Abstract
The structure of chromatin regulates the genetic activity of the underlying DNA sequence. We report here that the protein encoded by the proto-oncogene DEK, which is involved in acute myelogenous leukemia, induces alterations of the superhelical density of DNA in chromatin. The change in topology is observed with chromatin but not with naked DNA and does not involve dissociation of core histones from chromatin. Moreover, these effects require histone H2A/H2B dimers in addition to histone H3/H4. We additionally tested whether the DEK protein affects DNA-utilizing processes and found that the DEK protein substantially reduces the replication efficiency of chromatin but not of naked DNA templates.
Keywords: Chromatin structure, DEK, DNA topology, leukemogenesis
Experimental evidence has accumulated in recent years showing that chromatin is not a static structure, but rather is involved in the regulation of many nuclear processes such as transcription and replication (Felsenfeld 1996; Kingston et al. 1996; Peterson 1996; Svaren and Hörz 1996; Kornberg and Lorch 1999). In the eukaryotic nucleus, DNA is packaged into chromatin with its basic repeating motif, the nucleosome, consisting of 146 bp of DNA wrapped around a histone octamer in a left-handed superhelix (Luger et al. 1997). The structure of chromatin affects the accessibility of DNA to sequence-specific transcription factors and to DNA-dependent enzymes such as RNA and DNA polymerases. Chromatin structure can be modulated by different factors. It has been shown, for instance, that the acetylation state of the core histones is a critical determinant of transcriptional activity (Grunstein 1997; Struhl 1998). In addition, mobilization of the nucleosomes can be achieved by ATP-dependent chromatin remodeling factors such as the SWI/SNF and RSC complexes from yeast (Peterson and Tamkum 1995; Cairns et al. 1996), the NURF (Tsukiyama and Wu 1995), CHRAC (Varga-Weisz et al. 1997) and ACF (Ito et al. 1997) complexes from Drosophila, and related activities in human cells (Wang et al. 1996; LeRoy et al. 1998; Tong et al. 1998; Xue et al. 1998).
On the other hand, large regions of the genome can be kept transcriptionally inert by silencing mechanisms that restrict the access of factors to their target sequences (Pirrotta 1997, 1998; Ogbourne and Antalis 1998; Wallrath 1998). Factors that are involved in chromatin repression include the SIR (silent information regulator) proteins from yeast (Grunstein 1998), the heterochromatin binding protein HP1 (Wallrath 1998), and the Polycomb group (PcG) proteins from Drosophila, which are responsible for the maintenance of the inactive state of the homeotic genes (Pirrotta 1998).
We are interested in the influence of chromatin structure on chromatin replication (Alexiadis et al. 1997, 1998; Halmer and Gruss 1997; Halmer et al. 1998; Vestner et al. 1998). During these studies we identified a protein that affects chromatin structure and replication in vitro. This factor was found to be the product of the DEK proto-oncogene, which is involved in acute myeloid leukemia (AML) (von Lindern et al. 1992).
Results and Discussion
Identification of an activity that changes the linking number of DNA in chromatin
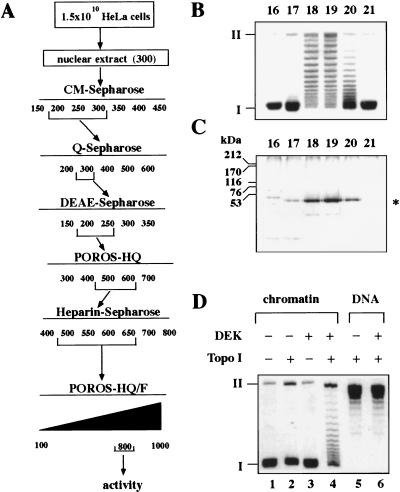
We prepared nuclear extracts from HeLa cells and separated proteins according to the chromatographic scheme outlined in Figure 1A. Individual fractions were incubated in the presence of a cytosolic S100 extract with SV40 minichromosomes (Alexiadis et al. 1998). The DNA was extracted from minichromosomes and investigated by agarose gel electrophoresis (Fig. 1B). We identified an activity that causes a drastic reduction in the number of negative supercoils introduced by the wrapping of DNA around histone octamers. Closed circular DNA with a reduced number of supercoils was visible as numerous topoisomers between fully superhelical form I and relaxed form II DNA (Fig. 1B). By using two-dimensional gel electrophoresis in the presence of chloroquine (Krude and Knippers 1991), we found that the average linking number of nucleosomal SV40 DNA was changed in the presence of this activity from −25 to −12 (data not shown). Analysis of the most purified active fractions by SDS-PAGE revealed a prominent band with an apparent molecular mass of 50 kD (Fig. 1C). The change in chromatin topology (Fig. 1B) was correlated with the amounts of the 50-kD protein. The 50-kD polypeptide was excised from a Coomassie-stained SDS–polyacrylamide gel, subjected to in-gel tryptic cleavage, and analyzed by nanoelectrospray mass spectrometry (Wilm et al. 1996 ). From the total mixture of tryptic peptides, nine were sequenced, which identified the protein as DEK.
