Abstract
The pre-mRNA splicing factor U2AF (U2 snRNP auxiliary factor) has an essential role in 3′ splice site selection. U2AF binds the intron pyrimidine tract between the branchpoint and the 3′ splice site and recruits U2 snRNP to the branch site at an early step in spliceosome assembly. Human U2AF is a heterodimer composed of large (hU2AF65) and small (hU2AF35) subunits. Both subunits contain a domain enriched in arginine–serine dipeptide repeats termed an RS domain. The two U2AF RS domains have been assigned essential and independent roles in spliceosome assembly in vitro—the hU2AF65 RS domain is required to target U2 snRNP to the branch site and the hU2AF35 RS domain is necessary for protein–protein interactions with constitutive and alternative splicing factors. We have investigated the functional requirements for the RS domains on the Drosophila U2AF homolog in vivo. In sharp contrast to its essential role in U2 snRNP recruitment in vitro, the RS domain on the Drosophila large subunit homolog (dU2AF50) was completely dispensable in vivo. Prompted by this unexpected result, we analyzed the RS domain on the Drosophila small subunit homolog (dU2AF38). Despite its requirement for enhancer-dependent splicing activity in vitro, the dU2AF38 RS domain was also inessential in vivo. Finally, we have tested whether the Drosophila U2AF heterodimer requires any RS domain. Flies mutant for both the small and large subunits could not be rescued by dU2AF50ΔRS and dU2AF38ΔRS transgenes. Therefore, in contrast to the separate roles assigned to the U2AF RS domains in vitro, our genetic data suggest that they may have redundant functions in vivo.
Keywords: Fruitfly, pre-mRNA splicing, RNA-binding proteins, RS domains, SR proteins
The generation of functional mRNAs in eukaryotes requires the accurate removal of noncoding sequences (introns) from pre-mRNAs by a process termed pre-mRNA splicing (Moore et al. 1993; Sharp 1994; Kramer 1996). Pre-mRNA splicing takes place in the spliceosome, a dynamic RNA–protein complex composed of small nuclear ribonucleoprotein particles (snRNPs) and extrinsic (non-snRNP) protein factors. The earliest steps in spliceosome assembly involve recognition of the 5′ splice site by U1 snRNP and the branchpoint-3′ splice site by U2 snRNP. Targeting of U2 snRNP to the branch site requires the extrinsic splicing factor U2AF (U2 snRNP auxiliary factor) (Ruskin et al. 1988). U2AF binds specifically to the intron pyrimidine tract located between the branchpoint and the 3′ splice site and recruits U2 snRNP to the branch site at an early step in spliceosome assembly (Ruskin et al. 1988; Zamore et al. 1992; Staknis and Reed 1994). Regulation of 3′ splice site choice, both positive and negative, can be realized by influencing the pyrimidine tract binding of U2AF (Tian and Maniatis 1993; Valcárcel et al. 1993; Reed 1996).
Human U2AF is a heterodimer composed of a 65-kD large subunit (hU2AF65) and a 35-kD small subunit (hU2AF35) (Zamore and Green 1989). Both subunits are highly conserved across species (Zamore and Green 1991); U2AF homologs have been identified in Drosophila melanogaster (Kanaar et al. 1993; Rudner et al. 1996), Schizosaccharomyces pombe (Potashkin et al. 1993; Wentz-Hunter and Potashkin 1996), and Caenorhabditis elegans (Zorio et al. 1997; T. Blumenthal, pers. comm.). The Drosophila U2AF large (dU2AF50) and small (dU2AF38) subunit homologs are 50 and 38 kD, respectively (Kanaar et al. 1993; Rudner et al. 1996). The large subunit contains three RNA recognition motifs (RRMs) and an amino-terminal arginine–serine-rich (RS) domain (Zamore et al. 1992). The small subunit contains a highly degenerate (pseudo-) RRM (Birney et al. 1993), two putative Zn2+ binding motifs (Worthington et al. 1996), and a carboxy-terminal RS domain and glycine-rich region (Zhang et al. 1992).
Biochemical studies of U2AF using extracts depleted of U2AF activity lead to some confusion as to the requirement for the large and small subunits in splicing. Depending on the substrate used and method of U2AF depletion (poly(U)–sepharose or immunoaffinity chromatography), different requirements for the large or large and small subunits were observed (Zamore and Green 1991; Zamore et al. 1992; Kanaar et al. 1993; Valcárcel et al. 1996; Zuo and Maniatis 1996; Gama-Carvalho et al. 1997). Both Drosophila U2AF subunits are required for viability suggesting that both subunits are necessary for splicing in vivo (Kanaar et al. 1993; Rudner et al. 1996).
Although both U2AF subunits contain RS domains, these domains have been assigned independent roles in spliceosome assembly. Consistent with a direct role in U2 snRNP recruitment, deletion of the RS domain from hU2AF65 (hU2AF65ΔRS) had no effect on pyrimidine tract binding yet it completely abolished the ability to restore splicing to U2AF-depleted extracts (Zamore et al. 1992; Valcárcel et al. 1996). Additionally, fusion of a synthetic RS domain containing seven RS dipeptides [(RS)7] (or any dipeptide repeat that possesses a net positive charge [(RA)7, (RG)7, (KS)7, but not (RD)7] to hU2AF65ΔRS was sufficient to restore splicing activity (Valcárcel et al. 1996). Based on the sole requirement for a net positive charge, it was proposed that the essential role of the hU2AF65 RS domain is to facilitate annealing of the U2 snRNA and the branch site sequence through charge shielding of the RNA phosophodiester backbones (Valcárcel et al. 1996).
Whereas the large subunit RS domain is thought to promote RNA–RNA interactions in U2 snRNP recruitment, the small U2AF subunit RS domain has been implicated in protein–protein interactions with constitutive and alternative splicing factors that serve to stabilize binding of hU2AF65 to intron pyrimidine tracts. A role for the small subunit in bridging constitutive and alternative splicing factors and hU2AF65 was first suggested by protein–protein interaction studies (Wu and Maniatis 1993; Amrein et al. 1994). These studies revealed that hU2AF35, but not hU2AF65, specifically interacts with the SR family of general splicing factors as well as the Drosophila alternative splicing factors transformer (TRA) and transformer2 (TRA2).
The SR proteins are a family of conserved splicing factors with similar domain structure and partially overlapping biochemical activities (Fu 1995; Manley and Tacke 1996). SR proteins contain at least one RRM-type RNA-binding domain and a serine–arginine-rich (SR or RS) domain that has been implicated in protein–protein interactions in vitro (Wu and Maniatis 1993; Amrein et al. 1994; Kohtz et al. 1994; Xiao and Manley 1997). In vivo, the Drosophila SRp55/B52 gene and the mammalian ASF/SF2 gene, including its RS domain, are essential for viability (Ring and Lis 1994; Wang et al. 1996). SR proteins are required at an early stage in mammalian spliceosome assembly and can promote U1 snRNP and U2AF binding to pre-mRNA in the earliest known mammalian spliceosomal complex (E complex) (Staknis and Reed 1994). In fact, SR proteins can simultaneously interact with both the U1 snRNP 70-kD protein, U1–70K, and with hU2AF35 in the yeast two-hybrid assay (Wu and Maniatis 1993). The RS domain on both U1–70K and hU2AF35 have been implicated in these protein–protein interactions (Wu and Maniatis 1993; Kohtz et al. 1994).
SR proteins also bind exonic enhancer elements located downstream from weak 3′ splice sites (Lavigueur et al. 1993; Sun et al. 1993; Wang et al. 1995; Tacke et al. 1997). Addition of SR proteins to nuclear extract promotes U2AF binding to pre-mRNA substrates containing these enhancer elements. Consistent with a role for hU2AF35 in bridging SR proteins bound to enhancers and hU2AF65 bound to weak pyrimidine tracts, reconstitution of enhancer-dependent splicing in U2AF-depleted extracts requires the addition of recombinant hU2AF35 (Zuo and Maniatis 1996). Addition of hU2AF35 lacking its RS domain (hU2AF35ΔRS) is insufficient for enhancer-dependent splicing, further implicating the RS domain in these critical protein–protein interactions (Zuo and Maniatis 1996).
