Abstract
The highly conserved protein kinase casein kinase II (CKII) is required for efficient Pol III transcription of the tRNA and 5S rRNA genes in Saccharomyces cerevisiae. Using purified factors from wild-type cells to complement transcription extracts from a conditional lethal mutant of CKII we show that TFIIIB is the CKII-responsive component of the Pol III transcription machinery. Dephosphorylation of TFIIIB eliminated its ability to complement CKII-depleted extract, and a single TFIIIB subunit, the TATA-binding protein (TBP), is a preferred substrate of CKII in vitro. Recombinant TBP purified from Escherichia coli is phosphorylated efficiently by CKII and, in the presence of a limiting amount of CKII, is able to substantially rescue transcription in CKII-deficient extract. Our results establish that TBP is a key component of the pathway linking CKII activity and Pol III transcription and suggest that TBP is the target of a CKII-mediated regulatory mechanism that can modulate Pol III transcription.
Keywords: Transcription, RNA polymerase III, TFIIIB, TBP, casein kinase II
The genes transcribed by RNA polymerase III (Pol III), like many transcribed by RNA Pol II, are strictly regulated according to cell cycle position, growth rate, and developmental phase (Hoeffler et al. 1988; Tower and Sollner-Webb 1988; Wolffe and Brown 1988; White et al. 1989, 1995a,b; Hartl et al. 1993; Reynolds 1993; Gottesfeld et al. 1994; Sethy et al. 1995). Pol III transcription also responds to a number of pathological conditions. For example, oncogenic transformation is generally associated with increased Pol III transcription (discussion in White et al. 1990, 1996). The regulation of class III gene expression operates principally at the level of the transcription initiation machinery, which includes three fundamental protein components, RNA Pol III, TFIIIB, and TFIIIC (for review, see Geiduschek and Kassavetis 1992; Willis 1993). TFIIIB is a complex of the highly conserved TATA-binding protein (TBP) with at least two other proteins generally referred to as TBP-associated factors or TAFs. There are two Pol III-specific TAFs in Saccharomyces cerevisiae (TFIIIB70/Brf1p and TFIIIB90/Tfc5p; gene identification summarized in Kumar et al. 1997), and one of these, Brf1p, is conserved in human (Wang and Roeder 1995). TFIIIC, composed of six subunits in yeast, is the principal sequence-specific DNA-binding component of the Pol III machinery (for review, see Geiduschek and Kassavetis 1992; Willis 1993).
TFIIIB and TFIIIC are the primary targets of the regulatory mechanisms that affect Pol III transcription. For example, TFIIIB is the target of the mechanism that represses Pol III transcription in metaphase extracts from Xenopus eggs (Hartl et al. 1993; Gottesfeld et al. 1994; Leresche et al. 1996) and mammalian tissue culture cells (White et al. 1995b). In mammalian cells and yeast both TFIIIB and TFIIIC are limiting in stationary phase (Tower and Sollner-Webb 1988; Sethy et al. 1995) and TFIIIC is limiting early in S phase in mammalian cells (White et al. 1995a). In the case of TFIIIB, the regulatory functions of individual subunits are becoming clear. The TBP subunit of Xenopus TFIIIB may be repressed in metaphase extracts (Leresche et al. 1996), and TBP is limiting for Pol III transcription in Drosophila tissue culture cells (Trivedi et al. 1996). In mammalian cells the metaphase silencing of Pol III transcription results from repression of one or more of the Pol III TAFs (White et al. 1995b), and a decline in Brf1p expression partly accounts for decreased TFIIIB activity in the stationary phase of yeast (Sethy et al. 1995). Considering these results, there has been intense interest in identifying the signaling pathways that impinge on the Pol III transcription machinery. A substantial body of evidence suggests that these signaling pathways involve protein phosphorylation events. As a result, protein kinases and phosphatases involved in the regulation of Pol III transcription are actively being sought.
One highly conserved S/T/Y protein kinase implicated in the regulation of Pol III transcription is casein kinase II (CKII; for review, see Pinna 1990; Tuazon and Traugh 1991; Litchfield and Lüscher 1993; Allende and Allende 1995). The CKII holoenzyme, a heterotetramer of two catalytic α subunits (αα′) and two noncatalytic β subunits (2β or ββ′), is generally implicated in transcriptional regulation by the observation that RNA synthesis is severely inhibited when a temperature-sensitive mutant of yeast CKIIα′ is shifted to the restrictive temperature (Hanna et al. 1995). We further reported that efficient Pol III transcription of the yeast 5S rRNA and tRNA genes requires CKII, and we have presented evidence suggesting that CKII activity is required for efficient initiation (Hockman and Schultz 1996). Taking advantage of the ability to prepare a CKII- and Pol III-deficient extract from the yeast temperature-sensitive mutant of CKII, we have pursued a biochemical approach aimed at identifying the component of the Pol III transcription machinery that responds to CKII inactivation. We show that the transcription factor TFIIIB is specifically defective in extracts depleted of CKII and that TBP, the central transcription factor in the nucleus, is the target of CKII among the subunits of TFIIIB.
Results
Transcription in CKII-deficient extract is rescued by TFIIIB
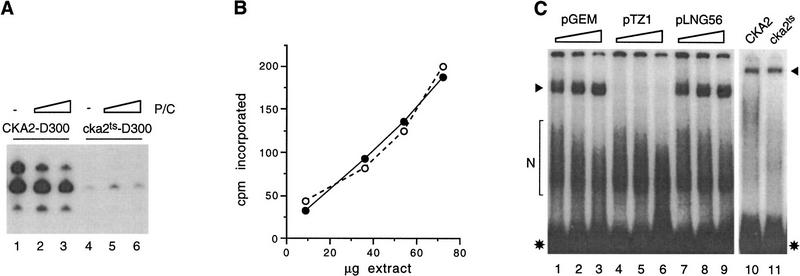
Two isogenic strains, designated CKA2 and cka2ts (temperature-sensitive), were used in this study. These strains differ only in the sequence of the CKA2 gene (encoding CKIIα′), which is wild type in CKA2 but carries mutations that confer a temperature-sensitive–lethal phenotype in strain cka2ts (Hanna et al. 1995). Whether the starting material is lysed cells (Hockman and Schultz 1996) or isolated nuclei (not shown), CKII activity in transcription extracts from strain cka2ts is ∼20-fold lower than wild type. To identify the CKII-responsive component of the Pol III transcription machinery, active transcription factors purified from wild-type extract were added to cka2ts extract with low CKII activity and impaired specific transcription. Initially we prepared two complementing fractions from wild-type cells, one enriched in Pol III/TFIIIC (fraction P/C) and another enriched in TFIIIB (Fig. 1A). Both wild-type and cka2ts extracts were responsive to TFIIIB, although the magnitude of the response differed significantly between the extracts. TFIIIB slightly stimulated transcription in wild-type extract (Fig. 1B, lanes 1–4), in agreement with previous reports (see Sethy et al. 1995). In contrast, TFIIIB strongly stimulated transcription in cka2ts extract; the purified factor was able to restore activity in the mutant to that of wild type supplemented with TFIIIB (Fig. 1B, lanes 5–8). Based on this result we propose that the defect in cka2ts extract results primarily from inactivation of TFIIIB.
Figure 1.
