Abstract
Recent reports show that ER stress plays an important role in diabetic retinopathy (DR), but ER stress is a complicated process involving a network of signaling pathways and hundreds of factors, What factors involved in DR are not yet understood. We selected 89 ER stress factors from more than 200, A rat diabetes model was established by intraperitoneal injection of streptozotocin (STZ). The expression of 89 ER stress-related factors was found in the retinas of diabetic rats, at both 1- and 3-months after development of diabetes, by quantitative real-time polymerase chain reaction arrays. There were significant changes in expression levels of 13 and 12 ER stress-related factors in the diabetic rat retinas in the first and third month after the development of diabetes, Based on the array results, homocysteine- inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1(HERP), and synoviolin(HRD1) were studied further by immunofluorescence and Western blot. Immunofluorescence and Western blot analyses showed that the expression of HERP was reduced in the retinas of diabetic rats in first and third month. The expression of Hrd1 did not change significantly in the retinas of diabetic rats in the first month but was reduced in the third month.
1. Introduction
Diabetic retinopathy (DR) is one of the severe complications of diabetes leading to loss of vision. Although the pathogenic mechanism of DR has been investigated for many years and a number of theories have been proposed [1, 2], the mechanism of DR remains unknown and needs further exploration.
Some diabetic patients are susceptible to DR, while others are quite resistant or develop minimal pathological changes [3]. It may be supposed that such DR-resistant patients are protected genetically. The existence of a DR-resistant gene was proposed, and a comparative study was performed of the gene expression between susceptible and resistant DR patients [4]. It was found that many endoplasmic reticulum (ER) stress-related factors are highly expressed in non-DR diabetic patients.
In our earlier work, we found that P58IPK/DNAJC3, an ER stress-related factor, binds to the ER transmembrane protein PERK (protein kinase RNA-activated- (PKR-) like ER kinase), which is normally activated by the ER stress/unfolded protein response. By binding to PERK, P58IPK thereby inhibits its phosphorylation of the α-subunit of eukaryotic translation initiation factor 2 (eIF-2α) and thus compromises eIF2/EIF2S3's mediator role in the translation of mRNA [5]. In this way, P58IPK inhibits ER stress in the endothelial cells of human retinal vessels. P58IPK also downregulates the expression of vascular endothelial growth factor (VEGF), which is associated with regulation of the pathology of DR [6]. VEGF plays a key role in DR [7, 8] and is regulated at the transcriptional level by the unfolded protein response pathway [9]. Recent reports also show that ER stress plays an important role in DR [10, 11]. Li et al. [12] demonstrated that multiple ER stress markers, including 78 kDa glucose-regulated protein (GRP78), phosphoinositol-requiring transmembrane kinase (IRE)1α, and phosphor-eIF2α were significantly upregulated in the retinas of animal models of type 1 diabetes and oxygen-induced retinopathy. Our recent work suggests that early progression of DR may be mediated by ER stress, but probably does not involve changes in activating transcription factor (ATF)4 or GRP78 [13]. Together, these studies suggest that although ER stress is involved in the development of DR, its specific pathogenesis is not yet understood.
ER stress is a complicated process involving a network of signaling pathways and hundreds of factors that function by triggering the PERK, IRE1 and ATF6 signaling pathways [14–16]. In order to delve into the effects of these ER stress-related factors on DR, we classified them into 11 categories according to function (Figure 1, Table 3), based on Jonikas et al. [17]. We selected 89 ER stress factors from more than 200, based on our work and that of others (Table 4) [13, 17–21]. These factors contain the 11 categories of ER stress. Expression of these factors in the retinas of diabetic rats was determined by quantitative real-time PCR (Q-PCR) arrays to find the specific factors and the ER stress signaling pathways that may play a key role in the pathogenesis of DR.
Figure 1.
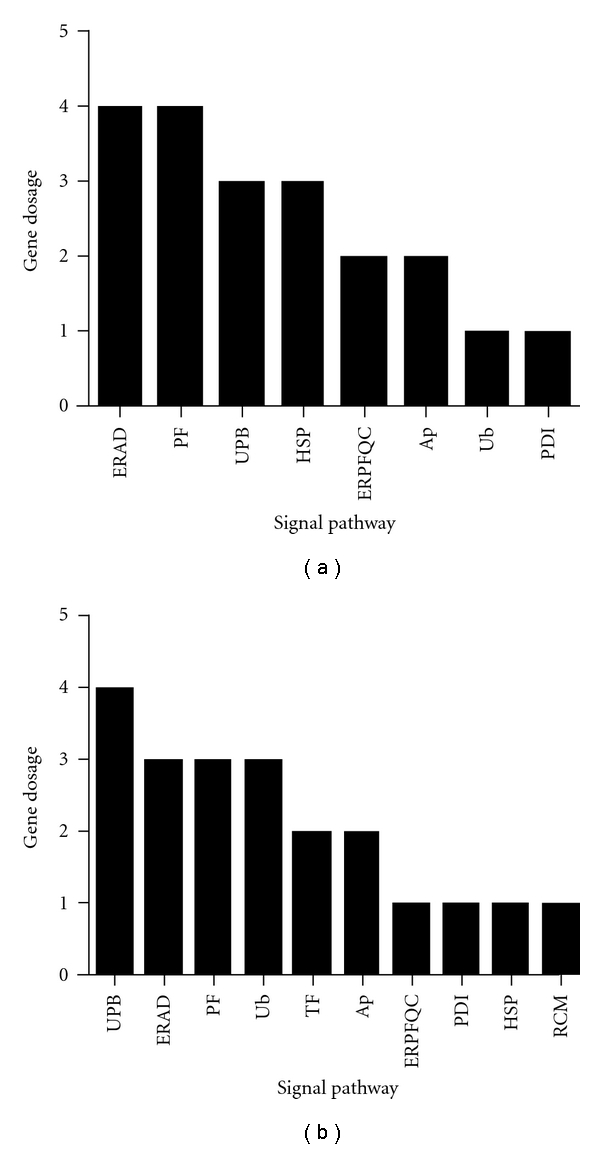
Assessment of the expression of ER stress-related factors in diabetic retinas in the first and third months after the development of diabetes by Q-PCR arrays. (a) the histogram of the expression of different genes in 11 signaling pathways related to ER stress after the first month; (b) the histogram of the expression of different genes in 11 signaling pathways related to ER stress after the third month. Unfolded Protein Binding: UPB, ER Protein Folding Quality Control: ERPFQC, Regulation of Cholesterol Metabolism: RCM, ER-associated degradation: ERAD, Ubiquitination: Ub, Transcription Factors: TF, Protein Folding: PF, Protein Disulfide Isomerization: PDI, Heat Shock Proteins: HSP, Apoptosis: Ap°.