Figure 1.
A 50-kD protein reduces the superhelical density of chromatin. (A) Purification scheme of the activity. Numbers indicate NaCl concentration (mm). (B) SV40 chromatin was incubated with the fractions (16–21) from the POROS-HQ/F column in the presence of a cytosolic S100 replication extract. Purified DNA was investigated by agarose gel electrophoresis and ethidium bromide staining. (I) Supercoiled; (II) relaxed closed circular and nicked DNA. (C) Protein composition of the POROS-HQ/F-fractions. Proteins of the fractions (16–21) were separated by SDS-PAGE and visualized by silver staining. The position of size markers and the 50-kD protein (asterisk) is indicated. The faster-migrating minor band on the SDS-polyacrylamide gel most likely consists of a degradation product of the 50-kD protein, as its amount increased after prolonged incubation at 37°C. (D) Topoisomerase dependence of the reaction. SV40 chromatin (lanes 1–4) or relaxed SV40 DNA (lanes 5,6) were incubated in the absence or presence of topoisomerase I and purified DEK protein. Purified products were analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining.
DEK, a 43-kD protein, was initially discovered as a fusion partner of the CAN nucleoporin in a specific subtype of AML (von Lindern et al. 1992). DEK often acts as a major immunoreactive antigen in patients with autoimmune diseases such as juvenile rheumatoid arthritis or lupus erythemathosis. However, despite these significant disease associations, the physiological function of DEK has yet to be determined.
Purified DEK changed the linking number of nucleosomal DNA in the presence of either topoisomerase I (Fig. 1D) or topoisomerase II (not shown). No additional proteins were necessary for the activity, suggesting that the cytosolic extract contributed topoisomerase activity to the reaction shown in Figure 1B. Significantly, the linking number of relaxed naked DNA was not changed by DEK in the presence of topoisomerase I (Fig. 1D), indicating that the changes in DNA topology only occur with chromatin templates (Fig. 1D). Increasing the amounts of purified protein did not result in a complete relaxation of minichromosomal DNA, but produced a wide range of topoisomers between the negatively supercoiled base state and relaxed DNA (data not shown). At a ratio of three molecules of DEK/nucleosome (100 ng DEK/100 ng chromatin), no further change in the topoisomer ladder was observed.
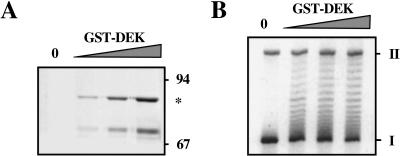
To demonstrate that the change in topology of minichromosomal DNA was caused by the DEK protein, we used recombinant DEK and found that increasing amounts of GST–DEK protein (Fig. 2A) change the topology of chromatin with the same efficiency as the purified native protein preparation (Fig. 2B). In addition, we incubated chromatin with increasing amounts of NAP-1, a protein known to interact with chromatin (Chang et al. 1997). Even with the highest amount of NAP-1, we could not detect any changes in chromatin topology (data not shown), demonstrating that the effect of DEK on DNA topology is specific.
Figure 2.
Recombinant GST–DEK changes the superhelical density of DNA in chromatin. (A) Increasing amounts of GST–DEK were separated by SDS-PAGE and visualized by silver staining. The positions of size markers and GST–DEK (*) are indicated. Lower band consists of a degradation product. (B) DEK assay with GST–DEK. SV40 chromatin was incubated with increasing amounts of GST–DEK in the presence of topoisomerase I. Purified DNA was investigated by agarose gel electrophoresis and ethidium bromide staining. (I) Supercoiled; (II) relaxed closed circular and nicked DNA.
