SUMMARY
The nucleotide-binding domain and leucine-rich repeat containing (NLR) proteins regulate innate immunity. Although the positive regulatory impact of NLRs is clear, their inhibitory roles are not well defined. We showed Nlrx1−/− mice exhibited increased expression of antiviral signaling molecules IFN-β, STAT2, OAS1 and IL-6 after influenza virus infection. Consistent with increased inflammation, Nlrx1−/− mice exhibited marked morbidity and histopathology. Infection of these mice with an influenza strain that carries a mutated NS-1 protein, which normally prevents IFN induction by interaction with RNA and the intracellular RNA sensor RIG-I, further exacerbated IL-6 and type I IFN signaling. NLRX1 also weakened cytokine responses to the 2009 H1N1 pandemic influenza virus in human cells. Mechanistically, Nlrx1 deletion led to constitutive interaction of MAVS and RIG-I. Additionally, an inhibitory function is identified for NLRX1 during LPS-activation of macrophages where the MAVS-RIG-I pathway was not involved. NLRX1 interacts with TRAF6 and inhibits NF-κB activation. Thus, NLRX1 functions as a checkpoint of overzealous inflammation.
INTRODUCTION
A hallmark of the innate immune response to influenza virus infection is the induction of antiviral cytokines and inflammation. However, an overzealous immune response is deleterious to the host when left unchecked. Host viral recognition is facilitated by three families of pattern recognition receptors (PRRs) that sense pathogen associated molecular patterns (PAMPs): the RIG-I like helicase (RLH) family, the Toll-like receptor (TLR) family, and the nucleotide binding domain (NBD) and leucine-rich-repeat-containing (NLR) family. Members of the NLR family are indispensable for both in vivo and cellular responses to pathogens, including the influenza A virus (Allen et al., 2009; Thomas et al., 2009). Previous in vitro studies have shown that the NLR, NLRX1, functions as an attenuator of RLH-mediated interferon- β (IFN-β) signaling (Moore et al., 2008). NLRX1 was found localized in the mitochondrial outer membrane and interacts with the mitochondrial antiviral signaling (MAVS) protein to disrupt virus-induced RLH-MAVS interactions (Moore et al., 2008).
After influenza virus infection, single stranded viral RNA is detected by RIG-I (Le Goffic et al., 2007; Rehwinkel et al., 2010), which then interacts with MAVS leading to ubiquitin ligases TRAF3 and TRAF6-dependent transcription of type I IFN (IFN-I) and NF-κB responsive genes, respectively (Kawai et al., 2005; Seth et al., 2005). Influenza employs its nonstructural protein 1 (NS1) to subvert the RIG-I-pathway (Guo et al., 2007; Pichlmair et al., 2006). The mammalian genome also encodes several negative regulators of MAVS signaling, although the in vivo relevance of most of these molecules has not been validated (Jia et al., 2009; Jounai et al., 2007; Moore et al., 2008; Xu et al., 2009; Yasukawa et al., 2009; You et al., 2009).
NLRX1 represents an NLR that functions as a modulator of PAMP receptors, rather than an actual receptor or sensor. Here we showed that NLRX1 is a negative in vivo regulator of the innate immune response to viral infection. We generated a Nlrx1−/− mouse strain and demonstrated that NLRX1 functions to down-regulate specific components of the IFN-I response and IL-6 signaling during influenza virus infection. Furthermore, we extend the relevancy of these findings by establishing that NLRX1 down-regulates several interferon response genes and IL-6 in human cells infected with the 2009 pandemic influenza virus A virus. Mechanistically, our data shows that NLRX1 functions as an attenuator of inflammation by intersecting the MAVS and TRAF6 pathways.
RESULTS
NLRX1 functions as a negative regulator of IFN-β production
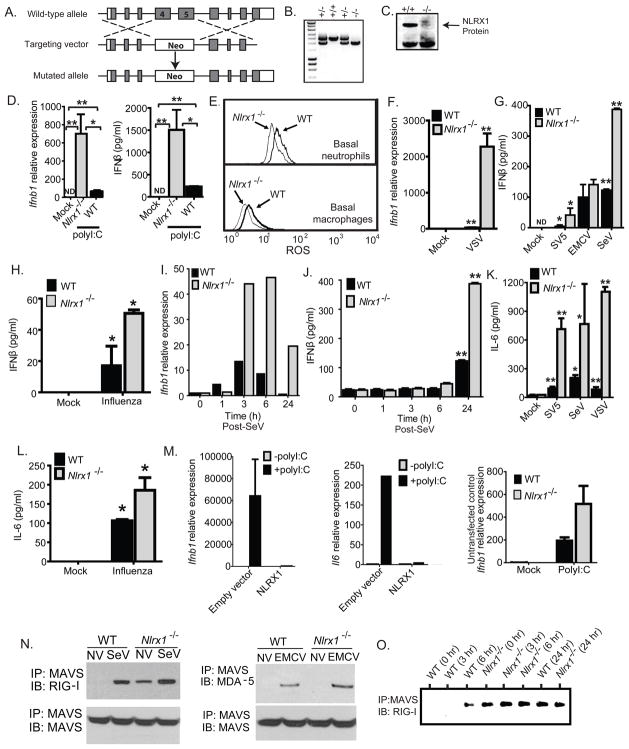
To study the role of NLRX1 in vivo, we generated a mutant mouse by disrupting the nucleotide binding domain (NBD) with a neomycin cassette (Figure 1A–B; Supplemental Figure S1A–C). Fibroblasts isolated from Nlrx1−/− mice were completely devoid of NLRX1 protein (Figure 1C). Our earlier in vitro analysis in cell lines concluded that NLRX1 functions as an attenuator of viral-induced MAVS-mediated IFN-I production (Moore et al., 2008). Therefore, we assessed Ifnb1 expression in Nlrx1−/− mouse fibroblasts. Constitutive Ifnb1 expression was equivalent between unstimulated wild-type (WT) and Nlrx1−/− fibroblasts (Supplemental Figure S1D), indicating that NLRX1 does not affect basal IFN expression. However, transfection of the synthetic RNA analog polyI:C into cells caused significantly more Ifnb1 transcript and IFN-β protein in Nlrx1−/− fibroblasts relative to WT controls (Figure 1D). These data suggest that NLRX1 functions as an attenuator of IFN-β production in response to dsRNA, which is an RLH agonist.
Figure 1. NLRX1 functions as a negative regulator of IFN-I production.