Table 3.
Q-PCR arrays showed that the expression of the ER stress factor had significant differences in the first and the third month in diabetic rat retina: the ER stress factor of significant differences belongs to different ER stress signaling pathways.
| Signaling pathway | First month | Third month |
|---|---|---|
| Unfolded protein binding | Cctd, ERdj4, Hspt70-3 | ERdj4, OMI/PARK13, CYPA, HUGT1 |
| ER protein folding quality control | GluII, PDIA10 | HUGT1 |
| Regulation of cholesterol Metabolism | — | SREBP1 |
| Regulation of translation | — | — |
| ERAD | Herp, NUC, Os9, ADO15 | Herp, OMI/PARK13, Hrd1 |
| Ubiquitination | Herp | Herp, UfD1, TGT |
| Transcription factors | — | ATF4, SREBP1 |
| Protein folding | Cctd, ERdj4, APG-1, PDIA10 | ERdj4, Ero1l, CYPA |
| Protein disulfide isomerization | PDIA10 | SREBP1 |
| Heat shock proteins | ERdj4, P58IPK, Hspt70-3 | ERdj4 |
| Apoptosis | JNK/JNK1, Casp12 | OMI/PARK13, NIPK/Trib3 |
Table 4.
Q-PCR array gene table. We selected 89 ER stress-related factors, and other 6 genes as a quality control a total; of 96 genes were detected in Q-PCR arrays.
| A01 | Rn.107561 | XM_341644 | AMFR | Autocrine motility factor receptor | AMFR |
| A02 | Rn.161941 | NM_001108183 | ARMET | Arginine rich, mutated in early-stage tumors | ARMET |
| A03 | Rn.2423 | NM_024403 | ATF4 | Activating transcription factor 4 (tax-responsive enhancer element B67) | CREB-2/CREB2 |
| A04 | Rn.222130 | NM_001107196 | ATF6 | Activating transcription factor 6 | ATF6A |
| A05 | Rn.18179 | NM_001002809 | ATF6B | Activating transcription factor 6 beta | CREB-RP/CREBL1 |
| A06 | Rn.42932 | NM_021702 | ATXN3 | Ataxin 3 | AT3/ATX3 |
| A07 | Rn.10668 | NM_017059 | BAX | BCL2-associated X protein | BCL2L4 |
| A08 | Rn.974 | NM_022399 | CALR | Calreticulin | CRT/RO |
| A09 | Rn.1762 | NM_172008.2 | CANX | Calnexin | CNX/IP90 |
| A10 | Rn.97889 | NM_182814.2 | CCT4 | Chaperonin containing TCP1, subunit 4 (delta) | CCT-DELTA/Cctd |
| A11 | Rn.62267 | NM_001106603.1 | CCT7 | Chaperonin containing TCP1, subunit 7 (eta) | CCT-ETA/Ccth |
| A12 | Rn.6479 | NM_024125.4 | CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | C/EBP-beta |
| B01 | Rn.104043 | NM_001013092.1 | CREB3 | CAMP responsive element binding protein 3 | LUMAN/LZIP |
| B02 | Rn.20059 | NM_001012115.1 | CREB3L3 | CAMP responsive element binding protein 3-like 3 | CREB-H/CREBH |
| B03 | Rn.11183 | NM_001109986 | DDIT3 | DNA-damage-inducible transcript 3 | CEBPZ/CHOP |
| B04 | Rn.110990 | NM_001014202.1 | DERL1 | Der1-like domain family, member 1 | DER-1/DER1 |
| B05 | Rn.11209 | NM_031627 | CHOP | Rattus norvegicus nuclear receptor subfamily 1, group H, member 3 | LXRalpha/Nr1h3 |
| B06 | Rn.40780 | NM_001109541 | DNAJB2 | DnaJ (Hsp40) homolog, subfamily B, member 2 | HSJ1/HSPF3 |
| B07 | Rn.29778 | NM_012699 | DNAJB9 | DnaJ (Hsp40) homolog, subfamily B, member 9 | DKFZp564F1862/ERdj4 |
| B08 | Rn.8642 | NM_001106486 | DNAJC10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | DKFZp434J1813/ERdj5 |
| B09 | Rn.162234 | NM_022232 | DNAJC3 | DnaJ (Hsp40) homolog, subfamily C, member 3 | HP58/P58 |
| B10 | Rn.91398 | NM_001013196 | DNAJC4 | DnaJ (Hsp40) homolog, subfamily C, member 4 | DANJC4/HSPF2 |
| B11 | Rn.107459 | NM_001033909 | Elf2 | E74-like factor 2 | Elf2 |
| B12 | Rn.81078 | NM_130422 | Casp12 | Caspase 12 | Casp12 |
| C01 | Rn.198593 | NM_001109339 | eIF2A | Eukaryotic translation initiation factor 2A, 65 kDa | CDA02/EIF-2A |
| C02 | Rn.24897 | NM_031599 | EIF2AK3 | Eukaryotic translation initiation factor 2-alpha kinase 3 | DKFZp781H1925/HRI |
| C03 | Rn.19198 | NM_001037208 | CRELD2 | cysteine-rich with EGF-like domains 2 | Creld2 |
| C04 | Rn.218563 | XM_344959.3 | ERN2 | Endoplasmic reticulum to nucleus signaling 2 | Ern2 |
| C05 | Rn.64648 | NM_138528 | ERO1L | ERO1-like (S. cerevisiae) | Ero1l |
| C06 | Rn.22325 | NM_144755 | TRB3 | Tribbles homolog 3 | NIPK/Trib3 |
| C07 | Rn.2459 | NM_001008317 | ERP44 | Thioredoxin domain containing 4 (endoplasmic reticulum) | PDIA10/TXNDC4 |
| C08 | Rn.57325 | NM_138917 | FBXO6 | F-box protein 6 | FBG2/FBS2 |
| C09 | Rn.99241 | NM_001106334 | GANAB | Glucosidase, alpha; neutral AB | G2AN/GluII |
| C10 | Rn.23744 | NM_001145840 | GANC | Glucosidase, alpha; neutral C | MGC138256 |
| C11 | Rn.4028 | NM_053523 | HERPUD1 | Homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | Sup |
| C12 | Rn.1950 | NM_212504 | HSPA1B | Heat shock 70 kDa protein 1B | HSP70-1B/HSP70-2/Hsp72 |
| D01 | Rn.187184 | NM_212546 | HSPA1L | heat shock protein 1-like | Hsp70-3/MGC112562/MGC114222 |
| D02 | Rn.211303 | NM_021863 | HSPA2 | Heat shock protein 2 | Hspt70/Hst70/MGC93458 |
