Abstract
Signals from the apical ectodermal ridge (AER) of the developing vertebrate limb, including fibroblast growth factor-8 (FGF-8), can maintain limb mesenchymal cells in a proliferative state. We report here that a specific CD44 splice variant is crucial for the proliferation of these mesenchymal cells. Epitopes carried by this variant colocalize temporally and spatially with FGF-8 in the AER throughout early limb development. A splice variant containing the same sequences expressed on model cells binds both FGF-4 and FGF-8 and stimulates mesenchymal cells in vitro. When applied to the AER, an antibody against a specific CD44 epitope blocks FGF presentation and inhibits limb outgrowth. Therefore, CD44 is necessary for limb development and functions in a novel growth factor presentation mechanism likely relevant in other physiological and pathological situations where a cell surface protein presents a signaling molecule to a neighboring cell.
Keywords: CD44, FGF, limb development, apical ectodermal ridge
The growth and differentiation of the developing vertebrate limb are initiated and maintained by interactions between proliferating mesenchymal cells and their overlying ectoderm (Harrison 1918). Mesenchymal cells contributing to the limb originate from the lateral plate mesoderm and their initial proliferation is stimulated by signals from the embryonic kidney (Stephens and McNulty 1981; Strecker and Stephens 1983; Geduspan and Solursh 1992). In birds and mammals, the accumulation of these mesenchymal cells then induces the ectoderm along the anterior to posterior rim to elongate and form a specialized structure called the apical ectodermal ridge (AER). Upon differentiation, the AER maintains the proximodistal outgrowth of the limb bud by keeping adjacent mesenchymal cells in an undifferentiated proliferating state (Zwilling 1955; Rubin and Saunders 1972; Summerbell et al. 1973). These proliferating cells constitute the so-called progress zone of the limb bud.
Particular members of the fibroblast growth factor (FGF) family can substitute for the growth-promoting functions of the AER and could be involved in the initiation of limb development (for review, see Yamaguchi and Rossant 1995; Cohn and Tickle 1996). These polypeptide growth factors affect cell motility, proliferation, survival, and differentiation in numerous cell types by binding to high affinity FGF receptors (for review, see Mason 1994). Among the known FGF proteins, FGF-2 (Savage et al. 1993; Dono and Zeller 1994), FGF-4 (Niswander and Martin 1992; Suzuki et al. 1992), and FGF-8 (Heikinheimo et al. 1994; Ohuchi et al. 1994; Crossley and Martin 1995; Mahmood et al. 1995; Crossley et al. 1996; Vogel et al. 1996; Ohuchi et al. 1997) are expressed in the AER. Heparin-coated beads loaded with any of these proteins can initiate the development of ectopic limbs when inserted into the flank of chick embryos, and can sustain limb development when placed on limb buds lacking an AER (Niswander et al. 1993; Niswander and Martin 1993; Fallon et al. 1994; Cohn et al. 1995; Mahmood et al. 1995; Crossley et al. 1996; Vogel et al. 1996; Ohuchi et al. 1997). However, only FGF-8 is expressed in prelimb ectoderm and later becomes restricted to the entire AER.
The binding of FGF proteins to their high affinity receptors may require several steps (for review, see Vlodavsky et al. 1996). For example, the association of FGF proteins with cell–surface heparan sulfate proteoglycans (HSPGs) is a prerequisite for high affinity FGF receptor activation (Heath et al. 1991; Rapraeger et al. 1991; Yayon et al. 1991; Ornitz et al. 1992; Spivak-Kroizman et al. 1994; Venkataraman et al. 1996). There is also growing evidence that additional HSPGs in the extracellular matrix can sequester and store secreted FGF proteins, and protect them from proteolytic degradation (Folkman et al. 1980; Gospodarowicz and Cheng 1986; Vlodavsky et al. 1987; Saksela et al. 1988; Cardon-Cardo et al. 1990; Gonzalez et al. 1990; Ishai-Michaeli et al. 1992). Therefore, it is likely that HSPGs in the developing limb have one or more of these functions and are crucial for the regulated activities of FGF proteins during limb morphogenesis.
In the present study, we have investigated the role of CD44 proteins in early limb development. Members of this family of transmembrane glycoproteins are expressed by cells of the limb bud, including those of the AER (Wheatley et al. 1993; Yu et al. 1996). Distinct CD44 variants are generated from a single gene by the alternative RNA splicing of up to 10 variant (“v”) exons and by extensive post-translational modifications. The amino acid sequences encoded by these variant exons are located in the extracellular portion of the protein near the transmembrane domain (Screaton et al. 1992; Tölg et al. 1993). A standard form of CD44 (CD44s) lacking these variant sequences is expressed by numerous cell types and is the smallest CD44 protein. It carries no variant exon sequences. Splice variants are only expressed in a limited number of tissues and in certain tumors (for review, see Sherman et al. 1996; Naor et al. 1997).
Particular CD44 proteins are likely to have distinct molecular properties. Our interest in these proteins stems from the finding that specific CD44 splice variants can contribute to the growth and metastasis of tumor cells (Günthert et al. 1991; for reviews, see Sherman et al. 1996; Naor et al. 1997). CD44 proteins have also been implicated in cell–cell adhesion (Jalkanen et al. 1987; St. John et al. 1990; DeGrendele et al. 1996), cell migration and invasion (Thomas et al. 1992; Lamb et al. 1997), and cell–matrix interactions, including binding to glycosaminoglycans such as hyaluronate, chondroitin sulfate, and heparan sulfate (Wayner and Carter 1987; Aruffo et al. 1990; Culty et al. 1990; Lesley et al. 1990; Jalkanen and Jalkanen 1992; Sleeman et al. 1997). Furthermore, certain CD44 proteins can be modified by glycosaminoglycans, which enable them to bind growth factors such as FGF-2 (Brown et al. 1991; Faassen et al. 1992; Tanaka et al. 1993; Jackson et al. 1995; Bennett et al. 1995a). For these reasons it is conceivable that CD44 may play one or more of these roles in the developing limb bud, where numerous cell–cell and cell–matrix interactions occur and where tightly regulated cellular signaling between the AER, limb ectoderm, and mesenchyme is required. It is also possible that the mechanisms used by CD44 in the embryonic limb are recapitulated during certain pathological processes, including tumor progression.
We report here that a CD44 variant carrying the alternatively spliced sequences encoded by variant exons v3 and v6 are required during early limb development. This variant characterized by v3- and v6-encoded epitopes is expressed initially by prelimb ectoderm and later becomes restricted to the AER. It is coexpressed temporally and spatially with FGF-8 in many embryonic tissues and colocalizes with FGF-8 on the surfaces of AER cells. Transfected cells bearing CD44v3–v10 can bind FGF-8 (as well as FGF-4) and present it to limb mesenchymal cells in vitro. (CD44v3–v10 indicates a CD44 protein that carries the amino acid sequences encoded by exons v3–v10 in addition to the constant CD44 structure.) CD44v5 would be a protein with only the v5 sequence in addition to the constant sequences. Furthermore, antibodies recognizing v3 and v6 epitopes inhibit FGF-8 presentation and limb bud outgrowth. These data indicate that CD44 is crucial for maintaining the growth-promoting functions of the AER and reveal a novel growth factor presentation mechanism, whereby a cell surface protein presents FGF to receptors on opposing cells.
Results
CD44 variants are expressed in prelimb ectoderm and in the AER
CD44 proteins were detected previously throughout the developing mouse limb bud using an antibody recognizing the common amino terminus of all CD44 proteins (Wheatley et al. 1993). To characterize which CD44 proteins are expressed by the different structures of the developing limb, we examined CD44 expression in rat embryos between embryonic days (E) 10.5 and 12.5 using whole mount immunohistochemistry with either an antibody that recognizes all CD44 proteins (5G8, epitope encoded by exon 15; Sleeman et al. 1996) or one that recognizes only CD44 proteins containing sequences encoded by CD44 exon v6 (1.1ASML; Günthert et al. 1991). Compared to total CD44 expression (Fig. 1A), which was identical to that reported in the mouse (Wheatley et al. 1993), CD44v6 epitope expression was highly restricted in E11.5 (Wainwright et al. 1996) and E12.5 rat embryos, with the strongest staining occurring in the AER (Fig. 1B). Similar results were obtained with an antibody against a v3 epitope and by in situ hybridization using v2 and v4 sequences as riboprobes (not shown). Some CD44v6 epitope staining was also evident in the vitelline vein and branchial arches, and, upon sectioning, in the developing forebrain (data not shown). In contrast to total CD44 expression, the v6 epitope could not be detected in the notochord or somites. In E10.5 embryos, CD44v6 was strongest in the ectoderm of the branchial arches (Fig. 1C) and in the putative forelimb ectoderm, between somites 7 and 14 (Fig. 1C,D). No CD44v6 was detected in the mesenchyme of either the branchial arches or early limb buds.
Figure 1.
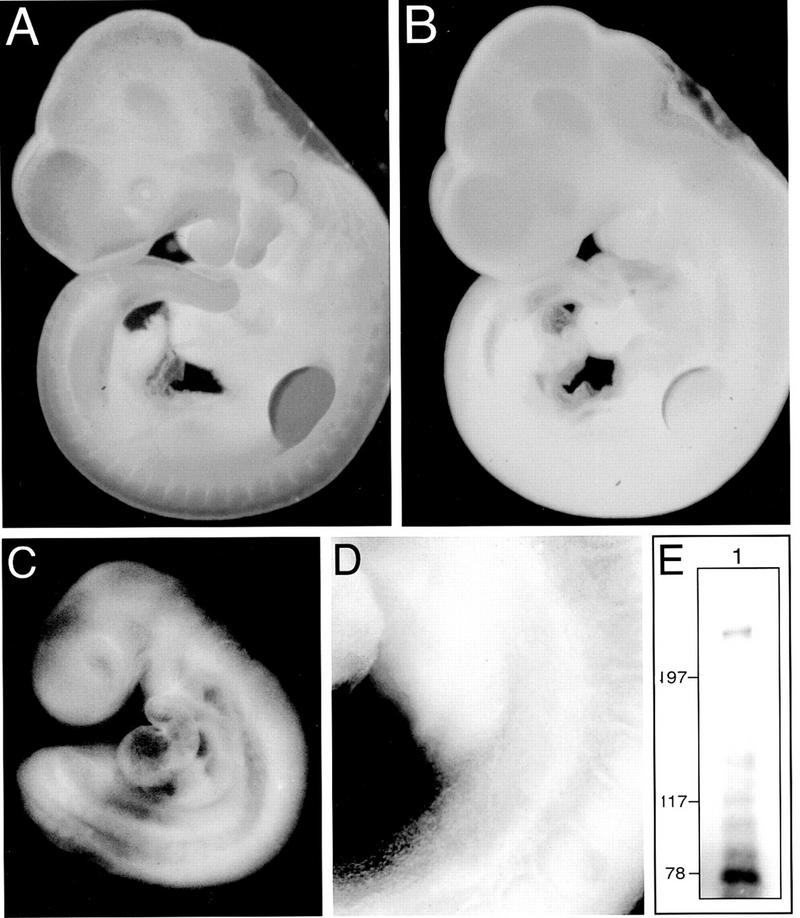
Splice variants of CD44 are expressed in putative limb ectoderm and become localized to the AER. Whole mount immunohistochemistry of E12.5 (A,B) and E10.5 (C,D) rat embryos showing expression of all CD44 proteins (A) and CD44 proteins containing the v6 epitope (B–D). Note that, compared to total CD44, the CD44v6 epitope is highly restricted during embryogenesis and is most prominent in the AER by E12.5 (B). Note also that CD44v6 expression can be detected in prelimb ectoderm (C, and at higher magnification in D). (E) Western blot of CD44 proteins immunoprecipitated from E12.5 rat limbs. In addition to CD44s (∼78 kD), the predominant CD44 variant protein has a molecular mass in excess of 200 kD. Several other smaller CD44 variant proteins are also visible. Marker proteins are indicated in kD.