One of the best characterized examples of enhancer-dependent splicing involves the sex-specific, alternative splicing of doublesex (dsx) in the sex determination pathway in Drosophila. The alternative splicing factors TRA and TRA2 are required for the female-specific, alternative splicing of dsx (Baker and Wolfner 1988; Cline and Meyer 1996). TRA and TRA2 activate a weak, female-specific, 3′ splice site in the dsx pre-mRNA. The resulting mRNA encodes a DSX isoform required for somatic female differentiation. TRA2 has an RRM and both TRA and TRA2 have RS domains. The RS domains on TRA and TRA2 have been implicated in protein–protein interactions with SR proteins and hU2AF35 (Wu and Maniatis 1993; Amrein et al. 1994). Biochemical analysis of the alternative splicing of dsx has revealed that TRA and TRA2 bind to exonic enhancer elements downstream of the regulated dsx intron and recruit SR proteins to form a splicing enhancer complex (Hedley and Maniatis 1991; Tian and Maniatis 1993). This complex promotes U2AF binding to the weak pyrimidine tract of the female-specific 3′ splice site (Zuo and Maniatis 1996). Reconstitution of female-specific dsx splicing requires both U2AF subunits as well as TRA and TRA2.
We have undertaken a molecular genetic analysis of the Drosophila U2AF homolog in vivo. To define a functional RS domain on the Drosophila U2AF large subunit, dU2AF50, we analyzed deletions and substitution mutations of the dU2AF50 RS domain. Surprisingly, in sharp contrast to the requirement for the hU2AF65 RS domain in U2 snRNP recruitment in vitro, we found that the dU2AF50 RS domain was completely dispensable in vivo. This unexpected result prompted an analysis of the Drosophila small subunit RS domain. Like the dU2AF50 RS domain, the dU2AF38 RS domain was completely dispensable in vivo, indicating that neither RS domain is necessary for splicing. Significantly, diplo X flies lacking the dU2AF38 RS domain were 100% viable and phenotypically female. Therefore, in vivo, female-specific, enhancer-dependent splicing of dsx was unaffected by the absence of the dU2AF38 RS domain. To determine whether the Drosophila U2AF heterodimer requires any RS domain, complementation tests were performed with dU2AF50, dU2AF38 double mutant flies. Whereas the combination of two wild-type transgenes could rescue the double mutant flies, the combination of dU2AF50ΔRS and dU2AF38ΔRS transgenes could not. Fusion of a synthetic RS domain containing seven RS dipeptides onto dU2AF50ΔRS was not sufficient to complement the double mutant in combination with dU2AF38ΔRS. Therefore, at least one RS domain on U2AF is required in vivo and a simple RS dipeptide repeat will not serve as a substitute. In contrast to the separate roles assigned to the U2AF RS domains in vitro, our genetic data suggest that the RS domains have redundant functions in vivo.
Results
The dU2AF50 RS domain is dispensable in vivo
We have shown previously that a mutation in the Drosophila U2AF large subunit gene is fully penetrant recessive lethal and can be rescued by a genomic transgene that contains dU2AF50 (Kanaar et al. 1993). The presence of intervening sequences in and around the amino-terminal RS domain of dU2AF50 prohibited a deletion analysis of the dU2AF50 RS domain using the rescuing genomic transgene. To facilitate our analysis of the dU2AF50 RS domain, we created an in vivo dU2AF50 expression vector. The dU2AF50 gene in the genomic clone was replaced with an oligonucleotide linker containing unique restriction sites and an improved translation initiation sequence (Cavener and Ray 1991) (see Materials and Methods). A transgene containing the wild-type dU2AF50 cDNA inserted into this expression vector rescued a dU2AF50 recessive lethal allele as efficiently as the original genomic transgene (Fig. 1A).
Figure 1.
The dU2AF50 RS domain is inessential in vivo. (A) Wild-type (WT) and dU2AF50 deletion and substitution derivative transgenes were tested for complementation of a dU2AF50 recessive lethal allele. A schematic diagram of the dU2AF50 domains is shown. The RS domain (RS), the dU2AF38 interaction domain (int), and the three RNA recognition motifs (RRM) are indicated. The percentages of rescue are from representative transgene lines. (B) Amino acid sequence comparison of the RS domains from dU2AF50 and hU2AF65. Identities and similarities are shown in black and gray boxes, respectively. Dashes denote gaps. Amino acid positions are shown on the right. The caret below the sequence indicates the dU2AF50 RS domain deletion site.
To define the functional requirements for the dU2AF50 RS domain in vivo, the RS domain from the dU2AF50-coding sequence (amino acids 1–34; Fig. 1B) was deleted (dU2AF50ΔRS) or replaced with a synthetic RS domain containing seven RS dipeptides [dU2AF50(RS)7] and inserted into the dU2AF50 expression vector. Germ-line transformants containing dU2AF50ΔRS and dU2AF50(RS)7 transgenes were generated and tested for their ability to complement a recessive lethal dU2AF50 allele. Balanced dU2AF50 mutant virgin females were crossed to males carrying a dU2AF50 transgene. Hemizygous, dU2AF50 mutant male progeny carrying the dU2AF50 transgene were scored and their percent viability was determined by comparison with their heterozygous mutant sisters. Surprisingly, both dU2AF50ΔRS and dU2AF50(RS)7 transgenes efficiently rescued the recessive lethal dU2AF50 allele (Fig. 1A). The high degree of sequence similarity between the RS domains on dU2AF50 and hU2AF65 (Fig. 1B) suggests that the inessential nature of the large subunit RS domain observed in vivo will not be specific to Drosophila.
A single RS dipeptide was present in the dU2AF50ΔRS rescuing transgene (Fig. 2A). To rule out the possibility that this single RS dipeptide was sufficient for dU2AF50 activity in vivo, the serine residue was deleted to create dU2AF50ΔRStrue. Similar to the findings with dU2AF50ΔRS, the dU2AF50ΔRStrue transgene also efficiently rescued the dU2AF50 mutant allele (Fig. 2A). Therefore, dU2AF50 does not require any RS dipeptides to support viability.
Figure 2.
