Abstract
The nuclear matrix antigen recognized by the monoclonal antibody (mAb) B1C8 is a novel serine (S) and arginine (R)-rich protein associated with splicing complexes and is named here SRm160 (SR-related matrix protein of 160 kD). SRm160 contains multiple SR repeats, but unlike proteins of the SR family of splicing factors, lacks an RNA recognition motif. SRm160 and a related protein SRm300 (the 300-kD nuclear matrix antigen recognized by mAb B4A11) form a complex that is required for the splicing of specific pre-mRNAs. The SRm160/300 complex associates with splicing complexes and promotes splicing through interactions with SR family proteins. Binding of SRm160/300 to pre-mRNA is normally also dependent on U1 snRNP and is stabilized by U2 snRNP. Thus, SRm160/300 forms multiple interactions with components bound directly to important sites within pre-mRNA. The results suggest that a complex of the nuclear matrix proteins SRm160 and SRm300 functions as a coactivator of pre-mRNA splicing.
Keywords: Serine–arginine-rich splicing factor, small nuclear ribonucleoprotein particle, spliceosome, coactivator, nuclear matrix
Precursor mRNA (pre-mRNA) splicing occurs within the spliceosome, a ∼60S complex composed of four small nuclear ribonucleoprotein particles (U1, U2, U4/U6, U5 snRNPs) and multiple non-snRNP factors (for review, see Sharp 1994; Krämer 1996). Although a wealth of biochemical and genetic data have provided detailed information on the role of specific spliceosomal components, it is not well understood how intron–exon segments are discriminated in vivo. To gain insights into this question, it will be important to determine how the splicing process is physically organized within the cell nucleus.
In situ hybridization and immunocytochemistry studies have localized in the nucleus individual components involved in pre-mRNA synthesis and processing. The majority of splicing factors are both widely distributed throughout the nucleus, excluding nucleoli, and are also concentrated in two distinct types of foci, referred to as “speckles” and coiled bodies (for review, see Lamond and Carmo-Fonseca 1993; Lawrence et al. 1993; Spector 1993). Mammalian nuclei typically contain 20–50 speckles and 1–5 coiled bodies. Both of these structures concentrate all four snRNPs, but only speckles are enriched for the largest subunit of RNA polymerase II (Pol II LS; Bregman et al. 1995; Mortillaro et al. 1996), poly(A)+ RNA (Carter et al. 1991; Huang et al. 1994), and essential non-snRNP splicing factors of the serine-arginine (SR)-family (Fu and Maniatis 1990; Spector et al. 1991). Fluorescence in situ hybridization studies have demonstrated that pre-mRNAs and spliced mRNAs of several specific cellular transcripts are located at the periphery or within speckles, indicating that a subset of pre-mRNAs are processed at these sites (Xing et al. 1993, 1995). However, detection of nascent RNA synthesis by a brief pulse with (BrdU) or 3H uridine reveals that the majority of nascent RNA synthesis, and by inference cotranscriptional splicing, occurs at foci separated from speckles (Fakan and Puvion 1980; Wansink et al. 1993; Fay et al. 1997). These and other studies indicate that speckles are structures from which splicing factors can be recruited to sites of nascent pre-mRNA transcription (Huang and Spector 1996; Misteli et al. 1997), in addition to functioning directly in splicing.
The SR family of proteins so far comprises a group of eight factors (SRp20, SRp30a/ASF/SF2, SRp30b/SC35, SRp30c, 9G8, SRp40, SRp55, and SRp75), which function in constitutive and regulated pre-mRNA splicing (for review, see Fu 1995; Manley and Tacke 1996; Reed 1996; Valcárcel and Green 1996). This family is highly conserved in metazoans and shares a similar structural organization, consisting of one or two amino-terminal RNA recognition motifs (RRMs) and an extensively phosphorylated carboxy-terminal domain that is rich in serine/arginine dipeptides (RS domain). The RRMs of SR family proteins bind RNA with some sequence specificity, whereas the RS domains function primarily in protein–protein interactions with other SR proteins. RS domains also participate in the targeting of SR proteins to speckles and may be important for the structural integrity of these subnuclear structures (Li and Bingham 1991; Gui et al. 1994; Hedley et al. 1995; Colwill et al. 1996; Misteli and Spector 1997; Cáceres et al. 1997).
Interactions between SR proteins are critical for the initial recognition and pairing of splice sites and also during subsequent steps in spliceosome formation. For example, binding of the SR family proteins SC35 and ASF/SF2 to the U1 snRNP 70-kD protein and factors associated with U2AF–65 kD (including U2AF–35 kD and Urp) is thought to facilitate the recognition and pairing of 5′ and 3′ splice sites, respectively (Wu and Maniatis 1993; Kohtz et al. 1994; Tronchère et al. 1997). U1–70 kD, U2AF–35 kD, and Urp proteins contain short RS domains involved in these interactions. SR proteins also promote exon inclusion by associating with specific sequences within exons called enhancers (for review, see Berget 1995; Black 1995; Chabot 1996). Besides these early roles, SR family proteins may also function at subsequent steps in pre-mRNA processing (Blencowe et al. 1994, 1995; Roscigno and Garcia-Blanco 1995; Alzhanova-Ericsson et al. 1996).
A subset of splicing factors in speckles remain spatially associated with these structures after the removal of chromatin and soluble material (Volgelstein and Hunt 1982; Spector et al. 1983, 1991; Verheijen et al. 1986; Smith et al. 1989; Nickerson et al. 1992; Blencowe et al. 1994; Wan et al. 1994). The underlying “nuclear matrix” (NM) remaining after nuclear extraction is composed of protein and the majority (>70%) of nuclear RNA, including most hnRNA (for review, see Mattern et al. 1997; Penman et al. 1997; Pederson 1998). Pulse-labeling studies indicate that newly synthesized transcripts associate with the matrix (Jackson and Cook 1985). Evidence that splicing occurs on the matrix was provided by Zeitlin et al. (1987, 1989), who demonstrated that processing of a β-globin pre-mRNA occurs with rapid kinetics in association with an isolated matrix fraction, supplemented with soluble nuclear factors. Despite increasing evidence for a role of the NM in pre-mRNA processing, the precise structural and functional nature of this nuclear entity is not well understood.
To gain insights into the structure and function of integral matrix components involved in pre-mRNA processing, we have screened a bank of monoclonal antibodies (mAbs) to the NM (anti-NM mAbs) for the recognition of NM proteins associated with splicing (Blencowe et al. 1994, 1995). Three distinct high molecular mass proteins (B1C8, 160–180 kD; H1B2, 240 kD; and B4A11, 300 kD) were identified as candidate antigens that are concentrated in NM speckles and that are also associated with splicing complexes assembled in vitro. All three of these antigens were also found to share properties of the SR family proteins: B1C8 and H1B2 share common phosphoepitopes and all three antigens share the SR protein property of precipitation in 20 mm MgCl2. The three anti-NM mAbs preferentially immunoprecipitated complexes containing exon sequences (pre-mRNA, intermediates, and mRNA product), but not the complex containing the excised intron lariat, indicating that the cognate antigens may function in splicing (Blencowe et al. 1994, 1995).
In this study, the role of B1C8 and B4A11 NM proteins in splicing has been investigated. Cloning of a cDNA encoding B1C8 reveals that it is a novel SR protein, and is renamed here as SRm160 (SR-related matrix protein of 160 kD). A complex of SRm160 and the B4A11 nuclear matrix antigen (now SRm300) functions as a pre-mRNA-specific splicing coactivator by forming interactions with SR family proteins and snRNPs.
Results
Isolation of a cDNA encoding SRm160, formerly the B1C8 nuclear matrix antigen
B1C8 (160 kD), renamed below as SRm160, was partially purified by a three-step procedure using conventional chromatography and magnesium precipitation, as previously described (see Fig. 4 in Blencowe et al. 1995). The protein was recovered from an SDS–polyacrylamide gel, digested with lys-C, and five resulting peptides were microsequenced. Searches of databases with the microsequences did not identify a known protein, although two of the microsequences were contained within a partial human cDNA sequence in the database of expressed sequence tags (EST 78873). A fragment (bases 1–1424) of EST 78873 was used to screen a λ cDNA library derived from human U937 cells and several positive plaques were isolated. Sequence analysis of the longest cDNA (3.7 kb) isolated from one of the plaques, and of several overlapping EST clones identified by database searches, revealed a single ORF of 820 amino acid residues (Fig. 1A). Sequence analysis of additional U937 λ cDNAs, EST cDNAs, genomic clones, as well as RT–PCR products, revealed the existence of variant SRm160 mRNAs with additional short exon sequences (B.J. Blencowe, G. Bauren, E. Reifenberg, and P.A. Sharp, unpubl.). These data indicate that multiple SRm160 protein isoforms may be expressed in mammalian cells.
Figure 4.