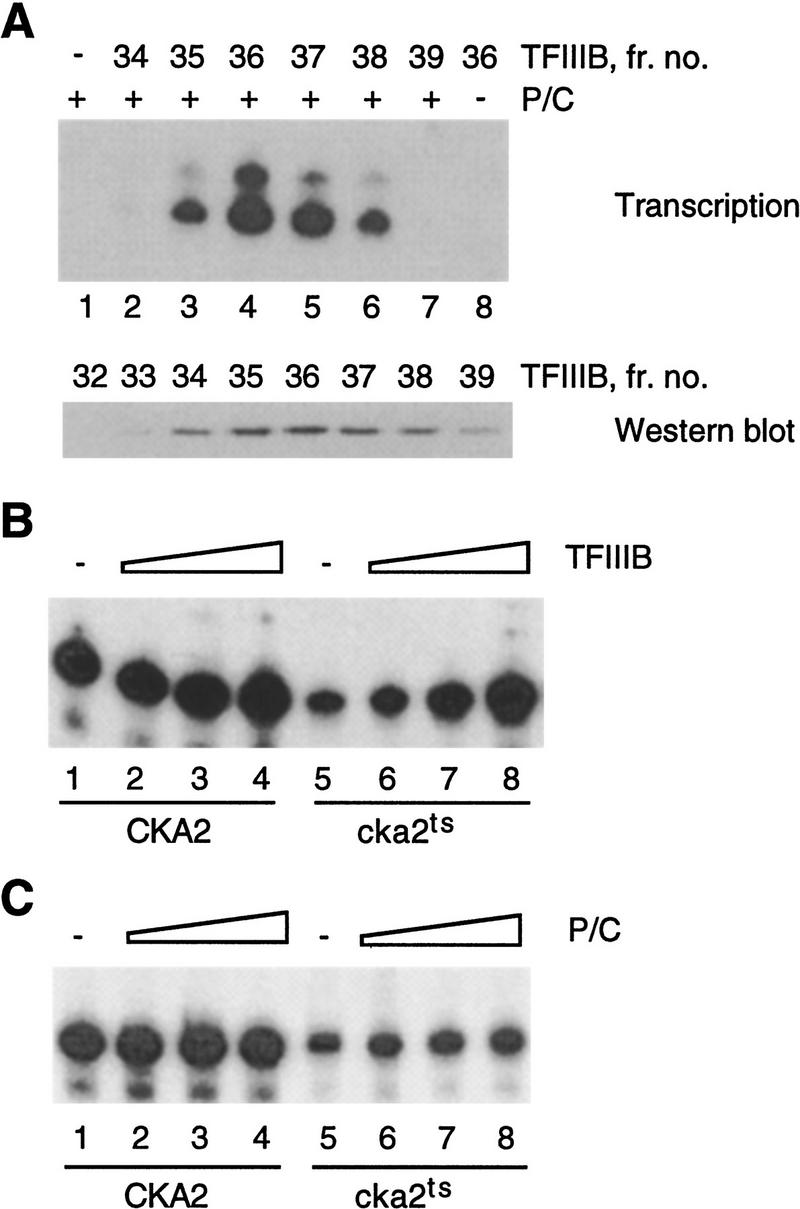
Pol III transcription in CKII-deficient whole cell extract is rescued by TFIIIB. (A) (Top) The P/C and TFIIIB fractions reconstitute Pol III transcription. tRNA transcription (20 ng template/reaction) was assayed using 4 μl of P/C, either added to 10-μl aliquots of fractions from the hydroxyapatite column used to purify TFIIIB (lanes 1–7), or on its own (lane 8). (Bottom) The peak of TFIIIB activity obtained by hydroxyapatite chromatography corresponds to the peak of TBP. Immunoblot of TFIIIB fractions (12-μl aliquots) using antiserum raised against recombinant TBP. (B) TFIIIB (0, 0.4, 1, and 2.5 μl) slightly stimulates 5S rRNA transcription in CKA2 extract (lanes 1–4) and fully restores transcription in cka2ts extract (lanes 5–8). (C) The P/C fraction (0, 0.8, 2 and 5 μl) does not significantly stimulate 5S rRNA transcription in CKA2 or cka2ts extract. The reactions in B and C used 60 μg of extract and plasmid pY5S at 400 ng/reaction.
This conclusion is supported by complementation experiments using the P/C (Pol III/TFIIIC) fraction. Titration of fraction P/C into wild-type extract was either without effect (not shown) or very slightly stimulated transcription (Fig. 1C, lanes 1–4). The P/C fraction slightly stimulated cka2ts extract, but it did not restore transcription to the wild-type level (Fig. 1C, lanes 5–8). This result suggests that neither RNA Pol III nor TFIIIC is significantly affected by the loss of CKII activity.
The Pol III transcription machinery can be partially purified by DEAE chromatography (Riggs and Nomura 1990). This procedure provides a 300 mm KCl cut from DEAE, referred to as fraction D300, in which the difference in transcription capacity between wild-type and cka2ts extracts is accentuated (Fig. 2A). In view of the results in Figure 1, we reasoned that a cka2ts–D300 fraction that is severely impaired compared to the wild type fraction (prepared in parallel) would be highly sensitive to the addition of TFIIIB. TFIIIB massively stimulates transcription in cka2ts–D300 (Fig. 2B). To show that this stimulatory activity corresponds to TFIIIB we supplemented cka2ts–D300 with aliquots of fractions from the hydroxyapatite column used for TFIIIB purification. The activity that stimulates cka2ts–D300 occurs in fractions 34–38, with the peak in fraction 36 and no activity in fraction 39 (Fig. 2C). This profile exactly corresponds to the profile of TFIIIB activity that complements the wild-type P/C fraction (Fig. 1A).
Figure 2.
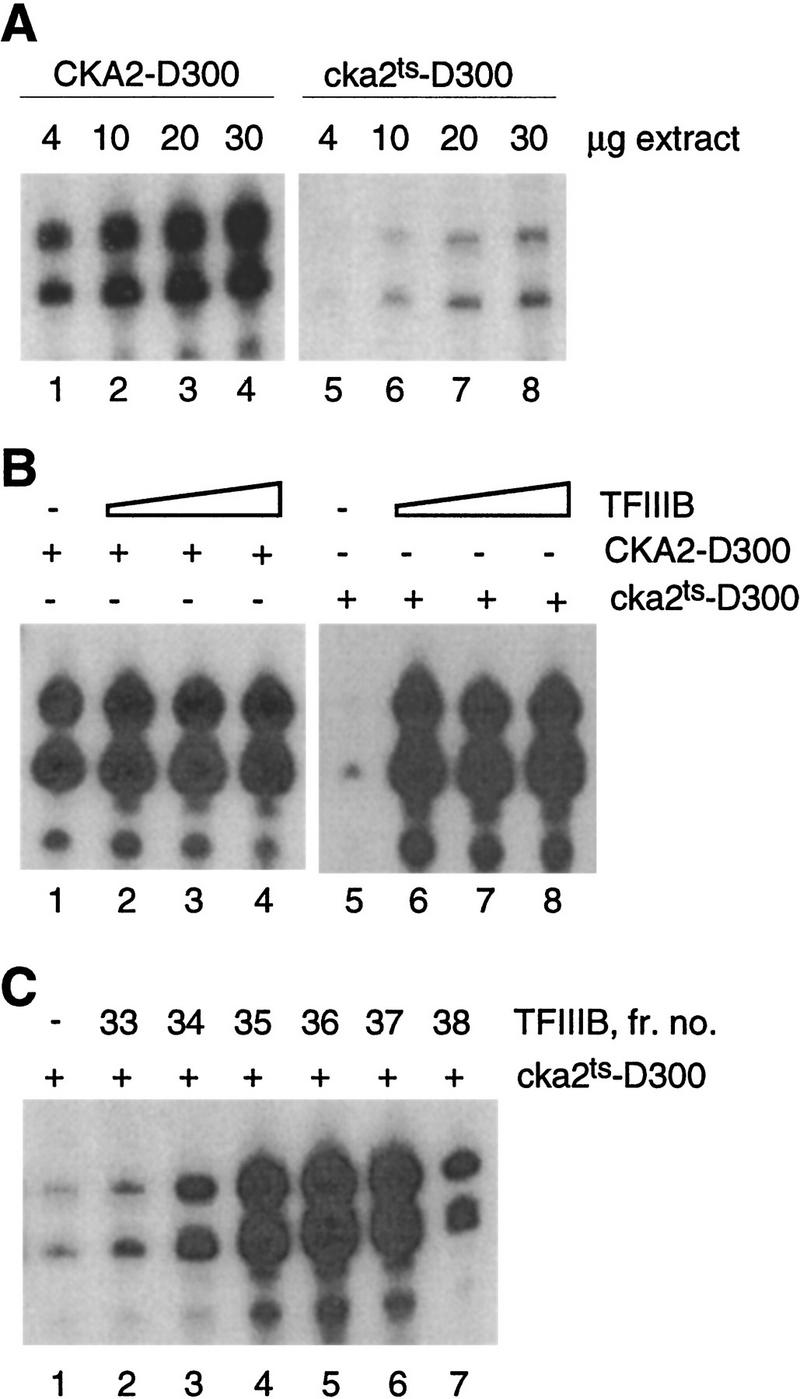
The transcription defect in CKII-depleted extract is preserved after partial purification of the Pol III transcription machinery and is fully rescued by TFIIIB. (A) tRNA transcription in the 300 mm KCl DEAE fraction from CKA2 and cka2ts extracts. (B) TFIIIB marginally stimulates tRNA transcription in CKA2–D300 (lanes 1–4) and fully restores transcription in cka2ts–D300 (lanes 5–8). The reactions received 0, 2, 5, or 10 μl of hydroxyapatite TFIIIB and 13 μg of D300 fraction. (C) The peak of TFIIIB activity (fraction 36, Fig. 1A) corresponds to the peak of activity capable of rescuing transcription in cka2ts–D300 (lane 5). Transcription was assayed using 13 μg of cka2ts–D300 added to 10-μl aliquots of fractions from the hydroxyapatite column used to purify TFIIIB. The template was at 20 ng/reaction.
