Abstract
SUMMARY
The Alx gene family is implicated in craniofacial development and comprises two to four homeobox genes in each vertebrate genome analyzed. Using phylogenetics and comparative genomics, we show that the common ancestor of jawed vertebrates had three Alx genes descendent from the two-round genome duplications (Alx1, Alx3, Alx4), compared with a single amphioxus gene. Later in evolution one of the paralogues, Alx3, was lost independently from at least three different vertebrate lineages, whereas Alx1 and Alx4 were consistently retained. Comparison of spatial gene expression patterns reveals that the three mouse genes have equivalent craniofacial expression to the two chick and frog genes, suggesting that redundancy compensated for gene loss. We suggest that multiple independent loss of one Alx gene was predisposed by extensive and persistent overlap in gene expression between Alx paralogues. Even so, it is unclear whether it was coincidence or evolutionary bias that resulted in the same Alx gene being lost on each occasion, rather than different members of the gene family.
INTRODUCTION
Homeobox genes are characterized by possession of a particular DNA sequence encoding a recognizable protein domain (Bürglin 1994). More than 200 homeobox genes have been identified in the human genome which can be categorized into 11 classes and around 100 gene families (Holland et al. 2007). The two major classes of homeobox gene are the ANTP and PRD classes, each of which includes many genes implicated in developmental patterning or cellular differentiation (Galliot et al. 1999; Nam and Nei 2005; Ryan et al. 2006;). The ANTP class includes Hox genes, ParaHox genes, NK genes, plus the Emx, Dlx, En, and Evx gene families among others. The PRD class includes other well-known developmental genes including Otx, Otp, Gsc, Pax2/5/8, Pax3/7, Pax4/6, Pitx, and Dmbx.
The Alx homeobox gene family belongs to the PRD class. Humans and mice possess three Alx homeobox genes, namely Alx1, Alx3, and Alx4. The Alx1 gene was initially named Cart1, but the nomenclature of this gene has been revised to recognize its paralogous relationship. Originally, the Alx genes derived their name from a Drosophila gene, aristaless, (aristaless homeobox) (Qu et al. 1997a), although the sequence similarity is not high. Indeed, there are several distinct groups of vertebrate genes, notably the Arx and Phox (Arix) gene families that show some similarity to aristaless (Beverdam and Meijlink 2001). It was shown later that aristaless itself does not belong to the Alx gene family, and that the Drosophila ortholog of this family has been secondarily lost in evolution (Ryan et al. 2006). The Alx gene family is certainly ancient, and single representatives have been reported from the sea anemone Nematostella (Ryan et al. 2006) and the sea urchins Strongylocentrotus and Lytechinus (Ettensohn et al. 2003) although the terminology Alx1 for the latter genes do not reflect one-to-one orthology with vertebrate Alx1.
The vertebrate Alx genes have a variety of important developmental roles, being implicated in neural tube closure (Zhao et al. 1996), limb development (Qu et al. 1997b), and most especially aspects of craniofacial development. The three mouse Alx genes show similar developmental expression patterns in the cranial regions of embryo. From early stages (E8.5–E9.5), all three are prominently expressed in frontonasal mesenchyme, and later in the first and second pharyngeal arches (Zhao et al. 1996; Qu et al. 1997a; ten Berge et al. 1998; Beverdam and Meijlink 2001;). Furthermore, gene targeting studies have shown the three Alx genes to have critical roles in craniofacial development in mice (Zhao et al. 1996; Qu et al. 1997b; Beverdam et al. 2001;). In humans, mutations in each of the three Alx genes have been reported to be associated with congenital craniofacial malformation (Wu et al. 2000; Wuyts et al. 2000; Mavrogiannis et al. 2001; Twigg et al. 2009; Uz et al. 2010;), which in each case are closely related to the phenotypes of the mouse mutants.
Although the developmental roles of Alx genes and their molecular interactions with each other have been well studied in mice and humans (Qu et al. 1999; Beverdam et al. 2001;), there remain large gaps in our understanding of the Alx gene family. In particular the evolution of this gene family within the chordates, and, more specifically, the vertebrates has not been scrutinized. Here we report the identification of Alx homeobox genes from amphioxus and from major classes of vertebrates. We have compared embryonic expression patterns between species, and related this to comparative genomic analyses in which we uncover an interesting evolutionary feature: repeated independent loss of the Alx3 gene.
MATERIALS AND METHODS
Cloning of Alx cDNAs
A Branchiostoma floridae embryonic cDNA library, kindly provided by Dr. J. Langeland, was screened using an AmphiDmbx probe (Takahashi and Holland 2004) under reduced stringency conditions. Two independent clones both containing a full coding region of AmphiAlx gene were isolated and sequenced (GenBank: JF460798, JF460799). Xenopus tropicalis Alx1 and Alx4 cDNAs (GenBank: JF460800, JF460801) were isolated by RT-PCR amplification from mRNA of stage 35 embryos. Primers used were based on genomic sequence from the Joint Genome Institute draft genome sequence (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html). Gallus gallus Alx1 and Alx4 genes were cloned by RT-PCR from chick embryo mRNA. Primers were based on predicted coding sequence.