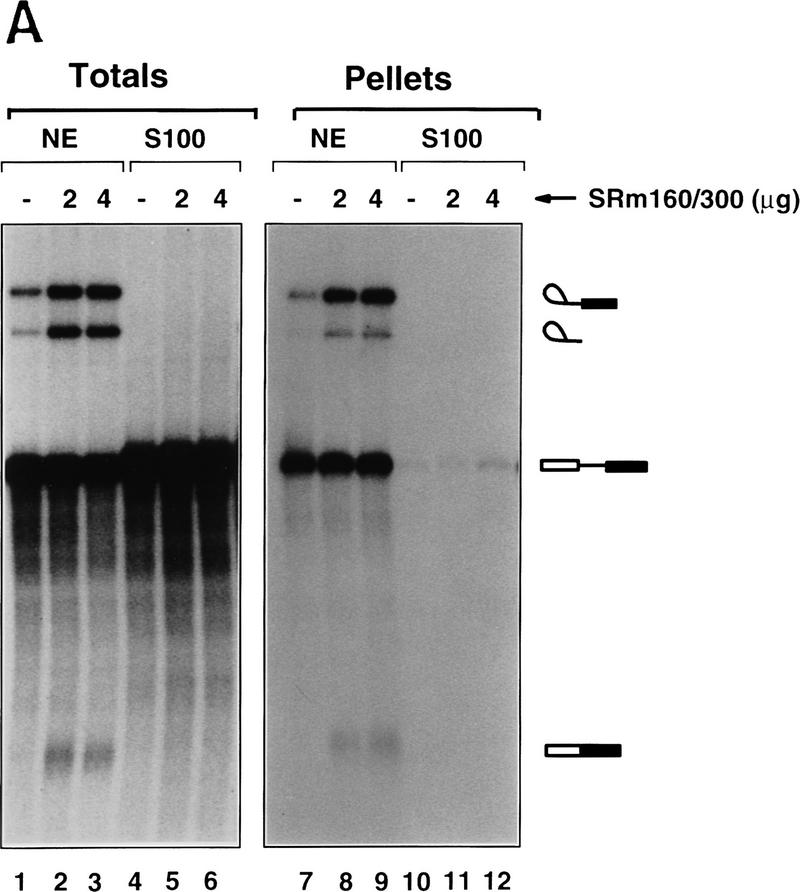
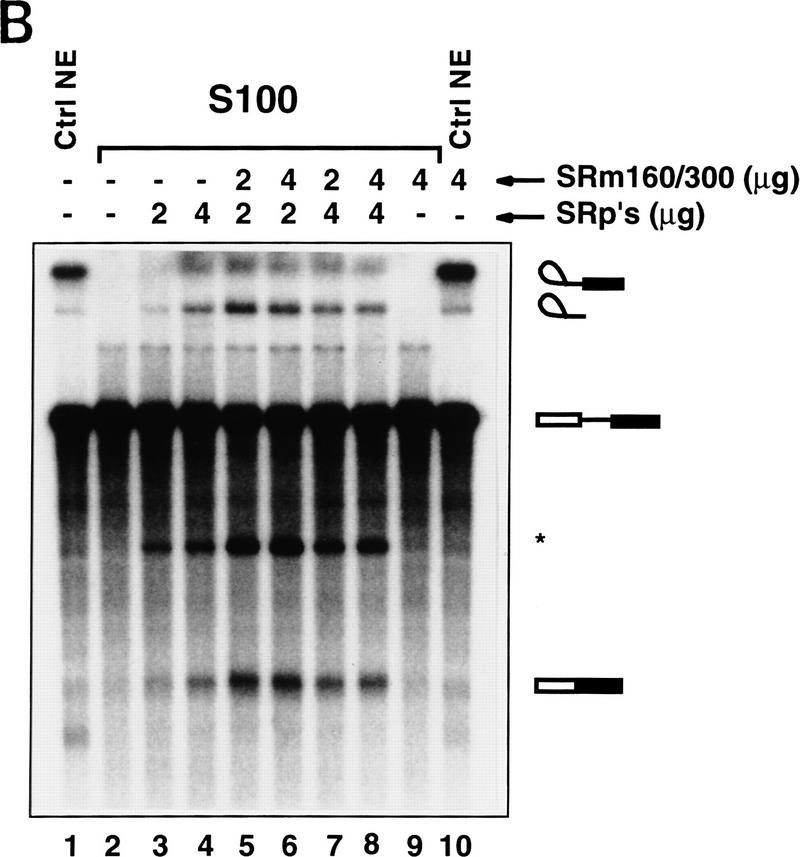
Pre-mRNA binding and promotion of splicing by SRm160/300 requires SR family proteins. (A) Immunoprecipitations were performed from splicing reactions incubated with PIP85A pre-mRNA for 40 min. The splicing reactions contained nuclear (lanes 1–3,7–9) or S100 cytoplasmic (lanes 4–6,10–12) extract, with (lanes 2,3,5,6,8,9,11,12) or without (lanes 1,4,7,10) purified SRm160/300 proteins added; the preparation of SRm160/300 is as described in Fig. 4, Blencowe et al. (1995). Fifty percent of the total RNA recovered directly from each splicing reaction (Totals) was loaded in lanes 1–6), whereas all of the RNA recovered after immunoprecipitation with mAb B1C8 (Pellets) was loaded in lanes 7–12. (B) S100 splicing reactions (lanes 2–9) containing β-globin pre-mRNA and varying amounts of SRm160/300 and SR family proteins, as indicated, were incubated for 1 hr. Control splicing reactions incubated in parallel contained nuclear extract (lane 1) or S100 extract (lane 2), without added proteins, and a reaction containing nuclear extract and 4 μg SRm160/300 (lane 10). The asterisk (*) indicates an RNA fragment protected from endogenous nuclease activity.
Figure 1.
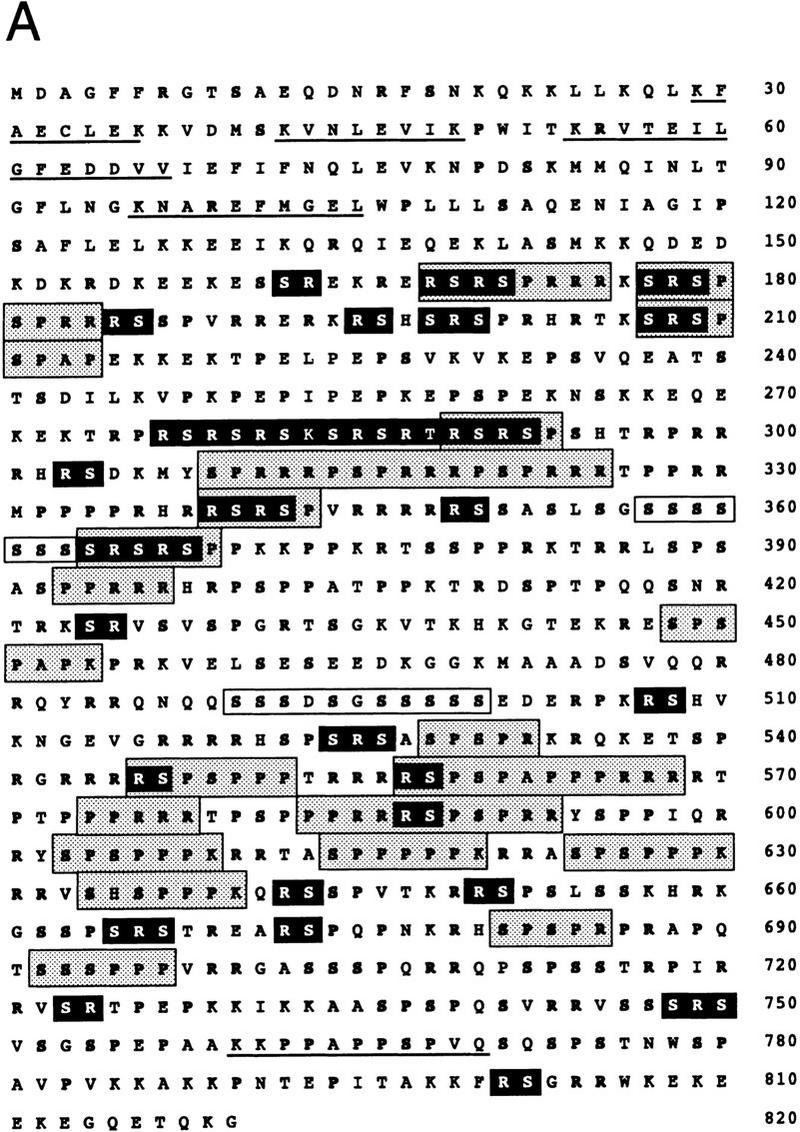

The SRm160 ORF and related sequences. (A) Amino acid sequence of the SRm160 ORF. Arginine (R), serine (S), and proline (P) amino acid residues are highlighted in bold-face type. SR/RS dipeptides are highlighted in black boxes. Serine-rich domains are highlighted in white boxes. Other repeat motifs containing P, R, or S residues are highlighted in gray boxes (see Table 1). The latter represent only the most frequently occurring repeats and are a subset of the total repeats identified as statistically significant by the statistical analysis of protein sequence (SAPS) algorithm (Brendel et al. 1992). Microsequences of lys-C peptides obtained from purified SRm160 protein are underlined. The SRm160 cDNA sequence has been deposited in GenBank (accession no. AF048977). (B) Multiple alignment of sequences homologous to the amino-terminal 150 amino acids of human SRm160. Partial cDNA ORF sequences from mouse (Mus musculus), fly (D. melanogaster), and three namatode species (C. elegans, A. simplex, and B. malayi) identified by BLAST searches were aligned using the Clustal algorithm. Sequences included in the alignment correspond to the following GenBank accession numbers: M. musculus, W67104; D. melanogaster, AA567455, AA440655, AA539347; C. elegans, D76066, C43686; A. simplex, S51495; B. malayi, AA089428. Residues identical to the majority sequence (top line) are shaded in black. Amino-terminal residues not present in the EST cDNAs are indicated by a blank space; gaps are indicated by a dash, and undefined residues are indicated by an X.
Northern analysis of polyadenylated RNA isolated from different human tissues with a cDNA probe (corresponding to ORF amino acids 1–473) detected a major ∼3.8 kb transcript and a second transcript of ∼6.0 kb (B.J. Blencowe and P.A. Sharp, unpubl.). The ∼3.8-kb transcript is ubiquitous, although variably expressed, whereas the 6.0-kb species appears to be expressed only in a subset of human tissues. The ∼3.8-kb transcript is approximately the same length as the ORF plus the 3′ UTR of the SRm160 sequence represented in Figure 1A. Although no cDNA sequence that was analyzed in λ phage, EST clones, or RACE PCR products contained >5 nucleotides of predicted 5′ UTR, corresponding genomic clones that were isolated contained in-frame stop codons and no consensus 3′ splice site upstream of the first ATG in the cDNA sequence (data not shown; GenBank accession no. AF048977). Highly homologous Drosophila melanogaster and Caenorhabditis elegans sequences, identified from searches of EST databases also did not contain ORFs upstream of the conserved first methionine residues and immediately diverged from the human sequence in this region (Fig. 1B; see below). The first methionine residue in these sequences is contained within consensus Kozak sequences. A Flag epitope-tagged SRm160 protein (corresponding to SRm160 ORF amino acids 1–820), expressed by transient transfection in HeLa cells and detected with an anti-FLAG antibody, precisely comigrated with the endogenous SRm160 protein (G. Bauren, B.J. Blencowe, and P.A. Sharp, unpubl.). Together, these analyses provide strong evidence that the first methionine residue in the predicted ORF is the site of initiation of translation and that the ORF in Figure 1 represents a full-length SRm160 protein. The data also indicate that, similar to previously characterized S/R-rich proteins that contain post-translational modifications (see below), the predicted molecular mass of the SRm160 protein (93.5 kD) is markedly different from the 160 kD observed in SDS–polyacrylamide gels.
SRm160 is a novel SR-related protein that is highly conserved in metazoans
The ORF of SRm160 contains a remarkably high content of Ser (S = 17.3%), Arg (R = 17.2%), and Pro (P = 16.2%) residues (Fig. 1A). The majority of the S, R, and P residues are located within the carboxy-terminal three-fourths of the protein and are concentrated within several types of reiterated motifs, including multiple SR repeats, two S-rich domains, and multiple R/P-rich motifs (Fig. 1A; Table 1). Potential serine and threonine phosphorylation sites for many different serine/threonine kinases are located throughout the S/R/P-rich region. In addition, the SRm160 ORF contains a consensus tyrosine phosphorylation site at amino acid 308, multiple proline-rich regions that include consensus sites for proline-directed kinases, and motifs that resemble binding sites for proteins with Src homology 3 (SH3) domains. The two S-rich domains, which are also potential phosphorylation sites, resemble similar sequences in previously identified nuclear phosphoproteins (Meier and Blobel 1992; Bourquin et al. 1997). The presence of multiple SR repeats in the ORF is consistent with previous biochemical and immunological evidence indicating that SRm160 is an SR-related protein, for example, its property of precipitation in 20 mm MgCl2 and reactivity with several mAbs that recognize SR family proteins. Moreover, the presence of multiple potential phosphorylation sites is consistent with previous evidence that SRm160 is highly phosphorylated (Blencowe et al. 1994, 1995).
Table 1.
Repetitive sequences in the SRm160 ORF
| Repeat consensus
|
Location
|
|---|---|
| RSRSP | 168, 289, 339, 365 |
| (S)PSPRR | 179, 314, 320, 528, 588, 681 |
| SPRRR | 171, 181, 309, 315, 321 |
| RSPSP | 178, 208, 546, 557, 587 |
| SPSPPPK | 209, 448, 547, 558, 603, 614 624, 634, 681, 692 |
| PPRRR | 393, 564, 574, 583 |
The SRm160 ORF sequence was analyzed for statistically significant repetitive sequences using the SAPS program of Brendel et al. (1992). Many repetitive sequences scored as significant, consisting of separated, tandem, or periodic repeats; only the repeats occurring four or more times are listed above and are highlighted in gray boxes in Figure 1A.