To identify the splice composition of the CD44 transcripts expressed in limb buds, we analyzed poly(A)+ RNA from either dissected rat E12.5 AER tissue or from distal limb mesenchyme and ectoderm using an RT–PCR-based assay (van Weering et al. 1993; Wainwright et al. 1996). We found that the AER expresses a CD44 splice variant containing all of the variant exon sequences (CD44v1–v10; see Fig. 2A), whereas the remainder of the limb bud expresses only CD44s (Fig. 2B). This result agrees with our in situ hybridizations and immunohistochemistry results described above. The PCR strategy used here does not allow for the identification of multiple transcripts with consecutive variant exon sequences all ending with exon v10. Such PCR products would be hidden underneath the laddering. Therefore, we further analyzed AER tissue using a different set of primers (not shown). Consistent with results from another laboratory (Yu et al. 1996), we identified additional CD44 splice variants with v3–v10, v4–v10, v6–v10, and v8–v10 sequences. CD44v3–v10 is the most prominent species. The same relative abundance of CD44v3–v10 is documented in the first lane of Figure 2A (primers outside the v region, lane FULL, expected size 1.25 kb), whereas a CD44v1–v10-derived PCR product is barely visible. Also a CD44s transcript was detected in the AER sample. Part or all of the CD44s signal may have originated from contaminating mesenchymal cells, as it was not possible to remove all of the mesenchyme from the dissected AER tissue. Western blot analysis of CD44 proteins immunoprecipitated from whole limb buds demonstrated the expression of one major CD44 variant protein with an apparent molecular mass of ∼230 kD and of several smaller CD44 variant proteins ranging in size from ∼90 to 140 kD (see Fig. 1E). Because PCR data localized variants exclusively to the AER, we assume that the 230-kD protein is carried by AER cells and represents the major CD44v3–v10 variant. These data indicate that splice variants of CD44 carrying v3 and v6 sequences are expressed by putative limb ectoderm during the initiation of limb bud formation and later become localized to the AER.
Figure 2.

CD44 transcripts expressed in the AER. RT–PCR analysis of RNA isolated from AER tissue (A) and the rest of the limb bud (B) of E12.5 rat embryos. After cDNA synthesis, the PCR protocol of van Weering et al. (1993) was followed (see Materials and Methods). In brief, CD44 sequences were first amplified using 5′ and 3′ primers (in the constant region of CD44), then this product was amplified further using a second 3′ primer and either a second 5′ primer located in the constant region (a probe of which is loaded in lane FULL) or variant exon-specific 5′ primers (v1–v10). The products of the second reaction were then analyzed on an agarose gel stained with ethidium bromide. (WATER) No cDNA; (−RT) without reverse transcriptase during the initial cDNA synthesis. The PCR product for CD44s is 421 bp in lane FULL. Note that the sizes in lanes v1–v10 are not comparable to the FULL lane as different primers were used (see also König et al. 1996).
A CD44v6-specific antibody blocks limb outgrowth
Because CD44 splice variants are localized to the AER throughout the course of early limb development, we postulated that these CD44 proteins could play a role in limb bud outgrowth. The CD44v6-specific antibody used above (1.1ASML) had been shown previously to inhibit the activity of CD44 splice variants in several biological systems (Reber et al. 1990; Arch et al. 1992; Seiter et al. 1993; Weiss et al. 1997). To test whether this antibody might similarly inhibit CD44 functions in the AER, we examined how 1.1ASML influenced limb development compared to the isotype-matched 5G8 antibody, which recognizes another CD44 epitope that is proximal to the membrane, and also to 1.1ASML, which had been preincubated with the v6 epitope peptide. The AERs of rat embryos isolated at a stage soon after AER formation (E11.5, equivalent to mouse limb stage 2; see Wanek et al. 1989), were removed aseptically from the developing right forelimbs using tungsten needles, incubated with antibodies, then grafted back onto the limb bud (scheme in Fig. 3). After 4 days in culture, limb buds from embryos that had received 1.1ASML-treated AERs (n = 14) were significantly shorter (by >50%) than unoperated control limbs (Fig. 3, cf. B with A, quantitation in C). In one set of experiments, the AER was removed and not grafted back onto the wounded limb bud. In each case (n = 3), limb bud outgrowth was inhibited to at least a similar degree as limbs that had received 1.1ASML-treated AER grafts. There were no significant differences in limb bud lengths between untreated limb buds and limb buds in embryos that had received AERs treated with 5G8 (n = 13) or 1.1ASML in combination with the CD44v6 peptide (n = 3) (Fig. 3C). Therefore, the effects of 1.1ASML were specific and not attributable to cytotoxicity. To further support that 1.1ASML treatment did not reduce the viability of AER cells upon binding, 5G8- and 1.1ASML-treated AER tissue was examined for FGF-8 RNA synthesis by in situ hybridization at 24 hr. In both cases similar levels of FGF-8 RNA were detected (not shown) indicating that 1.1ASML does not affect the viability of AER cells. This conclusion is consistent with trypan blue exclusion experiments, which showed marginal staining or no staining at all in all operated limbs (not shown). The limb bud from 1 of the 13 embryos treated with 5G8 demonstrated a partial malformation in the most anterior portion of the limb, where growth appeared to be arrested. This was likely, however, to be an artifact of the grafting procedure.
Figure 3.
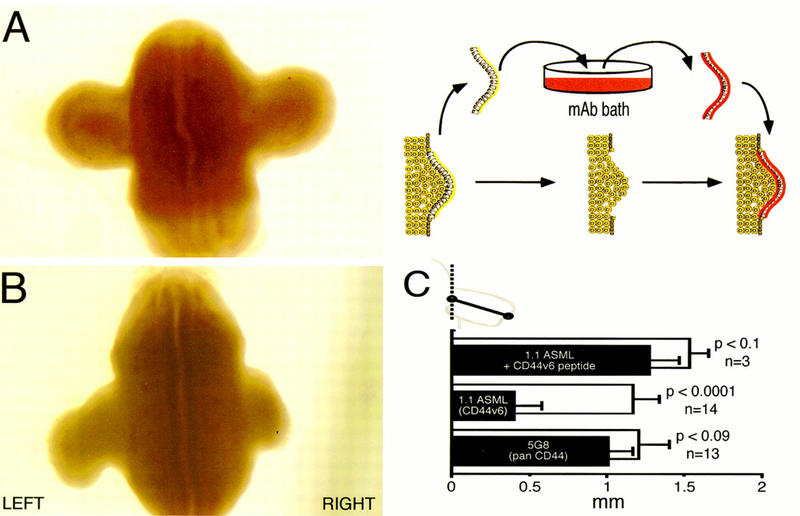
A CD44v6 epitope-specific antibody blocks limb bud outgrowth. AER tissue was dissected from the right limb bud, incubated with various antibodies, then grafted back onto the wounded limb. Left limbs remained unoperated. (A) AER treated with 5G8 (anti-CD44 antibody recognizing an epitope encoded by exon 15 = the exon 3′ of v10); (B) AER treated with 1.1ASML (CD44 exon v6 specific); (C) statistical analysis (paired t test) of grafting experiments demonstrating differences between treated (solid bars) and unoperated (open bars) limbs. Data are presented as means ± standard errors. Measurements were taken from the center of limb bud where they meet the flank, to the most distal point of the bud, as indicated in the cartoon.
To determine whether the 1.1ASML antibody has nonspecific effects on mesenchymal cell growth or whether it can influence AER function without being directly in contact with the basal surface of AER, embryos were cultured as described above except that the AERs were left intact. Instead, the mesenchyme subjacent to the AER (corresponding to the progress zone) on the right limb bud was exposed using a tungsten needle. These embryos were then cultured in medium lacking antibodies or in the presence of either 1.1ASML or 5G8. The mesoderm remained exposed to antibodies for several hours, after which time the overlying ectoderm healed. After 4 days in culture, no significant differences in limb bud development or length were observed between embryos grown in medium alone (n = 3; left, 1.03 ± 0.15 mm; right, 1.07 ± 0.06 mm) or in medium supplemented with 1.1ASML (n = 5; left, 1.06 ± 0.39; right, 0.92 ± 0.34) or 5G8 (n = 5; left, 1.12 ± 0.19; right, 1.1 ± 0.1). These data indicate that the 1.1ASML antibody can only inhibit limb outgrowth when directly in contact with the basal surface of the AER.
AER-induced limb bud mesenchymal cell proliferation is inhibited by a CD44v6-specific antibody in a coculture assay
To study the function of CD44 in the AER and the means by which the 1.1ASML antibody blocks limb bud outgrowth, we tested the effects of 1.1ASML on the proliferation-promoting ability of AER tissue when placed into coculture with dissociated primary limb bud mesenchymal cells. The AER and mesenchyme of E12.5 rat embryos were dissected from each other and the mesenchymal cells were dissociated and grown as a monolayer. After reaching ∼70% confluency, fragments of freshly isolated AER tissue were added either directly to the mesenchymal monolayers or after pretreatment with the CD44-specific antibodies 5G8, 1.1ASML, or 1.1ASML preblocked with its recognition peptide. After 16 hr of coculture, the cells were pulse-labeled for 4 hr with [3H]thymidine to measure their proliferative state. AER tissue in the absence of mesenchymal cells (Fig. 4, column 2) or upon removal from cocultures (not shown) did not incorporate [3H]thymidine. In the presence of AER tissue, mesenchymal cells incorporated significantly greater amounts of [3H]thymidine than mesenchymal cells cultured alone (Fig. 4, cf. column 3 with column 1). The stimulatory effects of the AER could be inhibited by the 1.1ASML antibody, but not by the 5G8 antibody or 1.1ASML that had been preincubated with a CD44v6 peptide (Fig. 4, columns 4,5,6). These results confirm that the 1.1ASML antibody can inhibit the growth-promoting function of the AER and suggest that CD44 proteins recognized by the antibody are required for mesenchymal cell proliferation.
Figure 4.