Phylogenetic analysis
Amino acid sequences of Alx homeodomain proteins from human and mouse were retrieved from NCBI Entrez Gene as follows: human Alx1 (NP_008913.2), Alx3 (NP_006483.2), Alx4 (NP_068745.2); mouse Alx1 (NP_766141.1), Alx3 (NP_031467.1), Alx4 (NP_031468.1). To identify chicken (G. gallus) and zebrafish (Danio rerio) sequences, we used reciprocal blast searching with human Alx proteins. Gene family members identified, with nomenclature defined after phylogenetic analysis, were: chicken Alx1 (XP_425445.2) and Alx4 (NP_989493.1); zebrafish Alx1 (NP_001038539.1), Alx3 (XP_695330.1), Alx4a (XP_001340966.1), and Alx4b (NP_001082826.1).
For lizard (Anolis carolinensis) and frog (X. tropicalis) sequences, tblastn searches using human Alx proteins against whole genome sequence in GenBank (lizard) or DOE Joint Genome Institute (frog) were conducted. Anolis sequences deduced were: Alx1 (AAWZ01006799.1–AAWZ01006800.1) and Alx4 (AAWZ01008092.1–AAWZ01008095.1). In Xenopus, Alx1 (protein model 196810) and a partial Alx4 (protein 173595) were readily identified in genomic sequence. However, part of the genomic region corresponding to the carboxyl-terminal end of the Alx4 protein is missing in the assembly around this gene, so we cloned the 3′-end of a cDNA by RT-PCR (GenBank: JF460801). We also predicted a further 14 amino acids at the N-terminus from scaffold_281|792714|792755. Echinoderm Alx protein sequences from Strongylocentrotus purpuratus, Paracentrotus lividus, and Lytechinus variegatus were used as outgroups for the analysis: StrpuAlx (NP_999809.1, originally reported as Alx1) (Ettensohn et al. 2003), LytvaAlx (AAP34699.1, originally reported as Alx1) (Ettensohn et al. 2003), and ParliAlx (ABG00197.1) (Rottinger et al. 2004).
The Alx protein sequences from all 10 species were aligned using ClustalW implemented in the BioEdit software package. The sequences were then manually trimmed of all sites that were not unambiguously aligned. For maximum-likelihood (ML) trees ProtTest 1.4 (Abascal et al. 2005) was used to determine the best protein substitution model and parameters for use in RAxML (Stamatakis et al. 2008). The JTT model of protein evolution was used with the proportion of invariable sites and gamma parameter estimated from the data, four categories of between-site rate variation; 100 bootstraps were used in the primary ML tree (final ML optimization likelihood: −4870.45565). The PHYLIP software package (http://evolution.genetics.washington.edu/phylip/) was used to create neighbor-joining (NJ) trees using the JTT model of protein evolution with 1000 bootstraps.
For identification of the Alx family genes in cartilaginous fish and agnathans, the genome data of the elephant shark (Callorhinchus milii; http://blast.fugu-sg.org/) (Venkatesh et al. 2007) and the sea lamprey (Petromyzon marinus, PMAR3; http://pre.ensembl.org/Petromyzon_marinus/Info/Index) were searched using the tblasn program with amphioxus and human Alx proteins. Orthology was tested using reciprocal blast searching to the human genome and by phylogenetic analysis (NJ and ML).
For identification of the duplicated Alx4 genes in teleosts other than zebrafish, genomic information from following sources was used: Takifugu rubripes (JGI v4.0; http://genome.jgi-psf.org/Takru4/Takru4.home.html); Tetraodon nigroviridis (http://www.ensembl.org/Tetraodon_nigroviridis/Info/Index); Oryzias latipes (http://www.ensembl.org/Oryzias_latipes/Info/Index); Gasterosteus aculeatus (http://www.ensembl.org/Gasterosteus_aculeatus/Info/Index).
Synteny analysis
Analysis of genomic regions was used to test hypotheses of gene loss. Genomic regions around Alx3 and its neighboring genes in zebrafish, zebra finch, mouse, and human were analyzed and compared with corresponding genomic regions of Xenopus, lizard, and chicken. Genome assemblies used were from NCBI (D. rerio Zv7, G. gallus Build 2.1, Taeniopygia guttata Build 1.1, Mus musculus Build 37.1, Homo sapiens Build 36.3; http://www.ncbi.nlm.nih.gov/mapview/), Joint Genome Institute (X. tropicalis genome assembly 4.1; http://genome.jgi-psf.org/Xentr4/Xentr4.home.html), and Broad Institute (A. carolinensis draft assembly AnoCar 1.0; http://genome.ucsc.edu/cgi-bin/hgGateway?db=anoCar1) (Kent et al. 2002). For genes that have not been annotated, the identity of genes was deduced by reciprocal blast searching to the human genome.
Whole mount in situ hybridizations
In situ hybridization of whole mount vertebrate embryos was carried out as described (Thompson et al. 2010). Probes for hybridization were synthesized by in vitro transcription using a DIG RNA Labeling Mix (Roche, Mannheim, Germany), following the supplier's instructions, and detection used alkaline phosphatase-conjugated anti-digoxigenin antibody followed by blue staining using NBT-BCIP (Roche).