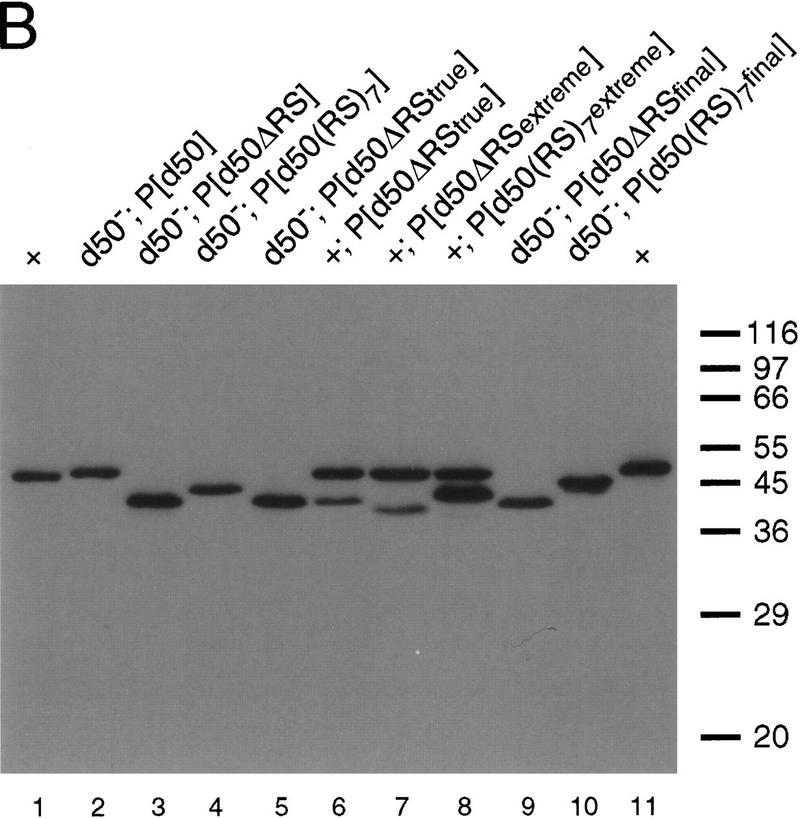
(A) Deletion analysis of the dU2AF50 RS domain. Wild-type dU2AF50 (WT) and deletion and substitution derivative transgenes were tested for complementation of a recessive lethal dU2AF50 allele. Amino-terminal amino acids spanning the RS domain and part of the dU2AF38 interaction domain are shown. The amino acids derived from the dU2AF50-coding sequence are shown in bold. The amino acids introduced through cloning or fused onto deletion derivatives are in roman type. Dots above the amino acid sequence indicate amino acids required for interaction with dU2AF38. The caret indicates the analogous position on dU2AF50 where the hU2AF65 RS domain was deleted for the in vitro reconstitution experiments (Valcárcel et al. 1996). The percentages of rescue shown are from representative transgene lines. (B) Protein expression levels of the dU2AF50 deletion derivatives. Immunoblot analysis of whole-fly extracts using an anti-dU2AF50 antibody. Whole-fly extracts are from w1118 (dU2AF50+) flies (lanes 1,11), dU2AF50 mutant flies carrying a wild-type (lane 2) or dU2AF50 derivative transgene as indicated above the gel (lanes 3,4,5,9,10) or w1118 flies carrying dU2AF50 derivative transgenes (lanes 6–8). Extracts from wild-type (w1118) or dU2AF50 mutant flies carrying the same dU2AF50ΔRStrue rescuing transgene are shown in lanes 4 and 5. The sizes of molecular mass markers are indicated in kD.
A positively charged domain is not required for dU2AF50 activity in vivo
In the in vitro reactivation experiments that demonstrated an essential requirement for the hU2AF65 RS domain, the amino terminus of the hU2AF65 protein was deleted up to a conserved proline repeat at amino acid 95 (amino acid 47 in dU2AF50, see Fig. 2A) (Valcárcel et al. 1993). In our dU2AF50ΔRStrue rescuing transgene the RS domain was deleted up to amino acid 37. This deletion left behind three positively charged residues (R37, R38, and K39; Fig. 2A). It was possible that the positively charged residues retained in dU2AF50ΔRStrue were sufficient for RS domain function. In fact, a hU2AF65 RS domain deletion that retained a few positively charged residues weakly reactivated splicing in a poly(U)-depleted extract in vitro (Valcárcel et al. 1993). In an attempt to directly correlate our in vivo complementation data with the published results from the in vitro reconstitution experiments, we deleted the dU2AF50 RS domain up to the conserved proline repeat (dU2AF50Δ1–46; see Fig. 2A) to create dU2AF50ΔRSextreme. We also fused a synthetic RS domain containing seven RS dipeptides onto this deletion creating dU2AF50(RS)7extreme. These dU2AF50 derivatives are analogous to the hU2AF65 RS deletion and synthetic RS domain fusion proteins used in the in vitro splicing reactivation experiments (Valcárcel et al. 1993). Independent transgenic lines (15–20) of each dU2AF50 derivative were generated and tested for complementation of the recessive lethal dU2AF50 allele. Consistent with the in vitro reactivation experiments, the dU2AF50ΔRSextreme transgene was not able to rescue the dU2AF50 mutant allele (Fig. 2A). In contrast to the ability of a synthetic RS domain to restore splicing activity to the analogous deletion in vitro, however, the dU2AF50(RS)7extreme transgene failed to rescue the dU2AF50 mutant allele (Fig. 2A). The inability of these mutant transgenes to complement the dU2AF50 recessive lethal allele was not a consequence of low protein expression levels. Whole-fly extracts from transgenic lines containing either dU2AF50ΔRSextreme or dU2AF50(RS)7extreme transgenes had mutant protein levels equal to or higher than rescuing dU2AF50 transgene lines as assessed by immunoblot analysis using anti-dU2AF50 antibodies (Fig. 2B, cf. lanes 7,8,9).
The inability of dU2AF50ΔRSextreme and dU2AF50(RS)7extreme to complement the recessive lethal dU2AF50 allele was probably attributable to disruption of the U2AF heterodimer. Recently, we have shown that a triple point mutation (W44A, D45A, and V46A) in dU2AF50 abolishes interaction with dU2AF38 completely in an Escherichia coli copurification assay and in Drosophila embryo extracts (Rudner et al. 1998). Furthermore, we have found that a dU2AF50 mutant lacking its RS domain efficiently associates with the small subunit (D.Z. Rudner, K.S. Breger, R. Kanaar, M.D. Adams, and D.C. Rio, in prep.). Consistent with a requirement for U2AF heterodimer formation, we have also shown that the dU2AF50 interaction mutant (W44A, D45A, V46A) is unable to complement the dU2AF50 recessive lethal allele (Rudner et al. 1998). All three of these critical residues were deleted in dU2AF50ΔRSextreme and dU2AF50(RS)7extreme (Fig. 2A) as well as in the analogous hU2AF65 mutant proteins (Valcárcel et al. 1996). Because the poly(U)-depleted extract does not require the small subunit for reactivation (Zamore et al. 1992), deletion of these conserved residues in hU2AF65 would not affect its activity in vitro. The requirement for heterodimer formation in vivo, however, complicates our molecular genetic analysis. We conclude that the inability of the dU2AF50 “extreme” derivatives to complement the dU2AF50 recessive lethal allele does not address the requirement for the remaining positively charged residues in dU2AF50ΔRStrue, but it does support the conclusion drawn from our previous study. Consistent with a requirement for both U2AF subunits for splicing in vitro, heterodimer formation is essential in vivo (Rudner et al. 1998).
To address the requirement for the remaining positively charged residues in dU2AF50ΔRStrue, a final set of dU2AF50 mutant transgenes were created. The three positively charged residues remaining at the amino terminus in the dU2AF50ΔRStrue transgene were deleted to create dU2AF50ΔRSfinal (Fig. 2A). A synthetic RS domain was fused to this final deletion mutant to create dU2AF50(RS)7final (Fig. 2A). Germ-line transformants were generated and tested for complementation of the recessive lethal dU2AF50 allele. Both the dU2AF50ΔRSfinal and dU2AF50(RS)7final transgenes efficiently rescued the dU2AF50 mutant allele (Fig. 2A). In contrast to the requirement for positive charges in the large subunit RS domain in vitro, the positively charged residues were not necessary in vivo. We conclude that the Drosophila U2AF large subunit RS domain is completely dispensable. These results indicate that the dU2AF50 RS domain is not required for U2 snRNP recruitment during spliceosome assembly in vivo.
The dU2AF38 RS domain and glycine-rich carboxyl terminus are dispensable in vivo
The lack of requirement for the dU2AF50 RS domain in vivo, prompted an analysis of the dU2AF38 RS domain. We have shown previously that a recessive lethal deletion mutation that disrupts the Drosophila U2AF small subunit gene can be rescued by a transgene containing a genomic clone that includes the gene encoding dU2AF38 (Rudner et al. 1996). The dU2AF38 gene is necessary for the observed rescue, as the genomic clone with a frameshift mutation in the dU2AF38-coding sequence is incapable of complementing the dU2AF38 null allele. We have used this rescuing genomic clone to analyze the dU2AF38 RS domain.