DEK is thought to be a member of a new family of site-specific DNA-binding factors involved in signal transduction and transcriptional regulation (Fu et al. 1997). Thus, we tested whether the DEK-induced change in chromatin topology is affected by different DNA sequences. To this end, SV40, puc18- and HK-plasmid DNA were assembled into chromatin in Drosophila S150 extracts and incubated in the presence of topoisomerase I and DEK. We found that DEK changes the linking number of all three chromatin templates to a similar extent, which indicates that the observed effect is independent of the underlying DNA sequence (data not shown).
DEK does not induce histone displacement
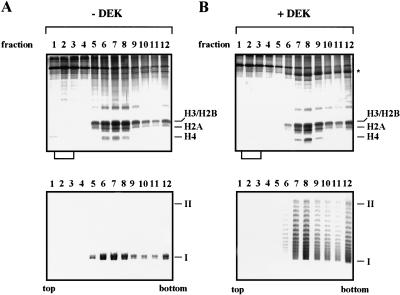
DEK-induced changes in DNA topology could be caused by displacement of nucleosomes. To test this possibility, we incubated chromatin with purified DEK protein and topoisomerase I and subjected the samples to glycerol gradient sedimentation (Fig. 3). Analysis by SDS-PAGE gave no evidence for a DEK-induced loss of histones, which should appear as free proteins close to the top of the gradients (Fig. 3, top). Furthermore, DEK was found in association with minichromosomes (asterisk in Fig. 3B, top, and confirmed by immunoblotting, not shown) and induced the expected change in DNA topology (Fig. 3B, bottom). These findings suggest that the change in linking number induced by the DEK protein is not caused by histone displacement.
Figure 3.
The DEK-induced change in linking number is not due to histone displacement. SV40 minichromosomes were incubated in the absence (A) or presence (B) of DEK and topoisomerase I and separated on glycerol gradients. Proteins of the gradient fractions (lanes 1–12) were analyzed by SDS-PAGE (top) and the DNA topology by agarose gel electrophoresis (bottom). The position of the core histones (H3, H2B, H2A, and H4) and supercoiled (I) and relaxed closed circular and nicked DNA (II) are indicated. Brackets below the SDS-PAGE show the position in which free histones are expected to appear. (Asterisk) Position of DEK protein.
DEK-mediated alteration of DNA topology in chromatin requires H2A/H2B
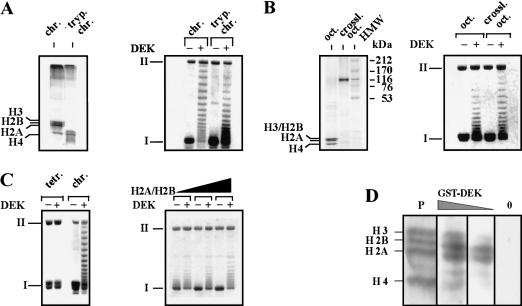
To investigate the components of the nucleosome that are necessary for interaction with DEK, we assembled chromatin with different histone compositions (Fig. 4A–C). First, we treated SV40 minichromosomes with trypsin, which removes the unstructured amino-terminal histone tails (Ausio et al. 1989; Quintini et al. 1996; Fig. 4A, left). Second, we cross-linked core histones within the histone octamer with dimethyl suberimidate (DMS) to prevent dissociation of histones from the octamer (Thomas 1989). Cross-linked histones migrate with an apparent molecular weight of around 100,000 dalton (Fig 4B, left). Third, we deposited H3/H4 tetramers onto DNA with purified H3/H4/N1 complexes (Zucker and Worcel 1990; Fig. 4C). These different substrates were incubated with and without DEK plus topoisomerase I. Agarose gel electrophoresis of extracted DNA revealed that DEK is active on chromatin substrates containing amino-terminally truncated histones (Fig. 4A, right) and cross-linked octamers (Fig. 4B, right). However, we observed that DEK was unable to change the topology of molecules containing only histone H3/H4 tetramers (Fig. 4C, left). Significantly, re-addition of H2A/H2B dimers to these templates fully restored DEK activity (Fig. 4C, right). These results demonstrate that the interaction of DEK with histone H2A/H2B dimers is necessary for the DEK-mediated change in topology of nucleosomal DNA. To provide evidence for a direct interaction of DEK with histone proteins, we performed a Far-Western blot analysis (Fig. 4D). We observed strong binding of DEK to histones H2A and H2B and a much weaker interaction with histones H3 and H4. No signal was obtained without the addition of DEK, excluding any cross-reactivities of histones with our antibody. By reducing the concentration of DEK in our assay (25 ng/cm2 filter), the binding was restricted mainly to H2A and H2B. The specificity of DEK for histones H2A and H2B explains the inability of DEK to change the topology of templates consisting of H3/H4 tetramers only.