RESULTS
The amphioxus Alx gene
Two amphioxus cDNA clones (GenBank: JF460798, JF460799) with high sequence similarity to vertebrate Alx genes were isolated. The two clones both include a complete open reading frame and are 98% identical to each other at the amino acid level. Using echinoderm Alx genes as outgroups (Ettensohn et al. 2003), phylogenetic analysis clearly shows that these clones encode the amphioxus ortholog of the vertebrate Alx gene family (AmphiAlx, Fig. 1). Our survey of the amphioxus genome (B. floridae; http://genome.jgi-psf.org/Brafl1/Brafl1.home.html) by blast revealed the same Alx gene, also identified by Takatori et al. (2008), suggesting that AmphiAlx is the single, polymorphic, amphioxus ortholog of vertebrate Alx genes. We discount a very similar sequence (proteinID 69456) neighboring the Alx gene (proteinID 119093) in the draft genome sequence as a probable “stutter” assembly artifact, as noted for other cases (Putnam et al. 2008). In any case, a local tandem duplication would not affect our use of the amphioxus gene as an outgroup for vertebrate sequences. In a previous study (Meulemans and Bronner-Fraser 2007), an EST clone (bfne089p08) was identified as amphioxus Alx; this EST corresponds to the 3′-UTR region of the AmphiAlx gene reported here. We also attempted to identify an ortholog in urochordates by blast searching the genomes of Ciona intestinalis (http://genome.jgi-psf.org/Cioin2/Cioin2.home.html) and Ciona savignyi (http://www.broadinstitute.org/annotation/ciona/). No ortholog was detected suggesting that the Alx gene was lost on the lineage leading to Ciona.
Fig. 1.
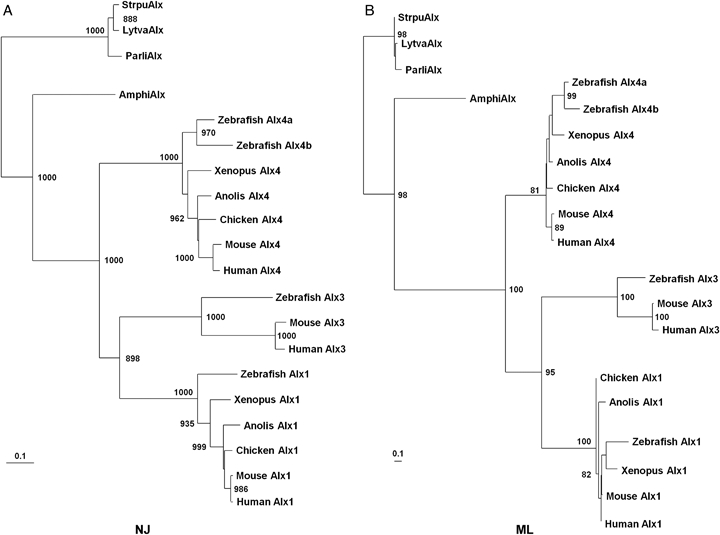
Molecular phylogenetic trees showing the relationships between deuterostome Alx proteins based on neighbor-joining (NJ, A) and maximum-likelihood (ML, B) analyses. Echinoderm Alx proteins were used as outgroups. Branch lengths are proportional to evolutionary distance corrected for multiple substitutions; the scale bar denotes 0.1 underlying amino acid substitutions per site. Figures on branches indicate robustness of each node (>70%), estimated from 1000 bootstrap replicates for NJ (A) and 100 replicates for ML (B). AmphiAlx (GenBank: JF460798) is clearly shown to be an ortholog of vertebrate Alx genes by both the NJ (100%) and ML (98%) methods. Furthermore, all extant bony vertebrates share at least two Alx gene duplication events: one that gave rise to the Alx4 and Alx1/3 paralogy groups (100% for both trees), and a second that gave rise to the Alx1 and Alx3 paralogy groups (89.8%, 95%). Further duplication of Alx4 genes is observed in zebrafish and other teleost fishes (see text).
Vertebrate Alx homeobox gene family
In order to examine orthology/paralogy relationships between the multiple Alx genes of different vertebrate species, phylogenetic analysis was performed. The single Alx genes of amphioxus and echinoderm species were ideal outgroups for this analysis. NJ (Fig. 1A) and ML (Fig. 1B) analyses, on amino acid sequence alignments, gave equivalent results.
The phylogenetic analysis confirms that all vertebrate Alx genes form a monophyletic group (NJ 100%, ML 100%), derived from a single ancestral gene. The vertebrate Alx proteins clearly divide into three lineages deduced to be three paralogous genes: Alx1, Alx3, and Alx4. It is also worth noting that the branch lengths for all Alx3 proteins are longer than those of other Alx proteins, suggesting that Alx3 genes may have rapidly evolved in comparison with Alx1 and Alx4.