| D03 | Rn.163092 | NM_153629 | HSPA4 | Heat shock protein 4 | Hsp110/ Hsp70/irp94 |
| D04 | Rn.144829 | NM_001106428 | HSPA4L | Heat shock protein 4-like | APG-1; MGC187594; OSP94 |
| D05 | Rn.11088 | NM_013083 | HSPA5 | Heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | BIP/GRP78 |
| D06 | Rn.37805 | NM_001011901 | HSPH1 | Heat shock 105 kDa/110 kDa protein 1 | DKFZp686M05240/HSP105 |
| D07 | Rn.107325 | NM_001106599 | HTRA2 | HtrA serine peptidase 2 | OMI/PARK13 |
| D08 | Rn.163330 | NM_001107321 | HTRA4 | HtrA serine peptidase 4 | FLJ90724 |
| D09 | Rn.772 | NM_022392 | INSIG1 | Insulin-induced gene 1 | CL-6 |
| D10 | Rn.16736 | NM_178091 | INSIG2 | Insulin-induced gene 2 | MGC26273 |
| D11 | Rn.9911 | NM_012806 | MAPK10 | Mitogen-activated protein kinase 10 | JNK3/JNK3A |
| D12 | Rn.4090 | XM_001056513 | MAPK8 | Mitogen-activated protein kinase 8 | JNK/JNK1 |
| E01 | Rn.9910 | NM_017322 | MAPK9 | Mitogen-activated protein kinase 9 | JNK-55/JNK2 |
| E02 | Rn.2362 | NM_053569 | MBTPS1 | Membrane-bound transcription factor peptidase, site 1 | PCSK8/S1P |
| E03 | Rn.212224 | NM_001035007 | MBTPS2 | Membrane-bound transcription factor peptidase, site 2 | S2P |
| E04 | Rn.144645 | NM_080577 | NPLOC4 | Nuclear protein localization 4 homolog (S. cerevisiae) | NPL4 |
| E05 | Rn.1492 | NM_053463 | NUCB1 | Nucleobindin 1 | DKFZp686A15286/NUC |
| E06 | Rn.1579 | NM_001007265 | OS9 | Osteosarcoma amplified 9, endoplasmic reticulum associated protein | OS-9 |
| E07 | Rn.11527 | NM_017319 | PDIA3 | Protein disulfide isomerase family A, member 3 | ER60/ERp57 |
| E08 | Rn.7627 | NM_001109476 | PFDN2 | Prefoldin subunit 2 | PFD2 |
| E09 | Rn.3401 | NM_001106794 | PFDN5 | Prefoldin subunit 5 | MM-1/MM1 |
| E10 | Rn.1463 | NM_017101 | PPIA | Peptidylprolyl isomerase A (cyclophilin A) | CYPA/CYPH |
| E11 | Rn.2232 | NM_133546 | PPP1R15A | Protein phosphatase 1, regulatory (inhibitor) subunit 15A | GADD34 |
| E12 | Rn.104417 | NM_001106806 | PRKCSH | Protein kinase C substrate 80K-H | AGE-R2/G19P1 |
| F01 | Rn.209127 | NM_001127545 | RNF139 | Ring finger protein 139 | HRCA1/RCA1 |
| F02 | Rn.209127 | NM_006913 | RNF5 | Ring finger protein 5 | RING5/RMA1 |
| F03 | Rn.4224 | NM_013067 | RPN1 | Ribophorin I | DKFZp686B16177/OST1 |
| F04 | Rn.99548 | NM_001100966 | SCAP | SREBF chaperone | KIAA0199 |
| F05 | Rn.98327 | NM_001034129 | SEC62 | SEC62 homolog (S. cerevisiae) | Dtrp1/HTP1 |
| F06 | Rn.24233 | NM_001107637 | SEC63 | SEC63 homolog (S. cerevisiae) | ERdj2/PRO2507 |
| F07 | Rn.20802 | NM_177933 | SEL1L | Sel-1 suppressor of lin-12-like (C. elegans) | IBD2/PRO1063 |
| F08 | Rn.4197 | NM_173120 | SELS | Selenoprotein S | AD-015/ADO15 |
| F09 | Rn.2119 | NM_030835 | SERP1 | Stress-associated endoplasmic reticulum protein 1 | RAMP4 |
| F10 | Rn.103851 | NM_199376 | SIL1 | SIL1 homolog, endoplasmic reticulum chaperone (S. cerevisiae) | BAP/MSS |
| F11 | Rn.221929 | XM_001075680 | SREBF1 | Sterol regulatory element binding transcription factor 1 | SREBP-1c/SREBP1 |
| F12 | Rn.41063 | NM_001033694 | SREBF2 | Sterol regulatory element binding transcription factor 2 | SREBP2/bHLHd2 |
| G01 | Rn.162486 | NM_001100739 | SYVN1 | Synovial apoptosis inhibitor 1, synoviolin | HRD1 |
| G02 | Rn.7102 | NM_012670 | TCP1 | T-complex 1 | CCT-alpha/CCT1 |
| G03 | Rn.20041 | NM_153303 | TOR1A | Torsin family 1, member A (torsin A) | DQ2/DYT1 |
| G04 | Rn.139603 | NM_001106380 | UBE2G2 | Ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) | UBC7 |
| G05 | Rn.106299 | NM_001007655 | UBE2J2 | Ubiquitin-conjugating enzyme E2, J2 (UBC6 homolog, yeast) | NCUBE2/PRO2121 |
| G06 | Rn.2022 | NM_001012025 | UBXN4 | UBX domain protein 4 | UBXD2/UBXDC1 |
| G07 | Rn.11946 | NM_053418 | UFD1L | Ubiquitin fusion degradation 1-like (yeast) | UFD1 |
| G08 | Rn.162227 | NM_133596 | UGCGL1 | UDP-glucose ceramide glucosyltransferase-like 1 | HUGT1 |
| G09 | Rn.107678 | NM_019381 | BI-1 | Transmembrane BAX inhibitor motif containing 6 | Tmbim6 |
| G10 | Rn.11790 | NM_001008301 | USP14 | Ubiquitin-specific peptidase 14 (tRNA-guanine transglycosylase) | TGT |
| G11 | Rn.98891 | NM_053864 | VCP | Valosin-containing protein | IBMPFD/TERA |
| G12 | Rn.101044 | NM_001004210 | XBP1 | X-box binding protein 1 | TREB5/XBP2 |
| H01 | Rn.973 | NM_001007604 | Rplp1 | Ribosomal protein, large, P1 | MGC72935 |
| H02 | Rn.47 | NM_012583 | Hprt | Hypoxanthine guanine phosphoribosyl transferase | Hgprtase/Hprt1 |
| H03 | Rn.92211 | NM_173340 | Rpl13a | Ribosomal protein L13A | Rpl13a |
| H04 | Rn.107896 | NM_017025 | Ldha | Lactate dehydrogenase A | Ldh1 |