Figure 4.
The DEK-induced change in topology depends on the presence of H2A/H2B dimers. (A, left) The amino-terminal histone domains of SV40 minichromosomes were removed by trypsin. Polypeptides were investigated by SDS-PAGE and visualized by silver staining. (Right) Non-treated chromatin (chr.) and trypsinized chromatin (tryp. chrom.) was used as substrate in the DEK assay. (B, left) Histone octamers (oct.) were cross-linked with DMS (crossl. oct.) and analyzed by SDS-PAGE stained with Coomassie brilliant blue. (HMW) High molecular weight marker in kilodaltons. (Right) SV40 DNA was reconstituted with control or cross-linked octamers and used as substrate in the DEK assay. (C, left) SV40 DNA was reconstituted with purified H3/H4 tetramers (tetr.) or histone octamers (chrom.). Chromatin was incubated in the absence or presence of DEK. (Right) H3/H4 containing chromatin was reconstituted with increasing amounts of H2A/H2B dimers and used as substrate in the DEK assay. (D) Far-Western blot analysis. Core histones were separated on an SDS–polyacrylamide gel, electroblotted, and renatured. The membrane was stained with Ponceau red (P) and incubated with decreasing amounts of GST–DEK (100 ng/cm2; 25 ng/cm2) and without DEK (0). The position of the core histones are indicated.
DEK reduces the replication efficiency of chromatin templates
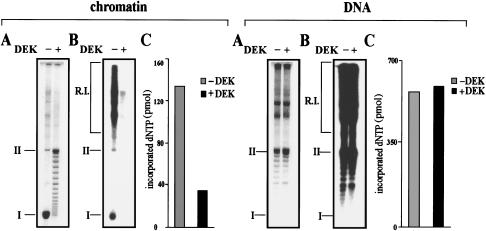
For genetic processes such as transcription or replication, initiation factors must gain access to their specific binding sites (Alexiadis et al. 1998). To determine whether DEK reduces the overall accessibility of chromatin, we treated SV40 minichromosomes with endonucleases. Both restriction endonuclease MboI (which cuts SV40 DNA at eight sites) and micrococcal nuclease (which attacks the linker DNA between nucleosomes) degrade DEK-treated minichromosomes with strongly reduced efficiency compared with untreated minichromosomes (not shown). More specifically, we investigated whether the DEK-mediated change in chromatin topology has an influence on chromatin replication by using DEK-treated minichromosomes that were preincubated with DEK as templates in the in vitro replication system (Fig. 5). Replication products were deproteinized and analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining (Fig. 5A, chromatin) and autoradiography (Fig. 5B, chromatin). Total incorporated nucleotides were determined by TCA precipitation (Fig. 5C, chromatin). We found that the replication efficiency of chromatin templates was significantly reduced in the presence of DEK (Fig. 5B,C), concomitant with a change in the topology (Fig. 5A), demonstrating that the altered chromatin structure inhibits the access of replication factors to the DNA template. In addition, we investigated the effects of DEK on the replication of protein-free DNA. However, no significant differences were detected in the replication efficiencies between protein-free DNA in the absence or presence of DEK (Fig. 5, DNA).
Figure 5.
DEK inhibits the replication of chromatin but not of protein-free DNA. (A) SV40 DNA and SV40 chromatin were preincubated with DEK and topoisomerase I and used as templates in the SV40 in vitro replication system. Purified DNA was separated on an agarose gel and visualized by ethidium bromide staining. (B) Autoradiography of the agarose gel shown in A. The position of replicative intermediates (R.I.) of supercoiled (I) and relaxed closed circular and nicked molecules (II) are indicated. (C) Nucleotide incorporation (pmole) of the samples shown in B, as determined by TCA precipitation.
Taken together, these data reveal that DEK specifically interacts with chromatin, alters the topology of nucleosomal DNA, and reduces the efficiency with which chromatin is replicated. Thus, DEK protein may have a global role in the structure and activity of chromatin. It is an interesting question whether the DEK–CAN fusion protein, expressed in AML (von Lindern et al. 1992), is also able to perform these changes and whether these effects contribute to the transforming potential of the protein.
