, mPER2, and mPER3 expression plasmids transfected with COS7 cells. In the mouse brain and mPER1-expressing COS7 cells, we detected a single band corresponding to the predicted molecular weight. Positive signals were not detected in mPER2 and mPER3-expressing COS7 cells. This antiserum was used for the SCN analysis.
The anti-mPER1 antiserum (Field et al. 1999; Hastings et al. 1999) used for MEFs and liver tissues was kindly provided by Dr. S.M. Reppert (Massachusetts General Hospital, Boston, MA).
Immunofluorescence of cultured cells
COS7 cells were grown on 24 × 24-mm cover glasses and transfected with 1 μg (total) of DNA by using of Lipofectamine-Plus (GIBCO BRL). Transfected cells were cultured in DMEM containing 10% FBS (12 hr) and then serum-free DMEM (12 hr). Serum shock was performed as described for indicated samples before fixation. Transfected cells were fixed in 4% paraformaldehyde in 0.1 m phosphate buffer. Fixed cells were permeabilized by 0.2% Triton-X in 0.1 m PBS for 5 min, and incubated with primary antibodies (anti-Flag M2 500×, anti-HA 1000×; for 16 hr at 4°C). After washing in PBS, FITC-conjugated anti-mouse IgG (Jackson Immuno Research) and/or Cy3-conjugated anti-rat IgG (Amersham) were used as the secondary antibodies in 1-hr incubations at room temperature. Cells were then washed and mounted with glycerol on a cover glass. Cells were observed by confocal laser microscopy (Bio-Rad).
Subcellular localization was assayed by examining immunoreactive cells that were classified into two groups, either cytoplasm dominant (C) in which the intensity of cytoplasmic staining was stronger than that in nucleus or the reverse (N nucleus dominant). We performed the above calculation in all 100 cells examined three to five times in each independent experiment.
Wild-type and mCry1/mCry2 double-mutant MEFs were cultured in DMEM/F10 with 10% FCS and seeded on glass coverslips 2 days before the immunohistochemistry. Cells were fixed in 0.1 m PBS containing 3% paraformaldehyde for 7 min, followed by a permeabilization step in 100% methanol for 20 min. After incubation with anti-mPER1 antibodies (16 hr, 4°C, 1:2000 dilution), washing, incubation with a secondary fluorescein-conjugated anti-rabbit antibody (1:100 dilution; DAKO) and washing, cells were analyzed under a fluorescence microscope. All steps, except incubation with anti-mPER1 antibodies, were performed at room temperature. Nuclei were counterstained with Hoechst solution at a dilution of 1:2000.
Immunohistochemistry of liver and brain
Animals were housed under 12-hr bright white light:12-hr dark condition. Zeitgeber time (ZT) was defined relative to lights on (ZT0) and lights off (ZT12). Animal experiments were performed under the approval of the Committee for Animal Research at Kobe University School of Medicine and the Animal Care and Use Committee at the Erasmus University Rotterdam.
For analysis of the liver, animals were decapitated at ZT12 and ZT24. Livers were immediately perfused with 0.1 m PBS, followed by overnight fixation in 4% paraformaldehyde, and dehydrated. After dehydration, samples were embedded in paraffin and cut into 7-μm sections. Endogenous peroxidase was inhibited by hydrogen peroxide-sodium azide treatment (0.6% H2O2 and 0.125% sodium azide in PBS for 30 min). Endogenous biotin was blocked by subsequent incubation with avidin and biotin (10 min each; biotin blocking system, DAKO). Sections were incubated with a primary rabbit anti-mPER1 antibody (1:2000 dilution), followed by a secondary biotin-conjugated anti-rabbit antibody (1:500). After extensive washing, the mPER1 immunoreactivity was visualized using streptavidin-conjugated peroxidase (Zymed), followed by a DAB (diaminbenzidine chromogen) reaction. Nuclei were counterstained with hematoxilin for 30 sec.
For the SCN, animals were perfused with saline, followed by a fixative (2% paraformaldehyde and 0.2% picric acid in 0.1 m phosphate buffer) under deep ether anesthesia at ZT14, and then post-fixed with the same fixative for 2 hr, then 30-μm-thick sections were cut in a cryostat and processed for free-floating immunohistochemistry as described previously (Yagita et al. 1994). After an application of biotinylated anti-rabbit IgG (1:500), sections were incubated with avidin-biotin-peroxidase (1:500, Vector) and visualized with DAB. After immunoreaction, SCN sections were mounted onto chrome alum/gelatin-coated slides, dehydrated through a graded series of ethanol, and mounted with Entellan.