In contrast to SR family splicing factors (see introductory section), the predicted SRm160 ORF lacks an RRM or an RRM homology (RRM-H) domain. Furthermore, searches of the databases with the ORF sequence did not reveal highly significant similarities to characterized proteins or to any sequence in the Saccharomyces cerevisiae genome. However, candidate mouse, D. melanogaster, C. elegans, and Brugia malayi homologs were identified in the EST databases and a homologous sequence within a parasitic nematode (Anisakis simplex) cDNA sequence was identified in the GenBank database (Fig. 1B). These partial cDNA sequences share remarkable identity with the amino-terminal, nonrepetitive, region of the human ORF: mouse = 97%, D. melanogaster = 62%, C. elegans = 57%, A. simplex = 54%, and B. malayi = 62%. This analysis suggests that the predicted SRm160 ORF corresponds to a protein that is highly conserved in metazoans, but absent in yeast.
Localization of SRm160 in interphase and metaphase cells
To establish the relationship between the cDNA ORF and the SRm160 nuclear matrix antigen originally detected by mAb B1C8, a 153-amino-acid protein fragment (ORF amino acids 7–160) was expressed as a GST fusion and used to immunize rabbits for the production of polyclonal antisera. The affinity-purified serum (rAb–SRm160), but not a corresponding pre–immune serum, specifically recognized a 160-kD protein in total nuclear extract (Fig. 2A, lane 1). rAb–SRm160 also recognized specifically SRm160 in a partially purified fraction, as did mAb B1C8 (data not shown). Also, consistent with previous results with mAb B1C8 (Blencowe et al. 1994; Wan et al. 1994), rAb–SRm160 stained specifically nuclear speckle structures in interphase cells and the spindle apparatus at metaphase (Fig. 2B). However, in contrast to mAb B1C8, rAb–SRm160 stained many smaller foci in addition to the predominant speckles, indicating that a form of SRm160 that is not strongly bound by mAb B1C8 is concentrated in additional nuclear foci. Identical to mAb B1C8, rAb–SRm160, but not preimmune serum, immunoprecipitated splicing complexes assembled on different pre-mRNAs from in vitro reactions, enriching for complexes containing exon sequences but not the intron–lariat RNA (data not shown; Blencowe et al. 1994). These results provide strong evidence that the isolated SRm160 cDNA in Figure 1 encodes the B1C8–160-kD nuclear matrix antigen.
Figure 2.
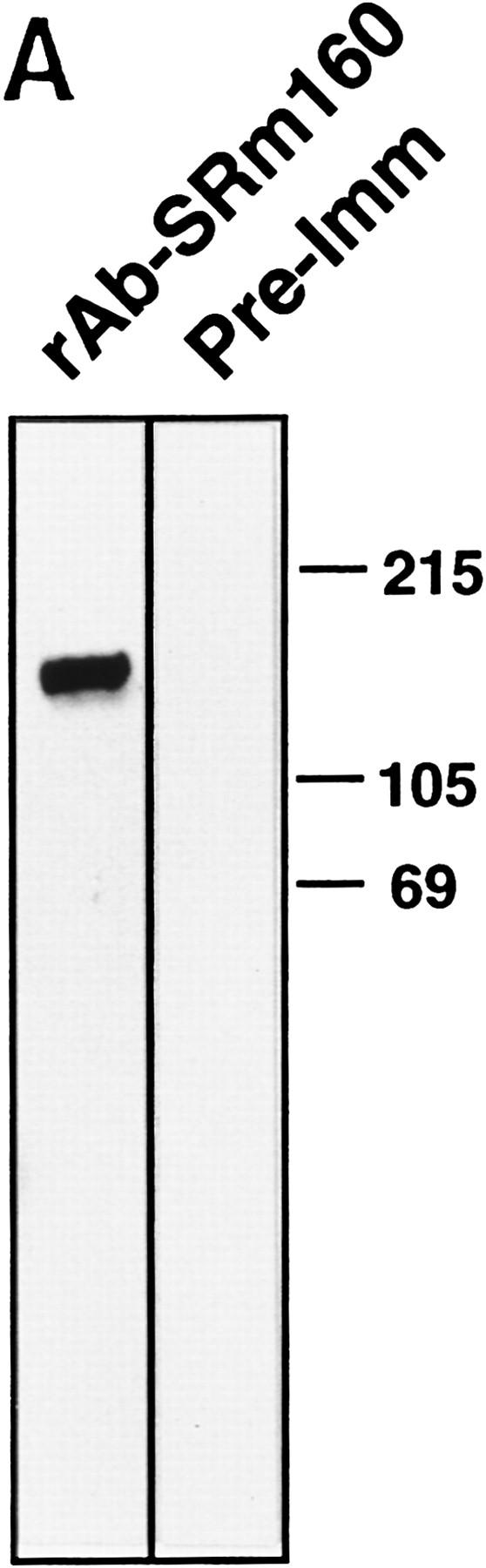
Association of SRm160 with interphase nucleoplasmic speckles, foci, and the mitotic spindle apparatus. (A) Total HeLa nuclear extract was separated on an SDS–polyacrylamide gel and immunoblotted with an affinity-purified polyclonal antiserum raised to a GST–SRm160 fusion protein containing SRm160 ORF amino acids 7–160 (rAb–SRm160, lane 1), and the corresponding preimmune serum (lane 2). (B) Interphase (top row) and metaphase (bottom row) human CaSki cells were double-immunolabeled with mAb B1C8 (red) and rAb–SRm160 (green) and visualized by confocal microscopy. The images were superimposed to reveal sites of overlap (yellow). The interphase image corresponds to a single confocal section proximal to the midline of the nucleus. The metaphase image corresponds to a stack of merged sections through the whole cell. Bar, 5 μm.
Association of SRm160 with SRm300 (the B4A11 nuclear matrix antigen) and specific SR family proteins
The absence of an RRM in SRm160 indicates that its association with splicing complexes could be mediated by protein–protein interactions. To identify proteins that bind to SRm160 and that may facilitate interactions with pre-mRNA, rAb–SRm160 immunoprecipitates were prepared from HeLa nuclear extracts and analyzed for coimmunoprecipitated proteins. Analysis of the total coimmunoprecipitated proteins revealed only four prominent species of ∼300, ∼220, 75, and 40 kD (data not shown). A candidate for the 300 kD species was SRm300 (previously the B4A11–300 kD nuclear matrix antigen), which shares similar properties to, and biochemically copurifies with, SRm160 (Blencowe et al. 1994, 1995, and in prep.). Because previous evidence suggests that RS domains can interact (Wu and Maniatis 1993; Amrein et al. 1994; Kohtz et al. 1994), candidates for the 75- and 40-kD species were SR family proteins of these sizes. To test this possibility, the immunoprecipitates were also probed with mAb 104, which detects a specific SR domain phosphoepitope shared between many SR proteins (Roth et al. 1990; Zahler et al. 1992).
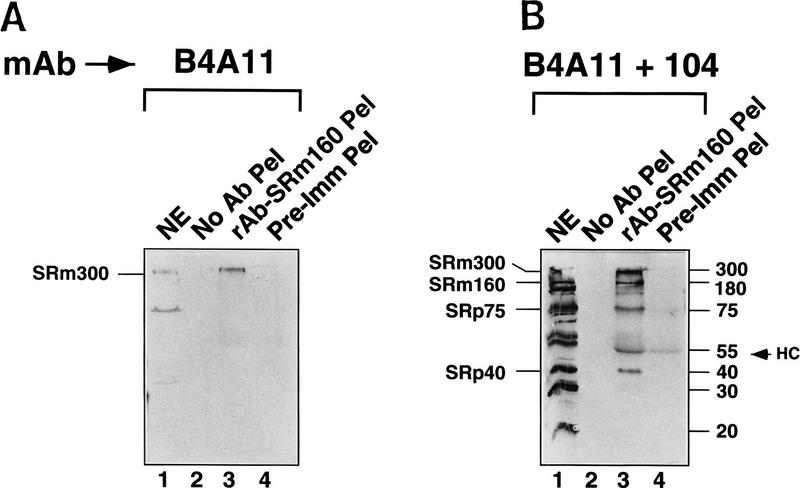
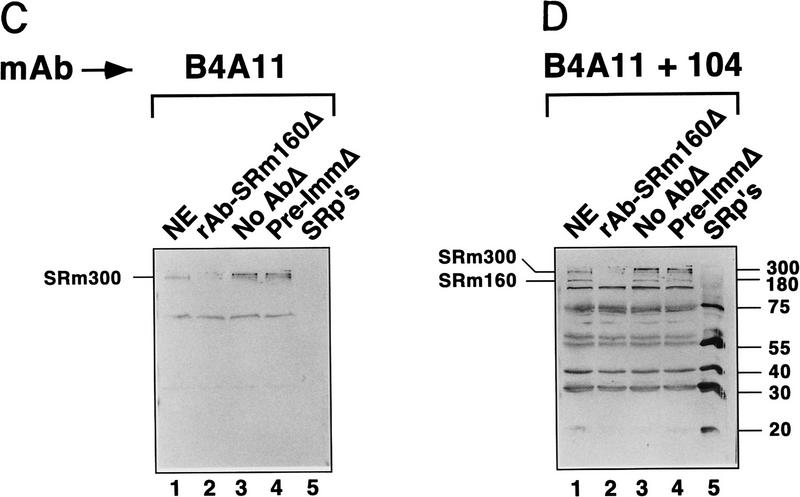
mAb B4A11 detected the SRm300 protein in the rAb–SRm160 immunoprecipitate (Fig. 3A,B, lane 3). Furthermore, mAb 104 detected proteins in the immunoprecipitate corresponding in size to SRp40 and SRp75, as well as detecting the SRm160 and SRm300 proteins (Fig. 3B, lane 3). The coimmunoprecipitation of these proteins by rAb–SRm160 was specific, as proteins corresponding in size to the other abundant SR–family proteins, including SRp20, SRp30, and SRp55, were not detected. Furthermore, no mAb 104 reactive proteins were immunoprecipitated by the preimmune serum (lane 4). Analysis of the supernatant fractions revealed that rAb–SRm160 essentially depleted all of the SRm160 protein and efficiently codepleted the SRm300 protein (Fig. 3C, 3D, lanes 2; data not shown). In contrast, only minor amounts of proteins corresponding in size to SRp40 and SRp75 were depleted as the majority of these proteins, as well as the other SR family proteins, remained in the supernatant (Fig. 3D, cf. lanes 2 and 4). These results indicate that essentially all of the SRm300 protein in nuclear extract is bound to SRm160 and that this SRm160/300 complex is specifically associated with a subfraction of SR proteins comigrating with SRp40 and SRp75.
Figure 3.