The results of adding P/C to whole cell extracts of CKA2 and cka2ts suggest that if either Pol III or TFIIIC is inactivated in cka2ts, the effect is minimal. This interpretation is supported by the results obtained when the P/C fraction is titrated into the respective D300 fractions: P/C produced a slight inhibition of transcription in CKA2–D300 (Fig. 3A, lanes 1–3) and marginally stimulated cka2ts–D300 (lanes 4–6). In further support of the conclusion that neither Pol III nor TFIIIC is significantly inhibited in cka2ts, we observed that bulk Pol III activity (elongation) and DNA binding by TFIIIC are similar in CKA2 and cka2ts extracts. Bulk Pol III activity, assayed using nonspecific duplex DNA as the template, was virtually identical in CKA2 and cka2ts whole cell extracts (Fig. 3B), as in nuclear extracts and the D300 fractions (data not shown). Based on previous work (Kassavetis et al. 1989) we established a DNA-binding assay for TFIIIC present in the D300 fraction (Fig. 3C, lanes 1–9). Using this assay we could not detect any difference in DNA binding by TFIIIC in CKA2 and cka2ts extracts (Fig. 3C, lanes 10,11). Taken together, the results in Figures 1, 2, 3 suggest that inactivation of CKII results in specific inactivation of the TFIIIB component of the Pol III transcription machinery.
Figure 3.
The transcription defect observed after partial purification of the Pol III transcription machinery from CKII-depleted extract is not due to inactivation of Pol III or TFIIIC. (A) The P/C fraction inhibits tRNA transcription in the 300 mm KCl DEAE fraction from CKA2 extract (lanes 1–4) and only marginally stimulates transcription in the corresponding fraction from cka2ts extract (lanes 5–8). All reactions received 13 μg of the D300 fraction, 20 ng of template, and 0, 4, or 8 μl of P/C. (B) Bulk Pol III activity in CKA2 (•) and cka2ts (○) whole cell extracts, expressed as counts per min (cpm) incorporated during a 25-min reaction at room temperature. (C) TFIIIC DNA-binding activity in the D300 fraction of wild-type extract (3.5 μg; lanes 1–9). The probe was an end-labeled restriction fragment from pTZ1 (4.4 ng/reaction). Specificity of binding was established using increasing amounts (700, 1000, 1700 ng) of nonspecific competitor (pGEM3), specific competitor (pTZ1), or a binding site mutant competitor (pLNG56). Lanes 10 and 11 compare TFIIIC binding in the D300 fractions from CKA2 and cka2ts cells. The specific TFIIIC shift (arrowheads) and free probe (asterisks) are indicated. Some nonspecific binding to probe DNA was observed in all experiments (N).
TFIIIB does not contain an activity that can restore the enzymatic function of temperature-sensitive CKII
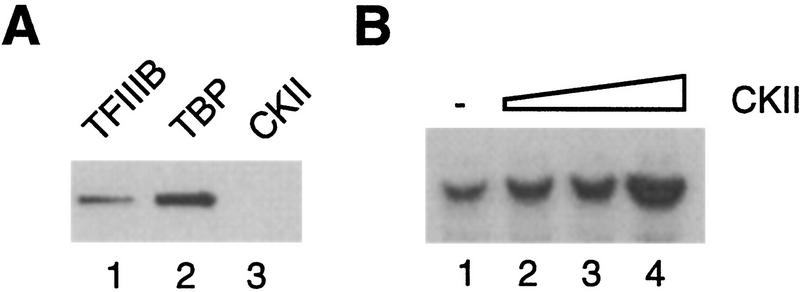
Although the fraction that stimulates cka2ts extract precisely coelutes with TFIIIB, our results do not rule out the possibility that the TFIIIB fraction contains a factor that restores CKII activity in cka2ts extract (or the D300 fraction). This factor could be CKII itself or a CKII activator (for review, see Tuazon and Traugh 1991; Allende and Allende 1995). We tested for CKII in the TFIIIB fraction using the peptide phosphorylation assay. CKII activity was measured in 3.2 μl of TFIIIB, an amount that fully rescues transcription in cka2ts–D300 (Fig. 2B). These measurements are compared to the CKII activity in 15 μg of wild-type D300 fraction (see Fig. 2A). As shown, TFIIIB contains <5% of the CKII activity measured in the CKA2–D300 fraction (Fig. 4A). The content of CKII in the TFIIIB fraction was also assessed by immunoblotting using an antiserum that specifically recognizes the β′ subunit of yeast CKII. We could not detect CKIIβ′ in amounts of TFIIIB similar to those used for the reconstitution of transcription (Fig. 4B), confirming the result obtained by assaying kinase activity. Therefore, CKII is substantially depleted in the fractions used for reconstitution of transcription.
Figure 4.
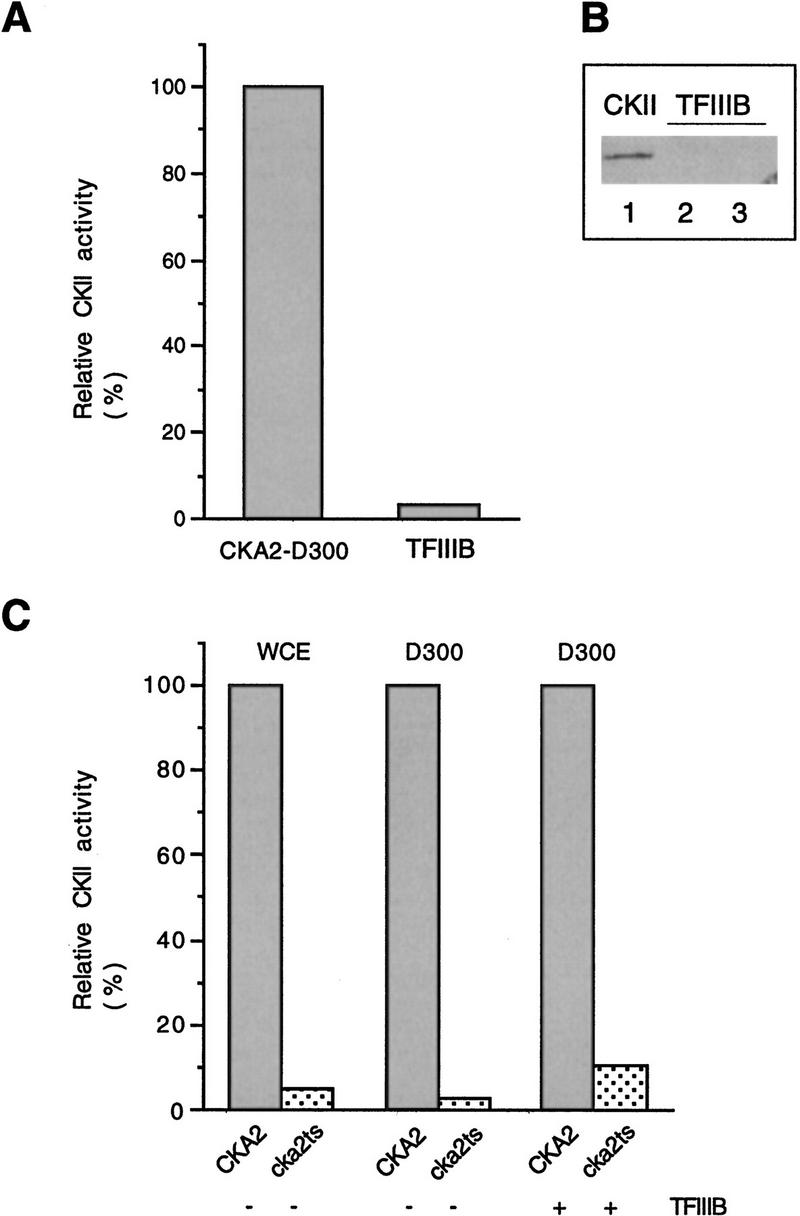
Rescue of transcription by TFIIIB does not involve restoration of CKII activity. (A) The TFIIIB fraction does not contain a significant amount of CKII activity. Bulk CKII activity is compared in 15 μg of CKA2–D300 extract (see Fig. 2A) and 4 μl of hydroxyapatite TFIIIB. CKII activity is expressed as a percentage of activity in 15 μg of CKA2–D300 (arbitrarily assigned the value of 100%). (B) The TFIIIB fraction does not contain a significant amount of the β′ subunit of CKII. Analysis of the TFIIIB fraction was performed by immunoblotting with a polyclonal antiserum raised against a carboxy-terminal peptide of yeast β′. (Lane 1) Purified CKII (1 μl); (lane 2) hydroxyapatite TFIIIB (15 μl); (lane 3) Cibachron blue TFIIIB (15 μl). (C) TFIIIB does not contain an activity that can restore the enzymatic function of temperature-sensitive CKII. Bulk CKII activity is expressed relative to wild type, which is arbitrarily assigned the value of 100%. The reactions used 6 μg of whole cell extract (WCE), 15 μg of D300, and 4 μl of hydroxyapatite TFIIIB.