The hU2AF35 RS domain has glycine-rich regions interdigitated with the RS dipeptide repeats, whereas the glycine-rich regions of dU2AF38 are distinct from its RS domain (Fig. 3B) (Rudner et al. 1996). To determine the in vivo requirement for the glycine-rich carboxyl terminus of dU2AF38, a stop codon was introduced at amino acid 216 in the dU2AF38-coding sequence in the rescuing genomic transgene to create dU2AF38ΔGly. This nonsense mutation eliminates the entire glycine-rich carboxyl terminus but leaves the RS domain intact (Fig. 3B). Germ-line transformants containing the dU2AF38ΔGly transgene were generated and tested for complementation of the dU2AF38 recessive lethal deletion mutation.The dU2AF38ΔGly transgene efficiently rescued the dU2AF38 null allele (Fig. 3A). The ability of the dU2AF38ΔGly transgene to rescue was not attributable to translational readthrough of the engineered stop codon as only dU2AF38 protein, of a size consistent with the deletion of the glycine-rich region, was detected by immunoblot analysis of whole-fly extracts from dU2AF38ΔGly-rescued flies (Fig. 4, cf. lanes 1, 2, and 3). Overexposure of the immunoblot or overloading the whole-fly extract failed to reveal any wild-type, full-length, dU2AF38 protein (data not shown). Therefore, this glycine-rich region including the carboxy-terminal run of 11 consecutive glycines, though rather distinctive and conserved from Drosophila to mammals, is not essential in vivo.
Figure 3.
dU2AF38 RS domain is inessential in vivo. (A) Wild-type (WT) and dU2AF38 deletion derivative transgenes were tested for complementation of a recessive lethal dU2AF38 null allele. A schematic diagram of the dU2AF38 domains is shown. Pseudo-RRM (ΘRRM), glycine-rich region (Gly), and RS domain (RS) are indicated. Gray lines indicate conserved cysteine and histidine residues in putative Zn2+-binding domains. Percentages of rescue shown are from representative transgene lines.(B) Amino acid sequence comparisons of the RS domain and glycine-rich region of the U2AF small subunit from human and Drosophila. Identities and similarities are shown in black and gray boxes, respectively. Gaps are denoted by dashes. Amino acid positions are shown on the right. The carets above the dU2AF38 sequences indicate the deletion points for dU2AF38ΔRS and are also the sites where stop codons were inserted for dU2AF38ΔRSGly and dU2AF38ΔGly. The caret below the hU2AF35 sequence indicates the deletion site in the hU2AF35ΔRS mutant used in the in vitro reconstitution experiments (Zuo and Maniatis 1996).
Figure 4.

Protein expression of the dU2AF38 deletion derivatives. Immunoblot analysis of whole-fly extracts using an anti-dU2AF38 antibody. Whole-fly extracts are from wild-type, dU2AF38+ flies (lane 1); homozygous ΔdU2AF38 (ΔE18) mutant flies carrying a wild-type dU2AF38 rescuing transgene (lane 2); homozygous ΔdU2AF38 carrying a dU2AF38ΔGly transgene (lane 3); homozygous ΔdU2AF38 carrying a dU2AF38ΔRS transgene (lane 4); homozygous ΔdU2AF38 carrying a dU2AF38 ΔRSGly transgene (lane 5). The size of molecular mass markers are indicated in kD.
To assess the in vivo requirement for the RS domain on dU2AF38, we deleted amino acids 189–213 by insertion of an oligonucleotide linker into the dU2AF38-coding sequence in the rescuing genomic transgene to create dU2AF38ΔRS. This in-frame deletion replaces the dU2AF38RS domain with a single glycine residue (Fig. 3B; see Materials and Methods). Germ-line transformants containing the dU2AF38ΔRS transgene were generated and tested for complementation of the dU2AF38 null allele. The dU2AF38ΔRS transgene completely rescued the dU2AF38 deletion mutation (Fig. 3A). We conclude that the U2AF small subunit RS domain is not required in vivo.
Because the glycine-rich regions are interdigitated with the RS dipeptide repeats in the hU2AF35RS domain, the hU2AF35RS deletion mutant used in the in vitro reconstitution experiments lacked both the RS domain and glycine-rich regions (Fig. 3B) (Zuo and Maniatis 1996). This deletion mutant was incapable of restoring enhancer-dependent splicing to the immunodepleted extracts. In an attempt to directly correlate our in vivo complementation data with the results of the in vitro reconstitution experiments, we inserted a stop codon at amino acid 189 in the dU2AF38-coding sequence of the rescuing genomic transgene to create dU2AF38ΔRSGly (Fig. 3B). This nonsense mutation eliminates the RS domain and glycine-rich carboxyl terminus of dU2AF38 and is a more extensive deletion than the hU2AF35 RS deletion used in vitro (Fig. 3B). Germ-line transformants containing the dU2AF38ΔRSGly transgene were generated and tested for complementation of the dU2AF38 recessive lethal deletion mutation. Surprisingly, the dU2AF38ΔRSGly transgene also completely rescued the dU2AF38 null allele (Fig. 3A). In contrast to the requirement for the hU2AF35 RS domain for splicing in vitro, the dU2AF38RS domain and glycine-rich region were completely dispensable in vivo. The ability of the dU2AF38ΔRSGly transgene to complement the dU2AF38 null mutant is not a consequence of translational readthrough of the engineered stop codon, as the dU2AF38 deletion mutant was the only protein detected by immunoblot analysis of a whole-fly extract from the rescued flies (Fig. 4, cf. lanes 1, 2, and 5).
Although the human and Drosophila small subunit RS domains differ in the placement of the glycine-rich regions, the overall sequence similarity (Fig. 3B) suggests that the lack of requirement for the small subunit RS domain in vivo is not specific to Drosophila. In fact, the recent identification of the U2AF small subunit homolog from S. pombe (pU2AF23) reveals substantial amino acid conservation in the amino terminus (77%) but the complete absence of a carboxy-terminal RS domain (Wentz-Hunter and Potashkin 1996). We conclude that the essential function of the U2AF small subunit does not reside in the RS domain and is therefore not necessary for constitutive splicing in vivo.
The dU2AF38 RS domain is not required for enhancer-dependent splicing of doublesex in vivo
If the dU2AF38 RS domain is required for enhancer-dependent, female-specific splicing of the dsx pre-mRNA, then female flies lacking the dU2AF38 RS domain (dU2AF38ΔRS or dU2AF38ΔRSGly) should be incapable of efficient female-specific dsx splicing. Inefficient female-specific splicing of dsx would cause partial or complete sexual transformation of female flies, a phenotype observed in TRA or TRA2 mutant females (Nagoshi et al. 1988; Nagoshi and Baker 1990). Dimorphic body parts on the diplo X, homozygous dU2AF38 mutant flies carrying the dU2AF38ΔRS or dU2AF38ΔRSGly transgenes were analyzed for sexual transformation. The rescued flies were phenotypically female and fully fertile (data not shown). Therefore, in contrast to previous biochemical studies using human splicing extracts and human U2AF35 (Zuo and Maniatis 1996), the dU2AF38 RS domain is not required for enhancer-dependent alternative splicing of dsx in vivo. These results are consistent with previous genetic analysis of a semi-lethal (hypomorphic) allele of dU2AF38 (Rudner et al. 1996). In these studies, no genetic interactions were observed between TRA, TRA2, and the dU2AF38 hypomorphic mutation.
To confirm that the splicing of dsx RNA is unaffected in the flies lacking the dU2AF38 RS domain, we analyzed dsx transcripts molecularly. Total RNA from w1118 (dU2AF38+) and dU2AF38 mutant flies carrying the dU2AF38ΔRSGly transgene was isolated. dsx splicing was analyzed by reverse-transcription PCR (RT–PCR) using primers specific for male and female dsx transcripts (Fig. 5B) (Amrein et al. 1994). No products were observed in the absence of reverse transcription (Fig. 5A, lanes 1,3,5,7). As expected, only male-specific dsx mRNA was observed in wild-type and dU2AF38ΔRSGly mutant males (Fig. 5A, lanes 2,6). Consistent with our phenotypic analysis of the dU2AF38ΔRSGly mutants, only female-specific dsx mRNA was observed in both wild-type and dU2AF38ΔRSGly mutant females (Fig. 5A, lanes 4,8). Therefore, in the absence of the dU2AF38 RS domain, dsx splicing enhancer function is normal.