A. Schematic of the generation of Nlrx1−/− mice through homologous recombination. B. PCR genotyping of offspring from heterozygous matings. C. Confirmation of the loss of NLRX1 protein in Nlrx1−/− offspring by immunoblot. D. Ifnb1 transcript and IFN-β protein were assessed in MEFs that were transfected with polyI:C. E. Neutrophils and macrophages were purified from the bone marrow of WT and Nlrx1−/− mice and were activated for 1 hour in the presence of DHR. Fluorescence intensity was used to measure ROS production. F. Ifnb1 transcription was measured in primary fibroblasts isolated from Nlrx1−/− mice following challenges with Vesicular Stomatitis Virus (VSV). G–H. IFNβ protein levels were determined by ELISA in the supernatants from Nlrx1−/− fibroblasts following stimulation with Simian Virus 5 (SV5), Encephalomyocarditis Virus (EMCV), Sendai Virus (SeV) or influenza A (PR/8/34) virus. I–J. Ifnβ gene transcription and IFNβ protein levels were measured over a timecourse SeV infection in primary ear fibroblasts isolated from Nlrx1−/− mice. K–L. IL-6 concentrations were assessed by ELISA in the supernatants from primary Nlrx1−/− MEFS following SV5, SeV, VSV and influenza virus infection. M. Nlrx1−/− MEFS were reconstituted with constructs containing full length human NLRX1 and assessed for Ifnb1 and Il6 gene transcription after polyI:C stimulation. N. The association between MAVS, RIG-I and MDA-5 were assessed by co-immunoprecipitation in primary ear fibroblasts isolated from WT and Nlrx1−/− mice. Data shown are at time 0 and 24 hours post-infection with either SeV (RIG-I) or EMCV (MDA-5). O. Primary ear fibroblasts were challenged with SeV and samples were harvested over a time course. The RIG-I-MAVS association was detected 6 hours after SeV challenge in WT fibroblasts and constitutive interaction was detected in Nlrx1−/− fibroblasts. *p < 0.05;**p < 0.005. See also Supplemental Figure S1.
Earlier reports found elevated reactive oxygen species (ROS) following overexpression of NLRX1 (Abdul-Sater et al., 2010; Tattoli et al., 2008). To assess this in a physiologically relevant model, we measured ROS in stimulated neutrophils and macrophages isolated from WT and Nlrx1−/− mice. We observed a small, quarter log reduction in basal ROS produced from Nlrx1−/− neutrophils and macrophages and in neutrophils stimulated with Pam3Cys (Figure 1E; Supplemental Figure S1E–F). However, ROS amounts were not markedly altered after PAMP stimulation with any other agonist in either cell type (Supplemental Figure S1E–F). Thus, NLRX1 likely plays a minimal role in ROS production.
NLRX1 inhibits the interaction between MAVS and RIG-I and functions as a negative regulator of virus infection
Previous studies utilizing fibroblasts isolated from either Mavs−/− or Ddx58−/− (RIG-I-deficient) mice revealed that loss of either protein resulted in significantly reduced amounts of IFN-β in response to Sendai Virus and Vesicular Stomatitis Virus (Kato et al., 2005; Sun et al., 2006). To assess the role of NLRX1, we challenged primary fibroblasts from our Nlrx1−/− mice with a panel of viruses designed to evaluate RIG-I and MDA5 signaling. These viruses included Simian Virus 5 (SV5), Encephalomyocarditis Virus (EMCV), Sendai Virus (SeV), Vesicular Stomatitis Virus (VSV) and Influenza A virus (A/PR/8/34). We observed a significant increase in IFN-β production in Nlrx1−/− fibroblasts following challenges with SV5, SeV, VSV and influenza A virus (Figure 1F–H). However, the amount of IFN-β induced by ECMV was unaffected by NLRX1 deficiency (Figure 1G). Because a previous study showed VSV-induced IFN-β production in MEFs was RIG-I dependent, whereas EMCV-induced IFN-β production was MDA5 specific (Kato et al., 2006), these data suggest that NLRX1 negatively regulates MAVS signaling that relies on RIG-I, but not MDA-5, dependent recognition of viruses.
To observe the kinetics of IFN-β production in the context of NLRX1 deficiency, we assayed SeV-infected samples over a time-course. We utilized SeV because it is a well characterized virus and a known activator of RIG-I. Following SeV infection, Ifnb1 expression was markedly increased in the Nlrx1−/− fibroblasts between 3 and 24 hours after infection (Figure 1I). IFN-β protein levels lagged behind transcription and were markedly enhanced at 24 hours after infection in Nlrx1−/− fibroblasts (Figure 1J). In addition to IFN-β, previous studies have also shown that IL-6 production in MEFs after SeV infection is dependent upon MAVS signaling (Sun et al., 2006). Thus, we expanded our analysis by assessing IL-6 production in Nlrx1−/− MEFs after virus challenge. Nlrx1−/− MEFs produced significantly increased amounts of IL-6 in response to SV5, SeV, VSV and influenza virus infection (Figure 1K–L). To complement the virus studies and to further confirm the inhibitory nature of NLRX1, primary MEFs from Nlrx1−/− mice were stably transfected with a construct encoding full-length human NLRX1. After transfection with polyI:C, reconstituted MEFs were capable of attenuating Ifnb1 and Il6 gene transcription (Figure 1M). Together, these data suggest that NLRX1 functions as a negative regulator of both IFN-I signaling and IL-6 production in primary MEFs following challenges with RIG-I inducing viruses.
Previous data suggested that the introduction of exogenous NLRX1 can inhibit the association of RIG-I with MAVS (Moore et al., 2008). To test this ex vivo, we examined the association of endogenous RIG-I or MDA-5 with MAVS after SeV and EMCV infections in fibroblasts that were isolated from WT and Nlrx1−/− mice. The RIG-I-MAVS association occurred constitutively in Nlrx1−/− cells in the absence of viral infection, whereas this association was not seen until 6 hrs after infection in WT fibroblasts (Figure 1N–O). The MDA5-MAVS interaction required stimulation by EMCV and was increased 24 hours after infection in the absence of NLRX1 (Figure 1N). The enhanced association between RIG-I and MAVS could, at least partially, explain the robust nature of the virus-induced IFN-β secretion in Nlrx1−/− fibroblasts.