| H05 | Rn.94978 | NM_031144 | Actb | Actin, beta | Actx |
| H06 | N/A | U26919 | RGDC | Rat genomic DNA contamination | RGDC |
| H07 | N/A | SA_00104 | RTC | Reverse Transcription Control | RTC |
| H08 | N/A | SA_00104 | RTC | Reverse transcription control | RTC |
| H09 | N/A | SA_00104 | RTC | Reverse transcription control | RTC |
| H10 | N/A | SA_00103 | PPC | Positive PCR control | PPC |
| H11 | N/A | SA_00103 | PPC | Positive PCR control | PPC |
| H12 | N/A | SA_00103 | PPC | Positive PCR control | PPC |
2. Methods
2.1. Diabetic Rat Model
Two-month-old male Sprague Dawley rats weighing 150 to 200 g were obtained from the animal center of Huazhong University of Science and Technology. Care, use, and treatment of animals were approved by the laboratory animal center of Huazhong University of Science and Technology. Rats were randomly divided into diabetic and control groups (n = 30 per group). The diabetic model was created by intraperitoneal injection of a single dose of streptozotocin (STZ; 65 mg/kg in 0.01 M citrate buffer, pH 4.5) [22]. Nondiabetic rats (the control group) were injected with citrate buffer only. Fasting plasma glucose was examined 3 d after STZ injection, and diabetes was confirmed by a value ≥16.7 mmol/L using Touch Glucometer (Boehringer Mannheim Diagnostics, Indianapolis, IN). Our previous work [13] and that of others [23] have established that in the STZ-induced diabetes model, diabetic retinopathy develops within one month of the development of diabetes. Accordingly, one and three months after the STZ injection, the retinas were separated from the eyes of both the diabetic and control groups. RNA was extracted and assessed using Q-PCR arrays, with 9 rats in each group.
2.2. Quantitative Real-Time RNA Polymerase Chain Reaction (Q-PCR) Arrays
The mRNA levels of 96 factors (89 ER stress-related factors and 7 quality control factors) were measured using Q-PCR arrays. Total RNA was extracted from rat retinal tissue using Trizol reagent (Invitrogen, Carlsbad, USA) according to the manufacturer's instructions. RNA was treated with DNAase (Invitrogen, Carlsbad, USA) and purified using Rneasy MinElute Clean-up Kit (Quiagen, Hilden, Germany). The cDNA was then synthesized using a SuperScript III kit (Invitrogen, Carlsbad, USA). Removing the plate seal from the PCR Array (SABioscience, Frederick, USA) and adding the cocktails to the PCR Array, Q-PCR was performed by using the Hot Star polymerase kit (Qiagen, Venlo, The Netherlands) with SYBR Green technology (ABI, Tampa, FL). PCR reaction buffer was added to a 384-well PCR array plate which was then tightly sealed with an optical adhesive cover. The thermocycling program consisted of 95°C for 10 min, then 40 cycles at 95°C for 15 s, and 60°C for one minute, then compared the differential expression of gene between the two groups.
2.3. Immunofluorescence
Immunofluorescence was performed on 5 μm frozen sections. Briefly, retinal sections were incubated with a rabbit anti-HERP (Santa Cruz Biotechnology, Santa Cruz, Calif) or anti-Hrd1 (Biosynthesis Biotechnology, Beijing, China) antibody (1 : 200) at 4°C overnight. This was followed by the secondary antibody, fluorescein-conjugated goat antirabbit IgG (Antigene, Wu Han, China), for one hour. The slides were visualized and photographed under a fluorescence microscope (Olympus, Hamburg, Germany).
2.4. Western Blot
Total protein was extracted from rat retinal tissue in 300 μL lysis buffer (50 mM Tris pH 7.5, 0.5 M NaCl, 1% NP-40, 1% sodium deoxycholate monohydrate, 2 mM EDTA, and 0.1% SDS). After centrifugation at 1000 ×g for 3 min, protein extracts were diluted with sample buffer (126 mM Tris HCl pH 6.8, containing 20% glycerol, 4% SDS, 0.005% bromophenol blue, and 5% 2-mercaptoethanol) at a 1 : 1 ratio and boiled for 3 minutes. The samples were fractionated according to size on a 12.5% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane (Millipore, Billerica, Mass), and probed with polyclonal anti-HERP (Santa Cruz Biotechnology, Santa Cruz, Calif) or polyclonal anti-Hrd1 (Biosynthesis Biotechnology, Beijing, China) antibodies. A secondary antibody, goat antirabbit IgG (Biosynthesis Biotechnology, Beijing, China) diluted 1 : 1000, was applied, and the chemiluminescent signal was detected. The same membrane was reused to detect β-actin (the internal control) by incubating it with mouse antihuman β-actin antibody (Gene, Hong Kong, China). Bands observed on the photography films were analyzed by automatic image analysis. The integrated optical density of each protein band was normalized to that of the corresponding β-actin band from the same sample.
