Abstract
Purpose of review
To describe the rationale for searching for genes for schizophrenia, prior efforts via candidate gene association and genomewide linkage studies, and to set the stage for the numerous genomewide association studies that will emerge by the end of 2008.
Recent findings
Genomewide association studies have identified dozens of new and previously unsuspected candidate genes for many biomedical disorders. At least seven new studies of approximately 20,000 cases plus controls are expected to be completed by the end of 2008.
Summary
Current results have few implications for clinical practice or research, and it is possible that this recommendation could be dramatically different in a year.
Keywords: schizophrenia, genetics, genomewide association, genomewide linkage
Introduction
The major question as of this writing in 10/2007 is whether knowledge about the genetic basis of schizophrenia will be substantially different in a year. Will there be at last a hard lead that withstands rigorous scrutiny? There are currently multiple studies scheduled for completion in 2008 that have the potential dramatically to improve understanding of schizophrenia. These efforts are genomewide association studies (GWAS, “jē' wŏs”), a relatively new method to elucidate the genetic basis of complex human diseases, and usually have very large samples by historical standards. In 2007 alone, GWAS has identified dozens of very strong and replicated associations for multiple different complex traits of biomedical importance. This review summarizes: the rationale for searching for genes for schizophrenia, current knowledge of the genetic component to the etiology of schizophrenia, and outlines the prospects for a substantial advance in the next year.
The Rationale for Searching for Genes for Schizophrenia
The rationale for searching for genes for schizophrenia comes from genetic epidemiological studies (family, adoption, and twin studies). Table 1 summarizes this body of work1, 2. Taken together, these data suggest that schizophrenia is familial and that genetic effects are the predominant component of its familiality. In context, the heritability3 of schizophrenia is quite high in comparison to many other biomedical disorders. Schizophrenia is known as a “complex trait” in that its inheritance is complex and does not conform to a typical mode of inheritance (e.g., autosomal dominant, sex-linked, or mitochondrial). It is critical to note that these results are from quasi-experimental studies and do not constitute rigorous proof of the involvement of genetic variation in the etiology of schizophrenia. Moreover, these results reveal nothing about the locations of the relevant genomic regions much less their function or the mechanism by which schizophrenia might result.
Table 1.
Summary of studies of the genetic epidemiology of schizophrenia.
| Study Type | Conceptual basis | Studies | Findings |
|---|---|---|---|
| Family | Risk of schizophrenia in 1st degree relatives of cases with schizophrenia vs. controls | 11 |
|
| Adoption | Risk of schizophrenia in adoption cluster (offspring of one set of parents raised from early in life by unrelated strangers | 5 |
|
| Twin | Risk of schizophrenia in monozygotic vs. dizygotic twins | 12 |
|
Progress to Date in Locating Genes for Schizophrenia
The two major strategies employed prior to 2007 to localize genetic variation for schizophrenia were candidate gene association studies and genomewide linkage studies.
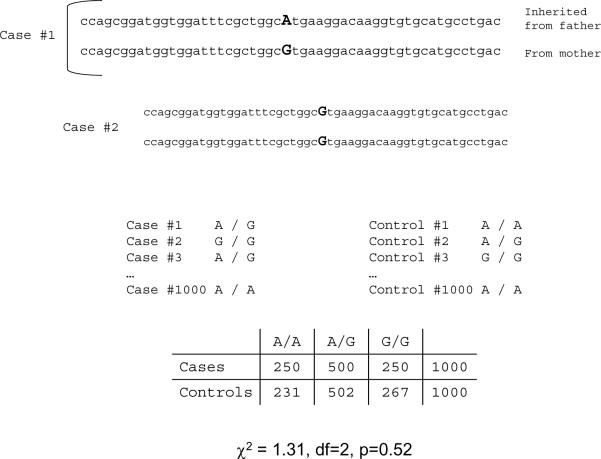
The conceptual basis of a candidate gene study is straightforward and entails the comparison of the genotype frequencies for a particular genetic marker in cases with schizophrenia to appropriately matched controls. An example is given in Figure 1 which shows how a test for association is created from genotype data. Candidate gene selection is usually based on knowledge of etiology and, for schizophrenia, most association studies have focused on the genes encoding proteins involved in some way with dopamine.