Figure 5.
The dU2AF38 RS domain is not required for enhancer-dependent dsx splicing in vivo. (A) RT–PCR analysis of dsx splicing. 32P-labeled RT–PCR products were subjected to electrophoresis through a native polyacrylamide gel and visualized by autoradiography. Total RNA isolated from w1118 (dU2AF38+) males (lanes 1,2) or females (lanes 3,4) flies or from dU2AF38 mutant males (lanes 5,6) or females (lanes 7,8) rescued by the dU2AF38ΔRSGly transgene was analyzed. The presence or absence of reverse transcriptase (RT) in the reaction is indicated above the lanes. Schematic diagrams of the female-specific (dsxf) and male-specific (dsxm) cDNA products are indicated on the left. The markers (M) are 32P-end-labeled MspI-cleaved pBR322 DNA. (B) Schematic diagram of dsx sex-specific alternative splicing. Boxes represent exons, lines represent introns. The male-specific splice and female-specific splice and polyadenylation site (A) are indicated. The primers used for the RT–PCR (Amrein et al. 1994) are shown schematically above the construct.
An RS domain is necessary on the U2AF heterodimer
To determine if any RS domain on the Drosophila U2AF heterodimer was required in vivo, the dU2AF38ΔRS and dU2AF50ΔRS transgenes were tested for the ability to rescue flies mutant for both the small and large subunits. Double mutant flies could be rescued by the combination of a wild-type dU2AF38 and a wild-type dU2AF50 transgene, but not by the combination of a dU2AF38ΔRS and a dU2AF50ΔRS transgene (Fig. 6; see Materials and Methods). We conclude that the presence of at least one RS domain on U2AF is required for viability.
Figure 6.
An RS domain on the U2AF heterodimer is required in vivo. Transgenes from wild-type and mutant derivatives of the two dU2AF subunits were tested for complementation of a dU2AF50, dU2AF38 double mutant. The combination of dU2AF rescuing transgenes used in each complementation cross are indicated on the left.
A synthetic RS domain is not sufficient for U2AF activity in vivo
Deletion of the small subunit RS domain provided a genetic background in which to investigate the functional requirements for the dU2AF50 RS domain in vivo. The dU2AF50(RS)7 and the dU2AF38ΔRS transgenes were tested for complementation of the dU2AF38, dU2AF50 double mutant. Even though fusion of an identical synthetic RS domain on hU2AF65ΔRS will restore splicing activity in vitro (Valcárcel et al. 1996), it did not rescue the double mutant in vivo (Fig. 6). We conclude that, in vivo, a simple RS dipeptide repeat is not equivalent to a U2AF RS domain.
Discussion
The results presented here demonstrate that the dU2AF50 RS domain and the dU2AF38 RS domain are completely dispensable in vivo. Although neither U2AF RS domain is required in vivo, we have found that at least one must be present on the U2AF heterodimer for biological activity. Finally, we have shown that a synthetic RS domain containing seven RS dipeptides is not equivalent to a U2AF RS domain.
The U2AF large subunit RS domain is dispensable in vivo
In contrast to the requirement for the hU2AF65 RS domain for U2 snRNP recruitment in vitro, our molecular genetic analysis indicates that this domain is not necessary for splicing in vivo. Resolution of these contradictory results is suggested by the synthetic lethality resulting from deletion of both U2AF RS domains. The requirement for at least one RS domain on U2AF indicates that the two domains might be functionally redundant. If the two RS domains can substitute for each other, a requirement for the dU2AF50 RS domain would have been masked by the presence of the dU2AF38 RS domain.
The biochemical analysis of hU2AF65 is consistent with functionally redundant U2AF RS domains. The in vitro reconstitution experiments analyzing the requirements for the hU2AF65 RS domain were performed in the absence of exogenous hU2AF35 and the RS deletion endpoint would have prohibited stable association with any hU2AF35 retained in the depleted extract (Valcárcel et al. 1996) (see Results). Under these conditions, in the absence of an associated hU2AF35 RS domain, a role for the hU2AF65 RS domain could have been revealed. In addition, fusion of the hU2AF35 RS domain onto hU2AF65ΔRS was sufficient to restore splicing activity to the poly(U)-depleted extracts (Valcárcel et al. 1996). This result indicates that the hU2AF35 RS domain is capable of U2 snRNP recruitment in vitro.
The results of our genetic assays demonstrate that a synthetic RS domain containing seven RS dipeptides is insufficient for U2AF activity in vivo indicating that a simple RS dipeptide repeat is not equivalent to a U2AF RS domain. Because this identical synthetic RS domain was sufficient for U2 snRNP recruitment in vitro (Valcárcel et al. 1996), it is possible that the synthetic RS domain is not sufficient for interaction with other splicing factors as might be required in vivo (see below). It is also possible there is a species-specific mechanistic difference between the Drosophila and human U2AF proteins.
The U2AF small subunit RS domain is dispensable for constitutive and dsx enhancer-dependent splicing in vivo
Although the biochemical analysis of hU2AF65 is consistent with functionally redundant RS domains, the experiments involving hU2AF35 are not. The protein–protein interaction studies that identified specific interactions between hU2AF35 and the constitutive and alternative splicing factors did not detect interactions between these splicing factors and hU2AF65 (Wu and Maniatis 1993). In these experiments, hU2AF65 was used as a negative control. The inability of hU2AF65 to interact with these splicing factors appears to be inconsistent with the U2AF RS domains having redundant functions.
The domain on hU2AF35 required for interaction with these splicing factors was found to include the hU2AF35 RS domain, but this interaction domain was not thoroughly mapped and might also require another part of hU2AF35 protein (Wu and Maniatis 1993). By analogy, it was shown recently that the RS domain on SRp30a (ASF/SF2) is not sufficient for interaction with the U1 snRNP specific protein U1–70K (Xiao and Manley 1997). If hU2AF35 required both its RS domain and another part of the hU2AF35 protein for interaction with constitutive and alternative splicing factors, then in the context of the U2AF heterodimer, it is possible that the hU2AF65 RS domain could satisfy the requirement for the hU2AF35 RS domain. Recently, it was found that the dU2AF50 RS domain can substitute for the TRA2 RS domain in somatic sex determination in vivo (W. Mattox, pers. comm.). This result indicates that the dU2AF50 RS domain can function in the protein–protein interactions in which the small subunit has been implicated.
Alternatively, it is possible that the protein–protein interactions between the small subunit and these splicing factors that have been observed in vitro and in the yeast two-hybrid assay are not relevant in vivo. Recently, a transgene containing the Drosophila U1 snRNP 70-kD (dU1–70K) gene lacking its RS domain was found to rescue a recessive lethal mutation in dU1–70K (S. Mount, pers. comm.). This result indicates that the U1–70K RS domain is also inessential in vivo and suggests that the protein–protein interactions observed between U1–70K and SR proteins in vitro are also not necessary for spliceosome assembly in vivo. Recent studies have shown, however, that the mammalian SR protein ASF/SF2 requires its RS domain for viability in vivo (Wang et al. 1996). Interestingly, the Saccharomyces cerevisiae U1–70K homolog (Smith and Barrell 1991), like the S. pombe U2AF small subunit, lacks an RS domain.