Acknowledgments
We thank Y. Takahashi (Kobe University) for technical advice, M. Muijtjens (Erasmus University) for establishing the MEF lines, Dr. S.M. Reppert (Massachusetts General Hospital, Boston, MA) for kindly providing mPER1 antibodies for analyses of liver and cultural MEFs, T. Kishimoto (Sumitomo Electric Industries) for donating HA-tagged mPer1, mPer2, and mPer3, Y. Minami (Kobe University) for donating the Flag-fused pcDNA3 vector, and Y. Shigeyoshi (Kinki University) for discussion. This work was supported by research grants to H.O. from the Special Coordination Funds of the Japanese Science and Technology Agency, the Grant-in-Aid for the Scientific Research on Priority Areas of the Ministry of Education, Science, Sports, and Culture of Japan, Mitsubishi Foundation, SRF and Uehara Memorial Foundation, to J.J.L. from the National Science Foundation (MCB-9307299), to J.C.D. and J.J.L. from the National Institute of Health (R01-MH 44651), to J.C.D. from the National Institute of Health (R37-GM 34985 and K05-MH01186), to J.H.J.H. and G.T.J.vd.H. from the Dutch cancer society. J.H.J.H. is recipient of the SPINOZA award of the Dutch Science Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL okamurah@kobe-u.ac.jp; FAX 81 78 382 5341.
References
- Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mPer1 and mPer2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Field MD, Maywood ES, O'brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Hastings MH. Central clocking. Trends Neurosci. 1997;20:459–464. doi: 10.1016/s0166-2236(97)01087-4. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: New insights into a core clock mechanism. J Neurosci. 1999;11:1–7. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang R-P, Toda T, Wei Y-F, Sancar A. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35:13871–13877. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Curtin KD, Rosbash M. PER protein interactions and temperature compensation of a circadian clock in Drosophila. Science. 1995;267:1169–1172. doi: 10.1126/science.7855598. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Luo C, Loros JJ, Dunlap JC. Nuclear localization is required for function of the essential clock protein FRQ. EMBO J. 1998;17:1228–1235. doi: 10.1093/emboj/17.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JHJ, van der Horst GTJ. Photic induction of mPer1 and mPer2 in Cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. Double-time is a new Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–96. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Reppert SM. A clockwork explosion. Neuron. 1998;21:1–4. doi: 10.1016/s0896-6273(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Saez L, Young MW. Regulation of nuclear entry of the drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Shimomura K, King DP, et al. Mammalian circadian autoregulatory loop: A timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Zylka MJ, Weaver DR, Kolakowski LFJ, Reppert SM. Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y, Sakakida Y, Okumura K, Takashima N, Okamura H. A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells. 1998a;3:167–176. doi: 10.1046/j.1365-2443.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, et al. A light independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998b;17:4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takumi T, Nagamine Y, Miyake S, Matsubara C, Taguchi K, Takekida S, Sakakida Y, Nishikawa K, Kishimoto T, Niwa S, et al. A mammalian ortholog of Drosophila timeless, highly expressed in SCN and retina, forms a complex with mPER1. Genes Cells. 1999;4:67–75. doi: 10.1046/j.1365-2443.1999.00238.x. [DOI] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- Todo T, Ryo H, Yamamoto K, Toh H, Inui T, Ayaki H, Nomura T, Ikenaga M. Similarity among the drosophila (6-4)photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- van der Horst GTJ, Muijtjens M, Kobayashi K, Takano R, Kanno S-I, Takao M, Wit JD, Verkerk A, Eker APM, Leenen DV, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- van der Spek PJ, Kobayashi K, Bootsma D, Takao M, Eker APM, Yasui A. Cloning, tissue expression, and mapping of a human photolyase homolog with similarity to plant blue-light receptors. Genomics. 1996;37:177–182. doi: 10.1006/geno.1996.0539. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Price JL, Sehgal A, Saez L, Young MW. Block in nuclear localization of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- Whitmore D, Sassone-Corsi P, Foulkes NS. PASting together the mammalian clock. Curr Opin Neurosci. 1998;8:635–641. doi: 10.1016/s0959-4388(98)80092-5. [DOI] [PubMed] [Google Scholar]
- Yagita K, Okamura H. Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 2000;465:79–82. doi: 10.1016/s0014-5793(99)01724-x. [DOI] [PubMed] [Google Scholar]
- Yagita K, Okamura H, Ibata Y. Rehydration process from salt-loading: Recovery of vasopressin and its coexisting galanin, dynorphin and tyrosine hydroxylase immunoreactivities in the supraoptic and paraventricular nuclei. Brain Res. 1994;667:13–23. doi: 10.1016/0006-8993(94)91708-6. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., S. Mitsui, L. Yan, K. Yagita, S. Miyake, and H. Okamura. 2000. Role of DBP in the Circadian Oscillatory Mechanism. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Young MW. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu Rev Biochem. 1998;67:135–152. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Levine JD, Jin X, Weaver DR, Reppert SM. Molecular analysis of mammalian timeless. Neuron. 1998a;21:1115–1122. doi: 10.1016/s0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homolog in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998b;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]