Figure 1.
Shows an example of an association study for one genetic marker. The genetic marker is the “famous” COMT val158met polymorphism located on chromosome 22 at base pair position 18,325,825. A “G” at this position codes for valine and an “A” codes for methionine. At the top of the Figure, DNA in the immediate vicinity of rs4680 is shown with the actual variant in larger font size. The DNA sequences on either side are the “keys” that allow the specific interrogation of rs4680 using PCR as the combination of the left and right sequences are found exactly once in the human genome. People usually have two copies of chr22, one from each parent. Each of the copies of chr22 could have either an A or a G at position 18,325,825. Thus, these two “alleles” make up a “genotype”. Case #1 inherited an A and a G – also know as alleles A and G and genotype A/G. Case #2 inherited a G from each parent and has two copies of allele G and genotype G/G. The basic test for an association between case/control status and rs4680 genotype is a 2 × 3 contingency table with the cells containing genotype counts. There is no significant association between case/control status and rs4680 genotype in this example.
There have been at least 1,240 studies of 649 candidate genes in schizophrenia† with COMT the most intensively studied (70 reports). Despite the tremendous amount of work that has gone into these studies, the candidate gene approach has not yielded replicable associations with schizophrenia that meet a high standard of proof. The performance of the candidate gene approach in schizophrenia research is not dissimilar to results from other complex traits of biomedical importance4, 5.
The conceptual basis of genomewide linkage studies is more complex, and is based on the correlations between genotypes and schizophrenia in families. These studies are based on families usually containing multiple affected individuals. Genetic markers for genomewide linkage studies (around 300 in the past but now totaling 10,000 or more) are selected to be relatively evenly spaced across the genome. These studies are not based on prior pathophysiological knowledge but rather can discover new knowledge about the locations of genes that might predispose or protect against the development of schizophrenia.
There have been genomewide linkage studies of 31 samples including 6,769 affected individuals. The results of these studies can be searched and investigated on the web‡. As discussed and depicted elsewhere2, these results do not appear to converge – no genomic region was identified by more than a few studies and an improbably large fraction of the genome (about half) was implicated by at least one study. Moreover, a meta-analysis of schizophrenia genomewide linkage studies6 was recently updated – genomic regions that previously met genomewide significance were considerably less impressive and no longer exceeded genomewide significance.
Comprehensive evaluation of the accumulated work to date is consistent with the following impressions. First, despite considerable effort, genetic variation that predisposes or protects against has not yet been identified with compelling replication. Second, evidence for several highly plausible candidate genes – e.g., NRG17, 8, DTNBP19, 10, or DISC111 – is suggestive but inconclusive and is dissimilar to the pattern of highly consistent and compelling replications found for other complex traits. For example, associations of genetic markers in FTO with body mass index were precisely replicated in 13 cohorts of over 38,000 individuals12. Finally, hindsight is the only perfect scientific tool and it is clear sample size requirements for adequate statistical power have almost always been underestimated in prior studies.
Genomewide Association Studies (GWAS)
In the past two years, technical advances combined with a fortuitous price war have made it possible to conduct GWAS using large samples. Genotyping for these studies is similar to that depicted in Figure 1 but scaled up massively. These studies routinely genotype each individual for 400,000–900,000 genetic variants, and sample sizes usually exceed 1,000 cases and 1,000 controls. A typical GWAS might contain over a billion genotypes (in contrast, a large genomewide linkage study in the past might have a million genotypes). Despite considerable skepticism prior to 2007, GWAS have identified strong evidence for ~70 new and previously unknown candidate genes for a variety of biomedical disorder in 2007. As an example, there were three candidate genes for type 2 diabetes mellitus identified prior to 2007 and there are now at least 10 with all new genes identified by GWAS13.