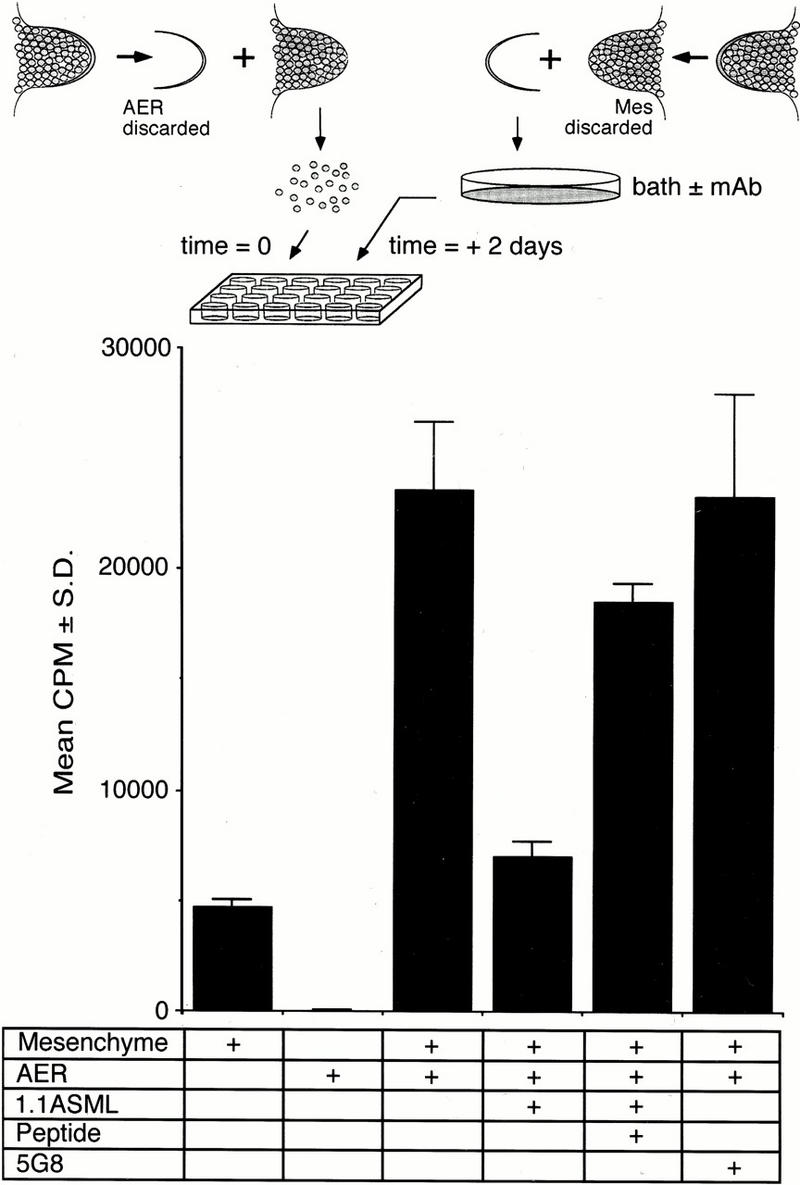
AER-mediated proliferation of primary mesenchymal cells in culture. Limb bud mesenchymal cells were put into culture as described in Materials and Methods. AER fragments were independently collected, treated either in PBS or in PBS containing 5G8, 1.1ASML, or 1.1ASML preblocked by its recognition peptide, washed, and placed on top of the mesenchymal cells. Within 2–3 hr the AER made adhesive contact to the mesenchymal cell layer. After 16 hr, thymidine incorporation was determined as a measure of mesenchymal cell proliferation. Plotted cpm represent the mean of triplicate samples ± s.d.
CD44 colocalizes with FGF-8 in the developing limb
There is no evidence to suggest that CD44 itself can function as a cell-bound growth factor. As described above, because FGF-8 is expressed throughout the AER and is likely to be one of the major signals maintaining limb bud mesenchymal cell proliferation, we reasoned that there could be a relationship between the functions of CD44 and FGF-8 in the developing limb. Therefore, we determined whether CD44 is coexpressed with FGF-8 in the developing rat embryo by in situ hybridization and immunohistochemistry. Using multiple CD44 variant exon-specific riboprobes and a probe for FGF-8, we found that CD44 and FGF-8 are coexpressed at E12.5 in the AER, branchial arches, and forebrain (data not shown). The CD44v6 epitope was also detected in prelimb ectoderm of E10.5 embryos (see Fig. 1D), at stages of development similar to when FGF-8 was reported to be expressed in mouse and chicken embryos (Heikinheimo et al. 1994; Crossley and Martin 1995; Mahmood et al. 1995; Crossley et al. 1996). To determine whether CD44 variants and FGF-8 proteins were colocalized in the same cellular compartments, we examined CD44v6 epitope and FGF-8 expression in frozen sections of limb buds from E12.5 embryos with double-labeling immunohistochemistry using 1.1ASML and a polyclonal FGF-8 antibody. As shown in Figure 5, the CD44v6 epitope and FGF-8 colocalize to the surfaces of AER cells. Therefore, CD44 variant proteins and FGF-8 are colocalized temporally and spatially in the AER and are expressed concomitantly at the onset of limb development.
Figure 5.
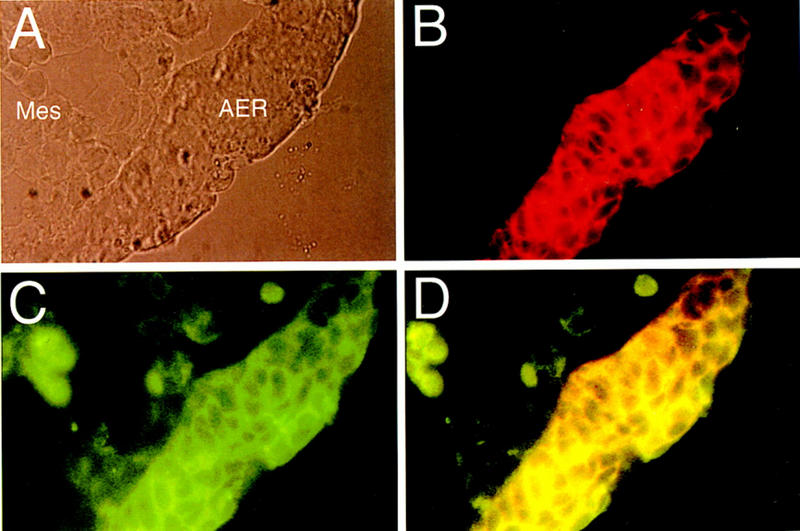
FGF-8 and v6 epitope containing CD44 variants colocalize in the AER. Immunohistochemical analysis of CD44 carrying the v6 epitope (B) and of FGF-8 (C) in the limb bud of an E11.5 rat embryo. A phase-contrast micrograph of the same section is shown in A. Double exposure (D) demonstrates CD44 and FGF-8 colocalization.
FGF-8 and FGF-4 bind to CD44v3–v10
As described in the introductory section, particular CD44 variants can bind to or be modified by glycosaminoglycans. For example, CD44 species carrying the v6 and v7 exon sequences have increased affinity for heparan sulfate and chondroitin sulfate (Sleeman et al. 1997). CD44 variants carrying the sequences encoded by exon v3 are modified covalently by heparan sulfate and can bind heparin-binding growth factors (Bennett et al. 1995a). Because the AER expresses CD44 proteins containing sequences encoded by exon v3, and as members of the FGF family bind to HSPGs, we explored whether CD44 proteins can bind FGF4 and FGF8. It is difficult to analyze directly CD44–FGF interactions in limb tissues because the AER itself produces several FGFs and CD44 variant proteins. Therefore, we used a cell culture system where we could easily manipulate CD44 expression. Namalwa Burkitt lymphoma cells, which do not express any CD44 (Sy et al. 1991) and do not produce detectable heparan sulfate (Fig. 6), had been stably transfected previously with either human CD44v3–v10 , CD44v8–v10, or CD44s. Human and rat CD44 amino acid sequences are highly homologous and many putative glycosylation and glycosaminoglycan addition sites are identical (Hofmann et al. 1991). Using an antibody that recognizes heparan sulfate, we found that Namalwa transfectants expressing CD44s (not shown) or CD44v8–v10 (Fig. 6A), did not carry detectable levels of heparan sulfate on their surfaces as measured by FACS analysis. Cells expressing CD44v3–v10, however, carried significant amounts of heparan sulfate (Fig. 6A). These findings show that Namalwa cells are capable of adding heparan sulfate to CD44, and that the appropriate CD44 proteins are the only major carriers of heparan sulfate on these cells.
Figure 6.
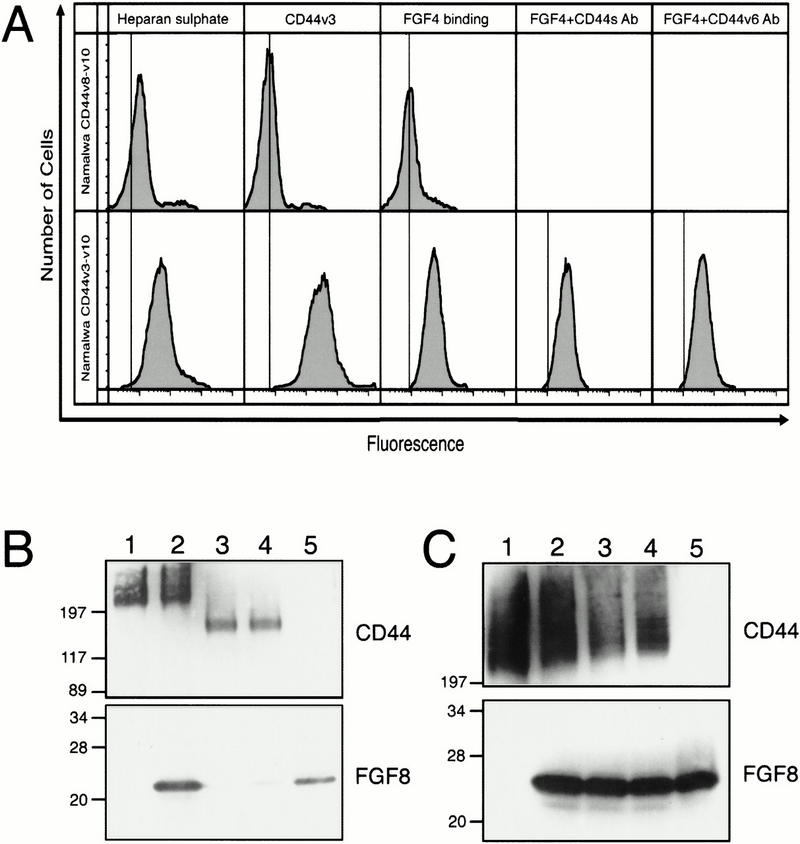
FGF binding to Namalwa cells expressing different CD44 splice variants. (A) Binding of FGF-4 to Namalwa cells expressing CD44v3–v10 or CD44v8–v10 measured by FACS analysis. The procedures of FACS analysis are described in Materials and Methods. The vertical line represents control maxima without primary antibody (in case of heparin and v3) or without FGF, but including both primary and secondary antibodies. The analyses indicate that most of the heparin sulfate is linked to the presence of CD44 carrying v3 sequences. FGF binding also depends on the presence of these CD44 proteins. The antibodies IM7 (recognizes an amino-terminal epitope on all CD44 proteins = CD44s antibodies) and VFF18 (CD44v6 epitope specific = CD44v6 antibodies) did not interfere with the binding. (B) Binding of FGF8 to Namalwa cells expressing CD44v3–v10 or CD44v8–v10 determined by Western blot analysis. Namalwa cells were incubated with FGF-8, washed extensively and lysed in sample buffer containing 2-mercaptoethanol. The lysate was resolved by SDS-PAGE and Western blotted with antibodies to FGF-8 and CD44. (Lane 1) Namalwa cells expressing CD44v3-10; (lane 2) Namalwa cells expressing CD44v3–v10 incubated with FGF-8; (lane 3) Namalwa cells expressing CD44v8–v10; (lane 4) Namalwa cells expressing CD44v8–v10 incubated with FGF-8; (lane 5) 50 ng FGF-8. (C) Western blot analysis showing that antibodies do not interfere with FGF-8 binding. Namalwa cells expressing CD44v3–v10 were incubated with 50 μg/ml IM7 or VFF18, washed and then tested for FGF-8 binding as described in B. (Lane 1) Namalwa cells alone; (lane 2) Namalwa cells plus FGF-8; (lane 3) Namalwa cells plus IM7 plus FGF-8; (lane 4) Namalwa cells plus VFF18 plus FGF-8; (lane 5) 50 ng FGF-8. Sizes of marker proteins are in kD.