NLRX1 is an attenuator of LPS-induced IFN-β and IL-6 production in macrophages
The essential role of MAVS in RIG-I mediated antiviral innate immunity is cell type specific (Kato et al., 2005; Sun et al., 2006). To address potential cell type differences in NLRX1 function, we assessed the role of NLRX1 in myeloid derived cells following polyI:C stimulation. There were no differences in Ifnb1 expression or IFN-β production from Nlrx1−/− and WT bone marrow derived macrophages stimulated with either polyI:C (MAVS) or 3′-ppp-RNA (RIG-I) (Figure 2A–B). We also generated plasmacytoid dendritic cells (pDCs), which are robust producers of IFN. There were no differences in IFN-β production from Nlrx1−/− pDCs stimulated with the known TLR7 agonist R848, TLR9 agonists CpG, or TLR3 agonist polyI:C (Supplemental Figure S1G). To determine if NLRX1 also contributes to bacteria induced IFN-β production, we challenged macrophages with either Legionella pneumophila or Listeria monocytogenes. Both of these bacteria are known to induce high amounts of IFN-β in macrophages. However, following in vitro infection, we did not observe a difference in Ifnb1 expression between WT and Nlrx1−/− macrophages (Supplemental Figure S1H). This indicates that NLRX1 does not play an obvious role in IFN-β induction by these bacteria.
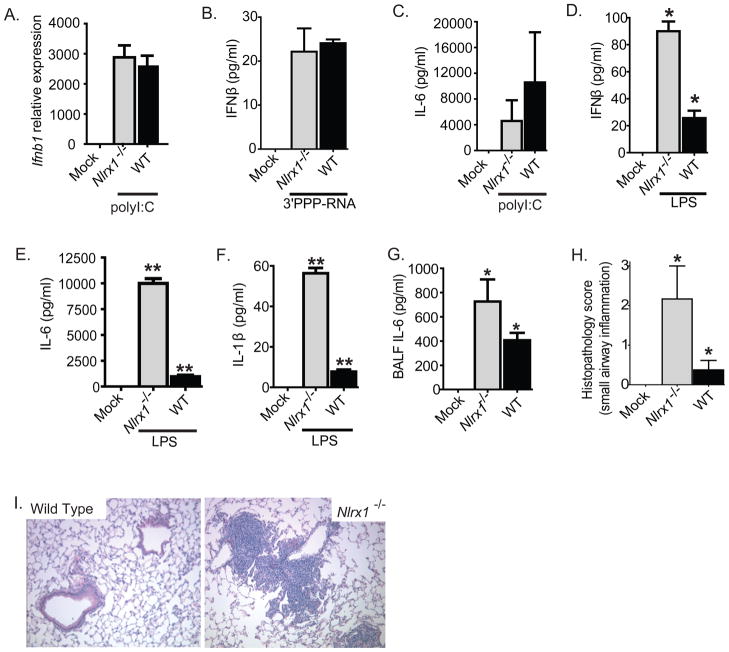
Figure 2. NLRX1 attenuates LPS mediated IFNβ and cytokine production in mouse macrophages.
A–B. Ifnβ transcript levels and IFNβ protein levels were measured in bone marrow macrophages that were isolated from WT and Nlrx1−/− mice following transfection with either polyI:C or 3′PPP-RNA. C. IL-6 production was measured in bone marrow macrophages isolated from WT and Nlrx1−/− mice following transfection with polyI:C. D–F. IFN-β, IL-6 and IL-1β protein levels were assessed in bone marrow derived macrophages that were challenged with LPS. G–I. Mice were challenged with LPS via i.t. instillation and monitored for changes in morbidity and innate immune responses. G. IL-6 levels were assessed by ELISA in BALF from WT and Nlrx1−/− mice, 24 hours following LPS challenge. H. Semiquantitative histopathological scoring was utilized to quantify lung histopathology 24 hours following LPS challenge. I. Peribroncholar and perivascular cuffing localized around the small/medium sized airways was a characteristic feature of the Nlrx1−/− histopathology.
In macrophages, the production of IFN-β and IL-6 after LPS stimulation occurs through a MAVS-independent mechanism (Sun et al., 2006). The data shown in Figure 1, suggests that RIG-I and MAVS are constitutively associated in Nlrx1−/− MEFs, which likely contributes to the increased amounts of IL-6 that we observed after virus stimulation in this cell type. Unlike the MEF data, a role for Nlrx1 in attenuating IL-6 was not observed in macrophages stimulated with transfected polyI:C, which is consistent with the literature (Figure 2C). In addition to polyI:C, we also examined the induction of IFN-β and IL-6 in macrophages stimulated by LPS. Under these conditions, Nlrx1−/− macrophages produced significantly more IFN-β and IL-6 after LPS stimulation (Figure 2D–E). We also assessed the production of IL-1β and observed a significant increase in IL-1β production in the Nlrx1−/− macrophages (Figure 2F). This suggests that the function of NLRX1 is not specific to IL-6. In addition to LPS, we did not observe a significant difference between WT and Nlrx1−/− macrophages stimulated with either TNF-α or Pam3CSK4 (Supplemental Figure S1I–J). LPS is an agonist for the TLR4 receptor, where signal transduction is facilitated through MyD88, TRAF3 and TRAF6 to mediate IFN-I production and canonical NF-κB initiated cytokine production. Together, these data suggest that NLRX1 functions as a negative regulator of this pathway in macrophages.
To assess the physiological relevance of these findings, mice were also subjected to an in vivo airway challenge with LPS, where LPS was delivered to the lungs via intratracheal (i.t.) instillation and the mice were harvested 24 hours after challenge. Consistent with the ex vivo macrophage data, LPS induced a significant increase in IL-6 production in the Nlrx1−/− mice (Figure 2G). Histopathology assessments revealed a significant increase in lung inflammation, which was especially concentrated around the small airways (Figure 2H–I). Together, these data suggest that NLRX1 functions as an attenuator of the TLR4 pathway in vivo in addition to its roles in attenuating MAVS-dependent IFN-I induction.