A complex of the SRm160 and SRm300 nuclear matrix proteins (SRm160/300) associates with SR family proteins comigrating with SRp40 and SRp75. (A,B) Proteins immunoprecipitated from HeLa nuclear extract by rAb–SRm160 were separated by SDS-PAGE, blotted, and probed with mAb B4A11 (A), followed by mAb104 (B). Protein immunoprecipitated from ∼0.5 mg of nuclear extract was loaded in lanes 2–4 and ∼0.15 mg of total nuclear extract was loaded in lane 1. Immunoprecipitations were performed with protein A beads alone (lane 2), rAb–SRm160 (lane 3), and preimmune serum (lane 4). Sizes of SR proteins SRm160 and SRm300 are indicated in kD. (HC) immunoglobulin heavy chain. (C,D) Supernatant fractions from the immunoprecipitations in A and B were separated and immunoblotted with mAbs B4A11 and 104, as in A and B. Total nuclear extract proteins are shown in lanes 1, and proteins recovered from the immunoprecipitation supernatant fractions prepared with rAb–SRm160, protein A beads alone and preimmune serum, are shown in lanes 2, 3, and 4, respectively. Approximately 0.15 mg of nuclear extract and of each supernatant fraction was loaded per lane. A sample of purified SR family proteins (∼4 μg) was separated as a marker in lane 5.
SRm160/300 associates with pre-mRNA and promotes splicing through interactions with SR family proteins
SRm160/300 could participate in splicing through interactions with SR family proteins. To investigate this possibility, a preparation of highly purified SRm160/300 proteins (see Materials and Methods; Fig. 4 in Blencowe et al. 1995) was assayed for splicing activity in S100 extracts, in conjunction with purified SR family proteins. “Cytoplasmic” S100 extracts of HeLa cells contain all factors required for splicing except SR family proteins, and are also deficient in SRm160 and SRm300 (Krainer et al. 1990; Zahler et al. 1992; data not shown). Addition of high levels of any one particular SR family protein to an S100 extract promotes splicing activity.
Addition of purified SRm160/300 to a S100 extract did not promote splicing activity or immunoprecipitation of PIP85A or β-globin pre-mRNA by mAb B1C8 (Fig. 4A, lanes 4–6, 10–12; data not shown). However, the same extract was active for splicing of either substrate upon addition of SR family proteins (Fig. 4B; data not shown). In contrast, addition of SRm160/300 to nuclear extract stimulated PIP85A splicing activity, and resulted in a corresponding increase in the level of immunoprecipitation of splicing complexes by mAb B1C8 (Fig. 4A, lanes 1–3, 7–9). These results demonstrate that SRm160/300 is functionally distinct from SR family proteins. It associates with complexes containing pre-mRNA and promotes splicing activity in nuclear extracts, yet, in the absence of SR family proteins, does not associate with pre-mRNA or restore splicing activity in S100 extracts.
To investigate whether the activity of SRm160/300 is mediated by interactions with the SR family of proteins, mixing experiments were performed in S100 reactions incubated with a β-globin pre-mRNA. Addition of varying amounts of SRm160/300 that were not active alone in an S100 extract (Fig. 4B, lane 9), together with limiting amounts of SR family proteins, resulted in a significant stimulation of splicing activity (cf. lanes 3 and 4 with lanes 5 and 6). The maximum level of stimulation was observed when 2 μg of SRm160/300 proteins was added to 2 μg of SR family proteins and was not increased upon addition of higher levels of SRm160/300 or SR family proteins (cf. lanes 5 and 6 with lanes 7 and 8). SRm160/300 also stimulated splicing when added to S100 reactions containing PIP85A supplemented with SR family proteins, but not when added alone (data not shown). Although SRm160/300 also stimulated β-globin splicing when added to nuclear extract, this level of stimulation was significantly lower than for the PIP85A substrate (lane 10, see below).
Further experiments were performed to determine whether the costimulatory effect of the SRm160/300 protein preparation was attributable to SRm160/300 proteins and not a potentially contaminating SR family protein activity. As in S100 assays, the SRm160/300 preparation alone did not restore splicing activity to U1 snRNP-depleted reactions incubated with β-globin pre-mRNA, which are, however, complemented by SR family proteins (Crispino et al. 1994; data not shown; see below and Fig. 7). The purified SRm160/300 preparation also did not promote intron–proximal splice site utilization in a pre-mRNA containing cis-competing 5′ splice sites, as did low levels of SR family proteins (data not shown).
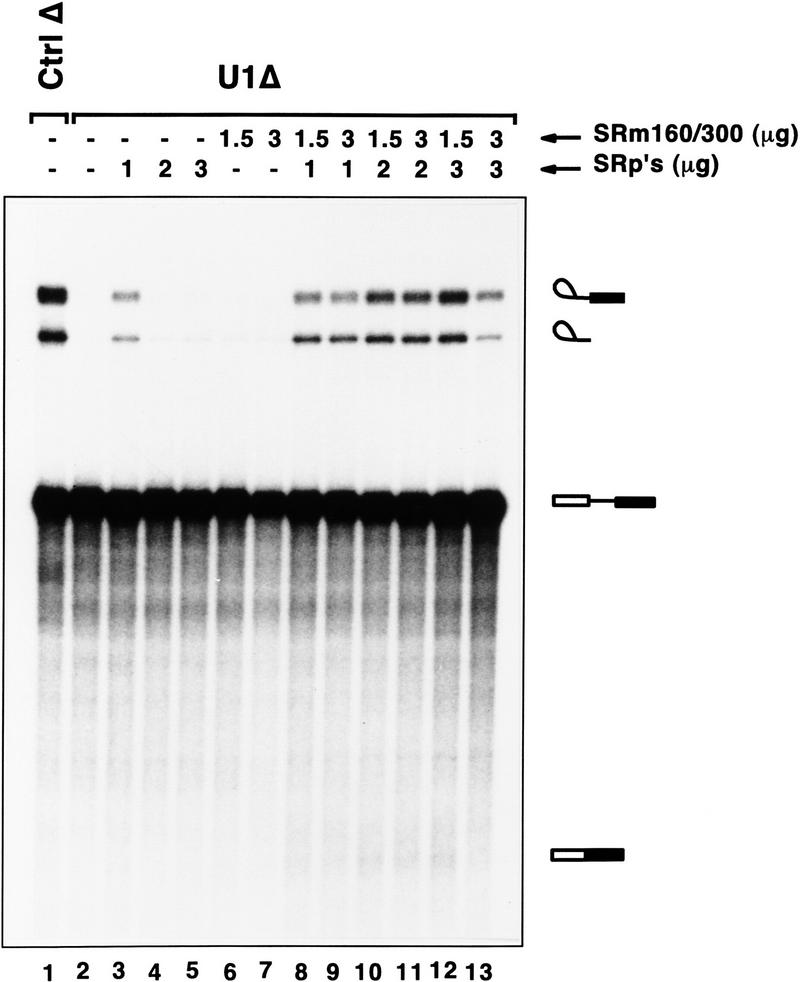
Figure 7.
SRm160/300 coactivates splicing in U1 snRNP-depleted reactions supplemented with SR family proteins. SRm160/300 proteins were added alone (lanes 6,7) or in combination with SR family proteins (lanes 8–13) to splicing reactions depleted of U1 snRNP. U1-depleted reactions containing excess SR proteins and no SRm160/300 are shown in lanes 3–5. Control reactions containing mock-depleted extract or U1-depleted extract with no added proteins are shown in lanes 1 and 2, respectively. The splicing reactions were incubated for 60 min before recovery of RNA.
The results in Figure 4B indicate that one or both of the SRm160/300 proteins promotes splicing activity in nuclear extracts and in S100 extracts supplemented with SR family proteins. Furthermore, they suggest that the ratio of SRm160/300 to SR family proteins is important for optimal splicing activity.
SRm160/300 is required for the splicing of specific pre-mRNAs
To determine whether SRm160/300 proteins are essential for splicing activity at endogenous concentrations of SR family proteins, nuclear extracts were immunodepleted of SRm160/300 proteins (see Fig. 3C,D, lanes 2) and tested for splicing activity using different pre-mRNA substrates (Fig. 5). Depletion of SRm160/300 prevented the first step of splicing of PIP85A pre-mRNA (Fig. 5A, lane 3). Splicing activity, however, was fully restored with the addition of purified SRm160/300 proteins (Fig. 5A, lanes 4–6). Restoration of activity, in the depleted extract was specific for SRm160/300 proteins, as the SRm160/300 protein buffer alone did not restore activity (lane 7), nor did addition of SRm160/300 proteins to nuclear extracts depleted of U2 snRNP (lane 8). Addition of high levels of SR family proteins to SRm160/300 depleted reactions resulted in a partial restoration of splicing activity, yet only addition of SRm160/300 proteins resulted in full restoration of activity (data not shown; see Discussion). These results indicate that, at endogenous concentrations of SR family proteins, SRm160/300 proteins are required for the first step of splicing of PIP85A pre-mRNA.
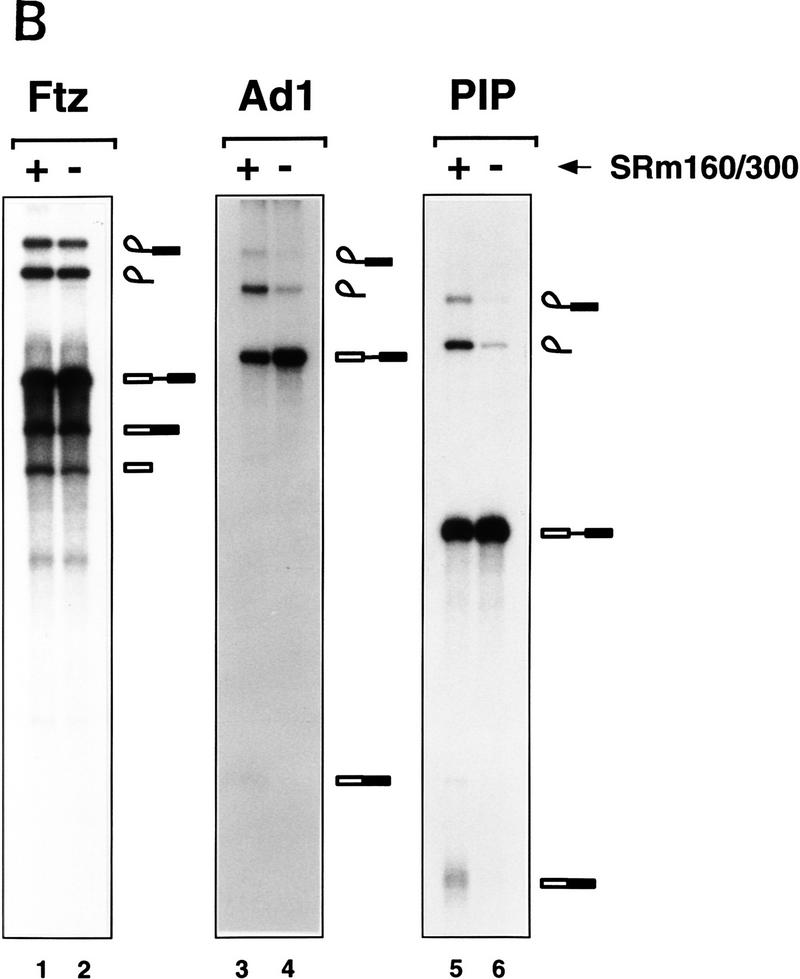
Figure 5.