As each of the three genes can be found in at least some species of actinopterygian and sarcopterygian vertebrates, we suggest that the three Alx genes were generated by the two-round whole genome duplications (2R-WGD) that took place in early vertebrate evolution (Dehal and Boore 2005; Holland et al. 2007;). Our analysis places Alx1 and Alx3 closer to each other than to Alx4 (NJ 89.8%, ML 95%). The first genome duplication will have generated two Alx genes, and we suggest that Alx1 and Alx3 were generated from one of these in the second genome duplication, whereas the other Alx gene gave rise to Alx4 and an “Alx2 ” gene that has not been detected. Subsequently, “Alx2 ” has been lost from all extant vertebrates examined to date.
For further elucidation of the early evolutionary history of vertebrate Alx genes, we also analyzed the genomic information of basal vertebrates available to date, namely cartilaginous fishes and agnathans. In chondrichthyes, draft genome sequence data from the elephant shark (C. milii) was analyzed. Despite its low genome coverage (1.4 ×), the genome sequences include three different sequences closely related to the homeodomain of human Alx proteins (GenBank: AAVX01553552.1, AAVX01326612.1, AAVX 01282984.1) or the carboxyl-terminal OAR domain of Alx proteins (Beverdam and Meijlink 2001) (GenBank: AAVX01514414.1, AAVX01274664.1, AAVX01004849.1). Our phylogenetic analysis using the OAR regions suggests these sequences are orthologs to Alx1, Alx3, and Alx4 of jawed vertebrates (NJ, ML, data not shown). This implies that the three distinct vertebrate Alx genes arose before the divergence of chondrichthyans and osteichthyans, compatible with origin during the 2R-WGD. We cannot deduce when loss of Alx2 occurred, however, from low coverage genome data. Analysis of draft genome sequence from the sea lamprey (P. marinus) revealed two contigs (Ensembl: Contig 7807, 24307) containing homeobox sequences from the Alx family (NJ, ML, data not shown). The lack of a genome assembly precludes further phylogenetic analysis.
Human and mouse have one each of the three Alx genes (Alx1, Alx3, Alx4), but quite different complements of genes are found in zebrafish, frog, chick, and lizard. For frog, chick, and lizard, only two genes were identified; phylogenetic analysis clearly shows these are Alx1 and Alx4. In the zebrafish genome, four Alx genes were identified. Our analysis shows that the protein denoted zgc: 162606 (GenBank: NP_001082826) is most closely related to the protein named zebrafish Alx4 (GenBank: XP_001340966). The implication is that zebrafish has two Alx4 genes, which we name Alx4a (GenBank: XM_001340930) and Alx4b (GenBank: NM_001089357). The duplication of Alx4 genes is also observed in the genomes of Takifugu (JGI: 598617, 579380), Tetraodon (GenBank: CAG08730, CAF99670), medaka (Ensembl: ENSORLT00000005949, ENSORLT00000008228), and stickleback (Ensembl: ENSGACT00000020637, ENSGACT 00000013078). This suggests that the duplication occurred in the teleost fish lineage after it had diverged from the other vertebrates. This conclusion is further supported by the observation that the two teleost Alx4 genes are located in duplicated chromosomal regions showing synteny to the human Alx4 genomic region (data not shown).
Independent losses of the Alx3 gene
The apparent absence of Alx3 in frog, chick, and lizard is intriguing. To test whether this reflects a true gene loss, or just incomplete data, we examined genome sequences for these species. Sequence searching using blast failed to identify orthologs of this gene in the draft genomes for any of the three species. More conclusive evidence came from examination of the genomic regions deduced to be syntenic to those harboring the Alx3 gene in human, mouse, and zebrafish (Fig. 2). In all three cases—frog, chick, and lizard—we found clear syntenic regions containing orthologs of many of the genes surrounding Alx3 in human, mouse, and zebrafish. Even the gene order is well conserved, apart from an inversion in zebrafish (Fig. 2). However, the Alx3 gene is clearly absent in frog, chick, and lizard from its expected position between Slc6a17 and Fam40a genes.
Fig. 2.
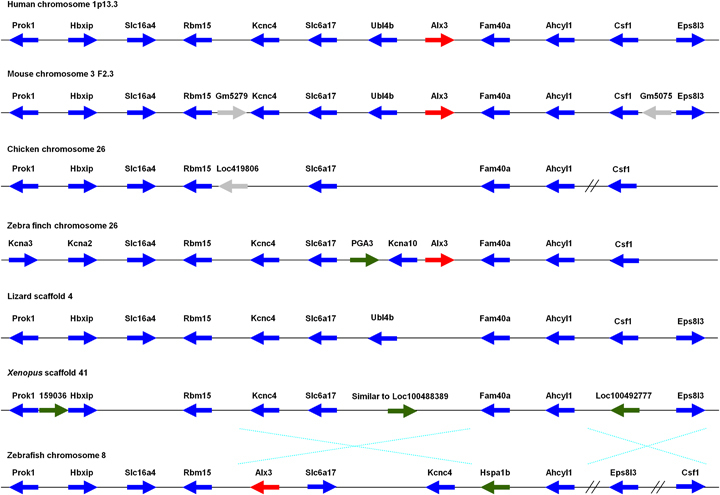
Schematic diagram showing the location of Alx3 and neighboring genes in syntenic regions of Homo sapiens, Mus musculus, Gallus gallus, Taeniopygia guttata, Anolis carolinensis, Xenopus tropicalis, and Danio rerio. Arrows indicate genes and their direction of transcription. The blue/black arrows represent loci for which syntenic conservation is confidently identified (zebra finch Kcna genes are denoted by blue arrows because their orthologs in other vertebrates are located immediately downstream of Prok1). Alx3 genes are highlighted with red arrows/gray shadows. The green/gray arrows denote loci for which orthologs are not located in the syntenic region; these are probably inserted secondarily. The gray arrows denote predicted pseudogenes. The dotted crosses indicate inversions within this region in zebrafish genome. The double oblique lines in the zebrafish and chicken genomic regions indicate presence of other genes between the loci shown.