To test the ability of each of these cell lines to bind FGFs, we used two different methods. Cells were preincubated with either recombinant FGF-8 or FGF-4 and then analyzed for FGF binding either by FACS analysis or by the resolution of total cell lysates in SDS-PAGE followed by Western blot analysis. We used the FGF-8b splice variant, which had been shown previously to be active in initiating and promoting limb outgrowth (Crossley et al. 1996; Vogel et al. 1996). Namalwa cells expressing human CD44v3–v10 bound both FGF-4 and FGF-8 (Fig. 6A,B, lane 2), whereas cells expressing CD44v8–v10 (Fig. 6A,B, lane 4) or CD44s (not shown) did not bind. The binding of FGFs could be reduced considerably when cells were pretreated with heparatinase (not shown). Therefore, we conclude that both FGF-4 and FGF-8, presumably like FGF-2 and other heparin-binding FGFs, can bind to CD44 proteins containing v3 sequences in the limb, most likely through heparan sulfate modifications to the sequence encoded by exon v3.
FGF-8 and FGF-4 can be presented to limb mesenchymal cells by CD44v3–v10
After establishing that CD44v3–v10 can bind the FGF proteins expressed in the AER and demonstrating that CD44 is crucial for limb bud outgrowth, we reasoned that CD44 proteins could be involved in presenting FGFs to mesenchymal cells adjacent to the AER. To address this possibility, we used the coculture assay described above using the Namalwa cells expressing different CD44 variants in place of AER tissue (see scheme in Fig. 7). We irradiated the Namalwa cells to ensure that they would not secrete cytokines that could influence limb bud mesenchymal cell proliferation. Each Namalwa cell line was added to cultures of primary limb mesenchymal cells in the presence and absence of limiting concentrations of either FGF-8 (Fig. 7) or FGF-4 (not shown), cocultured for 16 hr and then removed. The mesenchymal cells were assayed for [3H]thymidine incorporation. We found that in the presence of either FGF, limb mesenchymal cells that had been cocultured with CD44v3–v10-bearing Namalwa cells were stimulated to proliferate fourfold more than mesenchymal cells that had been cultured with the Namalwa cell line carrying CD44v8–v10 (Fig. 7, cf. column 4 with column 6). Neither FGF protein at the concentration used stimulated mesenchymal cells above background level (Fig. 7, cf. columns 1 and 2). The addition of either Namalwa transfectant alone also had no effect (Fig. 7, columns 3,5). We infer from these data that heparan sulfate-modified CD44 variants can act as functional low-affinity receptors for the FGF proteins expressed in the AER, and that cells expressing such variants can present FGF proteins to neighboring cells.
Figure 7.
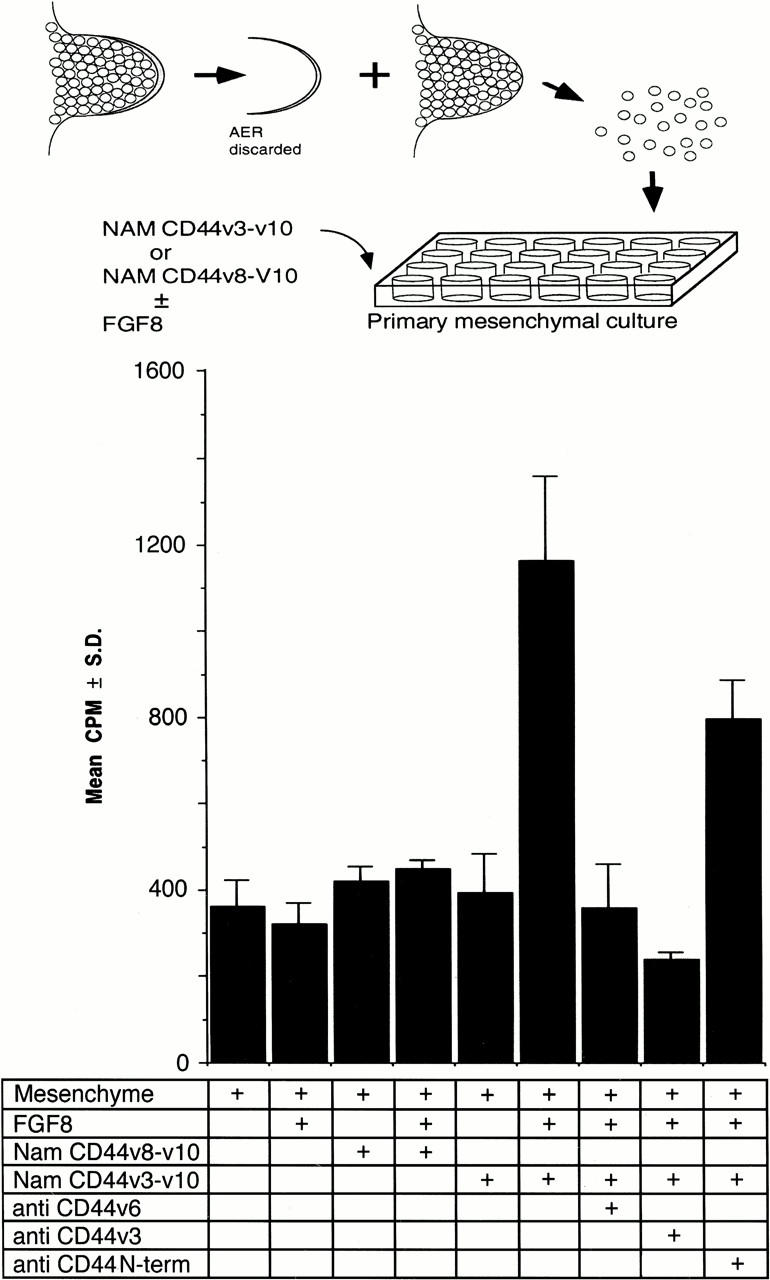
CD44v3–v10 can present FGF-8 to limb mesenchymal cells. Limb bud mesenchymal cells were cocultured with UV-irradiated Namalwa cells expressing either CD44v3–v10 or CD44v8–v10 in the presence or absence of FGF-8. The Namalwa cells were then removed and mesenchymal cell proliferation was monitored by [3H]thymidine incorporation. Note that FGF-8 only exerts a significant proliferative effect when added together with Namalwa cells transfected with CD44v3–v10. Antibodies used: VFF18 (anti CD44v6), rabbit polyclonal “antihoming receptor” (anti CD44v3) and IM7 (anti CD44 N-term). Plotted cpm are the mean values of triplicate samples ± standard deviation. The absolute incorporation was highly dependent on cell density and data can only be compared within one series and relative to mesenchymal cells grown alone.
CD44v6- and CD44v3-specific antibodies block FGF-8 and FGF-4 presentation, but not binding, by CD44
If the CD44 proteins in the AER bind and present FGF proteins to adjacent mesenchymal cells, then our finding that an anti-CD44v6 antibody can block limb outgrowth could be the result of the antibody preventing FGF proteins from binding to CD44. We tested this possibility by preincubating Namalwa cells expressing CD44v3–v10 with antibodies directed against either a human CD44v6 epitope (VFF18) or the amino-terminal domain of CD44 (IM7), and then incubating with either FGF-4 or FGF-8. We then analyzed FGF binding using FACS or Western blot analysis. The VFF18 antibody recognizes an epitope homologous to that recognized by the rat-specific 1.1ASML antibody used above (Dall et al. 1995). Both FGF-4 (Fig. 6A) and FGF-8 (Fig. 6C, lanes 3,4) still bound to CD44v3–v10 in the presence of either antibody. This indicates that interference with limb outgrowth was not attributable to prevention of FGF binding to CD44.
Another explanation for how a CD44v6-specific antibody influences the limb bud is that it prevents FGF proteins that are already bound to CD44 from stimulating mesenchymal cell proliferation. We tested this possibility by determining whether the VFF18 antibody could inhibit limb mesenchymal cell proliferation induced by FGF-4 or FGF-8 bound to CD44. The antibody inhibited the presentation of FGFs by Namalwa cells carrying CD44v3–v10; we found that [3H]thymidine incorporation was significantly reduced if Namalwa cells expressing CD44v3–v10 had been pretreated with VFF18 (Fig. 7, cf. columns 6 and 7). Interestingly, an antibody to exon v3, but not antibody IM7, also inhibited presentation (Fig. 7, columns 8,9). This finding indicates that the antibody used to block limb outgrowth functions by interfering with growth factor presentation in the rat limb bud, and suggests that growth factor presentation is an essential function of CD44 during limb morphogenesis.
Discussion
Our findings indicate that a particular CD44 splice variant plays a crucial role in limb development by helping to convey a proliferative signal from the AER to the mesenchyme. This splice variant is expressed in the AER and colocalizes with members of the FGF family, which are known to stimulate limb bud outgrowth. This CD44 variant can present FGF proteins to limb mesenchymal cells stimulating their proliferation. The function of CD44 is apparently crucial for limb development, as an antibody against the variant portion of CD44 can inhibit limb bud outgrowth.
The AER expresses several CD44 splice variants
We have demonstrated using immunohistochemistry, in situ hybridization, Western blot analysis, and RT–PCR that particular CD44 splice variants are expressed in the AER, whereas the remainder of the limb expresses only CD44s. This is in agreement with previous RT–PCR findings with RNA from mouse limb bud (Yu et al. 1996). At least two of these CD44 variants, CD44v1–v10 and CD44v3–v10, encode proteins that carry sequences for modification by heparan sulfate. Because the heparan sulfate-specific antibody does not work in Western blots, it was not possible to analyze directly heparan sulfate modifications on CD44 proteins expressed in the AER. One CD44 protein with a molecular mass of ∼230 kD predominates according to Western blot analysis (Fig. 1E). From its size and the AER RT–PCR data (Fig. 2A), it is likely that this protein is a glycosylated form of CD44v3–v10.
CD44 can mediate epithelial–mesenchymal interactions in the developing limb
Interactions between epithelia and mesenchyme are crucial throughout the course of embryogenesis (for review, see Birchmeier and Birchmeier 1993). The developing vertebrate limb is an example of such interactions, in which the AER and the limb mesenchyme are highly dependent on each other to establish limb outgrowth, differentiation, and patterning. When the AER is replaced by heparin beads carrying one of the FGFs produced by the AER, limb bud outgrowth progresses normally (Niswander et al. 1993; Niswander and Martin 1993; Fallon et al. 1994; Cohn et al. 1995; Mahmood et al. 1995; Crossley et al. 1996; Vogel et al. 1996). Soluble FGFs, even at high concentrations, cannot substitute, at least in vitro, for the AER. A low-affinity receptor carrying heparan sulfate expressed in the AER therefore, is likely to be necessary for presentation of these AER-derived growth-promoting factors. Our finding that a particular CD44 variant-specific antibody applied to the AER can severely inhibit limb outgrowth, indicates that CD44 is crucial for the communication of proliferative signals from the AER to the underlying limb mesenchymal cells. Because antibodies against CD44v6 and CD44v3 epitopes can block the presentation of FGF proteins to limb bud mesenchymal cells in vitro, and as FGFs are likely to be the chief AER-derived proliferative signals in the limb, we propose that CD44 is required for the presentation of FGFs to limb bud mesenchymal cells and for the maintenance of mesenchymal cell proliferation.
CD44 proteins (Wheatley et al. 1993; Nakamura et al. 1995; Yu and Toole 1997; C. Tölg, P. Gruss, and H. Ponta, unpubl.) are coexpressed with FGF-8 (L. Sherman, D. Wainwright, M. Ponta, and P. Herrlich, unpubl.) by a number of other epithelia that generate instructive signals to their associated mesenchymal cells, including the branchial arches and odontogenic placodes. It is possible, therefore, that CD44 variants are part of a general mechanism of epithelial–mesenchymal communication, whereby growth factors expressed by an epithelial cell are carried by CD44, then presented to an opposing mesenchymal cell.