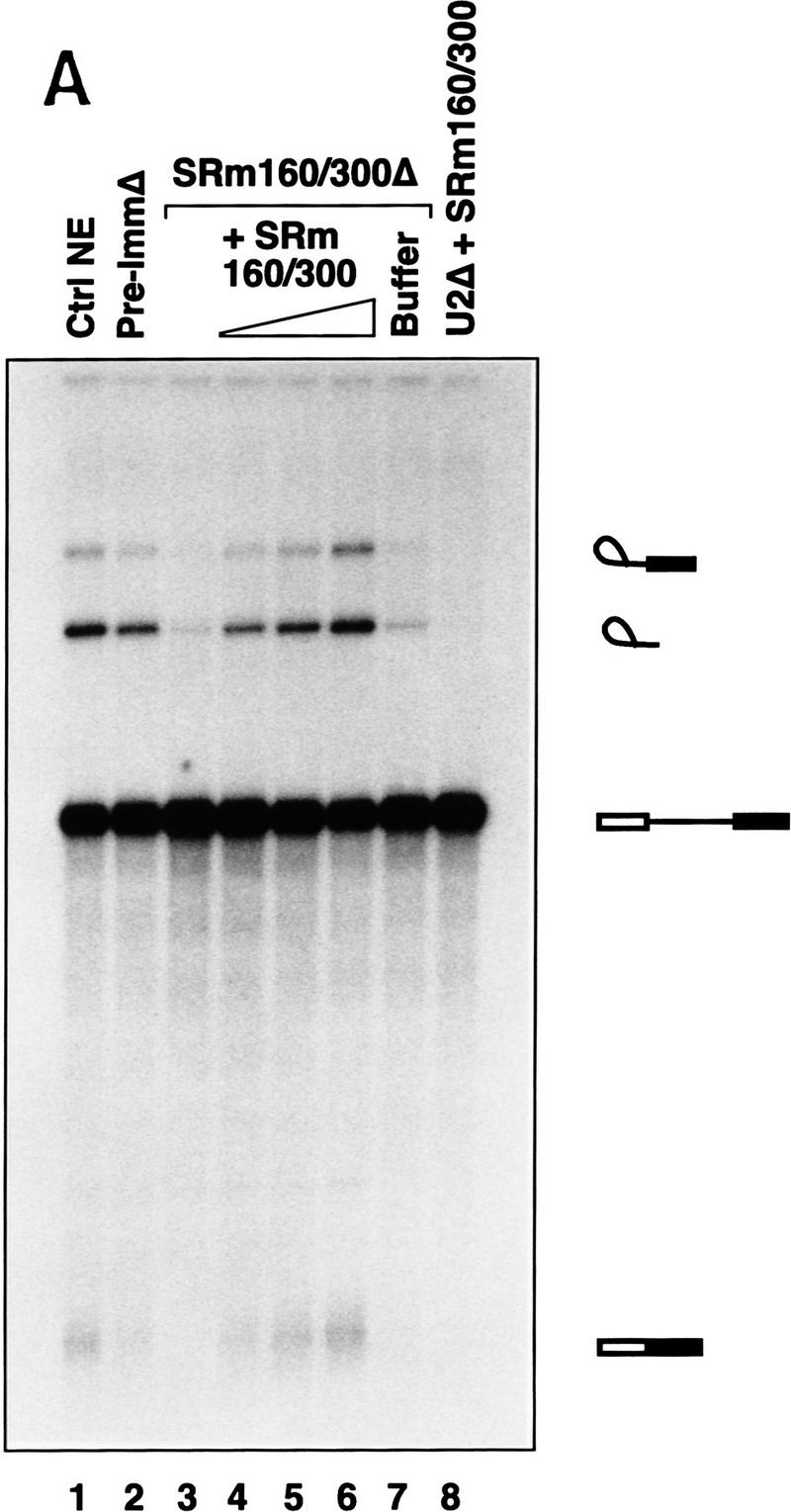
SRm160/300 is required for splicing of specific pre-mRNAs. (A) Depletion of SRm160/300 blocks the first step of splicing of PIP85A pre-mRNA. Splicing reactions containing nuclear extract depleted of SRm160/300 proteins (see Fig. 3C,D, lane 2) were incubated for 1 hr with PIP85A pre-mRNA in the presence (lanes 4–6), or absence (lanes 3,7), of purified SRm160/300 proteins. Control splicing reactions contained regular nuclear extract (lane 1), nuclear extract mock-depleted with preimmune serum (lane 2; see Fig. 3C,D, lane 4), SRm160/300-depleted extract plus protein buffer (lane 7), and U2 snRNP-depleted nuclear extract plus SRm160/300 proteins (lanes 8). Splicing reactions were performed in 30-μl reactions containing a range of 2–8 μg of SRm160/300 proteins in lanes 4–6 and 8 μg of SRm160/300 proteins in lane 8. (B) Comparision of the splicing activity of different pre-mRNAs in mock depleted (lanes 1,3,5) versus SRm160/300-depleted (lanes 2,4,6) nuclear extracts. Reactions were incubated for 1 hr with the Drosophila fushi tarazu (ftz, lanes 1,2), adenovirus major late (Ad1; lanes 3,4), and PIP85A (PIP; lanes 5,6) pre-mRNAs.
Surprisingly, depletion of SRm160/300 did not prevent splicing of the β-globin pre-mRNA substrate (data not shown). This was unexpected as antibodies to both SRm160 and SRm300 proteins immunoprecipitate β-globin splicing complexes (Blencowe et al. 1994), and excess SRm160/300 proteins stimulated β-globin pre-mRNA splicing in nuclear extract and in S100 reactions supplemented with SR proteins (see Fig. 4). Furthermore, it was found previously that mAbs to both SRm160 and SRm300 inhibited splicing of β-globin pre-mRNA (Blencowe et al. 1994; data not shown). However, consistent with the immunodepletion results, addition of rAb–SRm160 to nuclear extracts prevented splicing of PIP85A pre-mRNA but not β-globin pre-mRNA (data not shown). The most likely interpretation of these results is that the splicing of β-globin pre-mRNA may be promoted by two distinct pathways, only one of which is dependent on SRm160/300. Moreover, the previously observed inhibition of β-globin pre-mRNA splicing by mAbs SRm160 and SRm300 was probably attributable to one or more indirect effects of antibody binding.
Additional pre-mRNAs were tested for activity in the SRm160/300-depleted nuclear extracts. A pre-mRNA from the Drosophila fushi tarazu gene (Ftz), like the β-globin pre-mRNA, was spliced with similar efficiency in the depleted extract and extract mock-depleted with preimmune serum (Fig. 5B, lanes 1, 2). In contrast, a substrate derived from the major late region of adenovirus (Ad1), from which PIP85A pre-mRNA was derived, was spliced poorly in the depleted extract (lanes 3,4). Although inefficient, the level of Ad1 splicing in the SRm160/300-depleted extract was higher than that of PIP85A pre-mRNA (cf. lanes 3 and 4 with lanes 5 and 6). These results demonstrate that specific pre-mRNAs differ with respect to their dependence on SRm160/300 proteins for splicing.
When compared with previous findings, the above results indicate a correlation between SRm160/300 dependence and a requirement for U1 snRNP in splicing. The β-globin pre-mRNA is spliced efficiently in U1 snRNP-depleted reactions supplemented with excess SR family proteins, whereas, under the same conditions, the Ad1 pre-mRNA is spliced with reduced efficiency and the PIP85A pre-mRNA is spliced poorly (Crispino et al. 1994, 1996). Ftz pre-mRNA is also spliced efficiently in the absence of U1 snRNP, but without the addition of exogenous SR family proteins (Crispino et al. 1996). These correlative results indicate that pre-mRNAs that are not spliced in the absence of U1 snRNP, even when SR family proteins are in excess, have a stronger requirement for SRm160/300. This correlation was further strengthened by an analysis of PIP85A/β-globin chimeric pre-mRNAs, in which it was determined that sequences within the 3′ half, but not the 5′ half of the β-globin pre-mRNA, when transferred to PIP85A, conferred efficient splicing both in the absence of SRm160/300 or U1 snRNP when SR family proteins were in excess (Crispino et al. 1996; data not shown). Therefore, it is possible that SRm160/300 and U1 snRNP cooperate in promoting stable interactions leading to the formation of productive splicing complexes on some substrates. To investigate this possibility, SRm160/300 and U1 snRNP were tested for functional interdependence.
The association of SRm160/300 with pre-mRNA normally requires U1 snRNP and is stabilized by U2 snRNP
Under regular splicing conditions, binding of SRm160/300 to pre-mRNA is dependent on U1 snRNP. Immunoprecipitation of pre-mRNA by mAb B1C8 was very inefficient in splicing reactions depleted of U1 snRNP (Fig. 6A, lane 8), but was only slightly reduced in reactions depleted of U2 snRNP (lane 7). Identical immunoprecipitation results were obtained with rAb–SRm160 (data not shown). The lack of pre-mRNA immunoprecipitation resulting from depletion of U1 snRNP, and the reduction resulting from depletion of U2 snRNP, was not attributable to nonspecific losses as full levels of splicing activity and splicing complex immunoprecipitation were obtained in reactions containing an equal mix of the U1- and U2-depleted extracts (lane 9). Furthermore, these differences were not attributable to degradation as pre-mRNA was recovered intact in the supernatant fractions of the immunoprecipitations (data not shown). These results indicate that under regular splicing conditions, the association of SRm160/300 with pre-mRNA is greatly stimulated by the presence of U1 snRNP. In contrast, U2 snRNP is not strongly required but likely stabilizes the SRm160/300 association.
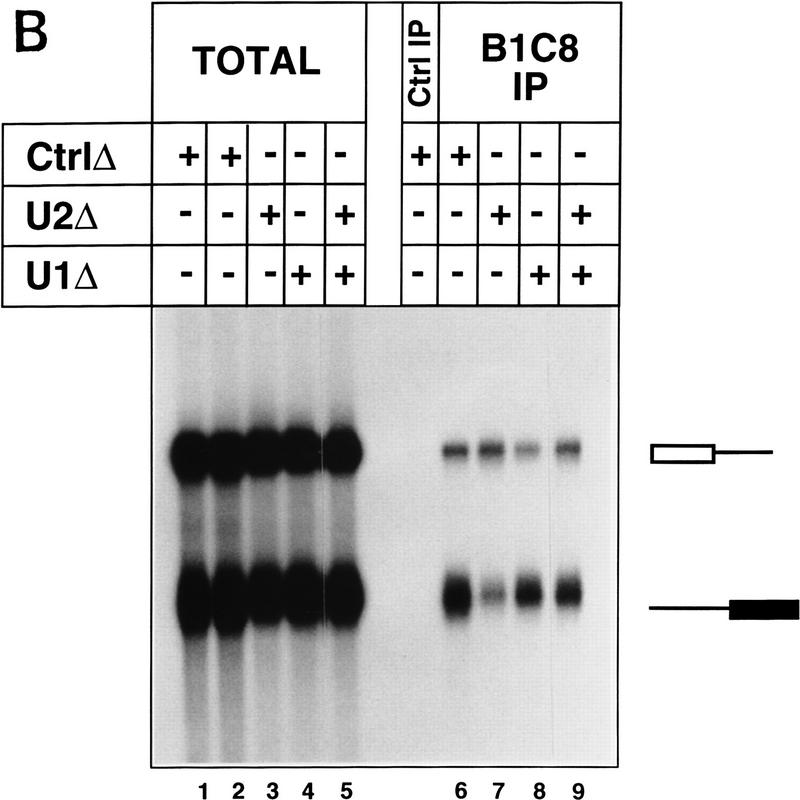
Figure 6.