We further extended our survey of the Alx3 genomic region to a larger number of vertebrate species. The Alx3 gene was found to be present in all mammalian and fish species for which there was adequate genome coverage (not shown). More surprisingly, we found the Alx3 gene in the zebra finch (T. guttata) genome, located in the appropriate syntenic region (Fig. 2).
It is clear, therefore, the Alx3 gene has been lost in evolution from the genomes of frog (X. tropicalis), lizard (A. carolinensis), and chicken (G. gallus). The known phylogenetic relationships between species possessing Alx3 and species lacking Alx3 reveal that these have been independent losses.
Consequences of gene loss on Alx gene expression
To determine whether the loss of Alx3 in evolution is associated with differences in expression of Alx1 and Alx4, we compared Alx gene expression in the developing head of mouse, chick, and frog embryos of comparable stages (Fig. 3). This region of the embryo is the major site of expression of Alx genes and displays phenotypic alterations in mice and humans carrying Alx mutations.
Fig. 3.
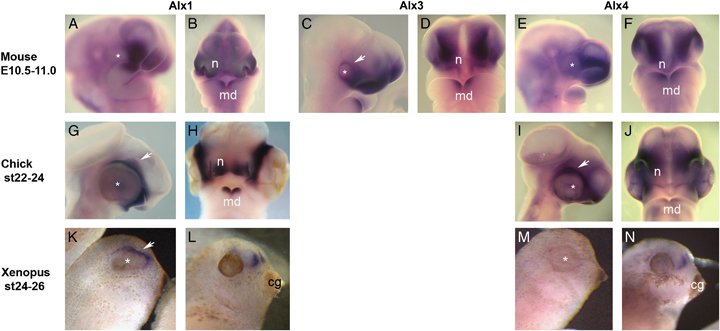
Alx expression in the developing head of mouse, chick, and frog embryos. Expression of mouse, chick, and Xenopus Alx genes during craniofacial development was compared using whole mount in situ hybridization. Lateral (A, C, E, G, I, K, L, M, N) and frontal (B, D, F, H, J) views of the developing head are shown. At E10.5-11 in mouse, Alx1 (A, B), Alx3 (C, D), and Alx4 (E, F) are expressed in the mesenchyme of the facial prominences, particularly around the nasal region (n), and at the distal tip of the mandible (md). At this stage, Alx3 is additionally expressed in the mesenchyme (C, white arrow) encircling the eye. ( ). At stages 22–24 of chick development, Alx1 (G, H) and Alx4 (I, J) are also expressed in the facial prominences, again with strong expression around the nasal region (n), and at the distal tip of the mandible (md). In the chick, however, both Alx1 (G) and Alx4 (I) are expressed in the periocular mesenchyme (white arrows). Periocular expression of Alx1 is observed in frog embryos at the stage of 24 (K, white arrow). At this stage, Alx4 is not expressed in frog embryos (M) but later at stage 35, both Alx1 (L) and Alx4 (N) are expressed in frontal mesenchyme above the cement gland (cg).
). At stages 22–24 of chick development, Alx1 (G, H) and Alx4 (I, J) are also expressed in the facial prominences, again with strong expression around the nasal region (n), and at the distal tip of the mandible (md). In the chick, however, both Alx1 (G) and Alx4 (I) are expressed in the periocular mesenchyme (white arrows). Periocular expression of Alx1 is observed in frog embryos at the stage of 24 (K, white arrow). At this stage, Alx4 is not expressed in frog embryos (M) but later at stage 35, both Alx1 (L) and Alx4 (N) are expressed in frontal mesenchyme above the cement gland (cg).
Consistent with previous reports, we find expression of mouse Alx1, Alx3, and Alx4 in the craniofacial mesenchyme of embryos at E10.5, with expression domains that largely overlap (Fig. 3, A–F). All three genes are expressed in the forming facial prominences and in the distal tip of the mandible. There is particularly strong expression around the nasal region. At this stage, mouse Alx3 is expressed in the periocular mesencyme and can be seen to circumscribe the eye (Fig. 3C, white arrow).