To test whether the TFIIIB fraction contained an activity that could stimulate the temperature-sensitive CKII, we assayed bulk CKII activity in the D300 fractions with and without added TFIIIB (Fig. 4C). Increased CKII activity can be measured when TFIIIB is added to cka2ts–D300; however, the activity remains 10-fold lower than in the equivalent reaction in which TFIIIB is added to CKA2-D300. Furthermore, transcription in cka2ts–D300 is not stimulated when purified CKII is used to raise the bulk CKII activity to the level observed upon addition of TFIIIB (not shown). We conclude that the transcription machinery responds directly to CKII depletion and that TFIIIB is the component of the transcription machinery that is inactivated when CKII is depleted.
TFIIIB must be phosphorylated to restore transcription in cka2ts extract
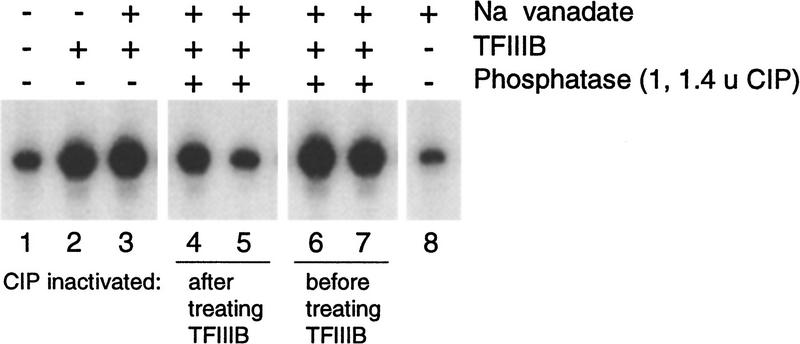
The results presented above demonstrate that TFIIIB responds to changes in CKII activity. This raises the possibility that direct CKII phosphorylation of TFIIIB is important for the activity of TFIIIB. We therefore tested whether dephosphorylation of TFIIIB affects its ability to stimulate transcription in cka2ts extract (Fig. 5). TFIIIB was treated with phosphatase in the presence or absence of the phosphatase inhibitor sodium vanadate and added to cka2ts nuclear extract (in which CKII activity is <2.5% of wild type; data not shown). TFIIIB retained its capacity to stimulate cka2ts extract when incubated with phosphatase in the presence of sodium vanadate (cf. lanes 1,3,6,7). In contrast, TFIIIB treated in turn with phosphatase and then sodium vanadate was unable to stimulate cka2ts extract (cf. lanes 1,3,4,5). Because sodium vanadate does not affect transcription on its own (lane 8), we conclude that TFIIIB must be phosphorylated to restore transcription to cka2ts extract.
Figure 5.
TFIIIB must be phosphorylated to restore transcription in cka2ts extract. TFIIIB was treated with phosphatase (CIP) in the presence or absence of sodium vanadate and then added to cka2ts nuclear transcription extract. The final composition of each reaction is indicated in the panel above the autoradiograph; the timing of CIP inactivation by sodium vanadate is indicated below the lane numbers. Plasmid pY5S was used as the template at 400 ng/reaction.
A single subunit of TFIIIB, the TBP, is phosphorylated by CKII
Four observations suggest that phosphorylation of TFIIIB by CKII can be limiting for transcription in wild-type extracts: (1) TFIIIB is limiting in wild-type extracts (Fig. 1B, lanes 1–4); (2) TFIIIB activity declines when CKII activity declines (Fig. 1B, lanes 5–8); (3) TFIIIB loses transcriptional activity when it is dephosphorylated (Fig. 5); (4) CKII stimulates transcription in some whole cell extracts from CKA2 cells, although our CKII preparations do not support transcription on their own (not shown) and do not contain TFIIIB as assayed by immunoblotting (Fig. 6). These observations suggest that CKII phosphorylation of a TFIIIB subunit is limiting for TFIIIB activity and, therefore, that a component of the wild-type TFIIIB fraction may be a substrate for in vitro phosphorylation by CKII. Likely substrates would include the TFIIIB subunits TBP, Brf1p, and TFc5p, respectively of approximate molecular masses 29, 70, and 90 kD. To examine the CKII phosphorylation of TFIIIB, the Cibachron blue fraction was incubated with CKII in the presence of [γ-32P]GTP as the phosphate donor (Fig. 7A). GTP was chosen because it is an efficient phosphate donor in reactions catalyzed by CKII but not most other protein kinases. The reaction products were analyzed by immunoblotting using a chemiluminescence detection system. The short exposures appropriate for the chemiluminescence assay were insufficient to detect the signal from the 32P-labeled proteins. Following immunodetection the membrane was stripped of antibody and then exposed to film to detect the 32P signal.
Figure 6.
(A) TFIIIB does not substantially copurify with CKII. Analysis of TFIIIB by immunoblotting using a polyclonal antiserum raised against recombinant TBP. (Lane 1) Five microliters of hydroxyapatite TFIIIB; (lane 2) 1 μl of purified TBP; (lane 3) 15 μl of purified CKII. (B) CKII can stimulate 5S rRNA transcription in whole cell extract (30 μg) from log phase cells. Plasmid pY5S was used as the template at 400 ng/reaction.
Figure 7.