CD44 is part of a novel growth factor presentation mechanism
Previous studies have supported the notion that FGFs must bind to HSPGs on the surfaces of target cells before activating their high-affinity receptors (for review, see Vlodavsky et al. 1996). We have shown here that CD44 variant proteins can act as HSPGs on the surface of cells, and have provided the first evidence that CD44 is involved in a novel growth factor presentation mechanism. The novelty of this mechanism lies in the fact that the growth factor secreted by one cell (here FGF-8 from an AER cell) is carried by an HSPG on this same cell (heparan sulfate-modified CD44) and presented to an opposing target cell (here limb bud mesenchymal cell; see Fig. 8) to stimulate its proliferation. It may suffice that FGF-8 is presented to high-affinity receptors on only the immediately adjacent mesenchymal cells. Because FGF-8 can maintain expression of FGF-10 in the apical mesenchyme of early limb buds (Ohuchi et al. 1997), it is possible that these cells respond by expressing FGF-10, which might then keep other neighboring mesenchymal cells in a proliferating state. Alternatively, the AER-derived signal may be passed on through gap junctions in the mesenchyme. Interestingly, FGF-4 increases mesenchymal gap junctional communication (Makarenkova et al. 1997).
Figure 8.

A model for FGF-8 presentation by CD44 in the developing limb. It is proposed that FGF-8 binds to the heparan sulfate side chains of a CD44 variant protein containing exon v3 sequences and that both FGF-8 and CD44 are inserted into the basal lamina as it is synthesized by AER cells. The FGF-8 is then presented to high-affinity FGF receptors (FGFR) on the surface of mesenchymal cells, possibly as the mesenchymal cells degrade the basal lamina. The binding of FGF-8 to its high-affinity receptor subsequently leads to mesenchymal cell proliferation.
The AER and limb mesenchyme are separated from one another by a basal lamina that is deposited by the AER, and which, according to dye exclusion assays, prevents passage of molecules as large as FGF proteins (Kelley and Fallon 1976; Wilcox and Kelley 1993). The basal lamina is likely to be a dynamic structure that is constantly renewed by the AER and proteolytically degraded by the mesenchymal cells. We propose that large CD44 splice variants and their bulky heparan sulfate side chains carrying FGF-8 are inserted into the basal lamina (Fig. 8). As the mesenchymal cells break down the basal lamina, heparan sulfate-modified CD44 proteins would presumably protect FGF-8 from degradation and keep it in a state suitable for presentation to either mesenchymal cell surface HSPGs or directly to high-affinity FGF receptors. It is possible that additional HSPGs in the limb bud could have similar functions. The only other HSPG known to be expressed in the AER is glypican (Niu et al. 1996). Glypican and related HSPGs are believed to serve in presenting FGF to high-affinity receptors on the same cell and therefore, may be involved in signaling within the AER (for review, see David 1993; Elenius and Jalkanen 1994; Niu et al. 1996; Steinfeld et al. 1996). Alternatively, CD44–FGF complexes could be shed into the basal lamina by the action of proteases. In this case, CD44 would act as a free matrix-associated HSPG. Although shedding of CD44 has been observed in many other systems (Campanero et al. 1991; Bazil and Horejsi 1992; Camp et al. 1993; Günthert et al. 1996), there is no evidence that free CD44 proteins are present in the basal lamina of the developing limb.
An important consideration is whether mice lacking CD44 would be deficient in limb outgrowth. There have been several approaches to abrogate CD44 expression, one of which has been the disruption of the CD44 locus. CD44 null mice are viable and do not demonstrate any obvious developmental deficiencies (see Results and F. Hilberg, unpubl.; Schmits et al. 1997). It is possible, however, that embryos can compensate for the lack of CD44 with other molecules. Interestingly, antisense CD44 expressed under the control of the keratin-5 promoter in transgenic mice reduces CD44 expression in mouse skin and causes a severe skin phenotype, including a proliferation defect and insensitivity to exogenously added growth factors (Kaya et al. 1997). This defect is not observed in the CD44 null mice. The keratin-5 promoter becomes active at E11.5 in the mouse embryo (Ramirez et al. 1994), a stage well after differentiation of the AER and early limb outgrowth. Both the antisense experiment and our antibody-blocking experiments abolish CD44 function acutely and after passage into unidirectional differentiation. Thus, in this situation, compensation for the lack of CD44 may no longer be possible in contrast to the choices that can be made earlier in development. Preliminary observations suggest that compensation in CD44 −/− limbs may be attributable to an increase by one order of magnitude in mesenchymal cell sensitivity to FGF-8.
CD44 may have additional functions in the developing limb
In agreement with previous observations in mouse limb buds (Yu et al. 1996), we found that CD44s is expressed by rat limb mesenchymal cells throughout the course of early limb development. One possible role for this CD44 protein may have to do with either adhesion to or recognition of hyaluronate. In the limb bud mesenchyme there is a proximal to distal gradient of hyaluronate (Kosher et al. 1981; Singley and Solursh 1981; Knudson and Toole 1985). The highest concentration of hyaluronate is among the cells opposing the ectoderm, except for the cells immediately beneath the AER where hyaluronate is not detectable (Yu et al. 1996). Therefore, CD44s could mediate hyaluronate adhesion within the mesenchyme or possibly hyaluronate degradation within the progress zone. Although binding to hyaluronate is up-regulated by the presence of variant exons (Sleeman et al. 1997), this binding does not appear to be relevant for the epithelial–mesenchymal cell interaction as the limb-blocking CD44v6-specific antibody does not interfere with this adhesion.
Our observation that both the CD44v6 epitope and FGF-8 are expressed throughout the AER and not just at the basal surface adjacent to the limb bud mesenchyme suggests that both proteins could function to maintain the AER itself.
Growth factor presentation may be a general function of CD44 splice variants
In addition to a role in presenting signals from epithelial cells to mesenchymal cells, the presentation of growth factors by CD44 may be of wider significance. Besides FGF-2, FGF-4, and FGF-8, heparan sulfate-modified CD44 proteins can bind a number of other heparin-binding growth factors (Tanaka et al. 1993; Bennett et al. 1995a). Furthermore, the presence of particular variant exon sequences in CD44 proteins can alter their affinity for various glycosaminoglycans (Bennett et al. 1995b; Sleeman et al. 1997). This altered affinity could permit CD44 to associate with a variety of cytokines and other substances that alter cellular proliferation and differentiation. Therefore, it is possible that the ability of some CD44 proteins to bind and sequester growth-promoting substances can explain the functions of CD44s in a number of normal and pathological systems.
Perhaps the most compelling putative role for CD44-mediated growth factor presentation is in the promotion of tumor cell growth and metastasis. Numerous experimental and clinical studies indicate that some CD44 variants play crucial roles in tumor progression (for review, see Günthert 1996; Sherman et al. 1996; Naor et al. 1997). Therefore, it is tantalizing to speculate that tumor cells use CD44 variants to sequester and present growth- or metastasis-promoting agents either to themselves, or, like the cells of the AER, to cells in their local microenvironment. In support of such a common mechanism of CD44 action, the CD44v6 antibody we used to block limb bud outgrowth also interferes with immune functions (Arch et al. 1992) and with tumor growth and metastasis (Reber et al. 1990; Günthert et al. 1991; Rudy et al. 1993; Seiter et al. 1993; Sleeman et al. 1996). The regulated presentation of growth factors is likely to be vital in each of these situations.
Materials and methods
Materials
The antibodies 5G8 (Sleeman et al. 1996), IM7 (Trowbridge et al. 1982), and 1.1ASML (Günthert et al. 1991) were affinity purified on protein G–Sepharose. VFF-11 and VFF-18 were obtained from Bender, Vienna, Austria. Hermes3 antibody was the kind gift of Dr. S. Jalkanen (University of Turku, Finland). The CD44v3-specific antibody was obtained from Novus Molecular, San Diego, CA (polyclonal rabbit “anti-homing receptor”). An anti-heparan sulfate antibody was obtained from Seikagaku Corporation, Tokyo, Japan. FGF-4 and FGF-8 antibodies were obtained from R&D systems. A polyclonal FGF-8 antibody was kindly donated by Dr. Clive Dickson (ICRF, London, UK). The peptide recognized by 1.1ASML (KWFENEWQGKNPPT) was kindly provided by Bender, Vienna. Biotinylated and HRP-conjugated secondary antibodies were from DAKO, streptavidin-conjugated phycoerythrin from PharMingen.
Cell culture
Namalwa Burkitt lymphoma cells and clones expressing CD44 were a gift of Dr. Steven Pals (Amsterdam, Netherlands) and were cultured as described (van der Voort et al. 1995). Primary cultures of limb bud mesenchymal cells were established from the forelimb buds of E12.5–E13.5 rat embryos. Timed pregnant BD-X rats (4–6 months of age, bred in the Institut für Genetik Animal Facility) were killed by CO2 inhalation. AER and ectoderm were removed from limb buds with tungsten needles. Mesenchymal cells from the distal third of limb were disaggregated by gentle trituration, washed, and resuspended in DMEM/10% FCS. Cells were plated in 24-well plates at a density of ∼1 × 105 cells/well and maintained in a humidified 5% CO2 atmosphere at 37°C.
Immunohistochemistry
For whole-mount immunohistochemistry, BD-X rat embryos were harvested between days E10.5 and E14.5 and processed essentially as described by Wheatley et al. (1993). As a blocking buffer we used PBS + 0.1% Triton X-100 + 10% FCS. Primary antibodies were used at 5 μg/ml. Immunostaining was detected by overnight incubations in 1 μg/ml of biotin-conjugated goat–anti-mouse IgG (DAKO) followed by streptavidin–peroxidase (DAKO). After the final washes, tissues were developed with 3,3-amino-9-ethyl carbazole (Sigma) in 50 mm sodium acetate (pH 4.7) for appropriate periods of time.
For double-labeling fluorescence immunohistochemistry, frozen coronal sections (10 μm) of forelimb buds from E12.5 embryos were fixed in ice cold methanol for 5 min, washed in PBS, and incubated with normal goat serum (10% in PBS) at room temperature for 30 min. Sections were then incubated in 1 μg/ml of 1.1ASML plus 1 μg/ml of a rabbit polyclonal anti-FGF-8 antibody in PBS for 1 hr. After washing three times for 5 min each, sections were incubated with a mixture of 1 μg/ml TRITC- and FITC-conjugated secondary antibodies for 30 min. Sections were then washed, mounted, and photographed. Background controls included exclusion of both primary antibodies and exclusion of one or the other primary antibody to ensure the specificity of each secondary antibody.
CD44 exon-specific RT–PCR analysis
Twenty whole limbs were isolated from E12.5 rats in ice cold PBS. The AERs were dissected from the rest of the limb using tungsten needles and both tissues were snap-frozen. Poly(A)+ RNA was then prepared using a Micro-FastTrack kit (InVitrogen). cDNAs were synthesized using reverse transcriptase and a CD44 primer from the 3′ nonvariant portion of rat CD44 (exon 19, positions 2433–2459; primer C2A in König et al. 1996; 5′-GGCACTACACCCCAATCTTC-3′), then amplified using PCR (25 cycles of 0.5 min at 95°C, 1.5 min at 54°C, and 1.0 min at 73°C, followed by incubation at 73°C for another 10 min) using a primer from the 5′ nonvariant portion of rat CD44 (exon 5, positions 621–640; C13 in König et al. 1996; 5′-AAGACATCGATGCCTCAAAC-3′) and another 3′ primer (exon 19, positions 2168–2189; 5′-CCAAGATGATAAGCCACTCTG-3′). From this PCR reaction, 2 μl was reamplified in the presence of individual variant exon-specific 5′ primers (as listed below) and 3′ primer 2168–2189. Conditions were the same as above except that the final incubation at 73°C was extended to 10 min. PCR products were analyzed by electrophoresis in 2.5% agarose and visualized by ethidium bromide staining.