The inability of hU2AF35ΔRS and wild-type hU2AF65 to reactivate splicing in the extracts depleted of U2AF by anti-hU2AF35 antibodies (Zuo and Maniatis 1996) is also inconsistent with the RS domains having redundant functions. It is possible that the recombinant hU2AF35ΔRS used in the reconstitution experiments was not active and could not interact with hU2AF65. Alternatively, the ability of the hU2AF65/hU2AF35ΔRS heterodimer to reactivate splicing might not have been possible to detect in this assay. Because recombinant hU2AF65 can associate with endogenous hU2AF35 retained in the immunodepleted extract to reactivate splicing activity, only a small range of hU2AF65/hU2AF35ΔRS protein concentrations could be tested (Zuo and Maniatis 1996). Reconstitution of splicing by hU2AF65/hU2AF35ΔRS might require protein concentrations outside this range. Therefore, the requirement for the hU2AF35 RS domain in the reconstitution experiments indicates that the small subunit is important for efficient enhancer-dependent splicing in vitro but cannot address whether it is essential.
Functional redundancy of U2AF RS domains in vivo
Because the U2AF proteins are complexed in a heterodimer and deletion of both RS domains results in synthetic lethality, it is reasonable to hypothesize that the RS domains are redundant and can functionally substitute for each other. An alternative interpretation of the synthetic lethality, however, is that the independent functions assigned to the two RS domains are both redundant with two other activities, one involved in U2 snRNP recruitment and the other in protein–protein interaction. Each redundant activity could individually support viability in the absence of one RS domain but deletion of both RS domains might be too great a burden for these two redundant activities resulting in the observed synthetic lethality. The (h)U2AF65 associated protein (UAP56), a human DEAD box protein required for U2 snRNP-branchpoint interaction, could be redundant with the hU2AF65 RS domain (Fleckner et al. 1997); and the novel set of bridging interactions between U1 snRNP and hU2AF65 suggested by the analysis of the branchpoint bridging protein (BBP) in yeast (Abovich and Rosbash 1997) and the recent identification of a (h)U2AF35 related protein (URP) in mammals (Tronchère et al. 1997), both qualify as potentially redundant with the hU2AF35 RS domain. Although this model is plausible, it does not account for the essential requirement for the individual U2AF RS domains observed in vitro. We favor the first model in consideration of parsimony.
Although we have detected modest splicing defects in dying dU2AF50 mutant larvae, it has not been possible to convincingly show that the cause of lethality in Drosophila U2AF subunit mutants is a splicing defect (D.Z. Rudner and D.C. Rio, unpubl.). This is likely attributable to the fact that the dying mutant larvae slowly run out of the U2AF protein and/or RNA that was maternally deposited in the mutant embryo. In metazoan nuclei, unspliced nuclear pre-mRNA may simply be degraded. In addition, the splicing of certain introns may be more sensitive to the level of U2AF than others, making detection of a defect in splicing nontrivial. Even with tight temperature-sensitive alleles in certain S. cerevisiae splicing factors, it is not always possible to detect a splicing defect in all introns at the nonpermissive temperature. The accumulated biochemical evidence demonstrating an essential requirement for U2AF in constitutive splicing in vitro (Zamore and Green 1991; Zuo and Maniatis 1996; Kanaar et al. 1993) and the requirement for the S. pombe U2AF large subunit homolog for splicing in vivo (Potashkin et al. 1993), however, makes it likely that the cause of death in the U2AF mutants in Drosophila is a defect in splicing. At this point in time, we cannot rule out the formal possibility that U2AF actually is dispensable for splicing in vivo and its essential function is in some unidentified capacity.
Our early view of the RS domains on U2AF consisted of two domains with highly specialized and independent roles in spliceosome assembly. The hU2AF35 RS domain stabilized hU2AF65 on the pyrimidine tract through protein–protein interactions with splicing factors bound to exonic enhancers and the hU2AF65 RS domain recruited U2 snRNP to the branch site sequence (Fig. 7A). The molecular genetic analysis of the Drosophila U2AF RS domains presented here provides a rather different view of the U2AF RS domains (Fig. 7B). Our analysis suggests that either of the RS domains can perform all the tasks assigned to the individual domains. Both RS domains can recruit U2 snRNP to the branch site and interact with constitutive or alternative splicing factors. This new model for the U2AF RS domains has both mnemonic and predictive value. Genetic experiments using the dU2AF deletion derivatives in combination with biochemical experiments with recombinant heterodimers employing assays that require both subunits will be invaluable in testing the predictions of the redundant function model for the U2AF RS domains.
Figure 7.
Old and new models for U2AF RS domains. (A) The two U2AF RS domains were originally assigned separate functions in spliceosome assembly. The large subunit (65) RS domain recruits U2 snRNP (U2) to the branch site “A” and the small subunit (35) RS domain interacts with the RS domains on constitutive and alternative splicing factors (SR) to stabilize the large subunit binding to the intron pyrimidine tract (py). (B) Both U2AF RS domains participate in all the tasks assigned to U2AF RS domains in the redundant function model. Both RS domains can target U2 snRNP to the branch site and interact with constitutive and alternative splicing factors.
Materials and methods
dU2AF50 in vivo expression vector and derivatives
pHSX (Jones and Rubin 1990) was cleaved with EcoRI and HindIII, treated with Klenow DNA polymerase and religated to create pdr1. The rescuing dU2AF50 genomic DNA fragment from pHSX–211S12 (Kanaar et al. 1993) was inserted into pdr1 between the BamHI and SalI sites to create pdr2. pdr2 was cleaved with SalI, treated with Klenow DNA polymerase and religated to create pdr3. Oligonucleotide linkers containing an improved Cavener translational initiation sequence (Schoner et al. 1990), a start codon and unique restriction enzyme sites (top strand: 5′-ATTAATTTTATTTAGCAACCAAAATGGCTAGCGGATCCGTCGACGAATTCGGTACCA-3′; bottom strand: 5′-GATCTGGTACCGAATTCGTCGACGGATCCGCTAGCCATTTTGGGTGCTAAATAAAATTAAT-3′) were annealed and inserted into pdr3 between the SspI and BamHI sites by partial cleavage to create pdr12. The dU2AF50-coding sequence from pdr6 (Rudner et al. 1998) was inserted into pdr12 between the BamHI and SalI sites to create pdr15. The dU2AF50 genomic DNA fragment pHSX–211S12 with an EcoRI site inserted 3′ to the stop codon was cleaved with BstEII and NotI and inserted into pdr15 by partial cleavage to create pdr76. pdr76 was cleaved with XbaI and BamHI, treated with Klenow DNA polymerase, and religated to create pdr113. pdr113 was cleaved with BamHI, treated with Klenow DNA polymerase, and religated to create pdr141. pdr141 is the wild-type dU2AF50 expression plasmid from which all deletion mutants are derived. The dU2AF50-coding sequence in pdr6 and all subsequent clones contain a single point mutation at nucleotide 745. This point mutation changes glycine 249 into serine. This mutation is in position 2 of the RNP1 octamer of the second RNA-binding domain. The G249S mutation had no detectable effect on dU2AF50 activity in vivo (see Fig. 1A).