There are at least seven on-going GWAS for schizophrenia with results expected by mid-2008. One small study has been published (whose main result is not replicating well per unpublished data)14, one has been submitted15, and the remainder are in various stages of completion. Taken together, these studies total approximately 10,000 cases and 10,000 controls. The major studies include the International Schizophrenia Consortium coordinated by Dr. Pamela Sklar at the MIT/Harvard Broad Institute (~7000 samples) and the Molecular Genetics of Schizophrenia group coordinated by Dr. Pablo Gejman of Northwestern University (~5,000 samples).
A welcome characteristic that has emerged in 2007 is the near-universal willingness of the investigators in the primary studies to participate in early data sharing. Nearly all investigators conducting GWAS of schizophrenia have agreed to participate in the Psychiatric GWAS Consortium (http://pgc.unc.edu) which will conduct high-quality meta-analyses of schizophrenia along with ADHD, autism, bipolar disorder, and major depressive disorder. Results of these meta-analyses will be made widely available to the research community as soon as possible. The total sample size across all these disorders should exceed 50,000 individuals with GWAS genotyping by the end of 2008.
Conclusion
It is now clear that GWAS can work in the sense of identifying highly replicable associations for human biomedical disorders of first-rank public health importance. Examples to date include T1DM, T2DM, Crohn's disease, cardiovascular disease, prostate cancer, breast cancer, body mass index, and height. The track record of GWAS in human genetics to date is exceptional and impressive. The key question currently is will it work for schizophrenia – will our knowledge of schizophrenia genetics be substantially different in one year? It is never optimal to conclude a review with uncertainty; however, in this instance, uncertainty is mitigated by the prospect of a convincing demonstration on the one year horizon of whether there is or is not common genetic variation relevant to the etiology of schizophrenia.
Footnotes
http://www.schizophreniaforum.org/res/sczgene/dbindex.asp (accessed 19OCT2007)
http://slep.unc.edu (accessed 19OCT2007).
REFERENCES
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of General Psychiatry. 2003;60:1187–92. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan PF. The genetics of schizophrenia. PLoS Medicine. 2005;2:614–618. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plomin R, DeFries JC, Craig IW, McGuffin P. Behavioral Genetics in the Postgenomic Era. 3rd ed. APA Books; Washington, DC: 2003. [Google Scholar]
- 4.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nature Genetics. 2003;33:177–82. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 5.Ioannidis JP. Commentary: Grading the credibility of molecular evidence for complex diseases. Int J Epidemiol. 2006;35:572–8. doi: 10.1093/ije/dyl003. [DOI] [PubMed] [Google Scholar]
- 6.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O'Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O'Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoega T, Helgason T. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. American Journal of Human Genetics. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. American Journal of Human Genetics. 2002;71:877–92. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Collier DA, He L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet. 2006;15:1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- 9.Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O'Neill FA, Walsh D, Kendler KS. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse Dysbindin gene, is associated with schizophrenia. American Journal of Human Genetics. 2002;71:337–48. doi: 10.1086/341750. Erratum in American Journal of Human Genetics 2002 Oct;72(4):1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mutsuddi M, Morris DW, Waggoner SG, Daly MJ, Scolnick EM, Sklar P. Analysis of high-resolution HapMap of DTNBP1 (Dysbindin) suggests no consistency between reported common variant associations and schizophrenia. Am J Hum Genet. 2006;79:903–9. doi: 10.1086/508942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Human Molecular Genetics. 2000;9:1415–23. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 12.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Davey-Smith G, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–62. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 14.Lencz T, Morgan TV, Athanasiou M, Dain B, Reed CR, Kane JM, Kucherlapati R, Malhotra AK. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol Psychiatry. 2007;12:572–80. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan PF, Lin D, Tzeng JY, van den Oord EJCG, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AC, Lieberman JA, Close SL. Genomewide association for schizophrenia in the CATIE study. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]