The expression of Alx genes in the developing chick head has not been reported previously. We find that expression of chick Alx1 and Alx4 is comparable to mouse Alx gene expression (Fig. 3, G–J). At stages 22–24 of development, both Alx1 and Alx4 are expressed in the facial prominences and the distal tip of the mandible and again there is strong expression around the nasal region. There is, however, an interesting difference in that chick Alx1 and Alx4 are expressed in the periocular mesenchyme encircling the eye in the chick at this stage (Fig. 3, G and I, white arrows). In mouse, this region expresses Alx3, but not Alx1 or Alx4.
In Xenopus, the Alx1 gene (Fig. 3K, white arrow) but not Alx4 (Fig. 3M) is expressed in the mesenchyme encircling the eye at stage 24. Later, at stage 35, expressions of both Alx1 and Alx4 are observed in frontal mesenchyme close to the hatching gland (Fig. 3, L and N). The slight difference in expression timing compared with amniotes is possibly due to the existence of the cement gland. Nevertheless, the sum of both spatial and temporal expression patterns of Xenopus Alx genes is largely equivalent to that seen for chick and mouse Alx genes.
DISCUSSION
Evolution of vertebrate Alx homeobox gene family
The accepted definition of a homeobox “gene family” is that set of genes descendent from a single gene in the common ancestor of bilaterian animals (Holland et al. 2007). In several invertebrates, including the sea anemone Nematostella (Ryan et al. 2006), sea urchin (Ettensohn et al. 2003), and amphioxus (Meulemans and Bronner-Fraser 2007; Takatori et al. 2008; and this study), only a single member of the Alx homeobox gene family has been identified. The implication is that the ancestral Alx gene has not duplicated in these lineages. In two Ciona species (tunicates), plus the nematode Caenorhabditis elegans and the insects Drosophila, Tribolium, and Apis, no Alx gene has been found. This implies secondary loss of the gene. In contrast, in vertebrates the Alx gene family includes three distinct genes—Alx1, Alx3, and Alx4—deduced to be descendent from the two WGDs that occurred in early vertebrate evolution (2R-WGD) (Holland et al. 2007; Putnam et al. 2008;) (summarized in Fig. 4). As Alx1 and Alx3 are more closely related to each other than either is to Alx4 (Fig. 1), it seems likely that the first round of genome duplication generated the common ancestor of Alx1 and Alx3 genes (Alx1/3 in Fig. 4) plus the ancestor of Alx4 gene (Alx2/4). The next duplication event gave rise to Alx1 and Alx3 genes from the Alx1/3 gene, and Alx4 and another Alx gene (“Alx2 ”) from the other (Fig. 4). This “Alx2 ” gene has not been identified in any vertebrate so far, and may have been lost soon after the 2R-WGD.
Fig. 4.
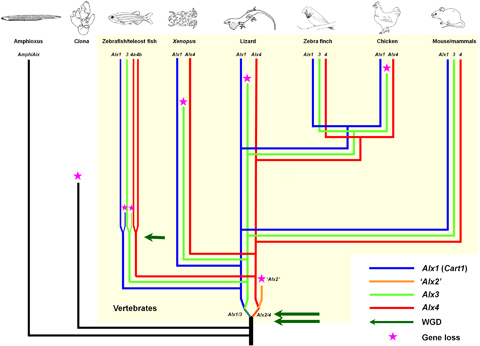
“Gene family tree” of vertebrate Alx homeobox genes in chordate evolution. After the two whole genome duplications (WGD, two green arrows at bottom of diagram) in early vertebrate evolution, four Alx homeobox genes were generated. The “Alx2” gene (yellow line) may have been swiftly lost, whereas the Alx3 genes were lost independently in at least three lineages (shown as the stars). Further modification in the teleost fish lineage followed the third WGD. The diagram depicts the phylogenetic location of each Alx3 gene loss, but note that the timing of loss within each lineage has not been determined.
In the teleost fish lineage, a further WGD occurred, subsequent to its divergence from basal actinopterygian fish (Jaillon et al. 2004). This will have generated duplicated Alx1, Alx3, and Alx4 genes, although our genomic analysis of five teleost species (zebrafish, Tetraodon, Takifugu, medaka, and stickleback) showed that only the duplicated Alx4 genes (Alx4a, Alx4b) were retained in these species. Duplicated Alx1 and Alx3 genes were lost after the third WGD in teleosts.
The enigma of vertebrate Alx3
The three principal members of the vertebrate Alx gene family (Alx1, Alx3, and Alx4) were established early in vertebrate evolution, and all were still present in the genome of the common ancestor of Chondrichthyes, Actinopterygii, and Sarcopterygii. However, database searching and analysis of syntenic regions reveal, definitively, that the Alx3 gene has been lost from the genome of at least three vertebrate species: the amphibian X. tropicalis, the squamate reptile A. carolinensis, and the bird G. gallus. These must all be independent losses as proved by the presence of the Alx3 gene in zebrafish, zebra finch, and mammals. Thus, the species which lack the Alx3 gene are on different evolutionary lineages, separated by lineages that possess Alx3 (Fig. 4).