NLRX1 attenuates inflammation through interactions with TRAF3 and TRAF6
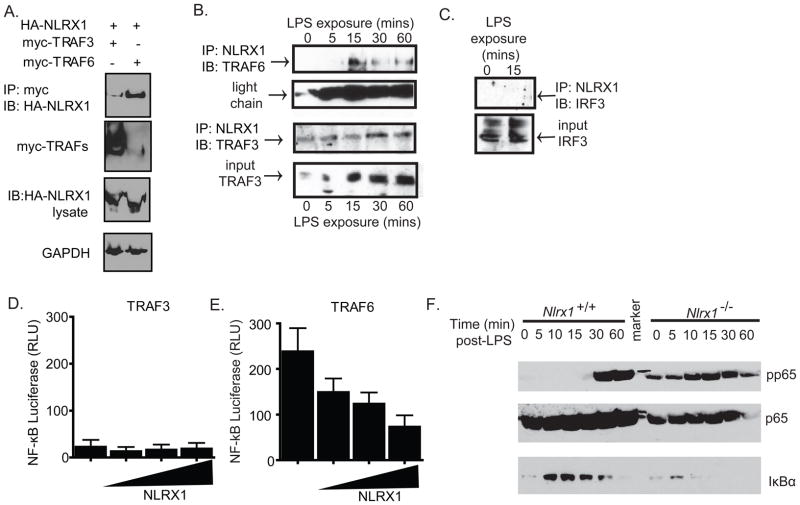
We next sought to explore the underlying mechanism associated with the negative regulatory functions of NLRX1 during LPS stimulation, which is not known to be mediated by RIG-I and MAVS. Based on the strong effect of NLRX1 on IL-6 induction, which is known to require TRAF and NF-κB activation, we first explored the functional and physical interactions of NLRX1 with TRAF3 and TRAF6. In an overexpression system, NLRX1 preferentially interacted with TRAF6 (Figure 3A, top panel) even though the input amount of TRAF6 was reduced relative to TRAF3 (Figure 3A, second panel). Interaction between over-expressed proteins can lead to artifacts; thus, we used LPS to activate endogenous TRAF3, TRAF6 and IRF3 in primary bone marrow derived macrophages. We detected NLRX1 associations with both TRAF3 and TRAF6 following activation with LPS (Figure 3B). In the case of TRAF6, the interaction with NLRX1 was detected within 15 mins of LPS stimulation. However, NLRX1 weakly associated with TRAF3 under both resting conditions and throughout the stimulation period (Figure 3B). We did not detect any interactions between NLRX1 and IRF3 in bone marrow macrophages following LPS exposure (Figure 3C).
Figure 3. NLRX1 functions as a novel inhibitor of TRAF6 and NF-κB.
A. Overexpression systems were utilized to assess NLRX1 interactions with TRAF6. HEK293T cells were transfected with HA-NLRX1 and either myc-TRAF-3 or myc-TRAF-6. Myc-TRAFs were immunoprecipitated and the blot was probed with HA-HRP antibody, myc-HRP and GAPDH-HRP. B–C. NLRX1 associations with TRAF6, TRAF3 and IRF3 were determined following LPS stimulation in primary bone marrow derived macrophages. D–E. NF-kB luciferase activity was determined in transfected 293T cells in the presence of increasing concentrations of input NLRX1 following stimulation with TRAF3 and TRAF6. F. Phosphorylation of p65 and levels of the inhibitor IκBα were assessed throughout a timecourse of LPS stimulation in MEFS. All studies were repeated a minimum of 3 times.*p < 0.05;**p < 0.005. See also Supplemental Figure S1.
We next utilized an overexpression system to assess the functional relevance of NLRX1 on TRAF6 mediated NF-κB activation. NLRX1 transfection dose dependently inhibited TRAF6 mediated induction of the NF-κB expression construct (Figure 3D–E). Under the resting state, the NF-κB heterodimer (p50-p65) is sequestered in the cytoplasm through its association with IκBα. Stimulation of cells results in the rapid phosphorylation, ubiquitination and subsequent degradation of IκBα, which results in the subsequent phosphorylation and activation of the p65 subunit and NF-κB activity. To evaluate the effect of NLRX1 on NF-κB, MEFs were generated from Nlrx1−/− mice and stimulated with LPS for up to one hour and the amounts of IκBα and phosphorylated p65 (pp65) were assessed. Following 30 minutes of LPS exposure, pp65 was markedly increased in the WT cells, but this process was accelerated in MEFS derived from Nlrx1−/− mice. A higher amount of pp65 was detected in naïve Nlrx1−/− MEFS and further increased by 10 minutes of LPS stimulation (Figure 3F). Consistent with these findings, the amounts of IκBα were reduced or not detected in Nlrx1−/− cells when compared to the WT controls. These results suggest that in addition to serving as an attenuator of IFN-I responses, NLRX1 also inhibits LPS induced TRAF6 activation of NF-κB. We also evaluated pp65 activation following TNFα stimulation. We observed a significant increase in pp65 within 5 mins of stimulation in cells from both sets of mice. However, no significant differences in pp65 levels were observed between WT and Nlrx1−/− in response to TNFR activation (Supplemental Figure S1K).
Nlrx1−/− mice showed enhanced viral clearance, but increased in lung pathology and IL-6 amounts after influenza virus infection
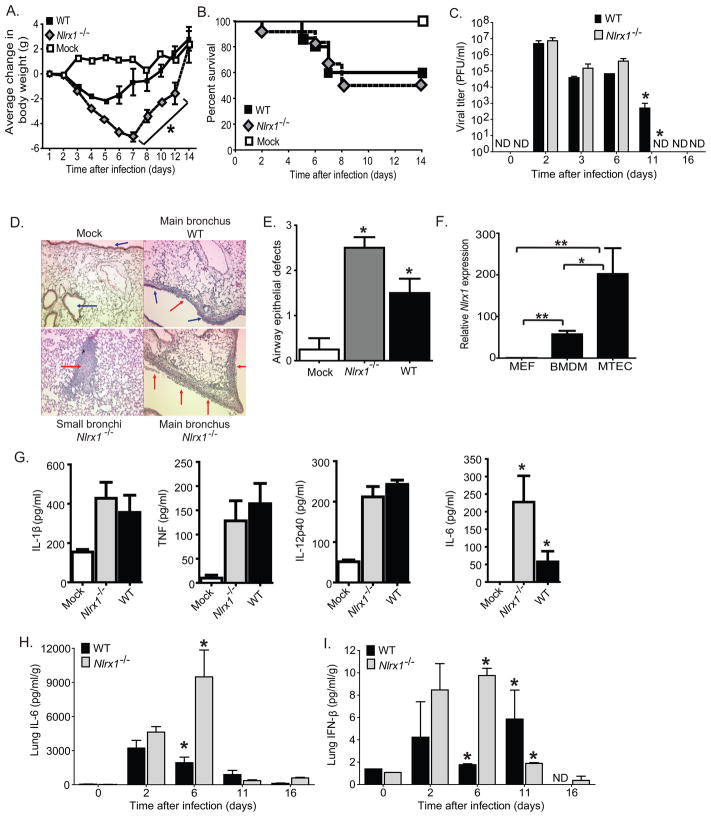
Because isolated fibroblasts from Nlrx1−/− mice exhibit a significant increase in viral induced IFN-I and IL-6 production ex vivo, we hypothesized that influenza virus infection of Nlrx1−/− mice would result in phenotypic and cytokine alterations in vivo. To test this hypothesis, we performed an intranasal exposure of WT and Nlrx1−/− mice with mouse-adapted influenza A/PR/8/34 (H1N1) virus and characterized disease progression. By 6 days post-infection (dpi), Nlrx1−/− mice exhibited significant morbidity as reflected by a rapid weight loss (Figure 4A). However, this increase in morbidity was not sufficient to cause an increase in mortality (Figure 4B). We utilized plaque assays from lung homogenates, which were collected over a 16 day time course, to determine the viral titer. We