2.5. Rat Retinal Capillary Endothelial Cell (RRCEC) Culture
RRCECs cultured in vitro were prepared as previously described [24]. Two-month-old male Sprague Dawley rats weighing 150–200 g (n = 60) were obtained from the animal center of Huazhong University of Science and Technology. After anesthesia, the eyes were removed, and the retinas harvested and homogenized by two gentle up-and-down strokes in a 15 mL homogenizer (Dounce; Bellco Glass, Vineland, NJ). The homogenate was filtered through an 88 μm sieve. The retentate was digested in 0.066% collagenase for 45 min at 37°C. The homogenate was centrifuged (1000 ×g for 10 min), and the pellet was resuspended in endothelial basal growth medium (Invitrogen-Gibco, Grand Island, NY), supplemented with 20% fetal bovine serum, 50 U/mL endothelial cell growth factor (Sigma-Aldrich, St. Louis, Mo), and 1% insulin-transferrin-selenium. RRCECs were cultured in fibronectin-coated dishes and incubated at 37°C in a humidified atmosphere containing 5% CO2.
Cultured endothelial cells were characterized by evaluating expression of factor VIII antigen (von Willebrand factor) and determining unchanged morphology under culture conditions by light microscopy. The expression of acetyl-LDL (Ac-LDL) receptors in endothelial cells was measured by adding fluorescence-labeled AC-LDL (Biomedical Technologies, Palatine, Il). Only cells from passages 3 to 7 were used in the experiments.
2.6. Cell Immunofluorescence
The RRCECs were grown in 24-well plates in human endothelial serum-free material basal growth medium containing 8.3 mM glucose. Upon attaining 80%, confluency cells were treated with medium containing 25 mM glucose for 2 d. Cells were then fixed with 4% formaldehyde for 15 min and permeabilized in 0.1% Triton X-100 for 10 min. Cells were incubated with primary antibody at 4°C overnight followed by secondary antibody for one hour. The slides were visualized and photographed under a fluorescence microscope (Olympus, Hamburg, Germany).
2.7. Statistical Analysis
Normally distributed data were compared using Student's independent samples t-test or one-way ANOVA where appropriate. When a significant difference was detected between groups, multiple comparisons of means were performed using the Bonferroni procedure, with type-I error rate at a maximum of 0.017 (0.05/3) adjustment. Statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) 15.0 software (SPSS, Chicago, IL). Data were presented as the mean ± standard deviation (SD). A probability (P) value <0.05 was considered statistically significant.
3. Results
3.1. Q-PCR Arrays
We detected 89 ER stress-related genes and found that the mRNA levels of 13 genes in the diabetic rats changed significantly during the first month (Table 1). We found that in the third month the levels of expression of 12 genes were changed significantly in these diabetic rats (Table 2). The changes in the expression levels of genes corresponded to 8 and 10 categories of signal pathways in the first and third months, respectively (Figure 1 and Table 3). The mRNA expressions of Erdj4 and HERP were lower both in the first and third months.
Table 1.
Q-PCR arrays showed that the expression of the ER stress factor had significant differences in the first and the third month in diabetic rat retina: The ER stress factor of differential gene expression in the first month.
| Symbol | Gene name | The average ratio of gene expression | t-test | |
|---|---|---|---|---|
| DR | control | P value | ||
| CCT4 | Cctd | 9.50E − 02 | 5.90E − 02 | 0.0134 |
| DNAJB9 | Erdj4 | 4.10E − 02 | 6.20E − 02 | 0.0125 |
| DNAJC3 | P58IPK | 1.70E − 02 | 3.00E − 02 | 0.0173 |
| Casp12 | Casp12 | 3.20E − 04 | 1.30E − 03 | 0.002 |
| ERP44 | Pdia10 | 2.10E − 02 | 2.50E − 02 | 0.0337 |
| GANAB | GluII | 8.10E − 02 | 1.40E − 01 | 0.045 |
| HERPUD1 | Herp | 1.50E − 01 | 2.60E − 01 | 0.0006 |
| HSPA1L | Hsp70-3 | 6.30E − 04 | 1.00E − 03 | 0.0183 |
| HSPA2 | Hspt70 | 7.30E − 03 | 1.30E − 02 | 0.0183 |
| MAPK8 | JNK | 1.10E − 02 | 2.00E − 02 | 0.0391 |
| NUCB1 | NUC | 2.80E − 02 | 5.60E − 02 | 0.0289 |
| OS9 | OS-9 | 9.10E − 02 | 1.50E − 01 | 0.0272 |
| SELS | AD-015 | 7.00E − 02 | 9.30E − 02 | 0.0486 |
Table 2.