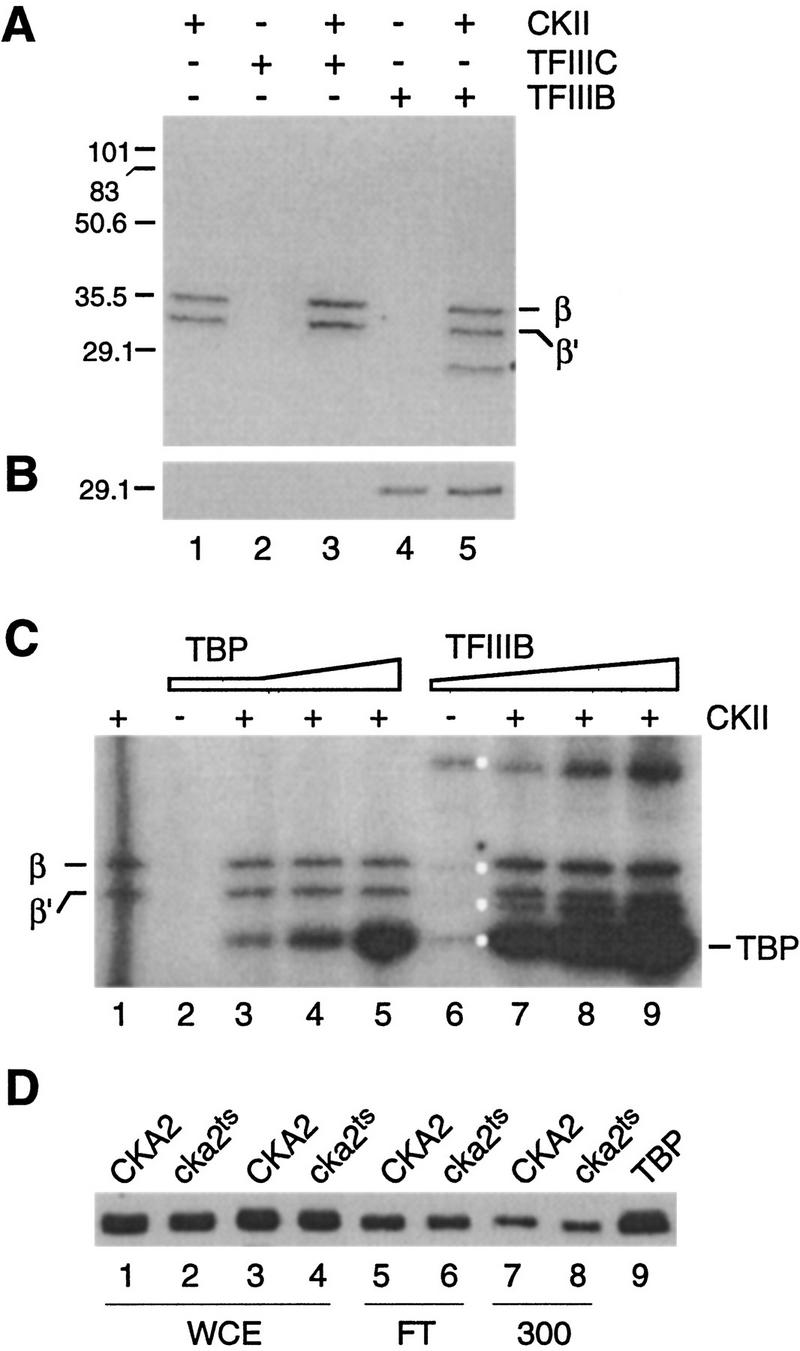
The TBP subunit of TFIIIB is preferentially phosphorylated by CKII. (A) Autoradiograph showing the labeled products resulting from in vitro phosphorylation of TFIIIB (10 μl) and affinity-purified TFIIIC (10 μl) with purified CKII (5 μl). The positions of the prestained molecular weight markers and the β subunits of CKII are shown on the left and right, respectively. (B) Immunoblotting analysis of the products in A using antiserum against TBP. As judged by the position of the markers, the immunoreactive band in lanes 4 and 5 exactly comigrates with the labeled 29-kD band in A, lane 5. (C) Recombinant TBP is phosphorylated by CKII in vitro. Reactions contained CKII on its own (lane 1; 0.5 μl), TBP on its own (0.1 μl; lane 2), CKII plus increasing amounts of recombinant TBP (0.1, 0.2, 0.5 μl; lanes 3–5), TFIIIB on its own (Cibachron blue fraction, 12 μl; lane 6), and CKII plus increasing amounts of TFIIIB (4, 8, 12 μl; lanes 7–9). Kinase/s in TFIIIB phosphorylate a number of proteins (white dots) that are not detected in shorter exposures (A, lane 4). (D) TBP is not depleted in whole cell extract or the DEAE fractions of cka2ts cells. Western blot analysis of whole cell extract (WCE; 42 μg per lane), the DEAE flowthrough (FT; 22 μg/lane), and 300 mm KCl fractions (300; 28 μg/lane) using TBP antiserum. Recombinant TBP (0.5 μl, lane 9) is the marker.
When incubated alone, yeast CKII autophosphorylates its β and β′ subunits (Fig. 7A, lane 1; Bidwai et al. 1994). TFIIIC does not contain significant GTP-dependent protein kinase activity on its own, and when used as a substrate for CKII only minor bands other than the CKII β and β′ are labeled (Fig. 7A, lane 2,3). Although some components of the TFIIIB fraction are labeled when TFIIIB is incubated on its own, these products are not detected (Fig. 7A, lane 4) except in longer exposures of the film (see below). On the other hand, when TFIIIB is used as a substrate for CKII, a single band in addition to the subunits of CKII is heavily labeled (Fig. 7A, lane 5). This band migrates at ∼29 kD, similar to the molecular mass of yeast TBP (yTBP). The possibility that the labeled 29-kD band is TBP was examined by immunoblotting using a yTBP antiserum (Fig. 7B). As expected, TBP is detected in the TFIIIB (lanes 4,5) but not the TFIIIC fraction (lanes 2,3). Furthermore, the protein labeled when TFIIIB is phosphorylated with CKII exactly comigrates with the band that reacts with the TBP antiserum. We also find that the labeled 29-kD band comigrates with recombinant TBP run in parallel and detected by silver staining or immunoblotting (not shown).
We were surprised to observe prominent labeling of only one polypeptide among those present in the TFIIIC and TFIIIB fractions, as our most purified TFIIIC and TFIIIB preparations contain ∼30 and 20 polypeptides, respectively, and many of the nine proteins comprising TFIIIC and TFIIIB contain CKII consensus sites (Hockman and Schultz 1996). Nonetheless, only TBP is significantly labeled by CKII. We conclude that the in vitro phosphorylation of TFIIIB-associated TBP by CKII is highly specific.
To extend the results obtained using cellular TFIIIB and purified CKII we tested whether recombinant TBP can be phosphorylated by CKII. Yeast TBP was expressed in Escherichia coli and purified to >95% homogeneity by ion exchange chromatography. Recombinant TBP, whether or not it was pretreated with phosphatase, was phosphorylated by CKII in the presence of [γ-32P]ATP as the phosphate donor [Fig. 7C, lanes 1–5; the same result was obtained with GTP (data not shown)]. Consistent with the idea that CKII is responsible for the observed phosphorylation of cellular and recombinant TBP, phosphorylation is completely blocked by heparin, a widely used inhibitor of CKII (not shown). This observation, combined with the results in Figure 7, A and B, demonstrates that in our experiments the only component of the wild-type Pol III transcription machinery that is a good substrate for CKII is the TBP subunit of TFIIIB.
Upon longer exposure of gels such as those in Figure 7 we detect a TFIIIB-associated activity that phosphorylates several proteins in the TFIIIB preparation, including TBP (Fig. 7C, lane 6; cf. with short exposure in A, lane 4). The level of TBP phosphorylation by the TFIIIB-associated kinase, however, is minor compared with that obtained when CKII is added to TFIIIB (lane 9). The molecular identity of the TFIIIB-associated kinase is not clear at the present time. It is unlikely to be a conventional form of CKII, as it is relatively insensitive to heparin (not shown) and is not associated with a significant amount of the β′ subunit (Fig. 4B).
Our results suggest that CKII phosphorylation of TBP increases the activity of TFIIIB. These results could be explained if the stability of TBP is affected by CKII. For example, the observation that transcription is severely impaired in cka2ts–D300 (Fig. 2A) could reflect loss of TBP during chromatography. To test whether changes in the amount of TBP could explain our results, we assessed the relative concentration of TBP in whole cell extract and various fractions from CKA2 and cka2ts cells by immunoblotting. We observe, in two independent sets of samples, that the concentration of TBP is virtually identical in CKA2 and cka2ts whole cell extracts and fractions from DEAE (Fig. 7D). We conclude that our results cannot be explained by an effect of CKII on the stability or chromatographic properties of TBP.
TBP and a limiting amount of CKII rescues Pol III transcription in CKII-deficient extract
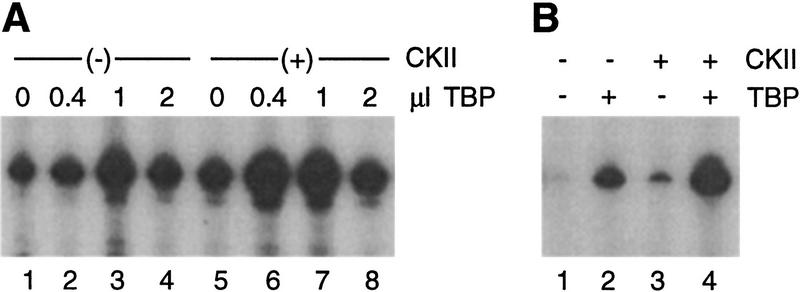
We next tested whether the activity of TBP as a Pol III transcription factor is influenced by CKII. This was done by supplementing nuclear transcription extract with recombinant TBP and purified yeast CKII, either alone or in combination (Fig. 8A). We observe significant stimulation of transcription in cka2ts extract by TBP alone (lanes 1–3). [Large amounts of TBP inhibit transcription (lane 4), perhaps by nonspecific occlusion of the template or by squelching.] The observed stimulatory effect of TBP on its own suggests that a TBP-dependent step in transcription is impaired as a consequence of CKII depletion. The effect of adding TBP and CKII together supports this idea. By comparing lanes 1–3 with 5–7 in Figure 8A, it is clear that the amount of TBP required to maximally stimulate cka2ts extract is significantly lowered when TBP is added along with a limiting amount of CKII. Importantly, under these conditions stimulation by CKII and TBP together is significantly greater than the total stimulation by each component added individually (lanes 1,2,5,6). The same result is obtained when TBP and CKII are added to the D300 fraction of cka2ts (Fig. 8B). These experiments demonstrate that the repression of TFIIIB resulting from inactivation of CKII can be overcome by bacterially expressed TBP in the presence of a limiting amount of CKII. Based on these results and the evidence presented in Figures 1, 2, 3, 4, 5, 6, 7 we propose that direct CKII phosphorylation of TBP is required for efficient Pol III transcription in yeast.