Why should Alx3 be lost repeatedly, but not Alx1 or Alx4? One relevant finding is that sequence analysis suggests that Alx3 and Alx1 are more closely related to each other than they are to Alx4. Consistent with this evolutionary inference, Alx1 and Alx3 (but not Alx4) have similar roles in neural tube closure (Zhao et al. 1996; Lakhwani et al. 2010;), whereas Alx4 is the only member involved in the axial patterning of limbs in chicken and mouse (Takahashi et al. 1998). This suggests that losing Alx3 or Alx1 could be more easily compensated for than losing Alx4. To assess why it is Alx3, not Alx1, that has been repeatedly lost, it is necessary to consider the expression of these genes.
Evolution of Alx gene expression and function in vertebrate embryos
The expression and function of mouse Alx family genes have been well studied (Zhao et al. 1996; Qu et al. 1997b, 1999; Beverdam et al. 2001; Lakhwani et al. 2010). All three genes are expressed in cephalic mesenchyme at early stages of mouse development and, consistent with this, mutations in each Alx gene show similar craniofacial phenotypes including defects of the facial bones. Clinical cases involving mutation of any one of the three human genes also display craniofacial deformity (Wu et al. 2000; Wuyts et al. 2000; Mavrogiannis et al. 2001; Twigg et al. 2009; Uz et al. 2010;). Although there are slight differences between phenotypes, and the exact developmental roles are not known, it is clear that each of the Alx genes plays an important role in craniofacial formation in both mouse and human.
We have analyzed the expression of Alx genes during head development in chick and frog embryos and compared expression with Alx genes of mouse. We find that expression is broadly similar between the three species. More specifically, the two Alx genes of chick or frog are expressed in patterns comparable to the sum of the three Alx genes of mouse. Interestingly, we note that chick Alx1 and Alx4, and Xenopus Alx1, are expressed in the embryonic periocular mesenchyme; in mouse neither Alx1 nor Alx4 shows specific expression in this region. Instead, it is mouse Alx3 that is expressed in the mesenchyme encircling the eye.
This finding may be relevant to understanding the loss of Alx3 in evolution. One possibility is that the loss of Alx3 in chick and frogs was tolerated in evolution because Alx1 or Alx4 expression compensates for the Alx role in periocular mesenchyme, and possibly other roles. Whether this is a reflection of pre-existing overlap of function (redundancy) before gene loss, or a subsequent change in gene expression following gene loss, is unclear. The former may seem more logical, but recent data from mouse mutants (Lakhwani et al. 2010) suggests the latter possibility should not be discounted. Although not noted explicitly by the authors, a recent analysis of Alx3−/− mutant mice showed a slightly altered expression pattern of Alx4, including an apparently novel ring of gene expression close to the eye region (Fig. 3I in Lakhwani et al. 2010). This suggests that Alx genes suppress each other in certain spatial domains, and that after gene loss a degree of functional compensation may occur. If the mechanism whereby Alx3 suppresses expression of Alx1 and Alx4 was established early in vertebrate evolution, then loss of Alx3 might have been readily compensated by activating expression of Alx paralogous genes. In this scenario no alteration of cis-regulatory elements was necessary, implying that Alx3 might be easily lost in evolution. A possible step toward testing this hypothesis would be to use morpholinos to suppress the action of Alx3 mRNA in zebrafish embryos.
If loss of Alx3 can be compensated by paralogous Alx expression, we should also ask why some species retain the Alx3 gene. One possibility is that Alx3 has evolved new roles in some vertebrates. For example, our phylogenetic trees show longer branch lengths leading to Alx3 proteins, suggesting more rapid evolution of peptide sequence. Mouse and human Alx3 proteins also contain discrete functional domains not possessed by other Alx proteins (Perez-Villamil et al. 2004). We suggest, therefore, that retention of Alx3 may be explained through the acquisition of new protein domains, interactions, and roles in certain vertebrate lineages.
These changing gene expression patterns, within and between species, together with multiple gene losses and changes to gene family composition, reveal a dynamic picture of Alx gene structure and function through vertebrate evolution. Similarly, craniofacial development is one of the most complicated aspects of vertebrate embryology, and it too has undergone drastic changes in vertebrate evolution. The relationship between the two is not straightforward, but the study of Alx genes has at least provided a glimpse into the complex relation between genome evolution and the developmental basis of craniofacial evolution.
Acknowledgments
The amphioxus cDNA library was kindly donated by James Langeland. We thank Fiona Sherwood, Shoko Ishibashi, and Enrique Amaya for their assistance with Xenopus work. We are grateful to Tatiana Solovieva and Hisano Takahashi for their help with animal drawings in Fig. 4. The lizard genome assembly was provided by the Broad Institute (http://www.broadinstitute.org/). The sea lamprey genome was sequenced at Washington University (http://genome.wustl.edu/). I. M. M. and A. G. are supported by the Wellcome Trust. This work was also funded in part by the Leverhulme Trust.