did not observe any significant differences in viral load between the WT and Nlrx1−/− mice through day 6 (Figure 4C). However, by day 11 the virus had been cleared from the Nlrx1−/− mice and was still present in the WT animals (Figure 4C). This suggests that the Nlrx1−/− mice are more efficient at viral clearance compared to the WT animals. We also examined histological sections through the main bronchiole of the left lung lobe for signs of morphological changes or inflammation at the 6 day time point. Airway epithelial cells showed substantial denuding and marked airway occlusion was observed throughout the lungs of Nlrx1−/− mice (Figure 4D). Furthermore, double blinded histological scoring (scale of 1–3) of lung sections were conducted and confirmed significantly increased airway epithelial injury (Figure 4E). We also found Nlrx1 to be highly expressed in primary mouse tracheal epithelial cell cultures (Figure 4F). Influenza virus infection typically induces airway inflammation and low levels of airway epithelial cell injury and denuding (Buchweitz et al., 2007). However, the excessive histopathology observed in the Nlrx1−/− mice is more consistent with features typically associated with a hyper-inflammatory response, increased cell death and is consistent with the increased production of interferons (Balachandran et al., 2000; Lechner et al., 2008). These data support the conclusion that NLRX1 is indeed functioning as an in vivo attenuator of influenza virus-induced inflammation and loss of this critical regulator results in increased lung injury and morbidity. However, a beneficial effect of this increased epithelial cell denuding and interferon response may be enhanced viral clearance in the Nlrx1−/− mice.
Figure 4. Characterization of the in vivo host immune response in Nlrx1−/− mice following influenza virus A/PR/8/34 infection.
A–B. Morbidity (weight loss) and mortality were measured for 14 days in Nlrx1−/− and WT mice following in vivo intranasal administration of influenza virus A/PR/8/34. C. Lungs were harvested at specific time points and homogenized to assess influenza viral titers, which were determined by standard plaque assay. D. Lungs were harvested 3 dpi and fixed by inflation and immersion in buffered PFA. Sections through the main bronchiole of the left lobe revealed increased airway epithelial cell denuding (red arrows) and small airway obstruction throughout the lungs of Nlrx1−/− mice. E. Histological scoring of H&E stained lung sections revealed a significant increase in airway epithelial defects. F. High basel levels of Nlrx1 transcription was observed in primary bone marrow derived macrophages (BMDM) and mouse tracheal epithelial cells (MTECs) from naïve WT mice. G. Serum was harvested from Nlrx1−/− and WT mice 3 dpi with influenza A/PR/8/34 virus. Influenza virus infection induced increased serum levels of IL-1β, TNFα, IL-12p40, and IL-6. H–I. Whole lungs were harvested from WT and Nlrx1−/− mice at specific time points after i.n. infection with influenza A/PR/8/34 virus. IL-6 and IFN-β levels were determined in cell free supernatants by ELISA. All studies were repeated a minimum of 3 times with 5–7 mice per group for each individual study.*p < 0.05;**p < 0.005. See also Supplemental Figures S2 and S3.
We next sought to assess the effects of NLRX1 on cytokines known to play roles in the host immune responses to influenza virus. IL-1β, TNFα, IL-12, and IL-6 facilitate a wide spectrum of immunologic functions during respiratory virus infection. No significant differences in serum levels of IL-1β, TNFα, and IL-12p40 were detected in the WT and Nlrx1−/− animals (Figure 4G). However, influenza virus-induced IL-6 expression has been linked to robust immunity to viral infection. Consistent with the differences in morbidity and lung damage, IL-6 was significantly elevated in the serum of Nlrx1−/− mice, compared to the WT animals (Figure 4G). In addition to serum, we also evaluated local cytokine concentrations in lung homogenates over the time course infection. We observed a significant increase in IL-6 and IFN-β levels in the Nlrx1−/− lungs (Figure 4H–I; Supplemental Figure S2). Together, these data are consistent with our ex vivo observations, which show that the removal of NLRX1 results in exacerbated IL-6 and IFN-β responses following in vivo influenza virus infection. We considered the possibility that the increase in IL-6 could be associated with the increase in airway epithelial cell denuding observed in the Nlrx1−/− mice. However, additional pharmacological experiments using an IL-6 receptor antagonist suggest that this pleiotropic cytokine does not contribute to the observed lung pathology, which might be caused by other cytokines or IFN-β (Supplemental Figure S3).