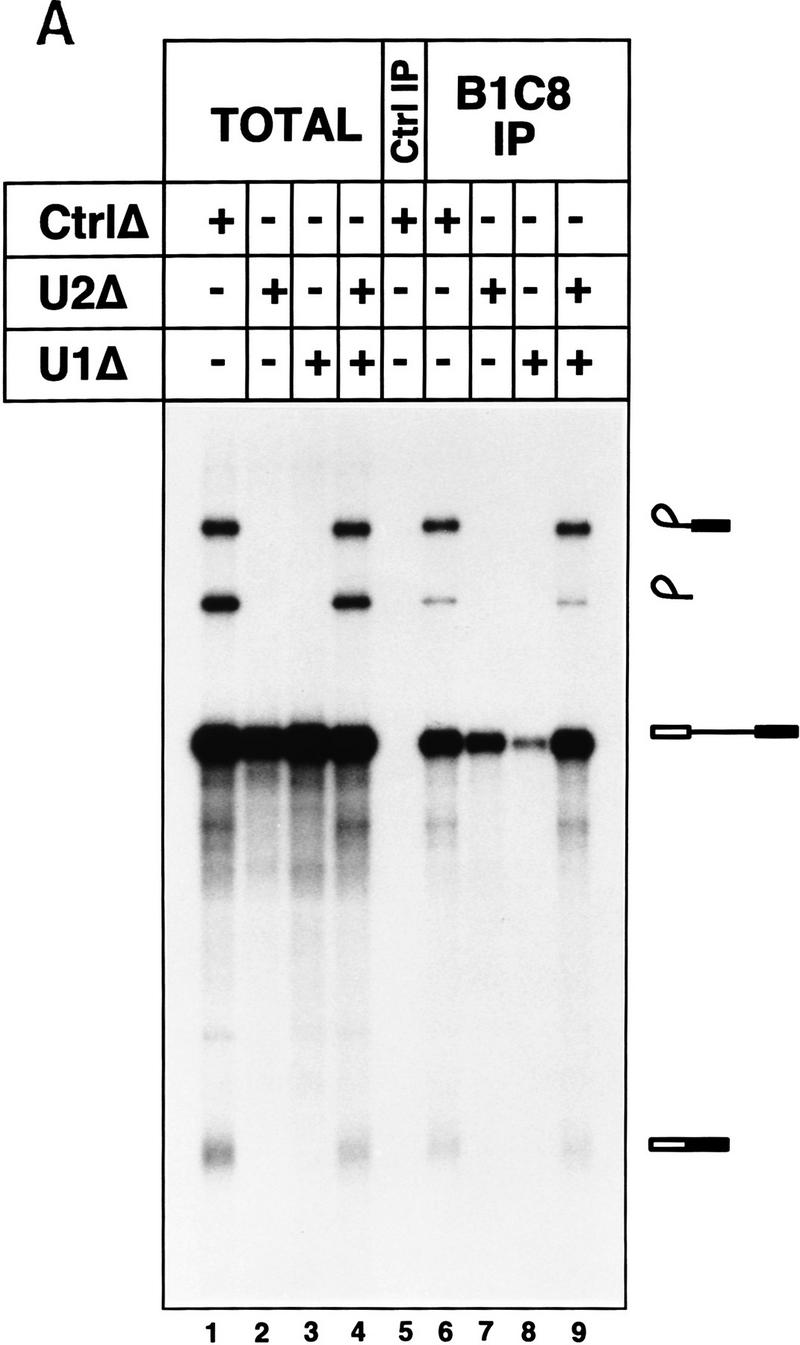
snRNP dependence for binding of SRm160/300 to pre-mRNA. (A) Splicing complexes were immunoprecipitated with mAb B1C8 from different snRNP-depleted and nondepleted reactions (lanes 6–9). Splicing reactions incubated for 40 min with PIP85A pre-mRNA contained mock-depleted (lane 1), U2-depleted (lane 2), U1-depleted (lane 3), or an equal mix of U1- and U2-depleted nuclear extracts (lane 4). RNAs recovered directly from the splicing reactions in lanes 1–4 represent 50% of the total sample, whereas all of the RNAs recovered from each immunoprecipitation were loaded in lanes 5–9. A control immunoprecipitation (lane 5) was performed with a nonspecific antibody from a splicing reaction containing mock-depleted nuclear extract, as in lane 1. (B) Complexes assembled on separate 5′ and 3′ halves of PIP85A pre-mRNA were immunoprecipitated with mAb B1C8 in the presence or absence of individual snRNPs (lanes 6–9). Equal amounts of the two half RNAs were incubated under splicing conditions for 40 min in mock-depleted (lanes 1,2), U2-depleted (lane 3), U1-depleted (lane 4), or an equal mix of U1- and U2-depleted nuclear extracts (lane 5). RNA recovered directly from the splicing reactions in lanes 1–5 represents 50% of the total sample, whereas all of the recovered from each immunoprecipitation was loaded (lanes 6–9); a control immunoprecipitation (lane Ctrl IP) was performed with a nonspecific antibody from a reaction containing mock-depleted nuclear extract (lane 1).
SRm160/300 associates independently with the 5′ and 3′ halves of pre-mRNA
The results in Figure 6A indicate that SRm160/300 may form multiple interactions with pre-mRNA dependent on snRNP binding. To further investigate this possibility, the ability of SRm160/300 to associate with separate 5′ and 3′ halves of PIP85A pre-mRNA was tested in the presence and absence of U1 and U2 snRNPs. mAb B1C8 immunoprecipitated both PIP85A 5′ and 3′ half RNAs incubated under splicing conditions in a mock-depleted nuclear extract (Fig. 6B, lane 6), although at a significantly lower efficiently than the full-length PIP85A pre-mRNA (cf. Fig. 6, cf. A with B). Moreover, the level of immunoprecipitation of the 5′ half RNA was approximately three- to fivefold lower than the 3′ half substrate (Fig. 6B, lane 6). Depletion of U2 snRNP significantly reduced the level of immunoprecipitation of the 3′ half RNA, but did not affect the level of immunoprecipitation of the 5′ half RNA (Fig. 6B, lane 7). Conversely, depletion of U1 snRNP resulted in a reduced level of immunoprecipitation of the 5′ half RNA, but did not significantly alter the level of immunoprecipitation of the 3′ half RNA (cf. lanes 8 and 9). Immunoprecipitation of the half RNAs was restored in a reaction containing an equal mix of the U1 and U2 snRNP-depleted extracts (lane 9). Similar immunoprecipitation results were obtained using rAb–SRm160 (data not shown).
The results indicate that SRm160/300 can associate independently with the 5′ and 3′ halves of PIP85A pre-mRNA and that these interactions are dependent on U1 and U2 snRNPs, respectively. Moreover, the results also indicate that the association of SRm160/300 with splicing complexes depends, in a cooperative manner, on the interactions of factors with multiple parts of the pre-mRNA. On an intact pre-mRNA substrate, the interaction with U1 snRNP is the most critical. When half RNAs are assayed, the interaction of U2 snRNP with the 3′ splice site is the most critical.
SRm160/300 can complement U1 snRNP-dependent splicing reactions supplemented with excess SR family proteins
The results in Figure 6 indicate that SRm160/300 forms multiple interactions with snRNP components bound at the 5′ and 3′ splice sites and that, under normal splicing reactions, SRm160/300 may function by a U1 snRNP-dependent assembly pathway. However, it is important to remember that endogenous levels of SRm160/300 are normally rate limiting for splicing in vitro (see Fig. 4) and, consequently, may be insufficient to promote critical interactions important for splicing when U1 snRNP is absent, even when SR family proteins are in excess. To test this possibility, purified SRm160/300 was added to U1 snRNP-depleted reactions in the presence or absence of excess SR family proteins (Fig. 7). Addition of SRm160/300 alone did not restore activity to U1-depleted reactions when assayed for splicing of PIP85A pre-mRNA (lanes 6,7). Remarkably, however, addition of SRm160/300 to U1-depleted reactions supplemented with excess SR family proteins restored splicing activity (lanes 8–12). The highest level of activity was obtained with the addition of 1.5 μg of SRm160/300 and 3 μg of SR family proteins (lane 12) and was saturated with the addition of more SRm160/300 (lane 13).
Addition of low levels (1 μg) of SR family proteins alone to the U1-depleted extract resulted in partial restoration of splicing activity to the PIP85A pre-mRNA substrate (lane 3), whereas higher levels (2–3 μg) did not promote splicing activity (lanes 4,5). These latter amounts of SR family proteins, however, did promote splicing of the β-globin pre-mRNA in U1 snRNP-depleted reactions (Crispino et al. 1994; data not shown). In contrast, no level of SRm160/300 tested restored splicing of PIP85A or β-globin pre-mRNA in the absence of U1 snRNP. It is possible that the low level of endogenous SRm160/300, which is required for splicing of PIP85A but not β-globin pre-mRNA, is titrated into nonfunctional complexes by addition of excess SR family proteins in the U1 snRNP-depleted reaction. These results provide further evidence for specific functional differences between SRm160/300 and SR family proteins and also indicate that not only the levels but ratios of these proteins is critical for promoting efficient splicing of PIP85A pre-mRNA. Taken together with the data in Figures 4 and 6, synergistic interactions between SRm160/300 and SR family proteins may play a critical role in the splicing of specific pre-mRNAs, by both U1-dependent and U1-independent mechanisms.
Discussion
SRm160/300 and SR family proteins have distinct but complementary functions
SRm160 is a novel protein that is distinct in structural organization and activity from previously characterized splicing factors. It contains an unusually high content of serine (S), arginine (R), and proline (P) residues. Although it contains multiple SR repeats, it lacks an RRM found in SR family proteins. SRm160 forms a stable complex with SRm300 (previously the B4A11 nuclear matrix antigen). Recent sequencing of cDNAs encoding SRm300 reveals that it also is a novel SR-related protein that lacks an RRM (B.J. Blencowe, R. Issner, and P.A. Sharp, in prep.). Biochemically purified SRm160/300 promotes splicing activity in nuclear extracts but, unlike SR family proteins, not in splicing-deficient cytoplasmic S100 extracts. However, coaddition of SRm160/300 and SR family proteins to S100 reactions significantly enhances splicing levels. Also in contrast to SR family proteins, SRm160/300 does not promote splicing activity when added in excess to U1 snRNP-depleted reactions. However, it significantly stimulates splicing in these reactions if added together with SR family proteins. Therefore, SRm160/300 cooperates with, but is functionally distinct from, SR family proteins.
Although SRm160/300 can promote splicing in the absence of U1 snRNP, under “normal” splicing reaction conditions containing endogenous levels of SR family proteins, SRm160/300 associates with pre-mRNA by a U1 snRNP-dependent pathway and its binding to pre-mRNA is further stabilized by U2 snRNP. SRm160/300 also associates preferentially with a subset of specific SR proteins comigrating with SRp40 and SRp75. We propose that the SRm160/300 complex fulfills the role of a “coactivator” of splicing. It interacts with multiple factors including SR proteins and snRNPs that bind directly to the precursor mRNA and, through these interactions, promotes splicing.
SRm160/300 functions as a pre-mRNA-specific splicing coactivator
Specific depletion of the SRm160/300 complex inactivates splicing of a subset of pre-mRNAs. However, addition of excess SRm160/300 to nuclear extracts, or to S100 reactions supplemented with SR proteins, stimulated splicing activity of all substrates tested. That SRm160/300 is rate limiting for general splicing is consistent with the general immunoprecipitation of splicing complexes by antibodies to SRm160 or SRm300. These results suggest that multiple pathways of interactions can result in splicing. Some splicing pathways are highly dependent on interactions involving SRm160/300, whereas other pathways use interactions that are less dependent, or are independent, of this coactivator.