Variant exon 5′ primers are as follows: v1, positions 785–804 (5′-GCCTCAACTGTGTACTCAAA-3′); v2, positions 916–935 (5′-GATGACTACCCCTGAAACAC-3′); v3, positions 1031–1050 (5′-ACGGAGTCAAATACCAACCC-3′); v4, positions 1156–1175 (5′-TGCAACTACTCCATGGGTTT-3′); v5, positions 1274–1293 (5′-TATAGACAGAAACAGCACCA-3′); v6, positions 1388–1407 (5′-TGGGCAGATCCTAATAGCAC-3′); v7, positions 1512–1531 (5′-CTGCCTCAGCCCACAACAAC-3′); v8, positions 1650–1663 (5′-CCAGTCATAGTACAACCCTT-3′); v9, positions 1757–1774 (5′-CAGAACTTCTCTACATTACC-3′); v10, positions 1841–1860 (5′-GGTCGAAGAAGAGGTGGAAG-3′). All nucleotide positions correspond to the sequence of rat CD44 cDNA (GenBank data bank, accession no. U96138).
Western blotting and immunoprecipitation
Proteins separated on SDS–polyacrylamide gels were transferred to Immobilon P (Millipore) membranes and detected using standard Western blotting procedures and chemiluminescence (ECL; Amersham).
For immunoprecipitations, 100 whole E12.5 rat limbs were lysed by trituration in 2 ml of RIPA buffer containing 2 mm PMSF. The lysate was cleared by centrifugation at 14,000 rpm and the supernatant pretreated with 50 μl of protein A + G agarose beads (Calbiochem) at 4°C for 30 min. The beads were removed and 5 μg/ml 5G8 antibody and 80 μl of protein A+G beads were added to the supernatant. After incubation at 4°C for 60 min, the beads were washed five times with fresh RIPA buffer and boiled in sample buffer containing 10% 2-mercaptoethanol for 10 min. The immunoprecipitates were resolved on a 7.5% SDS-PAGE gel. CD44 proteins were detected with the 5G8 antibody using Western blotting.
Limb bud mesenchymal cell coculture proliferation assays
Mesenchymal cell cultures were prepared in 24-well plates as described above. These cultures were grown to ∼70% confluency and then starved by serum withdrawal for 24 hr before the assay. The influence of AER fragments or of Namalwa cell clones (the latter in combination with FGFs) on proliferation was tested.
AERs were dissected from E12.5 rat embryos and immediately incubated for 45 min in PBS alone or PBS containing 50 μg/ml of 5G8, 1.1ASML, or 1.1ASML preincubated in a fivefold excess concentration of its recognition peptide. The AERs were washed twice and added to limb mesenchymal cell cultures. AER fragments from two limb buds were placed into each well. The AERs and mesenchymal cells were cocultured for 20 hr, after which the medium was replaced by medium containing 1 μCi/ml of [3H]thymidine (Amersham). After 4 hr, the cells were washed three times with PBS, solubilized in 10% SDS, and the incorporated radioactivity determined by scintillation counting. All samples were assayed in triplicate.
Namalwa cells (or clonal transfected lines expressing CD44 variants) were harvested, washed, resuspended in PBS, and then irradiated with 2000 J/m2 of UVC to prevent transcription, synthesis, and secretion of proteins that could potentially interfere with the assay. Approximately 1 × 105 Namalwa cells were added directly to limb mesenchymal cell cultures without or together with 50 ng/ml FGF-8 or FGF-4. In dose–response curves with similar sets of primary mesenchymal cell cultures and Namalwa CD44v3–v10 transfectants, 50 ng/ml proved to cause half-maximal stimulation. With nontransfected Namalwa or with mesenchymal cells alone we saw no stimulation by the addition of up to 200 ng/ml FGF. Where indicated in Figure 7, Namalwa cells were blocked with 50 μg/ml of the specified antibody in PBS for 30 min, washed twice and then added to the mesenchymal culture together with FGF. After 16 hr cultures were washed three times to remove Namalwa cells (which grow in suspension). The mesenchymal cells were pulse-labeled with 1 μCi/ml of [3H]thymidine for a further 4 hr. Incorporated radioactivity was determined by scintillation counting. All coculture assays were performed in triplicate.
FACS analyses and FGF-binding assays
For detection of cell surface expression of the CD44 splice variant carrying the v3 epitope and of heparan sulfate, cells were harvested, washed twice with ice-cold PBS, and resuspended in 3% FCS in PBS. After 15 min 1 μg/ml of primary antibody was added and the cells incubated for an additional 30 min. Cells were then washed twice and incubated with a biotinylated secondary antibody for 30 min. Cells were again washed twice and incubated with streptavidin-conjugated phycoerythrin for 30 min. Finally, the cells were washed three times and fluorescence detected using a FACS-Star Plus flow cytometer (Becton Dickinson).
For the detection of FGF-4 binding, Namalwa cells and clonal lines thereof expressing specific CD44 variants were washed twice in PBS and incubated on ice in 1% BSA, 0.01% NaN3 containing 0.5 μg/ml of human recombinant FGF-4 for 60 min. Bound FGF was detected with an FGF-4 antibody and then processed as above. To measure possible interference with FGF binding, cells were first treated with either 50 μg/ml of VFF18 or IM7 in 3% FCS in PBS on ice for 30 min or with 1 unit of heparinase II (Sigma) in PBS at 25°C for 3 hr, washed twice and then assayed as above for FGF binding.
For the detection of FGF8 binding, 5 × 106 Namalwa cells expressing CD44v3–v10 or CD44v8–v10 were harvested by centrifugation, washed three times in PBS, and then resuspended in 500 μl of ice cold PBS containing 3% BSA, 0.01% NaN3, and 0.2 μg/ml of recombinant mouse FGF-8b. The cells were then incubated on ice for 60 min, washed five times with 1 ml of PBS, and lysed in 50 μl of sample buffer containing 10% 2-mercaptoethanol. The samples were boiled for 10 min and centrifuged at 14,000 rpm for 30 sec to pellet insoluble material. The supernatant was loaded onto 12% (for FGF-8) or 6% (for CD44) SDS–polyacrylamide gels. FGF-8 was detected with a FGF-8b antibody and CD44 with Hermes3 (recognizes all CD44 proteins) by Western blot analysis. In antibody interference experiments, the cells were first incubated with 50 μg/ml IM7 or VFF18 for 30 min, washed three times with PBS, and analyzed for FGF-8 binding as above.
Embryo culture and experimental manipulation
Stage E11 rat embryos were dissected asceptically from the uterine decidua. The head, heart, and lower trunk were removed. Trunk sections were placed with dorsal side facing upward into 35-mm dishes containing 1 ml of ice cold PBS plus 1% methylcellulose. The AER was removed from the right limb of each embryo using a tungsten needle to gently peel the tissue away from the remainder of the limb. Any remaining fragments of the AER were dissected and discarded. Intact AER tissue was incubated at 4°C in 100 μl of PBS or PBS supplemented with 10 μg of either 1.1ASML, 1.1ASML that had been preincubated with its recognition peptide, or 5G8 for 30 min. AERs were then rinsed in PBS + methylcellulose and grafted back onto the operated limb by pushing the treated AER tissue into the wound where the mesenchyme was exposed. After 2 hr, medium was added to the embryo cultures, which were subsequently maintained in a humidified 5% CO2 atmosphere at 37°C. The medium used was a modification of that described by Ochiya et al. (1995) using 1% methylcellulose and no amphotericin B. After 4 days, limb buds were measured under a stereoscopic microscope.
Acknowledgments
We thank Martin Blum, William Scott, and Jonathan Sleeman for valuable discussions and their critical comments on this manuscript. We also thank Clive Dickson for the FGF-8 polyclonal antisera, Steven Pals for Namalwa cells and transfectants, and Norma Howells for assistance with animal breeding. This work was supported by grant He551/8-2 from the Deutsche Forschungsgemeinschaft, by Bender Wien (Boehringer-Ingelheim), and by Fonds der Chemischen Industrie.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL genetik@igen.fzk.de; FAX +49−7247−823354.