Q-PCR arrays showed that the expression of the ER stress factor had significant differences in the first and the third month in diabetic rat retina: The ER stress factor of differential gene expression in the third month.
| Symbol | Gene name | The average ratio of gene expression | t-test | |
|---|---|---|---|---|
| DR | control | P value | ||
| ATF4 | CREB-2 | 8.80E − 01 | 1.60E + 00 | 0.0178 |
| DNAJB9 | Erdj4 | 4.10E − 02 | 5.40E − 02 | 0.0106 |
| ERO1L | Ero1 | 9.50E − 03 | 1.20E − 02 | 0.0492 |
| TRB3 | Trib3 | 5.80E − 03 | 6.10E − 02 | 0.0024 |
| HERPUD1 | Herp | 7.20E − 02 | 2.80E − 01 | 0.0008 |
| HTRA2 | PARK13 | 2.20E − 02 | 4.00E − 02 | 0.0064 |
| PPIA | CYPA | 4.30E − 01 | 6.50E − 01 | 0.0238 |
| SREBF1 | SREBP1 | 2.10E − 02 | 2.80E − 02 | 0.0187 |
| SYVN1/Hrd1 | HRD1 | 7.90E − 02 | 1.20E − 01 | 0.0067 |
| UFD1L | UFD1 | 1.60E − 01 | 8.00E − 02 | 0.0463 |
| UGCGL1 | HUGT1 | 9.80E − 03 | 2.10E − 02 | 0.0833 |
| USP14 | TGT | 7.30E − 02 | 4.80E − 02 | 0.0405 |
3.2. Expression of HERP and HRD1 in the Retinas of Diabetic Rats
We detected HERP and Hrd1 protein expression levels in the retinas of diabetic rats by Western blot and immunofluorescence in the first and the third months of diabetes development. The Western blot suggested that the HERP expression decreased significantly in the first month (P = 0.004) and third month (P = 0.012) compared with the nondiabetic control group. No significant change in the expression level of Hrd1 was observed in the first month (P = 0.338), while it decreased significantly in the third month compared with the control group (P = 0.001; Figures 2 and 3).
Figure 2.
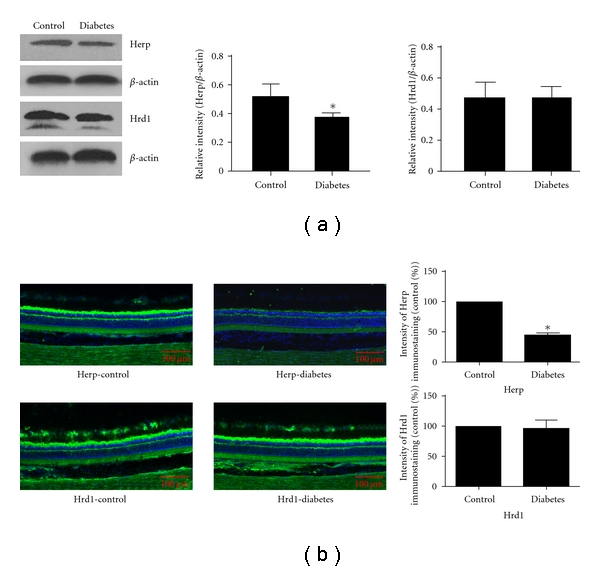
Western blot and immunofluorescence detected the expression of HERP and HRD1 in the first month after the development of diabetes: (a) Western blot detection of the expression of HERP and HRD1 in the first month. The expression of HERP in the diabetic group was less than that of the control group (P = 0.004); Hrd1 expression was similar in both groups (P = 0.338). (b) Immunofluorescence detection of the expression of HERP and HRD1 in the first month. The expression of HERP in the diabetic group was less than that of the control group (P = 0.008) Hrd1 expression was similar in both groups (P = 0.572).
Figure 3.
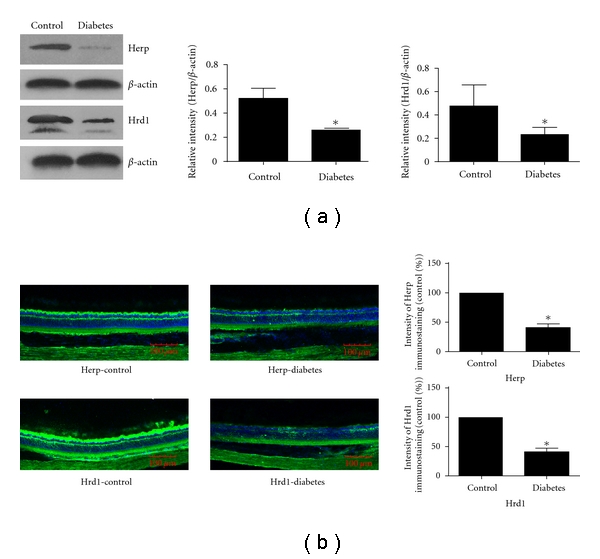
Western blot and immunofluorescence detected the expression of HERP and Hrd1 in the third month after the development of diabetes: (a) Western blot detection of the expression of HERP and Hrd1 in the third month. The expression of HERP and Hrd1in the diabetic group was less than that of the control group (P = 0.012 and P = 0.001, resp.). (b) immunofluorescence detection of the expression of HERP and HRD1 in the third month. The expression of HERP and Hrd1 in the diabetic group was less than that of the control group (P = 0.007 and P = 0.003, resp.).
The results of immunofluorescence were consistent with the Western blot. The protein level of HERP decreased significantly at both the first and third months (P = 0.008 and 0.007, resp.; Figures 2 and 3). There was no significant change in the expression of retinal HRD1 in the first month, while it decreased significantly in the third month (P = 0.572 and 0.003, resp. Figures 2 and 3).
3.3. Expression of HERP and HRD1 in RRCECs in the Presence of High-Glucose Concentration
The expression levels of HERP and HRD1 in RRCECs in vitro in the presence of high glucose concentration were decreased significantly compared to the control group (P = 0.013 and 0.024, resp.; Figure 4).
Figure 4.
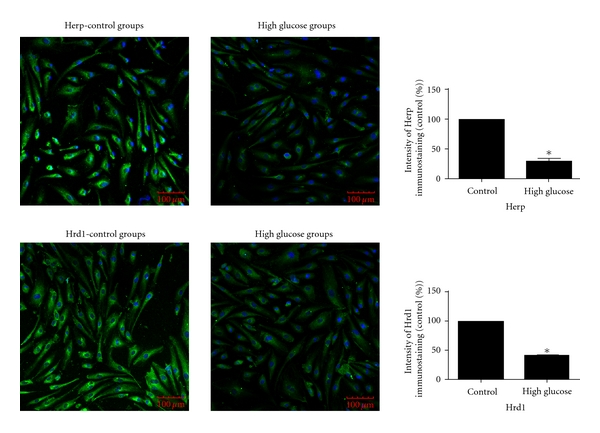
Immunofluorescence detection of the expression of HERP and HRD1 in RRCECs. The expression of HERP in the high glucose group was decreased compared to the control group, P = 0.013. The expression of HRD1 in the high glucose group was also decreased compared to the control group, P = 0.024.