NLRX1 functions in vivo as a negative regulator of the IFN-I response to influenza virus challenge
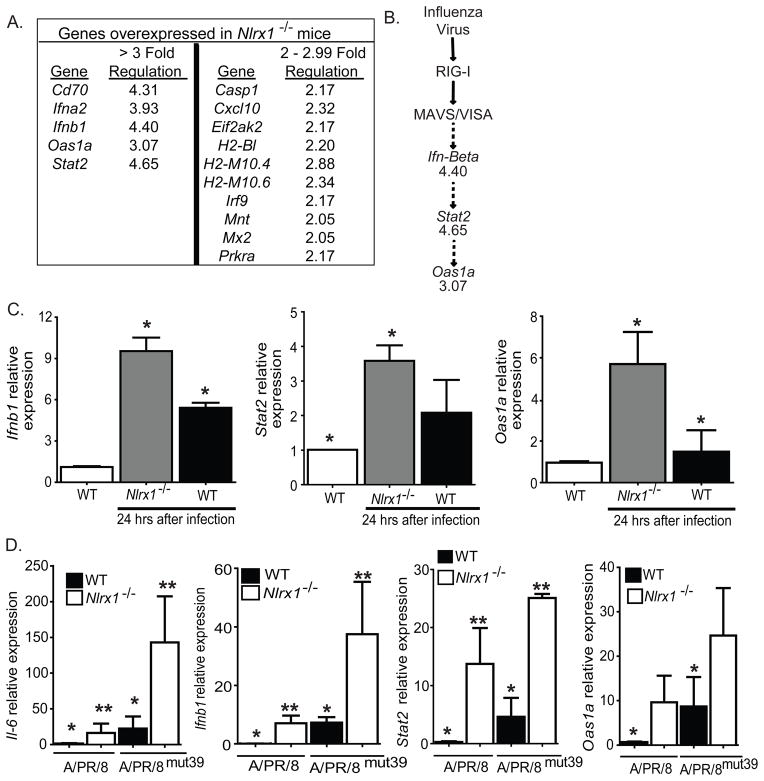
A direct assessment of IFN-I in animal tissues is difficult. However, downstream genes that are IFN-I responsive are frequently used as a surrogate marker for induction. To further investigate the specific cytokine milieu that is upregulated in the lungs of Nlrx1−/− mice, we performed expression profiling of a large subset of cytokines and interferon response genes in lung homogenates following influenza virus challenge (Supplemental Figure S4A). Consistent with its role as a negative regulator of IFN-I responses, we detected a greater than 3-fold upregulation of Ifna2, Ifnb1, Oas1a, and Stat2 gene expression in the lungs of Nlrx1−/− mice compared to WT animals. We also observed a greater than 4-fold increase in Cd70, a cytokine only known to be expressed by activated, but not resting, T and B lymphocytes (Figure 5A). Only 3 genes, H2-M10.1, H2-M10.6 and Oas2, were found to be greater than 2-fold upregulated in the lungs of naïve Nlrx1−/− mice compared to naïve WT animals (Supplemental Figure S4B–C). Ingenuity Pathway Analysis identified IFN-β, STAT2, and OAS1 as downstream effectors of influenza virus induced RIG-I and MAVS signaling (Figure 5B). These differences were confirmed by real-time PCR measurements (Figure 5C).
Figure 5. NLRX1 functions in vivo as a negative regulator of the IFN-I response to influenza virus challenge.
A. Gene expression was profiled for genes associated with IFN-I and viral innate immunity. Whole lungs were harvested from WT and Nlrx1−/− mice, 20 hours post-intratracheal (i.t.) inoculation with the influenza A/PR/8/34 virus. RNA was extracted and pooled in equal quantities for similarly treated mice from each genotype. Genes that demonstrated at least 2-fold changes in transcription between WT and Nlrx1−/− mice were considered significant. B. The expression profile data was analyzed using Ingenuity Pathways Analysis software, which revealed several genes in the RIG-I/MAVS pathway that were significantly up-regulated in the lungs of Nlrx1−/− mice. C. Realtime PCR was used to confirm increased Ifnβ, Stat2, and Oas1a gene expression in individual samples (un-pooled and distinct from the samples used in the array). D. NLRX1 and the viral NS1 protein have non-overlapping targets. WT and Nlrx1−/− mice were challenged with the influenza A/PR/8mut39 virus, which carries a mutant NS1 protein that attenuates the virus’s ability to inhibit the IFN-I response. IL-6, Ifnβ, Stat2, and Oas1a gene expression was assessed by realtime PCR. Expression profiling was repeated 2 times with samples pooled from 3 individual mice. All other studies were repeated 3 times with 3–7 mice per group for each individual study.*p < 0.05;**p < 0.005. See also Supplemental Figures S4 and S5.
The influenza virus encodes an NS1 protein that functions as a brake on host RIG-I mediated antiviral immunity (Guo et al., 2007; Pichlmair et al., 2006). We next sought to investigate the affects on lung cytokine production following infection of Nlrx1−/− mice with the influenza A/PR/8mut39 virus, which possesses a point mutation in the NS1 protein that attenuates the virus’s ability to inhibit IFN-I production (Hayman et al., 2007). Our initial in vivo studies with the influenza A/PR/8mut39 virus revealed no significant differences in mortality or viral titer in WT and Nlrx1−/− mice infected with the mutant virus compared to the control virus (Supplemental Figure S5A–B). Similar to the Nlrx1−/− mice that were challenged with the influenza A/PR/8 virus, WT mice that were infected with the influenza A/PR/8mut39 virus demonstrated a significant increase in lung histopathology, including a significant increase in airway epithelial cell denuding and small airway occlusion (Supplemental Figure S5C). Consistent with our hypothesis that the influenza virus NS1 protein and host NLRX1 protein target similar components of the RIG-I pathway, in vivo, we observed that infection of WT mice with the influenza A/PR/8mut39 virus resulted in significantly increased cytokine mRNA expression (Il6, Ifnb1, Stat2, and Oas1a) compared to the WT control virus (Figure 5D). Interestingly, we observed a significant increase in lung expression of Il6, Ifnb1, Stat2, and Oas1a when Nlrx1−/− mice were exposed to A/PR/8mut39, which extended beyond what was observed in WT animals (Figure 5D). Therefore, as illustrated in the model shown in Supplemental Figure S5D, releasing both the viral (NS1) and host (NLRX1) negative regulators of the RIG-I and MAVS pathway led to even further exacerbation of components of the host innate immune response.
NLRX1 attenuates the IFNβ-STAT2-OAS1 axis in response to the 2009 pandemic influenza A/California/08/09 virus
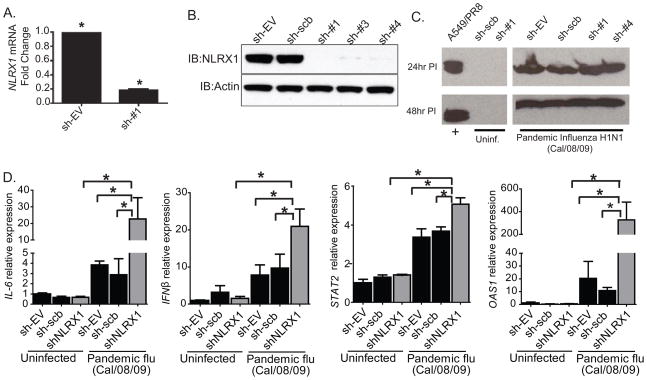
To evaluate the relevance of these findings in the context of human diseases, we evaluated the role of NLRX1 in the pandemic influenza A/California/08/09 (H1N1) virus. The pandemic virus was not mouse-adapted at the time of our studies and was restricted to use under BSL3 conditions. Hence, we did not use this virus in vivo. Instead, we chose a stable shRNA knockdown approach in human cells, which achieved a near-complete knockdown of NLRX1 mRNA and protein in 293T cells (Figure 6A–B). Infection with pandemic influenza A virus at an MOI=0.1 was verified by immunoblot for the influenza virus NS1 protein (Figure 6C). Congruent with our in vivo findings, we observed a significant increase in IL6, IFNB1, STAT2, and OAS1 mRNA in cells bearing shNLRX1 following infections with the pandemic influenza virus (Figure 6D). These data suggest that, similar to the other influenza viruses used in this study, NLRX1 also functions as an attenuator of the IFNβ-IL6 axis in response to influenza A/California/08/09 virus in human cells.