dU2AF50ΔRS was created by insertion of a linker (top strand: 5′-CTAGCGGCGCC-3′; bottom strand: 5′-TCGAGGCGCCG-3′) between the NheI and XhoI sites in pdr141 to create pdr134. dU2AF50(RS)7 was created by insertion of a linker (top strand: 5′-CTAGCCGCTCGCGTAGCCGCTCCCGGAGCCGCAGCCGTTCCCGC-3′; bottom strand: 5′-TCGAGCGGGAACGGCTGCGGCTCCGGGAGCGGCTACGCGAGCGG-3′) into pdr141 to create pdr137. pdr141 was cleaved with NheI and XhoI, treated with Klenow DNA polymerase, and religated to create dU2AF50ΔRStrue (pdr157). To create dU2AF50ΔRSextreme and dU2AF50(RS)7extreme a new XhoI site was inserted into pdr6 by site-directed mutagenesis using the oligonucleotide 5′-CGAATCCCGGCGGCGGTCTCGAGCAATAAAGCGACGGCTTGCG-3′ to create pdr169. pdr169 was cleaved with BamHI and BstEII and inserted into pdr141 to create pdr177. pdr177 was cleaved with NheI and XhoI, treated with Klenow DNA polymerase, and religated to create dU2AF50ΔRSextreme (pdr182a). The same oligonucleotide linker used to create dU2AF50(RS)7 was inserted between the NheI and XhoI sites in pdr177 to create dU2AF50(RS)7extreme (pdr179b). To create dU2AF50ΔRSfinal and dU2AF50(RS)7final the 5′ end of dU2AF50 was PCR amplified using the following 5′ primers: ΔRSfinal, 5′-CCGGATCCGCTAGCCCGTCGCTTTATTGGGATG-3′; and (RS)7final, 5′-CCGGATCCGCTAGCCGCTCGCGTAGCCGCTCCCGGAGCCGCAGCCGTTCCCGCTCGCCGTCGCTTTATTGGGATG-3′; and a 3′ primer downstream of the SphI site in the dU2AF50-coding sequence. The PCR products were cloned into pGem3Zf(+) (Promega) between the BamHI and SphI sites to create pdr219b and pdr220b. The NheI–SphI DNA fragments from pdr219b and pdr220b were ligated with an SphI–BstEII dU2AF50 fragment from pdr6 into pdr141 cleaved with NheI and BstEII in a three-way ligation to create pdr236 (dU2AF50ΔRSfinal) and pdr226 (dU2AF50(RS)7final). All PCR products, oligonucleotide linkers, and DNA fragments containing site-directed changes were confirmed by sequencing (U.S. Biochemical). NotI DNA fragments from the wild-type expression plasmid and all deletion or substitution derivatives were subcloned into a unique NotI site in the Drosophila transformation vector pw8 (Ashburner 1989).
dU2AF38 expression plasmids
The dU2AF38 genomic clone was subcloned into pHSX (Jones and Rubin 1990) between the EcoRI and ClaI sites to create pdr115. dU2AF38ΔGly was made by oligonucleotide linker insertion (top and bottom strand: 5′-GGATCCTTAGC-3′) into an XmaI site in the dU2AF38-coding sequence in pdr115 to create pdr160. dU2AF38ΔRS was made by oligonucleotide linker insertion (top strand: 5′-CTCTACGGC-3′; bottom strand: 5′-CCGGGCCGTAGAGGTAC-3′) between the KpnI and XmaI sites in the dU2AF38-coding sequence in pdr115 to create pdr162. dU2AF38ΔRSGly was created by oligonucleotide linker insertion (top and bottom strand: 5′-CTCTACTAACGGATCCGTTAGTAGAGGTAC-3′) into a KpnI site in the dU2AF38-coding sequence in pdr115 to create pdr192. All the cloning junctions and oligonucleotide linkers were sequenced. The dU2AF38 genomic clones from pdr160, pdr162, and pdr192 were subcloned into a unique NotI site in the Drosophila transformation vector pw8 (Ashburner 1989). Interestingly, we observed a reproducible 10-fold difference in transformation efficiency depending on the orientation of insertion into pw8. This phenomenon was also observed with the dU2AF50 transgenes.
Complementation analysis
Germ-line transformation of the wild-type dU2AF50 transgene and derivatives into w1118 embryos was as described (Spradling 1986). Independent transformant lines (10–30) were generated for each derivative. All autosomal insertions were tested for complementation of the dU2AF50 recessive lethal allele, 9–21XR15. y, w, 9–21XR15 f/Bins (y, w, sn, B) virgin females were mated to w/Y; P[w+; dU2AF50]/+ males. y, w, 9–21XR15, f/Y; P[w+; dU2AF50]/+ males were compared with their unbalanced y, w, 9–21XR15, f/w; P[w+; dU2AF50]/+ sisters. At least 150 progeny were scored in each complementation cross. At least one unbalanced stock (y, w, 9–21XR15, f/y, w, 9–21XR15, f; P[w+; dU2AF50]/+) was established for each of the rescuing dU2AF50 derivatives. For the dU2AF50 derivatives that did not rescue, we tested >15 independently isolated transgene lines to rule out the possibility of genomic position effect. No complementation was observed for any of these lines.
Two of the five dU2AF50 wild-type transgene lines tested complemented 9–21XR15. The percent rescue ranged from 75% to 123%. Isolate C8 28.1 is shown in Figures 1 and 2. Eight of the 12 dU2AF50ΔRS transgene lines tested complemented 9–21XR15. The percent rescue ranged from 52% to 150%. Isolate C18 9a is shown in Figures 1 and 2. Eight of the 11 dU2AF50(RS)7 transgene lines tested complemented 9–21XR15. The percent rescue ranged from 35% to 122%. Isolate C21 9b is shown in Figures 1 and 2. Three of the four dU2AF50ΔRStrue transgene lines tested complemented 9–21XR15. The percent rescue ranged from 56% to 104%. Isolate 5d is shown in Figure 2. Sixteen dU2AF50ΔRSextreme transgene lines were tested for complementation of 9–21XR15. No rescue was observed. dU2AF50(RS)7extreme transgene lines (24) were tested for complementation of 9–21XR15. No rescue was observed. Eleven of the 20 dU2AF50ΔRSfinal transgene lines tested complemented 9–21XR15. The percent rescue ranged from 11% to 100% (most were 80%–100%). Isolate C43 2a is shown in Figure 2. Seven of the 12 dU2AF50(RS)7final transgene lines tested complemented 9–21XR15. The percent rescue ranged from 61% to 157%. Isolate 2a is shown in Figure 2.
The 9–21XR15 dU2AF50 allele has not been characterized molecularly. It is fully penetrant recessive lethal and no endogenous dU2AF50 protein is detected by immunoblot in 9–21XR15 mutant flies (Fig. 3; our unpublished observations). dU2AF50ΔRS can also rescue a dU2AF50 deletion mutation, 9–21XR26. 9–21XR26 was not used in these studies because the deletion disrupts an adjacent essential gene complicating the genetic analysis.
Germ-line transformation of dU2AF38 mutant transgenes into w1118 embryos was as described previously (Spradling 1986). Independent transformant lines (5–10) were generated for each dU2AF38 derivative. All insertion lines on the X and third chromosomes were tested for complementation of the recessive lethal dU2AF38 null allele, ΔE18. w1118; ΔE18/Sm6β (Cy, Roi) virgin females were mated to w1118/Y; ΔE18/Sm6β (Cy, Roi); P[w+; dU2AF38]/+ males. Rescued, w1118; ΔE18/ΔE18; P[w+; dU2AF38]/+ progeny were scored and percent viability was determined by comparison with w1118; ΔE18/Sm6β; P[w+; dU2AF38]/+ siblings. At least 150 progeny were scored in each complementation cross. Unbalanced stocks (w1118; ΔE18/ΔE18; P[w+; dU2AF38]) were established with all three dU2AF38 deletion transgenes.
Two out of three dU2AF38 wild-type transgene lines tested complemented ΔE18. The percent rescue ranged from 82%–114%. Isolate 9a is shown in Figure 1. Both of the dU2AF38ΔGly transgene lines tested complemented ΔE18. The percent rescue ranged from 30%–90%. Isolate 1a is shown in Figure 1. Six of the seven dU2AF38ΔRS transgene lines tested complemented ΔE18. The percent rescue ranged from 55%–130%. Isolate 6b is shown in Figure 1. All eight of the dU2AF38ΔRSGly transgene lines tested complemented ΔE18. The percent rescue ranged from 70%–140%. Isolate 4a is shown in Figure 1.