Figure 8.
TBP and a limiting amount of CKII rescues pol III transcription in CKII-deficient extract. (A) Nuclear extract (16 μg) from cka2ts cells was supplemented with TBP alone (lanes 1–4) or increasing amounts of TBP in the presence of CKII (0.5 μl, lanes 6–8). The amount of CKII used in this experiment only slightly stimulated transcription on its own (cf. lanes 1 and 5). (B) The D300 fraction (11 μg) from cka2ts cells was supplemented with buffer (lane 1), TBP (2 μl, lane 2), CKII (2.5 μl, lane 3), and TBP plus CKII (lane 4). Plasmid pY5S was used as the template at 400 ng/reaction.
Discussion
We have identified TFIIIB as the component of the Pol III transcription machinery that responds to the inhibition of CKII activity. We show further that TFIIIB must be phosphorylated to restore transcription in a CKII-depleted extract and that a combination of TBP and a limiting amount of CKII rescues transcription in a CKII-depleted extract to a higher level than the sum of TBP and CKII added individually. We have therefore identified an effector (CKII) and target (TFIIIB/TBP) that may be components of a mechanism for regulating Pol III transcription in yeast.
Regulation of TFIIIB by phosphorylation
Previous studies using extracts from Xenopus eggs suggest that the phosphorylation status of TFIIIB is important for its activity (Hartl et al. 1993; Gottesfeld et al. 1994; Leresche et al. 1996). For example, the inactive form of TFIIIB from metaphase extracts can be activated by phosphatase treatment. Conversely, the active interphase form of TFIIIB can be inactivated by p34cdc2–cyclin B kinase. These results demonstrate that phosphorylation can inhibit TFIIIB. Our results confirm a role for phosphorylation in the regulation of TFIIIB; however, we find that CKII phosphorylation activates rather than inhibits yeast TFIIIB. Why might opposite effects of phosphorylation on the activity of TFIIIB have been detected in Xenopus and yeast? First, the different effects of phosphorylation may be species specific. We do not favor this possibility because p34cdc2 and CKII, the known protein kinases implicated in the regulation of TFIIIB, are highly conserved and are expressed in all cycling eukaryotic cells (for review, see Nurse 1990; Allende and Allende 1995), as are two potential targets of these kinases (the TBP and Brf1p subunits of TFIIIB: see introductory section). Furthermore, the unique and essential catalytic functions of p34cdc2 and CKA1/CKA2 are also highly conserved (Lee and Nurse 1987; Bidwai et al. 1992). The second and, in our view, more interesting possibility is that the regulation of TFIIIB is universally mediated by its phosphorylation and dephosphorylation and that the pattern of these events is more complex than suspected previously. For example, phosphorylation at some sites in TFIIIB might stimulate transcription, whereas phosphorylation at other sites (possibly on different subunits) might be inhibitory. The level of activation/inhibition may vary according to the site of phosphorylation, with some inhibitory sites possibly being dominant over sites required for maximal activity. The latter scenario has been described in another context; the p34cdc2 component of the p34cdc2–cyclin B complex is regulated by dominant inhibitory and stimulatory phosphorylation events (for review, see King et al. 1994). These considerations suggest the working hypothesis that TFIIIB is regulated by positive and negative phosphorylation events and that CKII is involved in the positive phosphorylation event.
The molecular target of CKII phosphorylation
Our evidence indicates that the regulation of TFIIIB by CKII occurs through direct CKII phosphorylation of the TBP subunit of TFIIIB. Specifically, the observation that CKII stimulates transcription in wild-type transcription extract correlates with the finding that only one polypeptide in our wild-type TFIIIB fraction, TBP, is efficiently phosphorylated by CKII in vitro. Furthermore, a limiting amount of CKII increases the stimulation of transcription in cka2ts extract by a fixed amount of TBP. Based on these results we propose that direct phosphorylation of TBP by CKII is required for efficient Pol III transcription in vitro. Consistent with this proposal, we note that yTBP contains three motifs that match the CKII consensus S/Txx acidic [the Y specificity motif is not present in yTBP (Wilson et al. 1997)]. Because the amino-terminal 60 residues of yTBP are dispensable for function in vivo (Reddy and Hahn 1991), potential CKII sites in this region are unlikely to play a significant role in Pol III transcription. The remaining 180 residues of TBP contain potential CKII sites at S128 and S183. Modeling studies using the VADAR program (Wishart et al. 1994) indicate that S183, which is buried in the crystal structure of TBP (Chasman et al. 1993), is highly unlikely to be solvent exposed. This residue therefore is unlikely to be available for phosphorylation by CKII. S128, on the other hand, is solvent exposed, highly conserved (Kim et al. 1993), and the CKII consensus sequence at S128 encompasses a small surface of TBP that is important for Pol III transcription in vivo (Cormack and Struhl 1993; Trivedi et al. 1996). Furthermore, mutation of S128 to G in yeast confers a temperature-sensitive phenotype that may reflect an impairment of Pol III transcription in vivo (Cormack and Struhl 1993). These considerations, in light of the recent report that TBP is limiting for transcription in Drosophila tissue culture cells (Trivedi et al. 1996), suggest the possibility that CKII phosphorylation of TBP is required for the maintenance of efficient Pol III transcription in vivo.
CKII and the regulatory mechanisms that impinge on TFIIIB
Our work potentially relates to the mounting evidence that TFIIIB is a key target of regulatory mechanisms that govern the output of Pol III genes. In mammalian cells the response of the Pol III transcription machinery to a wide variety of internal and external cues is governed by mechanisms that regulate TFIIIB (see introductory section). In Xenopus the cell cycle regulation of Pol III transcription operates at the level of TFIIIB, perhaps through differential phosphorylation of its 92-kD and TBP subunits (Leresche et al. 1996). The Drosophila Pol III transcription machinery is sensitive to the cellular concentration of TBP, and in yeast the silencing of transcription in stationary phase is mainly due to an effect on TFIIIB70/Brf1p (Sethy et al. 1995; Trivedi et al. 1996). In summary, TFIIIB is the target of a spectrum of regulatory effects that can act on distinct subunits of TFIIIB. The protein kinases involved in these diverse regulatory mechanisms remain to be fully characterized. Our results raise the possibility that CKII is one such kinase. Thus, the activity of CKII toward TBP may be regulated and therefore contribute directly to the regulation of Pol III transcription in vivo. On the other hand, CKII phosphorylation of TBP might be constitutive and ensure that the maximal level of Pol III transcription is achieved when the demand for Pol III products is at its peak. These hypotheses will be amenable to critical experimental analysis in yeast.
Materials and methods
Buffers
Buffers are described as follows: (YDBI) 50 mm KCl, 5 mm EGTA, 0.05 mm EDTA, 2.5 mm DTT, 0.4 mm PMSF, and 20% glycerol. (Buffer B) 1 mm EDTA, 1 mm DTT, 0.4 mm PMSF, and 20% glycerol. (Buffer L) 1 mm EDTA, 5 mm MgCl2, 2.5 mm DTT, 0.2 mm PMSF, 10% glycerol, and 0.05% NP-40. Buffers YDBI, B and L also contained 20 mm HEPES–KOH (pH 7.9) and 0.3 μg/ml of leupeptin. (Buffer K) 0.5 mm DTT, 0.4 mm PMSF, 0.6 μg/ml of leupeptin, 0.6 μg/ml of pepstatin, and 20% glycerol. (Buffer T) 20 mm Tris-HCl (pH 8), 1 mm EDTA, 1 mm DTT, 1 mm PMSF, 2 mm benzamidine-HCl, and 10% glycerol. The concentration of KCl (mm) in buffers B, K, L, and T is designated by the number after the letter abbreviation.