Figure 6. NLRX1 attenuates IL-6 and the IFNβ-STAT2-OAS1 axis in human cells in response to the 2009 pandemic influenza A/California/08/09 virus.
A–B. NLRX1 was efficiently knocked down by shRNA in 293T cells as shown by realtime PCR and western blot of three different shRNAs targeting NLRX1 (sh#1 to sh#3). C. Western blot for the influenza virus NS1 protein was used to confirm that cells were infected with the 2009 pandemic influenza A/California/08/09 virus at MOI=0.1. D. IL-6, IFNβ, STAT2 and OAS1A gene expression were assessed in 293T cells following the targeted knockdown of NLRX1 after infection with the 2009 pandemic influenza A/California/08/09 virus (MOI=0.1). Experiments were repeated 2 times in all three NLRX1 knockdown cell lines and scrambled control cell lines. *p < 0.05.
DISCUSSION
The observations based on the study of Nlrx1−/− mice solidify a role for NLRX1 as an in vivo checkpoint of IFN-I and IL-6 responses and additionally identifies a linkage between NLRX1, TRAF6 and NF-κB signaling. We have extended these findings to the influenza virus and identified an NLR that functions as an attenuator of host immune responses to this important human pathogen. Prior to this work, it was unclear if inhibitors of RIG-I-MAVS reduced antiviral immunity or attenuated hyperinflammatory responses in vivo. This report supports the hypothesis that NLRX1 functions as an in vivo cellular brake on both RIG-I-MAVS dependent and MAVS independent signaling pathways; thus, NLRX1 likely functions to prevent unwarranted or overwhelming host inflammatory responses to a variety of viruses. The finding that NLRX1 functions as a negative regulator of RIG-I-MAVS dependent signaling in response to viruses and dsRNA is consistent with the findings that Nlrx1−/− mice exhibited significant morbidity, lung injury, and inflammation following influenza virus challenge.
Several recent studies have described a role for NLR proteins in mediating innate immune responses to virus. Recent studies have shown that components of the NLRP3 inflammasome (NLRP3, Pycard and Caspase-1) function, in vivo, to mediate the innate immune response to influenza virus (Allen et al., 2009; Thomas et al., 2009). These studies demonstrate that NLRP3 mediates the posttranslational modification of proinflammatory cytokines (IL-1β and IL-18) in response to influenza virus infection. In addition to NLRP3, recent evidence has also suggested that the NLR, NOD2, also senses viral RNA and appears to interact with MAVS (Sabbah et al., 2009). The NOD2-MAVS interaction is dependent on the NBD and LRR domains of NOD2, which is similar to the mechanism of interaction that has been proposed for NLRX1-MAVS (Moore et al., 2008). Thus, it is interesting that the binding of NOD2 to MAVS appears to induce IFN-I production through IRF3 activation (Sabbah et al., 2009), whereas the binding of NLRX1 to MAVS attenuates this pathway (Moore et al., 2008).
One of the mechanisms by which NLRX1 attenuates MAVS activation is likely through direct competition with the RIG-I-MAVS interaction, shown previously in vitro (Moore et al., 2008). Our findings with Nlrx1−/− cells shows that MAVS and RIG-I constitutively interact with one another; however, this interaction is not sufficient to cause IFN-I induction. Instead, viral infection is needed to induce IFN-I, suggesting that in addition to the MAVS-RIG-I interaction, an additional viral-induced step is also required. The production of IFN-I following virus infection is dependent upon the RLH signaling pathway, which is tightly controlled through a complex series of both positive and negative regulators. One purpose of this regulation scheme is to protect the host from an overzealous and potentially detrimental innate immune response. A variety of molecules have recently been described as negative regulators of virus initiated RLH signaling and have been proposed to function through similar mechanisms as those described here for NLRX1(Moore et al., 2008). For instance, LGP2 is a RLH that is induced following viral infection and is functionally similar to RIG-I. However, LGP2 can function as a negative regulator by inhibiting RIG-I multimerization and competing with MAVS for binding with IKK-epsilon (Komuro and Horvath, 2006). Similar to the data shown here for NLRX1, LGP2 appears to have differential effects on RIG-I and MDA5 signaling. LGP2 has been shown to inhibit RIG-I signaling and augment MDA5 signaling in response to VSV and EMCV, respectively (Venkataraman et al., 2007). The Atg5-Atg12 conjugate is also a negative regulator of RLH signaling and has been shown to function through a mechanism that is similar to the one proposed for NLRX1. Atg5-Atg12 interferes with the CARD-CARD interactions between MAVS and RIG-I to inhibit IFN-I signaling (Jounai et al., 2007). Similar to NLRX1, in the absence of Atg5, MEFs stimulated with VSV and polyI:C were found to have significantly increased levels of IFN-I and IL-6 (Jounai et al., 2007).
A diverse range of molecules have been identified as negative regulators of inflammation. However, the concept that a subgroup of NLRs functions to attenuate inflammation is a relatively new finding and no previous work has analyzed this issue in vivo using gene deletion mice. One of the best characterized NLRs associated with a negative regulatory role is NLRP12. In vitro, NLRP12 has been shown to suppress inflammation by inhibiting components of the NF-κB pathway (Lich et al., 2007). Specifically, NLRP12 has been shown to associate with NF-κB-inducing kinase (NIK), which in turn facilitates proteasome mediated degradation of this kinase and inhibition of non-canonical NF-κB signaling (Lich et al., 2007). In addition to inhibiting non-canonical NF-κB signaling, which is typically associated with the activation of TNF family receptors, NLRP12 has also been shown to negatively regulate TLR signaling pathways by associating with activated forms of IRAK-1 (Williams et al., 2005). The association with IRAK-1 reduces the presence of hyperphosphorylated forms of IRAK-1. This results in the attenuated production of proinflammatory cytokines, including IL-6, following LPS stimulation (Williams et al., 2005). The NLR, NLRC5, has also been shown to be a negative regulator of NF-κB, AP-1 and IRF3 activation in response to LPS, polyI:C and VSV stimulation in overexpression and shRNA models (Benko et al., 2010; Cui et al., 2010). NLRC5 has been proposed to attenuate NF-κB signaling by interacting with IKKα and IKKβ to block phosphorylation and to inhibit RLH signaling by interacting with RIG-I and MDA5 (Cui et al., 2010). However, it should be noted that some of the negative regulatory functions associated with NLRC5 have not been observed by others (Kuenzel et al., 2010; Neerincx et al., 2010).