Consistent with complementary roles for SRm160/300 and SR family proteins on SRm160/300-dependent substrates, addition of excess SR family proteins partially restored splicing activity in SRm160/300-depleted reactions (B. Blencowe and P. Sharp, unpubl.). However, full restoration of activity required the addition of SRm160/300. Moreover, excess SR family proteins alone did not promote efficient splicing of PIP85A pre-mRNA in the absence of U1 snRNP; full restoration of activity also required the addition of SRm160/300. These findings indicate that one or more SR family proteins may have a parallel function with SRm160/300 on some substrates, but cannot fully substitute on other substrates. The results indicate that SRm160/300 and SR family proteins have overlapping but nonreciprocal functions and that cooperative interactions between these factors is critical for the splicing of specific pre-mRNAs.
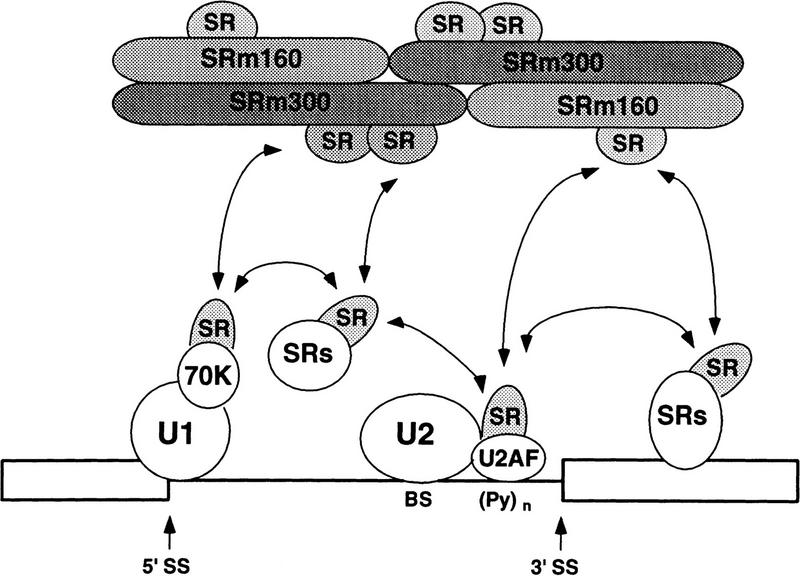
The above results suggest a model in which multiple interactions between U1 snRNP, SRm160/300, and SR proteins, under normal splicing conditions, function in the formation of splicing complexes (see Fig. 8). In this model, substrate-associated SR proteins and U1 snRNP bind SRm160/300. These interactions are further stabilized by binding of U2 snRNP to the branch site. This network of interactions could occur in conjunction with intron bridging interactions, which have been proposed recently (Abovich and Rosbash 1997). The model is consistent with earlier proposals for the involvement of an SR protein network (involving SC35 and ASF/SF2) in the formation of bridging interactions between U1-70K protein at the 5′ splice site and the U2 auxiliary factor 35-kD subunit and a related protein (Urp) at the 3′ splice site (Wu and Maniatis 1993; Kohtz et al. 1994; Tronchère et al. 1997). SRm160/300 may contribute to such an SR interaction network, or form a distinct network, possibly involving SR proteins of 40 and 75 kD, that is required for the recognition of pairs of exon–intron boundaries.
Figure 8.
Splicing coactivator model for the function of SRm160/300. SRm160/300 promotes splice-site pairing and splicing through multiple cooperative interactions with factors bound to pre-mRNA, including SR family proteins U1 and U2 snRNPs. A role for the RS domains of these factors in mediating an intron-wide network of interactions is emphasized. These early interactions are required for the splicing of specific pre-mRNAs. (BS) Branch site; [(Py)n] polypyrimidine tract; (SS) splice site.
SRm160/300 is reminiscent of splicing regulators that form cooperative interactions with SR family proteins on “enhancer” sequences. Splicing enhancers are typically located within exons where they stimulate the utilization of adjacent, weak splice sites. In a prototypic example, a multiprotein complex containing the Drosophila sex determination factors Tra and Tra2, both of which are SR proteins, and the RBP1 (SRp20) SR family protein, assembles on repeats of a specific enhancer sequence within exon 4 of doublesex pre-mRNA (Tian and Maniatis 1992, 1993; Heinrichs and Baker 1995; Lynch and Maniatis 1996). This complex promotes the utilization of an adjacent 3′ splice site resulting in exon 4 inclusion (female-specific splicing pattern) versus exon 4 exclusion (male-specific splicing pattern). Similar to SRm160, Tra is an SR protein that lacks an RRM and associates with other SR proteins that bind to RNA. Like Tra, it is possible that SRm160/300 also functions in association with specific splicing enhancers. However, the results of the present study indicate that Tra and SRm160/300 differ in that, under normal splicing conditions, binding of the latter to pre-mRNA requires U1 snRNP and is stabilized by U2 snRNP, whereas the former is not known to have this property. Thus, the SRm160/300 complex may function in an analogous manner to coactivators of transcription in that it promotes splicing by bridging between sequence-specific (SR family proteins) and basal (snRNP) factors of the spliceosome.
SRm160 phosphorylation
A remarkable property of SRm160 is its unusually high content of serine, arginine, and proline residues and multiple potential phosphorylation sites on serine, threonine, and tyrosine residues. Consistent with these sequence features, SRm160 collapses from 160 to 120 kD upon treatment with phosphatases, indicating that it is normally highly phosphorylated (B. Blencowe, R. Issner, and P. Sharp, unpubl.). Moreover, SRm160 is detected by three different mAbs (104, NM4, and NM22) that recognize distinct phosphoepitopes shared between SR family proteins and is a substrate for at least one SR-specific kinase (SRPK1; Gui et al. 1994), indicating that it is phosphorylated within its SR repeats (Blencowe et al. 1995, and unpubl.). Previous studies have demonstrated that changes in the phosphorylation state of splicing factors can have a profound effect on nuclear organization, splicing activity, and also splice site selection (for review, see Mermoud et al. 1994; Fu 1995; Misteli and Spector 1997). The abundance of potential phosphorylation sites and the extensive phosphorylation of SRm160 suggest that it may be an important target of kinases and phosphatases that regulate splicing.
SRm160/300 as an integral nuclear matrix component
SRm160 and SRm300 are integral phosphoproteins of the nuclear matrix, indicating that they may function in association with this substructure (Blencowe et al. 1994; Wan et al. 1994). The precise structural and functional nature of the nuclear matrix is not understood. Extensive morphological characterization indicates that the matrix is composed of a lattice of interconnecting filaments that enmesh nucleoli and nonnucleolar substructures, including speckles (Nickerson et al. 1997, and references therein). Previous studies support a role for the nuclear matrix in splicing. Specific pre-mRNAs, spliced mRNAs, splicing factors, and splicing have all been detected in association with nuclear matrix preparations (for review, see Mattern et al. 1997; Penman et al. 1997; Pederson 1998). The results in the present study support a previous model in which it was proposed that SR-related proteins associated with the nuclear matrix perform critical roles in splicing (Blencowe et al. 1994). As integral matrix proteins, it is also possible that SRm160 and SRm300 function in the nuclear organization of pre-mRNA splicing. Both proteins are highly concentrated within speckled domains in the matrix. Consistent with evidence that a significant component of cotranscriptional splicing may occur outside of speckles, SRm160 is also detected in many smaller foci separated from speckle domains. However, the detection of specific pre-mRNAs and corresponding spliced mRNAs at the periphery of or within speckles indicates that these structures are sites of processing of a subset of pre-mRNAs (Xing et al. 1993, 1995; Huang and Spector 1996). It is possible that the preferential localization of these transcripts to speckle domains is influenced by their dependence on specific combinations of pre-mRNA splicing factors, including SRm160/300.
Materials and methods
cDNA isolation and sequencing
Sequence analysis of four overlapping EST clones (78873, 113038, 121604, and 138500), and two cDNAs (pSRm160–1C1A and pSRm160–2C2B) isolated from a human myeloid cell line (U937) cDNA library (kind gift of Stefano Volinia, Univerista’ delgi Studi, Ferrara, Italy) in Lambda ZapII (Strategene), allowed the assembly of a 3.7-kb contig containing the SRm160 ORF (see Fig. 1). Sequences 5′ to the cDNA sequence in human genomic DNA were obtained from P1-derived artificial chromosome (PAC) clones, isolated by Genomic Systems, Inc., using a PstI fragment (bases 1–1424 in EST 78873) as a hybridization probe. cDNA clones were sequenced on both strands using dGTP and dITP reactions and sequenase (U.S. Biochemical). PAC clones were sequenced using ampliTaq polymerase (Roche), as per the manufacturer’s recommendations.
Antibodies
A cDNA fragment encoding SRm160 ORF amino acids 7–160 was generated by PCR amplification from EST 78873 and inserted into the pGEX–KG expression vector (Pharmacia) to generate pGST–SRm160. The GST–SRm160 fusion protein was expressed and purified from cell lysates on glutathione–Sepharose (Pharmacia), as per the manufacturer’s instructions. Female New Zealand rabbits were immunized with GST–SRm160 according to standard immunization protocols by Covance Co. (Denver, PA). Serum antibodies were affinity purified on immobilized SRm160–GST fusion protein and eluted with high and low pH buffers, as described by Harlow and Lane (1988). Total serum immunoglobulin was purified on protein A–Sepharose beads and eluted using Tris–glycine buffer (pH 3.0), as described by Harlow and Lane (1988).
Murine mAbs used in the study are B1C8 (Blencowe et al. 1994; Wan et al. 1994), B4A11 (Blencowe et al. 1994), and mAb104 (Roth et al. 1990).
Nuclear extracts
HeLa nuclear and cytoplasmic S100 extracts were prepared as described by Dignam et al. (1983). Nuclear extracts were depleted of individual snRNPs using the antisense affinity depletion (AAD) method (Barabino et al. 1990; Blencowe and Barabino 1995). Levels of snRNP depletion were analyzed and quantitated by Northern blotting; >99% removal of the targeted species was achieved in each case. Depletion of SRm160/300 proteins was performed using conditions essentially as described for snRNPs by the AAD method. Approximately 0.5 mg of total purified serum immunoglobulin was cross-linked to 0.2 ml (packed bead volume) of protein A–Sepharose CL4B beads (Pharmacia) using dimethylpilimidate (Harlow and Lane 1988). Immunodepletion was performed using two 45-min incubations of 0.1 ml of rAb–SRm160-linked beads (=packed bead volume) from 0.5 ml of a “high salt” nuclear extract (HSNE, Blencowe and Barabino 1995). Immunodepleted extracts were dialyzed into regular buffer D (Dignam et al. 1983) and characterized for the extent of depletion and splicing activity.
Protein purification
SR family proteins were purified as described by Zahler et al. (1992). SRm160 and SRm300 proteins were purified as described in Blencowe et al. (1995); 10–12 μg of the final DEAE-FT fraction containing SRm160 and SRm300 proteins was separated by SDS-PAGE, transferred to nitrocelluluse membrane, excised, and digested with lys-C. Five of the resulting lys-C peptides were microsequenced.
For functional studies of SRm160/300 proteins, the final DEAE-FT fraction, described in Figure 4 of Blencowe et al. (1995), was dialyzed against 3 × 1-liter changes of a buffer containing 5% glycerol, 100 mm NaCl, 15 mm HEPES (pH 7.6), 0.75 mm EDTA, 1.5 mm DTT, 2.5 mm KF, 2.5 mm β-glycerophosphate, and 0.2 mm Pefabloc (Boehringer). The dialyzed fraction (∼1 mg/ml) was added directly to splicing reactions.