Materials and methods
Purification of DEK
HeLa cells (1.5 × 1010; Computer Cell Culture Center, SA, Belgium) were resuspended in hypotonic buffer (20 mm HEPES-KOH at pH 7.4, 5 mm KCl, 1.5 mm MgCl2, 0.1 mm DTT) and homogenized in a Dounce homogenizer. Nuclei were pelleted for 10 min at 9000 rpm (SS34) at 4°C. Nuclear proteins were eluted for 45 min with 2 volumes of buffer A-300 (20 mm HEPES at pH 7.6, 300 mm NaCl, 10 mm sodium bisulfite, 1 mm EDTA) and centrifuged for 30 min at 9000 rpm (SS34) at 4°C. The supernatant was adjusted to 100 mm NaCl, 10% glycerol, 10 mm β-mercaptoethanol, and supplied with a protease inhibitor cocktail (Boehringer Mannheim) and stirred for 15 min on ice.
About 200 ml of nuclear extract were purified according to the flow chart outlined in Figure 1A. All columns were pre-equilibrated with buffer A-100 and eluted with the indicated salt steps. For purification, we used the following columns: a 20-ml CM Sepharose column (Pharmacia), a 3-ml Q Sepharose column (Pharmacia), a 1.2-ml DEAE–Sepharose column (Pharmacia), a 0.4-ml POROS/HQ column (Boehringer Mannheim), and a 0.5-ml heparin–Sepharose column (Pharmacia). Columns were eluted with the indicated salt steps with the following total volumes: 140, 30, 14.4, 6, and 10 ml, respectively. Finally, DEK-containing fractions were loaded onto a 0.8-ml POROS-HQ/F column (Boehringer Mannheim), washed with 5 volumes of buffer A-100, and eluted with a 12-ml linear gradient. From 1.5 × 1010 HeLa cells, ∼20 μg of DEK protein could be purified to near homogeneity.
DEK assay (reduction of the superhelical density of chromatin)
A total of 4 μl of the fractions (∼100 ng of DEK protein) were dialyzed on a Whatman Filter (Type VS, pore size 0.025 μm) against buffer A-100 in the presence of 1 μg/μl BSA (Pharmacia) for 90 min at 4°C and 100-ng salt-treated SV40 minichromosomes (Gruss and Knippers 1995) were incubated with the dialyzed fraction in the presence of 5 units of topoisomerase I (Promega) and 200 ng/μl BSA in 20 mm HEPES at pH 7.6, 120 mm NaCl, 1 mm EDTA, and 10 mm sodiumbisulfite for 30 min at 37°C. Purified DNA was separated on an 0.8% agarose gel in 0.5× TBE-buffer at 3.5 V/cm for 16 hr.
Sedimentation analysis
Salt-treated SV40 minichromosomes (8 μg) were incubated in the presence or absence of 8 μg of DEK with 30 units of topoisomerase I for 1 hr at 37°C in buffer A100/0.2 μg/μl BSA. Complexes were loaded on 4-ml 10%–35% glycerol gradients (20 mm HEPES at pH 7.6, 10 mm sodium disulfite, 1 mm EDTA, 100 mm NaCl) and run in a TLS55 rotor for 1 hr at 55,000 rpm at 4°C. TCA-precipitated proteins were separated on an 18% SDS–polyacrylamide gel and stained with silver. DNA was investigated by 0.8% agarose gel electrophoresis. In the absence of chromatin, the DEK protein was found in fractions 1–3 (not shown).
Far-Western blot analysis
Core histones (5 μg) were separated on an 18% SDS-PAGE and electroblotted to polyvinylidene difluoride membranes (Immobilon-P; Millipore). After transfer, the blots were stained with Ponceau red, and the positions of the four core histones were marked. Renaturation of the core histones was done in PBS-0.05% Tween 20 (PBST) for 2 hr at room temperature, and the membrane was blocked in PBS–BSA (PBS, 2% BSA, 0.5% NP40) for 2 hr at room temperature. The membrane was incubated with recombinant GST–DEK protein (250–100 ng/cm2 filter) in PBS–BSA for 2 hr at room temperature, and DEK binding was detected by antibody staining with ECL (Amersham).