4. Discussion
The STZ-induced rat diabetes model is an established animal model for studying DR. Although we did not verify the development of DR in this study, our previous studies and the publication from another group have demonstrated that DR develops within one month of STZ-induced diabetes [13, 22, 23]. Our results indicate that of 89 ER stress genes, the expression of 12 genes in the retinas of diabetic rats was downregulated by the third month of diabetes development, and the expression of CCT4 increased within the first month. We did not observe any change in the expression of AFT4 or GFP78 at either time point in our study, which is consistent with our earlier results [13].
The expression of genes belonging to 8 different categories of ER stress factors was altered in the first month, while those of 10 categories were changed by the third month, suggesting that with increasing time more categories of ER stress factors were involved in the pathogenic process of DR. The expression of a number of related factors of the ERAD signaling pathways was downregulated, indicating that the ERAD signaling pathway may play an important role in DR. The ERAD system is an important pathway of protein degradation in the ER [25, 26] and plays important physiological roles. The ER is the location of protein synthesis, and secretion [27, 28] and has strict quality control mechanisms which allow secretion of correctly folded protein into the cytoplasm. The wrongly folded protein will be degraded through ERAD. ERAD therefore is a quality control system of the ER.
Recent studies found that HRD1 plays a central role in the ERAD-luminal pathway [29] and that HERP coordinates and regulates HRD1-mediated ubiquitylation [28], so we selected HRD1 and HERP from the ERAD pathway for further study. HERP expression was downregulated significantly in the retinas of diabetic rats in the first and third months. HERP is a membrane-bound, ubiquitin-like protein that is located in the ER. It forms a complex with ubiquitinated proteins and with the 26S proteasome [30–33]. HERP functions to degrade wrongly folded nonglycosylated proteins by forming a protein-enzyme complex with Derlin-1, HRD1, and p97 [34]. In our study, HRD1 expression in the retinas of diabetic rats remained unchanged in the first month, while it decreased in the third month. HRD1 is an E3 ubiquitin ligase and a key factor of ERAD [35–37]. ERAD has three pathways in yeast [38]: ERAD-L, ERAD-M, and ERAD-C. Both ERAD-L and ERAD-M are the key enzymes of HRD1. In the mammalian ERAD, HRD1 plays a very broad role in the ubiquitination process of abnormal proteins in the ER. The ubiquitin ligase HRD1 is mainly involved in the degradation of glycosylation proteins [39–41].
The decreased expression of HERP and HRD1 at both the mRNA and protein levels could lead to a decrease in function of ERAD's ability to remove wrongly folded proteins in the cell. Misfolded protein accumulation in the ER induces ER stress and activates signaling pathways, including PERK, ATF6, and IRE1 [15]. Persistent ER stress leads to cell death and induction of inflammation [42–45]. An inflammatory milieu is instrumental in breaking down the blood-retinal barrier in DR [46, 47].
In conclusion, we have shown by in vivo and in vitro experiments that an elevated concentration of glucose leads to downregulation of the ERAD signaling pathway. Such downregulation may result in local inflammation and DR.
Acknowledgments
This research was supported by the National Nature Science Foundation (Grant no. 30872823), China. S. Yan, C. Zheng, and Z.-q. Chen contributed equally to this paper.
References
- 1.Frank RN. Diabetic retinopathy. The New England Journal of Medicine. 2004;350(1):48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 2.Cheung N, Wong TY. Diabetic retinopathy and systemic vascular complications. Progress in Retinal and Eye Research. 2008;27(2):161–176. doi: 10.1016/j.preteyeres.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Cejkova P, Novota P, Cerna M, et al. HLA DRB1, DQB1 and insulin promoter VNTR polymorphisms: interactions and the association with adult-onset diabetes mellitus in Czech patients. International Journal of Immunogenetics. 2008;35(2):133–140. doi: 10.1111/j.1744-313X.2008.00749.x. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Zhang HQ, Shi Y, et al. Overexpression of nuclear transport factor 2 may protect against diabetic retinopathy. Molecular Vision. 2009;15:861–869. [PMC free article] [PubMed] [Google Scholar]
- 5.Yan W, Frank CL, Korth MJ, et al. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(25):15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B, Li D, Li GG, Wang HW, Yu AX. P58(IPK) inhibition of endoplasmic reticulum stress in human retinal capillary endothelial cells in vitro. Molecular Vision. 2008;14:1122–1128. [PMC free article] [PubMed] [Google Scholar]
- 7.Hattori T, Shimada H, Nakashizuka H, Mizutani Y, Mori R, Yuzawa M. Dose of intravitreal bevacizumab (avastin) used as preoperative adjunct therapy for proliferative diabetic retinopathy. Retina. 2010;30(5):761–764. doi: 10.1097/IAE.0b013e3181c70168. [DOI] [PubMed] [Google Scholar]
- 8.Kant S, Seth G, Anthony K. Vascular endothelial growth factor-A (VEGF-A) in vitreous fluid of patients with proliferative diabetic retinopathy. Annals of Ophthalmology. 2009;41(3-4):170–173. [PubMed] [Google Scholar]
- 9.Ghosh R, Lipson KL, Sargent KE, et al. Transcriptional regulation of VEGF-A by the unfolded protein response pathway. PLoS One. 2010;5(3) doi: 10.1371/journal.pone.0009575. Article ID e9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oshitari T, Hata N, Yamamoto S. Endoplasmic reticulum stress and diabetic retinopathy. Vascular Health and Risk Management. 2008;4(1):115–122. doi: 10.2147/vhrm.2008.04.01.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAlpine CS, Bowes AJ, Werstuck GH. Diabetes, hyperglycemia and accelerated atherosclerosis: evidence supporting a role for endoplasmic reticulum (ER) stress signaling. Cardiovascular and Hematological Disorders—Drug Targets. 2010;10(2):151–157. doi: 10.2174/187152910791292529. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Wang JJ, Yu Q, Wang M, Zhang SX. Endoplasmic reticulum stress is implicated in retinal inflammation and diabetic retinopathy. FEBS Letters. 2009;583(9):1521–1527. doi: 10.1016/j.febslet.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, Wang HS, Li GG, Zhao MJ, Zhao MH. The role of endoplasmic reticulum stress in the early stage of diabetic retinopathy. Acta Diabetologica. 2011;48:103–111. doi: 10.1007/s00592-009-0170-z. [DOI] [PubMed] [Google Scholar]