REFERENCES
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Beverdam A, Brouwer A, Reijnen M, Korving J, Meijlink F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development. 2001;128:3975–3986. doi: 10.1242/dev.128.20.3975. [DOI] [PubMed] [Google Scholar]
- Beverdam A, Meijlink F. Expression patterns of group-I aristaless-related genes during craniofacial and limb development. Mech. Dev. 2001;107:163–167. doi: 10.1016/s0925-4773(01)00450-6. [DOI] [PubMed] [Google Scholar]
- Bürglin T. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the Homeobox Genes. Oxford: Oxford University Press; 1994. pp. 25–73. [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettensohn CA, Illies MR, Oliveri P, De Jong DL. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development. 2003;130:2917–2928. doi: 10.1242/dev.00511. [DOI] [PubMed] [Google Scholar]
- Galliot B, de Vargas C, Miller D. Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. Dev. Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhwani S, Garcia-Sanz P, Vallejo M. Alx3-deficient mice exhibit folic acid-resistant craniofacial midline and neural tube closure defects. Dev. Biol. 2010;344:869–880. doi: 10.1016/j.ydbio.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Mavrogiannis LA, et al. Haploinsufficiency of the human homeobox gene ALX4 causes skull ossification defects. Nat. Genet. 2001;27:17–18. doi: 10.1038/83703. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS One. 2007;2:e787. doi: 10.1371/journal.pone.0000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, Nei M. Evolutionary change of the numbers of homeobox genes in bilateral animals. Mol. Biol. Evol. 2005;22:2386–2394. doi: 10.1093/molbev/msi229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Villamil B, Mirasierra M, Vallejo M. The homeoprotein Alx3 contains discrete functional domains and exhibits cell-specific and selective monomeric binding and transactivation. J. Biol. Chem. 2004;279:38062–38071. doi: 10.1074/jbc.M400800200. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Qu S, Li L, Wisdom R. Alx-4: cDNA cloning and characterization of a novel paired-type homeodomain protein. Gene. 1997a;203:217–223. doi: 10.1016/s0378-1119(97)00497-6. [DOI] [PubMed] [Google Scholar]
- Qu S, et al. Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development. 1997b;124:3999–4008. doi: 10.1242/dev.124.20.3999. [DOI] [PubMed] [Google Scholar]
- Qu S, Tucker SC, Zhao Q, deCrombrugghe B, Wisdom R. Physical and genetic interactions between Alx4 and Cart1. Development. 1999;126:359–369. doi: 10.1242/dev.126.2.359. [DOI] [PubMed] [Google Scholar]
- Rottinger E, Besnardeau L, Lepage T. A Raf/MEK/ERK signaling pathway is required for development of the sea urchin embryo micromere lineage through phosphorylation of the transcription factor Ets. Development. 2004;131:1075–1087. doi: 10.1242/dev.01000. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 2006;7:R64. doi: 10.1186/gb-2006-7-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Takahashi M, et al. The role of Alx-4 in the establishment of anteroposterior polarity during vertebrate limb development. Development. 1998;125:4417–4425. doi: 10.1242/dev.125.22.4417. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Holland PW. Amphioxus and ascidian Dmbx homeobox genes give clues to the vertebrate origins of midbrain development. Development. 2004;131:3285–3294. doi: 10.1242/dev.01201. [DOI] [PubMed] [Google Scholar]
- Takatori N, et al. Comprehensive survey and classification of homeobox genes in the genome of amphioxus, Branchiostoma floridae. Dev. Genes Evol. 2008;218:579–590. doi: 10.1007/s00427-008-0245-9. [DOI] [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, el Bahi S, Guenet JL, Robert B, Meijlink F. Mouse Alx3: an aristaless-like homeobox gene expressed during embryogenesis in ectomesenchyme and lateral plate mesoderm. Dev. Biol. 1998;199:11–25. doi: 10.1006/dbio.1998.8921. [DOI] [PubMed] [Google Scholar]
- Thompson H, Blentic A, Watson S, Begbie J, Graham A. The formation of the superior and jugular ganglia: insights into the generation of sensory neurons by the neural crest. Dev. Dyn. 2010;239:439–445. doi: 10.1002/dvdy.22179. [DOI] [PubMed] [Google Scholar]
- Twigg SR, et al. Frontorhiny, a distinctive presentation of frontonasal dysplasia caused by recessive mutations in the ALX3 homeobox gene. Am. J. Hum. Genet. 2009;84:698–705. doi: 10.1016/j.ajhg.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uz E, et al. Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia. Am. J. Hum. Genet. 2010;86:789–796. doi: 10.1016/j.ajhg.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, et al. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YQ, Badano JL, McCaskill C, Vogel H, Potocki L, Shaffer LG. Haploinsufficiency of ALX4 as a potential cause of parietal foramina in the 11p11.2 contiguous gene-deletion syndrome. Am. J. Hum. Genet. 2000;67:1327–1332. doi: 10.1016/s0002-9297(07)62963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts W, Cleiren E, Homfray T, Rasore-Quartino A, Vanhoenacker F, Van Hul W. The ALX4 homeobox gene is mutated in patients with ossification defects of the skull (foramina parietalia permagna, OMIM 168500) J. Med. Genet. 2000;37:916–920. doi: 10.1136/jmg.37.12.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Behringer RR, de Crombrugghe B. Prenatal folic acid treatment suppresses acrania and meroanencephaly in mice mutant for the Cart1 homeobox gene. Nat. Genet. 1996;13:275–283. doi: 10.1038/ng0796-275. [DOI] [PubMed] [Google Scholar]