In vitro replication assay
The experiments were carried out as described previously (Gruss et al. 1993) using purified SV40 T-Ag as initiator protein and cytosolic HeLa S100 extract as source for replication enzymes. For in vitro replication, 400 ng of salt-treated SV40 minichromosomes and 100 ng of SV40 DNA were preincubated with 400 ng of DEK, 3 mm ATP, and 2 μg of T-Ag for 30 min at 37°C, and then used as templates exactly as described (Alexiadis et al. 1998). Purified replication products were run on an 0.8% agarose gel in TBE buffer (14 hr, 3.5 V/cm). One-tenth of the replication assay was precipitated with TCA to determine the incorporated nucleotides.
Acknowledgments
We thank R. Mettke for technical assistance, B. Vestner for cross-linked histone octamers, Wen Jiang for purified NAP1 protein, and G. Grosveld for DEK expression vectors. We are very grateful to Jim Kadonaga for critical comments on the manuscript. This work was supported by grants from DFG to C.G. Work in the Protein Interaction Laboratory is supported by the Danish National Research Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Claudia.Gruss@uni-konstanz.de; FAX 49 7531 884036.
References
- Alexiadis V, Halmer L, Gruss C. Influence of core histone acetylation on SV40 minichromosomes replication in vitro. Chromosoma. 1997;105:324–331. doi: 10.1007/BF02529747. [DOI] [PubMed] [Google Scholar]
- Alexiadis V, Varga-Weisz PD, Bonte E, Becker PB, Gruss C. In vitro chromatin remodelling by CHRAC at the SV40 origin of DNA replication. EMBO J. 1998;17:3428–3438. doi: 10.1093/emboj/17.12.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausio J, Dong F, van Holde KE. Use of selectively trysinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Chang L, Loranger SS, Mizzen C, Ernst SG, Allis CD, Annunziato AT. Histones in transit: Cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 1997;36:469–480. doi: 10.1021/bi962069i. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin unfolds. Cell. 1996;86:16–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- Fu GK, Grosveld G, Markovitz DM. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc Natl Acad Sci. 1997;94:1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- ————— Yeast heterochromatin: Regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Gruss C, Knippers R. The SV40 minichromosome. In: Adolph KW, editor. Methods in molecular genetics. Orlando, FL: Academic Press; 1995. pp. 101–113. [Google Scholar]
- Gruss C, Wu J, Koller T, Sogo JM. Disruption of the nucleosomes at the replication fork. EMBO J. 1993;12:4533–4545. doi: 10.1002/j.1460-2075.1993.tb06142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halmer L, Gruss C. Accessibility for topoisomerase I and II regulates the replication efficiency of SV40 minichromosomes in vitro. Mol Cell Biol. 1997;17:2624–2630. doi: 10.1128/mcb.17.5.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halmer L, Vestner B, Gruss C. Involvement of topoisomerases in the initiation of simian virus 40 minichromosome replication. J Biol Chem. 1998;274:34792–34798. doi: 10.1074/jbc.273.52.34792. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes & Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Chromatin-modifying and remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- Krude T, Knippers R. Transfer of nucleosomes from parental to replicated chromatin. Mol Cell Biol. 1991;11:6257–6267. doi: 10.1128/mcb.11.12.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Ogbourne S, Antalis TM. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998;331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL. Multiple switches to turn on chromatin. Curr Biol. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Tamkum JW. The SWI-SNF complex: A chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. PcG complexes and chromatin silencing. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- ————— Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- Quintini G, Treuner K, Gruss C, Knippers R. Role of amino-terminal histone domains in chromatin replication. Mol Cell Biol. 1996;16:2888–2897. doi: 10.1128/mcb.16.6.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Svaren J, Hürz W. Regulation of gene expression by nucleosomes. Curr Opin Genet Dev. 1996;6:164–170. doi: 10.1016/s0959-437x(96)80046-3. [DOI] [PubMed] [Google Scholar]
- Thomas JO. Chemical cross-linking of histones. Methods Enzymol. 1989;170:549–563. doi: 10.1016/0076-6879(89)70064-1. [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Vestner B, Bustin M, Gruss C. Stimulation of replication efficiency of a chromatin template by chromosomal protein HMG-17. J Biol Chem. 1998;273:9409–9414. doi: 10.1074/jbc.273.16.9409. [DOI] [PubMed] [Google Scholar]
- von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath LL. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- Wang W, Coté J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zucker K, Worcel A. The histone H3/H4 N1 complex supplemented with histone H2A–H2B dimers and DNA topoisomerase I forms nucleosomes on circular DNA under physiological conditions. J Biol Chem. 1990;266:14487–14496. [PubMed] [Google Scholar]