- 14.Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Structure and Function. 2008;33(1):75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 15.Lin JH, Li H, Yasumura D, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirasawa H, Jiang C, Zhang P, Yang FC, Yokota H. Mechanical stimulation suppresses phosphorylation of eIF2alpha and PERK-mediated responses to stress to the endoplasmic reticulum. FEBS Letters. 2010;584(4):745–752. doi: 10.1016/j.febslet.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 17.Jonikas MC, Collins SR, Denic V, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohno K. Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. Journal of Biochemistry. 2010;147(1):27–33. doi: 10.1093/jb/mvp196. [DOI] [PubMed] [Google Scholar]
- 19.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. Journal of Cell Biology. 2010;189(5):783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapoor A, Sanyal AJ. Endoplasmic reticulum stress and the unfolded protein response. Clinics in Liver Disease. 2009;13(4):581–590. doi: 10.1016/j.cld.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Hosoi T, Ozawa K. Endoplasmic reticulum stress in disease: mechanisms and therapeutic opportunities. Clinical Science. 2010;118(1):19–29. doi: 10.1042/CS20080680. [DOI] [PubMed] [Google Scholar]
- 22.Brucklacher RM, Patel KM, VanGuilder HD, et al. Whole genome assessment of the retinal response to diabetes reveals a progressive neurovascular inflammatory response. BMC Medical Genomics. 2008;13:1–26. doi: 10.1186/1755-8794-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Liu R, Cui Z, et al. Functional characterization of 58-kilodalton inhibitor of protein kinase in protecting against diabetic retinopathy via the endoplasmic reticulum stress pathway. Molecular Vision. 2011;17:78–84. [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Yin W, Hong X, et al. Remodeling retinal neovascularization by ALK1 gene transfection in vitro. Investigative Ophthalmology and Visual Science. 2008;49(10):4553–4560. doi: 10.1167/iovs.07-0995. [DOI] [PubMed] [Google Scholar]
- 25.Hoseki J, Ushioda R, Nagata K. Mechanism and components of endoplasmic reticulum-associated degradation. Journal of Biochemistry. 2010;147(1):19–25. doi: 10.1093/jb/mvp194. [DOI] [PubMed] [Google Scholar]
- 26.Ando H, Ichihashi M, Hearing VJ. Role of the ubiquitin proteasome system in regulating skin pigmentation. International Journal of Molecular Sciences. 2009;10(10):4428–4434. doi: 10.3390/ijms10104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma R, Tsuchiya M, Bartlett JD. Flouride induces endoplasmic reticulum stress and inhibits protein synthesis and secretion. Environmental Health Perspectives. 2008;116(9):1142–1146. doi: 10.1289/ehp.11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coe H, Michalak M. Calcium binding chaperones of the endoplasmic reticulum. General Physiology and Biophysics. 2008;28:F96–F103. [PubMed] [Google Scholar]
- 29.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143(4):579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kny M, Standera S, Hartmann-Petersen R, Kloetzel PM, Seeger M. Herp regulates Hrd1-mediated ubiquitylation in a ubiquitin-like domain-dependent manner. Journal of Biological Chemistry. 2011;286(7):5151–5156. doi: 10.1074/jbc.M110.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kokame K, Agarwal KL, Kato H, Miyata T. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. Journal of Biological Chemistry. 2000;275(42):32846–32853. doi: 10.1074/jbc.M002063200. [DOI] [PubMed] [Google Scholar]
- 32.Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Molecular Cell. 2007;28(4):544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chigurupati S, Wei Z, Belal C, et al. The homocysteine-inducible endoplasmic reticulum stress protein counteracts calcium store depletion and induction of CCAAT enhancer-binding protein homologous protein in a neurotoxin model of Parkinson disease. Journal of Biological Chemistry. 2009;284(27):18323–18333. doi: 10.1074/jbc.M109.020891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulze A, Standera S, Buerger E, et al. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. Journal of Molecular Biology. 2005;354(5):1021–1027. doi: 10.1016/j.jmb.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 35.Kanehara K, Xie W, Ng DT. Modularity of the Hrd1 ERAD complex underlies its diverse client range. Journal of Cell Biology. 2010;188(5):707–716. doi: 10.1083/jcb.200907055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. Journal of Cell Biology. 2010;188(2):223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shmueli A, Tsai YC, Yang M, Braun MA, Weissman AM. Targeting of gp78 for ubiquitin-mediated proteasomal degradation by Hrd1: cross-talk between E3s in the endoplasmic reticulum. Biochemical and Biophysical Research Communications. 2009;390(3):758–762. doi: 10.1016/j.bbrc.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126(2):361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 39.Carroll SM, Hampton RY. Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. Journal of Biological Chemistry. 2010;285(8):5146–5156. doi: 10.1074/jbc.M109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Molecular Cell. 2009;34(2):212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458(7237):453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 42.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140(6):900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida H. ER stress and diseases. FEBS Journal. 2007;274(3):630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verfaillie T, Garg AD, Agostinis P. Targeting ER stress induced apoptosis and inflammation in cancer. doi: 10.1016/j.canlet.2010.07.016. Cancer Letters. In press. [DOI] [PubMed] [Google Scholar]
- 46.Kern TS. Contributions of inflammatory processes to the development of the early stages of diabetic retinopathy. Experimental Diabetes Research. 2007;2007:14 pages. doi: 10.1155/2007/95103. Article ID 95103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myśliwiec M, Balcerska A, Zorena K, Myśliwska J, Lipowski P, Raczyńska K. The role of vascular endothelial growth factor, tumor necrosis factor alpha and interleukin-6 in pathogenesis of diabetic retinopathy. Diabetes Research and Clinical Practice. 2008;79(1):141–146. doi: 10.1016/j.diabres.2007.07.011. [DOI] [PubMed] [Google Scholar]