Strains and extract preparation
Extracts were prepared from S. cerevisiae strains CKA2, cka2ts, and the protease-deficient BJ5626 (Jones 1991) grown to an OD at 600 nm of 1–2 (see Hockman and Schultz 1996). CKA2 and cka2ts were grown at room temperature, and BJ5626 at 30°C. Whole cell extracts were prepared after breaking the cells under liquid nitrogen, using a motorized mortar grinder for large-scale extracts (Schultz et al. 1997). Nuclear extracts were prepared in parallel from strains CKA2 and cka2ts according to Dunn and Wobbe (1995).
Purification and assay of transcription factors from yeast
Large-scale whole cell extracts were fractionated according to Kassavetis et al. (1989), with modifications that take into account the presence of an inhibitor that is concentrated on Bio-Rex 70. The transcriptionally active 35%–70% ammonium sulfate cut (2.8 grams of protein) of whole cell extract (7 grams) was dialyzed to YDBI and applied to 140 ml of Bio-Rex 70 (Bio-Rad). This column was step-eluted with 100, 250, and 500 mm KCl in YDBI. The 500 mm KCl fraction contained all components of the Pol III transcription machinery, although it was transcriptionally inert because of an inhibitor that is separated from Pol III/TFIIIC on DEAE and from TFIIIB on hydroxyapatite. The Bio-Rex 500 mm KCl fraction (160 mg) was dialyzed to B100 and applied to a 40-ml DEAE–Sepharose Fast Flow (Pharmacia) column equilibrated in B100; fractions were collected from the 100, 300, and 600 mm KCl washes. TFIIIB (108 mg), eluted in the flowthrough, was dialyzed to buffer K containing 25 mm KPO4 (buffer K25). Fifty milligrams of this fraction was loaded onto a 20-ml Bio-Gel hydroxyapatite (Bio-Rad) column equilibrated in buffer K25. The column was washed with buffer K50 and developed with a 75-ml gradient (50–300 mm KPO4) that eluted TFIIIB at ∼200 mm KPO4. TFIIIB fractions (∼3 mg) were dialyzed against buffer B200 and applied to a 3-ml Cibacron blue–Sepharose column (Fluka). Following washing with buffer B200, TFIIIB was eluted with B1000. Pol III and TFIIIC (20 mg) eluted together in the 300 mm cut from DEAE. Ten milligrams of this fraction (P/C) was dialyzed to buffer L100, precleared with 0.6 mg of pGEM3, and applied to a 1.1-ml TFIIIC affinity column [synthetic box B+ 29-mer coupled in 10 repeat fragments (average) to Sepharose CL2B (Kassavetis et al. 1989)]. The column was washed with buffer L200, and TFIIIC was eluted with L1000. Bulk Pol III activity was measured according to Schultz and Hall (1976). TFIIIB was monitored during chromatography by immunoblotting with a polyclonal antiserum against TBP (1:10,000 dilution). TFIIIB was also monitored by its ability to complement fraction P/C, which is transcriptionally inactive and devoid of TBP but enriched in bulk Pol III activity and TFIIIC DNA-binding activity. TFIIIC was monitored by its DNA-binding activity according to the method of Kassavetis et al. (1989). The Pol III machinery was obtained in a single fraction (D300) by applying whole cell extract in YDBI to a DEAE–Sepharose Fast Flow column (Riggs and Nomura 1990). The column was washed with 90 mm KCl/YDBI, and the transcription machinery was then eluted with 300 mm KCl/YDBI.
Expression and purification of recombinant TBP
Yeast TBP was expressed in E. coli strain BL21(DE3) and purified according to a protocol developed by Steve Hahn (pers. comm.; see also Reddy and Hahn 1991). Briefly, cells containing plasmid pSH228 were induced with 0.4 mm IPTG for 2 hr. The soluble fraction resulting from sonication of cells in buffer T50 was applied to DEAE–Sepharose Fast Flow, and the flowthrough collected. This material was applied to Mono S (Pharmacia) and eluted with a 50–500 mm KCl gradient (in buffer T). TBP was recovered in the 250–300 mm KCl fractions.
Purification of CKII and assay by peptide phosphorylation
CKII was purified according to Bidwai et al. (1994) from cells broken under liquid nitrogen using a mortar grinder (Schultz et al. 1997). The heparin–agarose fraction of CKII (85% pure) was used for most experiments. Peptide phosphorylation was performed as described (Hockman and Schultz 1996), except that a TCA precipitation step was introduced to remove polypeptides from the reaction mix prior to spotting onto P81 paper. This step reduces the variability between replicate assays. Specifically, 25 μl reactions are stopped by adding TCA to 5%, incubated on ice for 10 min, and then spun for 5 min in a microcentrifuge. Twenty microliters of the supernatant is then spotted onto P81 paper and processed for scintillation counting.
Preparation and use of CKIIβ′ antiserum
A peptide (DLTKSGGFKT-3′) corresponding to the 10 carboxy-terminal residues of CKIIβ′ (amino acids 219–228; Reed et al. 1994) was coupled to keyhole limpet hemacyanin (KLH) at the peptide’s amino terminus and used to immunize New Zealand white rabbits. After collecting naive serum, the rabbits were injected with 1 mg of KLH-conjugated peptide. The injection was repeated 2, 5, 9, and 13 weeks after the first immunization; serum was screened by ELISA (Harlow and Lane 1988) using the target peptide coupled to BSA and peroxidase-conjugated AffiniPure goat anti-rabbit IgG (Jackson ImmunoResearch Labs). The reaction product was detected using a peroxidase substrate kit (Bio-Rad). A strong ELISA reaction (compared to preimmune serum) was observed using serum collected at 5, 9, and 13 weeks. The serum specifically detected CKIIβ′ at a 1:2500 dilution in immunoblots.
Immunoblotting
Proteins were resolved on 12% or 15% SDS–polyacrylamide gels and electroblotted to nitrocellulose membrane in Tris-glycine buffer. The membranes were incubated with antiserum in 20% horse serum in TBST (100 mm Tris-HCl at pH 7.5, 150 mm NaCl, 5 mm MgCl2, 0.1% Tween 20). The immunoreactive species were detected using horseradish peroxidase-conjugated anti-rabbit antibody and the ECL chemiluminescence detection system (Amersham).
In vitro kinase and phosphatase reactions
Purified CKII was incubated with potential substrates in 20 μl final volume containing 15 mm HEPES–KOH (pH 7.9), 130 mm KCl, 10 mm MgCl2, 2.5 mm EGTA, 0.2 mm EDTA, and 1 μl [γ-32P]GTP or [γ-32P]ATP (NEN; 3000 Ci/mmole). The reactions were performed for 15 min at 30°C, stopped with SDS-PAGE loading buffer, and the products resolved as described under immunoblotting. Hydroxyapatite TFIIIB was dephosphorylated by incubation for 20 min at room temperature with the calf alkaline intestinal phosphatase (Sigma) in a buffer containing 20 mm Tris-HCl (pH 8.5), 50 mm NaCl, 5 mm MgCl2, 0.1 mm ZnCl2, and 1 mm DTT. Sodium vanadate (Sigma) was used at 0.5 mm.
In vitro transcription reactions
Multiple round transcription reactions were performed according to Hockman and Schultz (1996) using pY5S for 5S rRNA transcription (Schultz et al. 1992) and pTZ1, which contains the SUP4 tRNATyr gene, for tRNA transcription (Kassavetis et al. 1989). Details are noted in the legends to Figs. 1, 2, 3, 5, 6, and 8. The final buffer composition of all reactions in an experiment was identical. Reconstitution of transcription in cka2ts extract with CKII and TBP was performed without preincubation of CKII and TBP, as CKII phosphorylation of TBP prior to mixing with cell extract usually dampens rather than enhances the stimulation of transcription (not shown). We speculate that in a kinase reaction containing only purified TBP and CKII, phosphorylation of TBP occurs at some sites that both are not available when TBP is assembled into TFIIIB and must be unmodified for TBP to participate in transcription.