References
- Arch R, Wirth K, Hofmann M, Ponta H, Matzku S, Herrlich P, Zöller M. Participation in normal immune responses of a splice variant of CD44 that encodes a metastasis-inducing domain. Science. 1992;257:682–685. doi: 10.1126/science.1496383. [DOI] [PubMed] [Google Scholar]
- Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- Bazil V, Horejsi V. Shedding of the CD44 adhesion molecule from leukocytes induced by anti-CD44 monoclonal antibody stimulating the effect of a natural receptor ligand. J Immunol. 1992;149:747–753. [PubMed] [Google Scholar]
- Bennett KL, Jackson DG, Simon JC, Tanczos E, Peach R, Modrell B, Stamenkovic I, Plowman G, Aruffo A. CD44 isoforms containing exon v3 are responsible for the presentation of heparin-binding growth factor. J Cell Biol. 1995a;128:687–698. doi: 10.1083/jcb.128.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KL, Modrell B, Greenfield B, Bartolazzi A, Stamenkovic I, Peach R, Jackson DG, Spring F, Aruffo A. Regulation of CD44 binding to hyaluronan by glycosylation of variably spliced exons. J Cell Biol. 1995b;131:1623–1633. doi: 10.1083/jcb.131.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W. Molecular aspects of mesenchymal–epithelial interactions. Annu Rev Cell Biol. 1993;9:511–540. doi: 10.1146/annurev.cb.09.110193.002455. [DOI] [PubMed] [Google Scholar]
- Brown TA, Bouchard T, St. John T, Wayner E, Carter WG. Human keratinocytes express a new CD44 core protein (CD44E) as a heparin-sulfate intrinsic membrane proteoglycan with additional exons. J Cell Biol. 1991;113:207–221. doi: 10.1083/jcb.113.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp RL, Scheynius A, Johansson C, Pure E. CD44 is necessary for optimal contact allergic responses but is not required for normal leukocyte extravasation. J Exp Med. 1993;178:497–507. doi: 10.1084/jem.178.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanero MR, Pulido R, Alonso JL, Pivel JP, Pimentel-Muinos FX, Fresno M, Sánchez-Madrid F. Down-regulation by tumor necrosis factor-alpha of neutrophil cell surface expression of the sialophorin CD43 and the hyaluronate receptor CD44 through a proteolytic mechanism. Eur J Immunol. 1991;21:3045–3048. doi: 10.1002/eji.1830211222. [DOI] [PubMed] [Google Scholar]
- Cardon-Cardo C, Vlodavsky I, Haimovitz-Friedman A, Hicklin D, Fuks Z. Expression of basic fibroblast growth factor in normal human tissues. Lab Invest. 1990;63:832–840. [PubMed] [Google Scholar]
- Cohn MJ, Tickle C. Limbs: A model for pattern formation within the vertebrate body plan. Trends Genet. 1996;12:253–257. doi: 10.1016/0168-9525(96)10030-5. [DOI] [PubMed] [Google Scholar]
- Cohn MJ, Izpisúa-Belmonte JC, Abud H, Heath JK, Tickle C. Fibroblast growth factors induce additional limb development from the flank of chick embryos. Cell. 1995;80:739–746. doi: 10.1016/0092-8674(95)90352-6. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf-8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Minowada G, MacArthur CA, Martin GR. Roles for FGF-8 in the induction, initiation, and maintenance of chick limb development. Cell. 1996;84:127–136. doi: 10.1016/s0092-8674(00)80999-x. [DOI] [PubMed] [Google Scholar]
- Culty M, Miyake K, Kincade PW, Silorski E, Butcher EC, Underhill C. The hyaluronate receptor is a member of the CD44 (H-CAM) family of cell surface glycoproteins. J Cell Biol. 1990;111:2765–2774. doi: 10.1083/jcb.111.6.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall P, Heider K-H, Sinn H-P, Skroch-Angel P, Adolf G, Kaufmann M, Herrlich P, Ponta H. Comparison of immunohistochemistry and RT-PCR for detection of CD44v-expression, a new prognostic factor in human breast cancer. Int J Cancer. 1995;60:471–477. doi: 10.1002/ijc.2910600408. [DOI] [PubMed] [Google Scholar]
- David G, Bai XM, Van der Schueren B, Marynen P, Cassiman JJ, Van den Berghe H. Spatial and temporal changes in the expression of fibroglycan (syndecan-2) during mouse embryonic development. Development. 1993;119:841–854. doi: 10.1242/dev.119.3.841. [DOI] [PubMed] [Google Scholar]
- DeGrendele HC, Estess P, Picker LJ, Spiegelman MH. CD44 and its ligand hyaluronate mediate rolling under physiologic flow: A novel lymphocyte-endothelial cell primary adhesion pathway. J Exp Med. 1996;183:1119–1130. doi: 10.1084/jem.183.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dono R, Zeller R. Cell type-specific nuclear translocation of fibroblast growth factor-2 isoforms during chicken kidney and limb morphogenesis. Dev Biol. 1994;163:316–330. doi: 10.1006/dbio.1994.1151. [DOI] [PubMed] [Google Scholar]
- Elenius K, Jalkanen M. Function of the syndecans—a family of cell surface proteoglycans. J Cell Sci. 1994;107:2975–2982. doi: 10.1242/jcs.107.11.2975. [DOI] [PubMed] [Google Scholar]
- Faassen AE, Schrager JA, Klein DJ, Oegema TR, Couchman JR, McCarthy JB. A cell surface chondroitin sulfate proteoglycan, immunologically related to CD44, is involved in type I collagen-mediated melanoma cell motility and invasion. J Cell Biol. 1992;116:521–531. doi: 10.1083/jcb.116.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon J, López A, Ros M, Savage M, Olwin B, Simandl B. FGF-2, apical ectodermal ridge growth signal for chick limb development. Science. 1994;264:104–107. doi: 10.1126/science.7908145. [DOI] [PubMed] [Google Scholar]
- Folkman J, Klagsbrun M, Sasse J, Wadzinski M, Ingber D, Vlodavsky I. A heparin-binding angiogenic protein—basic fibroblast growth factor—is stored within basement membrane. Am J Pathol. 1980;130:393–400. [PMC free article] [PubMed] [Google Scholar]
- Geduspan JS, Solursh M. A growth promoting influence from the mesonephros during limb outgrowth. Dev Biol. 1992;151:242–250. doi: 10.1016/0012-1606(92)90230-e. [DOI] [PubMed] [Google Scholar]
- Gonzalez A-M, Buscaglia M, Ong M, Baird A. Distribution of basic fibroblast growth factor in the 18-day rat fetus: Localization in the basement membranes of diverse tissues. J Cell Biol. 1990;110:753–765. doi: 10.1083/jcb.110.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospodarowicz D, Cheng J. Heparin protects basic and acidic FGF from inactivation. J Cell Physiol. 1986;128:475–484. doi: 10.1002/jcp.1041280317. [DOI] [PubMed] [Google Scholar]
- Günthert AR, Sträger J, von Reyher U, Henne C, Joos S, Koretz K, Moldenhauer G, Kramner PH, Möller P. Early detachment of colon carcinoma cells during CD95 (APO-1/Fas)-mediated apoptosis. I. De-adhesion from hyaluronate by shedding of CD44. J Cell Biol. 1996;134:1089–1096. doi: 10.1083/jcb.134.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U. CD44 in malignant disorders. Curr Top Microbiol Immunol. 1996;213:271–285. [PubMed] [Google Scholar]
- Günthert U, Hofmann M, Rudy W, Reber S, Zöller M, Haussmann I, Matzku S, Wenzel A, Ponta H, Herrlich P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65:13–24. doi: 10.1016/0092-8674(91)90403-l. [DOI] [PubMed] [Google Scholar]
- Harrison RG. Experiments on the development of the forelimb of Amblystoma, a self-differentiating equipotential system. J Exp Zool. 1918;25:413–461. [Google Scholar]
- Heath WF, Cantrell AS, Mayne NG, Jaskunas RS. Mutations in the heparin-binding domains of human basic fibroblast growth factor alter its biological activity. Biochemistry. 1991;30:5608–5615. doi: 10.1021/bi00236a039. [DOI] [PubMed] [Google Scholar]
- Heikinheimo M, Lawshé A, Shackleford GM, Wilson DB, MacArthur CA. Fgf-8 expression in the post-gastrulation mouse suggests roles in the development of the face, limbs and central nervous system. Mech Dev. 1994;48:129–138. doi: 10.1016/0925-4773(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Hofmann M, Rudy W, Zöller M, Tölg C, Ponta H, Herrlich P, Günthert U. CD44 splice variants confer metastatic behavior in rats: Homologous sequences are expressed in human tumor cell lines. Cancer Res. 1991;51:5292–5297. [PubMed] [Google Scholar]
- Ishai-Michaeli R, Svaln C-M, Chajek-Shaul T, Korner G, Ekre H-P, Vlodavsky I. Importance of size and sulfation of heparin in release of basic fibroblast growth factor from the vascular endothelium and extracellular matrix. Biochemistry. 1992;31:2080–2088. doi: 10.1021/bi00122a027. [DOI] [PubMed] [Google Scholar]
- Jackson DG, Bell JI, Dickinson R, Timans J, Shields J, Whittle N. Proteoglycan forms on the lymphocyte homing receptor CD44 are alternatively spliced variants containing the v3 exon. J Cell Biol. 1995;128:673–685. doi: 10.1083/jcb.128.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalkanen S, Jalkanen M. Lymphocyte CD44 binds the COOH-terminal heparin-binding domain of fibronectin. J Cell Biol. 1992;116:817–825. doi: 10.1083/jcb.116.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalkanen S, Bargatze RF, de los Toyos J, Butcher EC. Lymphocyte recognition of high endothelium: Antibodies to distinct epitopes of an 85–95 kD glycoprotein antigen differentially inhibit lymphocyte binding to lymph node, mucosal, or synovial endothelial cells. J Cell Biol. 1987;105:983–990. doi: 10.1083/jcb.105.2.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya G, Rodriguez I, Jorcano JL, Vassalli P, Stamenkovic I. Selective suppression of CD44 in keratinocytes of mice bearing an antisense CD44 transgene driven by a tissue-specific promoter disrupts hyaluronate metabolism in the skin and impairs keratinocyte proliferation. Genes & Dev. 1997;11:996–1007. doi: 10.1101/gad.11.8.996. [DOI] [PubMed] [Google Scholar]
- Kelley RO, Fallon JF. Ultrastructural analysis of the apical ectodermal ridge during vertebrate limb morphogenesis. I. The human forelimb with special reference to gap junctions. Dev Biol. 1976;51:241–256. doi: 10.1016/0012-1606(76)90141-x. [DOI] [PubMed] [Google Scholar]
- Knudson CB, Toole BP. Changes in the pericellular matrix during differentiation of limb bud mesoderm. Dev Biol. 1985;112:308–318. doi: 10.1016/0012-1606(85)90401-4. [DOI] [PubMed] [Google Scholar]
- Kosher RA, Savage MP, Walker KH. A gradation of hyaluronate accumulation along the proximodistal axis of the embryonic chick limb bud. J Embryol Exp Morphol. 1981;63:85–98. [PubMed] [Google Scholar]
- König H, Moll J, Ponta H, Herrlich P. Trans-acting factors regulate the expression of CD44 splice variants. EMBO J. 1996;15:4030–4039. [PMC free article] [PubMed] [Google Scholar]
- Lamb RF, Hennigan RF, Turnbull K, Katsanakis KD, MacKenzhie ED, Birnie GD, Ozanne BW. AP-1-mediated invasion requires increased expression of the hyaluronan receptor CD44. Mol Cell Biol. 1997;17:963–976. doi: 10.1128/mcb.17.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley J, Schulte R, Hyman R. Binding of hyaluronic acid to lymphoid cell lines is inhibited by monoclonal antibodies against Pgp-1. Exptl Cell Res. 1990;187:224–233. doi: 10.1016/0014-4827(90)90085-o. [DOI] [PubMed] [Google Scholar]
- Mahmood R, Bresnick J, Hornbruch A, Mahony C, Morton N, Colquhoun K, Martin P, Lumsden A, Dickson C, Mason I. A role for FGF-8 in the initiation and maintenance of vertebrate limb bud outgrowth. Curr Biol. 1995;5:797–806. doi: 10.1016/s0960-9822(95)00157-6. [DOI] [PubMed] [Google Scholar]
- Makarenkova H, Becker DL, Tickel C, Warner AE. Fibroblast growth factor 4 directs gap junction expression in the mesenchyme of the vertebrate limb bud. J Cell Biol. 1997;138:1125–1137. doi: 10.1083/jcb.138.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason IJ. The ins and outs of fibroblast growth factors. Cell. 1994;78:547–552. doi: 10.1016/0092-8674(94)90520-7. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Kimura S, Kenmotsu S, Sakai H, Saku T, Ozawa H. Immunolocalization of CD44 and heparan sulfate chains on the stratum intermedium and papillary layer in the rat enamel organ. Arch Histol Cytol (Japan) 1995;58:323–334. doi: 10.1679/aohc.58.323. [DOI] [PubMed] [Google Scholar]
- Naor D, Sionov RV, Ish-Shalom D. CD44: Structure, function and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- Niswander L, Martin GR. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- ————— FGF-4 and BMP-2 have opposite effects on limb outgrowth. Nature. 1993;361:68–76. doi: 10.1038/361068a0. [DOI] [PubMed] [Google Scholar]