Immunocytochemistry
CaSki cells (CRL 1550) were washed with Hank’s balanced salt solution at 37°C before extraction for 2 min at room temperature with cytoskeletal buffer [10 mm PIPES (pH 6.8), 300 mm sucrose, 100 mm NaCl, 3 mm MgCl2, and 1 mm EGTA, 20 mm vanadyl riboside complex, 1 mm 4-(2-aminoethyl-benzenesulfonyl fluoride] containing 0.5% Triton X-100. The samples were fixed in cytoskeletal buffer containing 4% (wt/vol) formaldehyde for 20–50 min at room temperature, washed three times in PBS containing 0.1% (vol/vol) Tween 20, blocked with 10% (vol/vol) normal goat serum in PBS-Tween, and incubated with antibodies in PBS–Tween containing 2% (vol/vol) normal goat serum for 1–2 hr. To localize SRm160 in mitotic cells, taxol was included at 1 μg/ml in the Hank’s balanced salt solution wash and at 2 μg/ml in the cytoskeletal buffer containing Triton to preserve spindle microtubules. rAb–SRm160 immunostaining was detected with a biotinylated goat anti-rabbit IgG with Extravidin–FITC (Sigma) and mAb B1C8 staining was detected with a rhodamine-coupled goat anti-mouse IgM. The samples were mounted in Vectashield (Vector Laboratories) and examined by epifluorescence microscopy and confocal microscopy using a Leica TCS system.
Immunoprecipitation
Immunoprecipitation of splicing complexes was performed as described by Blencowe et al. (1994). SRm160 associated proteins were immunoprecipitated with rAb–SRm160 cross-linked to protein A beads using the same conditions as described above for immunodepletion; the pellet samples shown in Figure 3A,B correspond to proteins recovered from protein A beads after the first round of SRm160 immunodepletion from HSNE. After immunoprecipitation, the beads were washed three times for 5 min in a buffer containing 350 mm KCl, 20 mm HEPES (pH 7.6), 0.05% NP-40, and 1 min in an identical buffer but containing 100 mm KCl. The beads were drained, resuspended in two volumes of Laemmli loading buffer, and boiled for 5 min. Eluted proteins were analyzed in 12.5% SDS–polyacrylamide gels.
Splicing assays
Unless stated otherwise in the figure legends, splicing assays were performed in 15-μl volumes containing 5 μl of extract, 1 μl of a concentrated splicing mix (30 mm MgCl2, 22.5 mm ATP, 75 mm creatine phosphate, 75 mm DTT for reactions containing PIP85A, Ad1, Ftz, and chimeric PIP-βGlo pre-mRNAs or 45 mm MgCl2, 22.5 mm ATP, 75 mm creatine phosphate, 75 mm DTT for reactions containing β-globin pre-mRNA), 1 μl of RNAsin (Promega), and 1 μl of pre-mRNA substrate (labeled with 6000Ci/mmole[ α-32P]UTP) diluted to ∼300 cps in H20. Splicing reactions containing PIP85A and Ftz pre-mRNAs were analyzed on 15% denaturing polyacrylamide gels and reactions containing Ad1 and β-globin pre-mRNAs on 10% and 8% gels, respectively.
SDS-PAGE and immunoblot analysis
SDS-PAGE and immunoblots were performed essentially as described by Harlow and Lane (1988). Immunoblots were developed using secondary antibodies conjugated to alkaline phosphatase (Promega), also as described by Harlow and Lane (1988).
Acknowledgments
We thank J. Borrow for reagents and advice on cDNA isolation and J. Crispino for providing snRNP-depleted extracts. We are also grateful to R. Arnaout and J. Augliera for assistance with DNA sequencing, to R. Cook and S. Densmore for peptide microsequencing, and to M. Lauro for oligonucleotide synthesis. S. Volinia (Univerista’ delgi Studi, Ferrara) generously provided the U937 cDNA library, and colleagues in the splicing community, including X.-D. Fu, A. Mayeda, A. Krainer, M. Roth, J. Steitz, and A. Zahler, generously provided antibodies and recombinant SR family proteins during the course of these studies. We thank G.M. Whitesides and R. Jackman for the use of their confocal microscope. We thank D. Chasman for advice on sequence analysis; E. Reifenberg, M. Beddal, M. Siafaca, and members of the laboratory in general for their support and helpful discussions; B. Davis, G. Buarén, L. Lim, P. McCaw, R. Pulak, and A. Simons for kindly reviewing the manuscript. This work was supported by U.S. Public Health Service grants R01-AI32486, RO1-GM34277, and RO1-CA42063 from the National Institutes of Health (NIH), by a cooperative agreement CDR-8803014 from the National Science Foundation to P.A.S., and, partially, by National Cancer Institute (NCI) Cancer Center Support (core) grant P30-CA14051. B.J.B. was supported by a Fellowship and Career Development Award from the U.S. Department of Defense Breast Cancer Research Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sharppa@mit.edu; FAX (617) 253-3867.
References
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Alzhanova-Ericsson AT, Sun X, Visa N, Kiseleva E, Wurtz T, Daneholt B. A protein of the SR family binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from gene to the nuclear pore. Genes & Dev. 1996;10:2881–2893. doi: 10.1101/gad.10.22.2881. [DOI] [PubMed] [Google Scholar]
- Amrein H, Hedley ML, Maniatis T. The role of specific protein-RNA interactions and protein-protein interactions in positive and negative control of pre-mRNA splicing by transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- Barabino SML, Blencowe BJ, Ryder U, Sproat BS, Lamond AI. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990;63:293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Black D. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Barabino SML. Antisense affinity depletion of RNP particles. Methods Mol Biol. 1995;37:67–76. doi: 10.1385/0-89603-288-4:67. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Nickerson JA, Issner R, Penman S, Sharp PA. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Issner R, Kim J, McCaw P, Sharp PA. New proteins related to the Ser-Arg family of splicing factors. RNA. 1995;1:852–865. [PMC free article] [PubMed] [Google Scholar]
- Bourquin JP, Stagljar I, Meier P, Moosmann P, Silke J, Baechi T, Georgiev O, Schaffner W. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1997;11:2055–2061. doi: 10.1093/nar/25.11.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman D, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V, Bucher P, Nourbakhsh I, Blaisdell B E, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Mistelli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KC, Taneja KL, Lawrence JB. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B. Directing alternative splicing: Cast and scenarios. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley J, Bell J, Duncan P. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Crispino JD, Blencowe BJ, Sharp PA. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science. 1994;265:1866–1869. doi: 10.1126/science.8091213. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Mermoud JE, Lamond AI, Sharp PA. Cis-acting elements distinct from the 5′ splice site promote U1-independent pre-mRNA splicing. RNA. 1996;2:664–673. [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebowitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Puvion E. The ultrastructural visualization of nuclear and extranucleolar RNA synthesis and distribution. Int Rev Cyt. 1980;65:255. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Fay F, Taneja K, Shenoy S, Lifshitz L, Singer R. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A) Exp Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gui J-F, Lane WS, Fu X-D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. New York, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hedley M, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc Natl Acad Sci. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs V, Baker BS. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck T, Ellisman M, Spector DL. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. Transcription occurs at the nucleoskeleton. EMBO J. 1985;4:919–925. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes & Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M. The coiled body. Trends Cell Biol. 1993;3:198–204. doi: 10.1016/0962-8924(93)90214-l. [DOI] [PubMed] [Google Scholar]
- Lawrence J, Carter K, Xing X. Probing functional organization within the nucleus: Is genome structure integrated with RNA metabolism? Cold Spring Harbor Symp Quant Biol. 1993;58:807–818. doi: 10.1101/sqb.1993.058.01.088. [DOI] [PubMed] [Google Scholar]
- Li H, Bingham PM. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- Lynch K, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes & Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- Manley J, Tacke R. SR proteins and splicing control. Genes & Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Mattern K, van Driel R, de Jong L. Composition and structure of the nuclear matrix. In: Curtis BR, editor. Nuclear structure and gene expression. New York, NY: Academic Press; 1997. pp. 87–110. [Google Scholar]
- Meier U, Blobel G. Nopp140 shuttles on tracks between nucleus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Mermoud J, Calvio C, Lamond AI. Uncovering the role of Ser/Thr protein phosphorylation in nuclear pre-mRNA splicing. Adv Prot Phosphatases. 1994;8:99–118. [Google Scholar]
- Misteli T, Spector DL. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 1997;7:135–138. doi: 10.1016/S0962-8924(96)20043-1. [DOI] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu HL, Du L, Warren SL, Sharp P, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson JA, Krockmalnic G, Wan KM, Turner CD, Penman S. A normally masked nuclear matrix antigen that appears at mitosis on cytoskeleton filaments adjoining chromosomes, centrioles and midbodies. J Cell Biol. 1992;116:977–987. doi: 10.1083/jcb.116.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson JA, Krockmalnic G, Wan KM, Penman S. The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc Natl Acad Sci. 1997;94:4446–4450. doi: 10.1073/pnas.94.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. 1998. Thinking about a nuclear matrix. J. Mol. Biol. (in press). [DOI] [PubMed]
- Penman S, Blencowe B, Nickerson J. Nuclear structure and gene expression. New York, NY: Academic Press; 1997. The nuclear matrix: Past and present; pp. 3–24. [Google Scholar]
- Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- Roscigno R, Garcia-Blanco M. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA. Nobel lecture: Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- Smith HC, Harris SG, Zillmann M, Berget SM. Evidence that a nuclear matrix protein participates in pre-mRNA splicing. Exp Cell Res. 1989;182:521–533. doi: 10.1016/0014-4827(89)90255-3. [DOI] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of snRNPs. Biol Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu X-D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992;256:237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- ————— A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- Tronchère H, Wang J, Fu X-D. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388:397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- Valcárcel J, Green MR. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- Verheijen R, Kuijpers H, Vooijs P, van Venrooij W, Ramaekers F. Distribution of the 70k U1 RNA-associated protein during interphase and mitosis: Correlation with other U RNP particles and proteins of the nuclear matrix. J Cell Sci. 1986;86:173–190. doi: 10.1242/jcs.86.1.173. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Hunt BF. A subset of small nuclear ribonucleoprotein particle antigens is a component of the nuclear matrix. Biochem Biophys Res Commun. 1982;105:1224–1232. doi: 10.1016/0006-291x(82)91099-3. [DOI] [PubMed] [Google Scholar]
- Wan K, Nickerson JA, Krockmalnic G, Penman S. The SRm160 protein is in the dense assemblies of the nuclear matrix and relocates to the spindle and pericentriolar filaments at mitosis. Proc Natl Acad Sci. 1994;91:594–598. doi: 10.1073/pnas.91.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson C, McNeil PM, Jr, Lawrence J. Nonrandom gene organization: Structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stalk JA, Roth MB. SR proteins: A conserved family of pre-mRNA splicing factors. Genes & Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zeitlin S, Parent A, Silverstein S, Efstratiadis A. Pre-mRNA splicing and the nuclear matrix. Mol Cell Biol. 1987;7:111–120. doi: 10.1128/mcb.7.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S, Wilson RC, Efstratiadis A. Autonomous splicing and complementation of in vivo assembled spliceosomes. J Cell Biol. 1989;108:765–777. doi: 10.1083/jcb.108.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]