To test the requirement for an RS domain on the dU2AF heterodimer, the third chromosome, rescuing dU2AF38 transgenes were P[w+; dU2AF38 9] and P[w+; dU2AF38ΔRS 2a]. Both transgenes fully complement the dU2AF38 recessive lethal allele, ΔE18. The third chromosome dU2AF50 rescuing transgene lines used were P[w+; dU2AF5028.1]; P[w+; dU2AF50ΔRS 9a] and P[w+; dU2AF50(RS)7 1b]. All three transgenes fully complement the dU2AF50 recessive lethal allele, 9–21XR15. y, w, 9–21XR15, f/Bins (y, w, sn, B); ΔE18/Sm6β (Cy, Roi); P[w+; dU2AF50]/+ virgin females were crossed to w/Y; ΔE18/Sm6β (Cy, Roi); P[w+; dU2AF38]/+ males. Rescued y, w, 9–21XR15, f/Y; ΔE18/ΔE18; P[w+; dU2AF50]/P[w+; dU2AF38] male progeny were scored and percent rescue was determined by comparison with y, w, 9–21XR15, f/w; ΔE18/Sm6β; P[w+; dU2AF50]/P[w+; dU2AF38] sisters. y, w, 9–21XR15, f/w; ΔE18/ΔE18; P[w+; dU2AF38] and y, w, 9–21XR15, f/Y; ΔE18/Sm6β; P[w+; dU2AF50] siblings were also scored to ensure individual transgenes could efficiently complement ΔE18 and 9–21XR15. Rescue of ΔE18 by both dU2AF38 transgenes was ∼100%. Rescue of 9–21XR15 by the dU2AF50 transgene was between 44%–72%. As a further control, complementation of ΔE18 and 9–21XR15 by P[w+; dU2AF50ΔRS 9a] and P[w+; dU2AF38 9] or P[w+; dU2AF50 28.1] and P[w+; dU2AF38ΔRS 2a] was analyzed. Rescue of the double mutant by P[w+; dU2AF50ΔRS 9a] and P[w+; dU2AF38 9] or P[w+; dU2AF50 28.1] and P[w+; dU2AF38ΔRS 2a] was 16% and 43%, respectively.
Immunoblot analysis
Whole-fly extract (1/8 fly equivalent per lane for dU2AF50 and 1/4 fly equivalent per lane for dU2AF38) was subjected to electrophoresis on a 10% SDS-polyacrylamide gel, transferred to nitrocellulose, blocked and probed with affinity-purified anti-dU2AF50 or anti-dU2AF38 polyclonal antibodies as described previously (Rudner et al. 1998).
RT–PCR analysis
RNA from 100 adult flies was isolated using guanidinium thiocyanate and a CsCl step gradient (Sambrook et al. 1989). dsx splicing was analyzed by RT–PCR. dsx primers specific for male and female RNA isoforms described by Amrein et al. (1994) were used. DNase I-treated total RNA (2 μg) was reverse transcribed using 25 pmoles of both male-specific and female-specific 3′ primers in the same vessel. Twenty percent of the reverse transcriptase reaction was amplified in a standard PCR reaction containing both 3′ primers and the common 5′ primer. The PCR reactions included 0.2 μCi of [α-32P]dCTP (800 Ci/mmole). Amplification products were analyzed on an 8% native polyacrylamide and visualized by autoradiography. Products were typically analyzed between cycles 20 and 24. To confirm the identity of the male and female products, unlabeled amplification reactions were separated on an 8% native polyacrylamide gel, electroblotted onto Hybond N+ membrane (Amersham) and hybridized with a 32P-labeled probe from the upstream exon common to both RNA isoforms.
Acknowledgments
We acknowledge members of the Rio and Cline laboratories for encouragement and support; Roland Kanaar for advice; Melissa Adams for advice and critical reading of the manuscript; Lissa T. Merritt for support; and Tom Cline for providing a home for the Drosophila genetics. This work was initiated with support from the ACS (DB112) and subsequently supported by the National Institutes of Health (RO1 HD28063).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL don𝔄rio@uclink4.berkeley.edu; FAX (510) 642-6062.
References
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:1–10. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Amrein H, Hedley ML, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: A laboratory handbook. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Baker BS, Wolfner MF. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes & Dev. 1988;2:477–489. doi: 10.1101/gad.2.4.477. [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;1921:5803–5316. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- Cavener DR, Ray SC. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline TW, Meyer BJ. Vive la difference: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcárcel J, Green M. U2AF65 recruits a novel human DEAD box protein required for assembly of U2 snRNP-branch point interaction. Genes & Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M, Krauss RD, Chiang L, Valcárcel, Green MR, Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley ML, Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell. 1991;65:579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- Jones KR, Rubin GM. Molecular analysis of no-on-transient, A, a gene required for normal vision in Drosophila. Neuron. 1990;4:711–723. doi: 10.1016/0896-6273(90)90197-n. [DOI] [PubMed] [Google Scholar]
- Kanaar R, Roche SE, Beall EL, Green MR, Rio DC. The conserved pre-mRNA splicing factor U2AF from Drosophila: Requirement for viability. Science. 1993;262:569–573. doi: 10.1126/science.7692602. [DOI] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lavigueur A, La Branche H, Kornblihtt AR, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes & Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes & Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- Nagoshi RN, Baker BS. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: Cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes & Dev. 1990;4:89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- Nagoshi RN, McKeown M, Burtis KC, Belote JM, Baker BS. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- Potashkin J, Naik K, Wentz-Hunter K. U2AF homolog required for splicing in vivo. Science. 1993;262:573–575. doi: 10.1126/science.8211184. [DOI] [PubMed] [Google Scholar]
- Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- Ring HZ, Lis JT. The SR protein B52/SRp55 is essential for Drosophila development. Mol Cell Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Kanaar R, Breger KS, Rio DC. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc Natl Acad Sci. 1996;93:10333–10337. doi: 10.1073/pnas.93.19.10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner, D.Z., R. Kanaar, K.S. Breger, and D.C. Rio. 1998. Interaction between subunits of the heterodimeric splicing factor U2AF is essential in vivo. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Ruskin B, Zamore PD, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual, second edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schoner BE, Belagaje RM, Schoner RG. Enhanced translational efficiency with two-cistron expression system. In: Goeddel DV, editor. Methods in enzymology. San Diego, CA: Academic Press; 1990. pp. 94–103. [DOI] [PubMed] [Google Scholar]
- Sharp PA. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- Smith V, Barrell BG. Cloning of a yeast U1 snRNP 70K protein homologue: Functional conservation of an RNA-binding domain between humans and yeast. EMBO J. 1991;10:2627–2634. doi: 10.1002/j.1460-2075.1991.tb07805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. P element-mediated transformation. In: Roberts DB, editor. Drosophila: A practical approach. Washington, D.C.: IRL Press; 1986. pp. 175–197. [Google Scholar]
- Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Mayeda A, Hampson RK, Krainer AR, Rottman FM. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes & Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: Creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- Tronchère H, Wang J, Fu X. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388:397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- Valcárcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA of branch point and promotion base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- Valcárcel J, Singh R, Zamore PD, Green MR. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- Wang J, Takagaki Y, Manley JL. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes & Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hoffmann HM, Grabowski PJ. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- Wentz-Hunter K, Potashkin J. The small subunit of the splicing factor U2AF is conserved in fission yeast. Nucleic Acids Res. 1996;24:1849–1854. doi: 10.1093/nar/24.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington MT, Amann BT, Nathans D, Berg JM. Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proc Natl Acad Sci. 1996;93:13754–13759. doi: 10.1073/pnas.93.24.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein-RNA interactions and is necessary for splicing. Genes & Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Green MR. Biochemical characterization of U2 snRNP auxiliary factor: An essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio AR, Lea K, Blumenthal T. Cloning of Caenorhabditis U2AF65: An alternatuvely spliced RNA containing a novel exon. Mol Cell Biol. 1997;17:946–953. doi: 10.1128/mcb.17.2.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes & Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]