- Niswander L, Tickle C, Vogel A, Booth I, Martin GR. FGF-4 replaces the apical ectodermal ridge and directs outgrowth and patterning of the limb. Cell. 1993;75:579–587. doi: 10.1016/0092-8674(93)90391-3. [DOI] [PubMed] [Google Scholar]
- Niu S, Antin PB, Akimoto K, Morkin E. Expression of avian glypican is developmentally regulated. Dev Dynamics. 1996;207:25–34. doi: 10.1002/(SICI)1097-0177(199609)207:1<25::AID-AJA3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Ochiya T, Sakamoto H, Tsukamoto M, Sugimura T, Terada M. Hst-1 (FGF-4) antisense oligonucleotides block murine limb development. J Cell Biol. 1995;130:997–1003. doi: 10.1083/jcb.130.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H, Yoshioka H, Tanaka A, Kawakami Y, Nohno T, Noji S. Involvement of androgen-induced growth factor (FGF-8) gene in mouse embryogenesis and morphogenesis. Biochem Biophys Res Commun. 1994;204:882–888. doi: 10.1006/bbrc.1994.2542. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Nakagawa T, Yamamoto A, Araga A, Ohata T, Ishimaru Y, Yoshioka H, Kuwana T, Nohno T, Yamasaki M, Itoh N, Noji S. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF-8, an apical ectodermal factor. Development. 1997;124:2235–2244. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Yayon A, Flanagan G, Svahn CM, Levi E, Leder P. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol Cell Biol. 1992;12:240–247. doi: 10.1128/mcb.12.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A, Bravo A, Jorcano J, Vidal M. Sequences 5′ of the bovine keratin 5 gene direct tissue and cell type-specific expression of a lac Z gene in the adult and during development. Differentiation. 1994;58:53–64. doi: 10.1046/j.1432-0436.1994.5810053.x. [DOI] [PubMed] [Google Scholar]
- Rapraeger AC, Krufka A, Olwin BB. Requirement of heparin sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- Reber S, Matzku S, Günthert U, Ponta H, Herrlich P, Zöller M. Retardation of metastatic tumor growth after immunization with metastasis-specific monoclonal antibodies. Int J Cancer. 1990;46:919–927. doi: 10.1002/ijc.2910460528. [DOI] [PubMed] [Google Scholar]
- Rubin L, Saunders JW., Jr Ectodermal-mesodermal interactions in the growth of limb buds in the chick embryo: Constancy and temporal limits of the ectodermal induction. Dev Biol. 1972;28:94–112. doi: 10.1016/0012-1606(72)90129-7. [DOI] [PubMed] [Google Scholar]
- Rudy W, Hofmann M, Schwartz-Albiez R, Zöller M, Heider K-H, Ponta H, Herrlich P. The two major CD44 proteins expressed on a metastatic rat tumor cell line are derived from different splice variants: Each one individually suffices to confer metastatic behavior. Cancer Res. 1993;53:1262–1268. [PubMed] [Google Scholar]
- St. John T, Meyer J, Idzerda R, Gallatin WM. Expression of CD44 confers a new adhesive phenotype on transfected cells. Cell. 1990;60:45–52. doi: 10.1016/0092-8674(90)90714-p. [DOI] [PubMed] [Google Scholar]
- Saksela O, Moscatelli D, Sommer A, Rifkin DB. Endothelial cell-derived heparin sulfate binds basic fibroblast growth factor and protects it from proteolytic degradation. J Cell Biol. 1988;107:743–751. doi: 10.1083/jcb.107.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage MP, Hart CE, Riley B, Sasse J, Olwin BB, Fallon JR. Distribution of FGF-2 suggests it has a role in chick limb bud growth. Dev Dyn. 1993;198:159–170. doi: 10.1002/aja.1001980302. [DOI] [PubMed] [Google Scholar]
- Schmits R, Filmus J, Gerwin N, Senaldi G, Kiefer F, Kundig T, Wakeham A, Shahinian A, Catzavelos C, Rak J, Furlonger C, Zakarian A, Simard JJ, Ohashi PS, Paige CJ, Gutierrez-Ramos JC, Mak TW. CD44 regulates hematopoietic progenitor distribution, granuloma formation, and tumorigenicity. Blood. 1997;90:2217–2233. [PubMed] [Google Scholar]
- Screaton GR, Bell MV, Jackson DG, Cornelis FB, Gerth U, Bell JI. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci. 1992;89:12160–12164. doi: 10.1073/pnas.89.24.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiter S, Arch R, Komitowski D, Hofmann M, Ponta H, Herrlich P, Matzku S, Zöller M. Prevention of tumor metastasis formation by anti-variant CD44. J Exp Med. 1993;177:443–455. doi: 10.1084/jem.177.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman L, Sleeman J, Dall P, Hekele A, Moll J, Ponta H, Herrlich P. The CD44 proteins in embryonic development and in cancer. Curr Topics Microbiol Immunol. 1996;213/I:249–269. doi: 10.1007/978-3-642-61107-0_15. [DOI] [PubMed] [Google Scholar]
- Singley CT, Solursh M. The spatial distribution of hyaluronic acid and mesenchymal condensation in the embryonic chick wing. Dev Biol. 1981;84:102–120. doi: 10.1016/0012-1606(81)90375-4. [DOI] [PubMed] [Google Scholar]
- Sleeman JP, Arming S, Moll JF, Hekele A, Rudy W, Sherman LS, Kreil G, Ponta H, Herrlich P. Hyaluronate-independent metastatic behavior of CD44 variant-expressing pancreatic carcinoma cells. Cancer Res. 1996;56:3134–3141. [PubMed] [Google Scholar]
- Sleeman J, Kondo K, Moll J, Ponta H, Herrlich P. Variant exons v6 and v7 together expand the repertoire of glycosaminoglycans bound by CD44. J Biol Chem. 1997;272:31837–31844. doi: 10.1074/jbc.272.50.31837. [DOI] [PubMed] [Google Scholar]
- Spivak-Kroizman T, Lemmon MA, Dikic I, Ladbury SE, Pinchasi D, Hwang J, Jaye M, Grumley G, Schlessinger J, Lax I. Heparin-induced oligomerisation of FGF molecules is responsible for FGF receptor dimerization, activation and cell proliferation. Cell. 1994;71:1015–1024. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Steinfeld R, Van Den Berghe H, David G. Stimulation of fibroblast growth factor receptor-1 occupancy and signaling by cell surface-associated syndecans and glypican. J Cell Biol. 1996;133:405–416. doi: 10.1083/jcb.133.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens TD, McNulty TR. Evidence for a metameric pattern in the development of the chick humerus. J Embryol Exp Morphol. 1981;61:191–205. [PubMed] [Google Scholar]
- Strecker TR, Stephens TD. Peripheral nerves do not play a trophic role in limb skeletal morphogenesis. Teratology. 1983;27:159–167. doi: 10.1002/tera.1420270204. [DOI] [PubMed] [Google Scholar]
- Summerbell D, Lewis JH, Wopert L. Positional information in chick limb morphogenesis. Nature. 1973;224:492–496. doi: 10.1038/244492a0. [DOI] [PubMed] [Google Scholar]
- Suzuki HR, Sakamoto H, Yoshida T, Sugimora T, Terada M, Solursh M. Localization of Hst-1 transcripts to the apical ectodermal ridge in the mouse embryo. Dev Biol. 1992;150:219–222. doi: 10.1016/0012-1606(92)90020-h. [DOI] [PubMed] [Google Scholar]
- Sy MS, Guo Y-J, Stamenkovic I. Distinct effects of two CD44 isoforms on tumor growth in vivo. J Exp Med. 1991;174:859–866. doi: 10.1084/jem.174.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Adams DH, Hubscher S, Hirano H, Siebenlist U, Shaw S. T-cell adhesion induced by proteoglycan-immobilized cytokine MIP-1β. Nature. 1993;361:79–82. doi: 10.1038/361079a0. [DOI] [PubMed] [Google Scholar]
- Thomas L, Byers HR, Vink J, Stamenkovic I. CD44H regulates tumor cell migration on hyaluronate-coated substrate. J Cell Biol. 1992;118:971–977. doi: 10.1083/jcb.118.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tölg C, Hofmann M, Herrlich P, Ponta H. Splicing choice from ten variant exons establishes CD44 variability. Nucleic Acids Res. 1993;21:1225–1229. doi: 10.1093/nar/21.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge IS, Lesley J, Schulte R, Hyman R, Trotter J. Biochemical characterization and cellular distribution of a polymorphic, murine cell-surface glycoprotein expressed in lymphoid tissues. Immunogenetics. 1982;15:299–312. doi: 10.1007/BF00364338. [DOI] [PubMed] [Google Scholar]
- van der Voort R, Manten-Horst E, Smit L, Ostermann E, van den Berg F, Pals ST. Binding of cell-surface expressed CD44 to hyaluronate is dependent on splicing and cell type. Biochem Biophys Res Comm. 1995;214:137–144. doi: 10.1006/bbrc.1995.2267. [DOI] [PubMed] [Google Scholar]
- van Weering DHJ, Baas PD, Bos JL. A PCR-based method for the analysis of human CD44 splice products. PCR Methods Appl. 1993;3:100–106. doi: 10.1101/gr.3.2.100. [DOI] [PubMed] [Google Scholar]
- Venkataraman G, Sasisekharan V, Herr AB, Ornitz DM, Waksman G, Cooney CL, Langer R, Sasisekharan R. Preferential self-association of basic fibroblast growth factor is stabilized by heparin during receptor dimerization and activation. Proc Natl Acad Sci. 1996;93:845–850. doi: 10.1073/pnas.93.2.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlodavsky I, Folkman J, Sullivan R, Fridman R, Ishai-Michaelli R, Sasse J, Klagsbrun M. Endothelial cell-derived basic fibroblast growth factor: Synthesis and deposition into subendothelial extracellular matrix. Proc Natl Acad Sci. 1987;84:2292–2296. doi: 10.1073/pnas.84.8.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlodavsky I, Miao H-Q, Medalion B, Danagher P, Ron D. Involvement of heparin sulfate and related molecules in sequestration and growth promoting activity of fibroblast growth factor. Cancer Metastasis Rev. 1996;15:177–186. doi: 10.1007/BF00437470. [DOI] [PubMed] [Google Scholar]
- Vogel A, Rodriguez C, Izpisúa-Belmonte J-C. Involvement of FGF-8 in initiation, outgrowth and patterning of the vertrebrate limb. Development. 1996;122:1737–1750. doi: 10.1242/dev.122.6.1737. [DOI] [PubMed] [Google Scholar]
- Wainwright D, Sherman L, Sleeman J, Ponta H, Herrlich P. A splice variant of CD44 expressed in the rat apical ectodermal ridge contributes to limb outgrowth. Ann NY Acad Sci. 1996;785:345–349. doi: 10.1111/j.1749-6632.1996.tb56305.x. [DOI] [PubMed] [Google Scholar]
- Wanek N, Muneoka K, Holler-Dinsmore G, Burton R, Bryant SV. A staging system for mouse limb development. J Exp Zoo. 1989;249:41–49. doi: 10.1002/jez.1402490109. [DOI] [PubMed] [Google Scholar]
- Wayner EA, Carter WG. Identification of multiple cell adhesion receptors for collagen and fibronectin in human fibrosarcoma cells possessing unique alpha and common beta subunits. J Cell Biol. 1987;105:1873–1884. doi: 10.1083/jcb.105.4.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JM, Renkl AC, Termeer C, Dittmar H, Taxis S, Howells N, Sleeman J, Hofmann M, Schöpf E, Ponta H, Herrlich P, Simon JC. An essential role for CD44 variant isoforms in epidermal Langerhans cells and blood dendritic cell function. J Cell Biol. 1997;137:1137–1147. doi: 10.1083/jcb.137.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley SC, Isacke CM, Crossley PH. Restricted expression of the hyaluronan receptor, CD44, during postimplantation mouse embryogenesis suggests key roles in tissue formation and patterning. Development. 1993;119:295–306. doi: 10.1242/dev.119.2.295. [DOI] [PubMed] [Google Scholar]
- Wilcox MJ, Kelley RO. Cell of the apical ectodermal ridge in quail limb buds are coupled to one another but are isolated from subjacent mesoderm. Prog Clin Biol Res. 1993;383A:339–349. [PubMed] [Google Scholar]
- Yamaguchi TP, Rossant J. Fibroblast growth factors in mammalian development. Curr Opin Gen Dev. 1995;5:485–491. doi: 10.1016/0959-437x(95)90053-j. [DOI] [PubMed] [Google Scholar]
- Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- Yu Q, Toole BP. Common pattern of CD44 isoforms is expressed in morphogenetically active epithelia. Dev Dynamics. 1997;208:1–10. doi: 10.1002/(SICI)1097-0177(199701)208:1<1::AID-AJA1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Yu Q, Grammatikakis N, Toole BP. Expression of multiple CD44 isoforms in the apical ectodermal ridge of the embryonic mouse limb. Dev Dynamics. 1996;207:204–214. doi: 10.1002/(SICI)1097-0177(199610)207:2<204::AID-AJA8>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Zwilling E. Ectoderm–mesoderm relationship in the development of the chick embryo limb bud. J Exp Zool. 1955;128:423–442. [Google Scholar]