Acknowledgments
We are grateful to Darren Hockman for expert technical assistance, especially in development of the CKII assay (according to helpful suggestions from Colin Rasmussen) and the preparation of antisera. Ron Reeder kindly provided the TBP antiserum, and plasmids pTZ1 and pLNG56 were supplied by Peter Guideschek. pSH228 was from Steve Hahn, who also advised on the purification of recombinant TBP. Max Cummings and Mike Ellison are acknowledged for the molecular modeling, and we thank Claiborne Glover III for helpful discussions and Charlotte Spencer for comments on the manuscript. Peptides were prepared by the Alberta Peptide Institute (Edmonton), and oligonucleotides were synthesized by the DNA Core Facility, Biochemistry Department, University of Alberta. This work was supported by an establishment grant from the Alberta Heritage Foundation for Medical Research and by operating grants from the Canadian National Cancer Institute and Medical Research Council (MRC). M.C.S. is a scholar of the MRC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL michael.schultz@ualberta.ca; FAX (403) 492-9556.
References
- Allende JE, Allende CC. Protein kinase CK2: An enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- Bidwai AP, Hanna DE, Glover CVC. Purification and characterization of casein kinase II (CKII) from Δcka1 Δcka2 Saccharomyces cerevisiae rescued by Drosophila CKII subunits. The free catalytic subunit of casein kinase II is not toxic in vivo. J Biol Chem. 1992;267:18790–18796. [PubMed] [Google Scholar]
- Bidwai AP, Reed JC, Glover CVC. Casein kinase II of Saccharomyces cerevisiae contains two distinct regulatory subunits, β and β′. Arch Biochem Biophys. 1994;309:348–355. doi: 10.1006/abbi.1994.1123. [DOI] [PubMed] [Google Scholar]
- Chasman DI, Flaherty KM, Sharp PA, Kornberg RD. Crystal structure of yeast TATA-binding protein and model for interaction with DNA. Proc Natl Acad Sci. 1993;90:8174–8178. doi: 10.1073/pnas.90.17.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Struhl K. Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science. 1993;262:244–248. doi: 10.1126/science.8211143. [DOI] [PubMed] [Google Scholar]
- Dunn B, Wobbe CR. Preparation of protein extracts from yeast. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York, NY: Greene/Wiley; 1995. pp. 13.13.1–13.13.9. [DOI] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA. RNA polymerase III transcription complexes. In: McKnight SL, Yamamoto K, editors. Transcriptional regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- Gottesfeld JM, Wolf VJ, Dang T, Forbes DJ, Hartl P. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science. 1994;263:81–84. doi: 10.1126/science.8272869. [DOI] [PubMed] [Google Scholar]
- Hanna DE, Rethinaswamy A, Glover CVC. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J Biol Chem. 1995;270:25905–25914. doi: 10.1074/jbc.270.43.25905. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D, editors. Antibodies. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartl P, Gottesfeld J, Forbes DJ. Mitotic repression of transciption in vitro. J Cell Biol. 1993;120:613–624. doi: 10.1083/jcb.120.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockman DJ, Schultz MC. Casein kinase II is required for efficient transcription by RNA polymerase III. Mol Cell Biol. 1996;16:892–898. doi: 10.1128/mcb.16.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffler WK, Kovelman R, Roeder RG. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988;53:907–920. doi: 10.1016/s0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- Jones EW. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Riggs DL, Negri R, Nguyen LH, Geiduschek EP. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- King RW, Jackson PK, Kirschner MW. Mitosis in transition. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Kumar A, Kassavetis GA, Geiduschek EP, Hambalko M, Brent CJ. Functional dissection of the B′′ component of RNA polymerase III transcription factor IIIB: a scaffolding protein with mutliple roles in assembly and initiation of transcription. Mol Cell Biol. 1997;17:1868–1880. doi: 10.1128/mcb.17.4.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- Leresche A, Wolf VJ, Gottesfeld JM. Repression of RNA polymerase II and III transcription during M phase of the cell cycle. Exp Cell Res. 1996;229:282–288. doi: 10.1006/excr.1996.0373. [DOI] [PubMed] [Google Scholar]
- Litchfield DW, Lüscher B. Casein kinase II in signal transduction and cell cycle regulation. Mol Cell Biochem. 1993;127/128:187–199. doi: 10.1007/BF01076770. [DOI] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Pinna LA. Casein kinase 2: An “eminence grise” in cellular regulation? Biochim Biophys Acta. 1990;1054:267–284. doi: 10.1016/0167-4889(90)90098-x. [DOI] [PubMed] [Google Scholar]
- Reddy P, Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991;65:349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- Reed JC, Bidwai AP, Glover CVC. Cloning and disruption of CKB2, the gene encoding the 32-kDa regulatory β′-subunit of Saccharomyces cerevisiae casein kinase II. J Biol Chem. 1994;269:18192–18200. [PubMed] [Google Scholar]
- Reynolds WF. The tyrosine phosphatase cdc25 selectively inhibits transcription of the Xenopus oocyte-type tRNAtyrC gene. Nucleic Acids Res. 1993;21:4372–4377. doi: 10.1093/nar/21.18.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs DL, Nomura M. Specific transcription of Saccharomyces cerevisiae 35S rDNA by RNA polymerase I in vitro. J Biol Chem. 1990;265:7596–7603. [PubMed] [Google Scholar]
- Schultz LD, Hall BD. Transcription in yeast: α-Amanitin sensitivity and other properties which distinguish between RNA polymerases I and III. Proc Natl Acad Sci. 1976;73:1029–1033. doi: 10.1073/pnas.73.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MC, Reeder RH, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- Schultz MC, Hockman DJ, Harkness TAA, Garinther WI, Altheim BA. Chromatin assembly in a yeast whole-cell extract. Proc Natl Acad Sci. 1997;94:9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethy I, Moir RD, Librizi M, Willis IM. In vitro evidence for growth regulation of tRNA transcription in yeast. A role for transcription factor (TF) IIIB70 and TFIIIC. J Biol Chem. 1995;270:28463–28470. doi: 10.1074/jbc.270.47.28463. [DOI] [PubMed] [Google Scholar]
- Tower J, Sollner-Webb B. Polymerase III transcription factor B activity is reduced in extracts of growth-restricted cells. Mol Cell Biol. 1988;8:1001–1005. doi: 10.1128/mcb.8.2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi A, Vilalta A, Gopalan S, Johnson DL. TATA-binding protein is limiting for both TATA-containing and TATA-lacking RNA polymerase III promoters in Drosophila cells. Mol Cell Biol. 1996;16:6909–6916. doi: 10.1128/mcb.16.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuazon PT, Traugh JA. Casein kinase I and II—Multipotential serine protein kinases: Structure, function, and regulation. Adv Second Messenger Phosphoprotein Res. 1991;23:123–164. [PubMed] [Google Scholar]
- Wang Z, Roeder RG. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Stott D, Rigby PW. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989;59:1081–1092. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]
- White RJ, Stott D, Rigby PW. Regulation of RNA polymerase III transcription in response to Simian virus 40 transformation. EMBO J. 1990;9:3713–3721. doi: 10.1002/j.1460-2075.1990.tb07584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Gottlieb TM, Downes CS, Jackson SP. Mitotic regulation of a TATA-binding-protein-containing complex. Mol Cell Biol. 1995a;15:1983–1992. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Gottlieb TM, Downes CS, Jackson SP. Cell cycle regulation of RNA polymerase III transcription. Mol Cell Biol. 1995b;15:6653–6662. doi: 10.1128/mcb.15.12.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Trouche D, Martin K, Jackson SP, Louzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;383:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- Willis IM. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- Wilson LK, Dhillon N, Thorner J, Martin GS. Casein kinase II catalyzes tyrosine phosphorylation of the yeast nucleolar immunophilin Fpr3. J Biol Chem. 1997;272:12961–12967. doi: 10.1074/jbc.272.20.12961. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Willard L, Richards FM, Sykes BD. VADAR: A comprehensive program for protein structure evaluation. Version 2.1. Edmonton, Canada: University of Alberta; 1994. [Google Scholar]
- Wolffe AP, Brown DD. Developmental regulation of two 5S ribosomal RNA genes. Science. 1988;241:1626–1632. doi: 10.1126/science.241.4873.1626. [DOI] [PubMed] [Google Scholar]