In addition to intersecting with the RIG-I-MAVS pathway, NLRX1 also attenuates NF-κB activation through its interaction with components of the TRAF6 pathway. TRAF6 is known to regulate a diverse range of innate immune signals and cytokines, including NF-κB and IL-6. Indeed, we demonstrate increased NF-κB signaling in Nlrx1−/− cells following LPS stimulation. This identifies TRAF6 as an interacting partner for NLRX1. The interaction of NLRX1 with TRAF6 was also found in a companion submission (Xiaojun, 2010). Together, our data are in contrast with an earlier report, which showed that overexpressed NLRX1 synergized ROS-mediated NF-κB activation (Tattoli et al., 2008). The data generated from our Nlrx1−/− mice shows that NLRX1 regulation of NF-κB occurs through a MAVS-independent pathway and is inhibitory, which is a finding that is also supported by the companion submission (Xiaojun, 2010). These findings suggest that NLRX1 attenuates IFN-I and NF-κB, either partially or wholly, through separate pathways in a cell type- and pathogen specific manner.
From a therapeutic standpoint, it has been proposed that targeted knockdown of negative regulators of MAVS could enhance antiviral immune responses. This report highlights that removal of just one of the many reported brakes of RIG-I-MAVS signaling can indeed result in elevated responses to influenza A virus. However, this comes at a price of increased systemic exposure to proinflammatory cytokines, such as IL-6. Further elucidation of the role of NLRX1 and other molecular brakes on antiviral signaling and the likely bifurcation of downstream transcriptional activation may allow for safe and targeted approaches to increasing host responses to viral infection. Considering the attenuating effects of NLRX1 on pulmonary inflammation shown here, this protein is likely to have a broad and complex role in the control of inflammatory diseases beyond viral infection.
EXPERIMENTAL PROCEDURES
Mouse Generation
The Nlrx1−/− mice were generated as illustrated in Figure 1A. Exons 4 and 5 (NBD domain) of Nlrx1 were replaced with a Neo cassette. The targeting vector was confirmed by restriction analysis and by sequencing. The linearized targeting vector was electroporated into IC1 C57BL/6 embryonic stem (ES) cells (InGenious targeting Labs, New York). Homologous recombinants were verified and injected into Balb/c blastocysts. The resulting chimeras were mated with female C57BL/6J mice and the resulting heterozygotes were crossed to produce homozygous Nlrx1−/− mice.
Ex vivo primary cell challenges
Primary fibroblasts (MEFs and ear fibroblasts), bone marrow derived macrophages and pDCs were isolated and cultured using standard procedures. To assess PAMP stimulation, cells were challenged with either polyI:C or 3′PPP-RNA (transfected with Lipofectamine 2000 (Sigma)), LPS (1μg/ml; Escherichia coli, sterile serotype 0111:B4(Sigma)), TNF-α or Pam3CSK4. Unless otherwise stated, all cells were treated with 30 MOI of each virus or 1 MOI of each bacterium, except Legionella, which was used at 0.01 MOI. For assessments of ROS, neutrophils and macrophages were purified from bone marrow using a discontinuous Percoll gradient. The leukocytes were resuspended and incubated with dihydrorhodamine-123 (DHR) (ANAspec) and the respective stimuli for 1hr. Cells were then washed and analyzed for fluorescence intensities using a CyAn ADP high resolution cytometer (DAKO). Alternatively, blood was collected and incubated with DHR and the respective stimuli, after red cell lysis, and fluorescence intensities were acquired using flow cytometry. In this case, high side scatter and high forward scatter cell populations were selected for analysis.
In vivo influenza A Virus Infection
Animals were anesthetized and challenged by either intratracheal (i.t.) administration of LPS (1 mg/ml) or intranasal (i.n.) administration of 6×104 PFU/ml of influenza virus A/PR/8/34 or A/PR/8/34mut39. Mice were observed and weight was assessed daily for up to 14 dpi. Mice were euthanized either 3 or 7 dpi, and serum was collected following cardiac puncture. Total cytokine levels were determined by ELISA. For histopathologic examination, lungs were fixed by inflation and immersion in buffered formalin and subjected to H&E staining. Lung sections were scored based on assessments of mononuclear and polymorphonuclear cell infiltration, perivascular and peribronchiolar cuffing, and estimates of the percent of lung involved with the disease, while blinded to genotype and treatment. Epithelial cell defects were determined based on the severity and extent of damage to the epithelial cell layer.
Expression Profiling
Mice were challenged by intrachael (i.t.) administration of 6×104 PFU/ml of influenza virus A/PR/8/34 and sacrificed 16–20 hrs post-infection. Total RNA was isolated from whole lungs. RNA from 3 different animals was pooled together prior to the cDNA reaction. Pooled samples were analyzed using RT2 Profiler PCR Arrays (SABiosystems) following the manufacturers protocols or by Taqman rtPCR analysis (ABI). Array results were confirmed on total RNA from individual mice that were challenged independently of those used for array profiling. The array data was analyzed utilizing Ingenuity Pathways Analysis software (Ingenuity Systems).
Lentivirus-based delivery of NLRX1-targeting shRNA
The NLRX1-targeting shRNA sequences and scrambled control sequences were cloned into the lentiviral vector FG12 according to standard molecular cloning protocols and NLRX1 expression was reduced in HEK293T cells. The shRNA hairpin sequences used were: sh-scrambled 5′-GCACATCGCTTCTCGGGATTCTCTTGAAATCCCGAG AAGCGATGTGC-3′, sh-NLRX1, 5′-GCACATCTTCCGTCGGGATTCTCTTGAAAT CCCGACGGAAGATGTGC-3′. The altered sequences in the control scrambled construct are underlined.
Influenza Virus A/Californina/08/09 Infection of NLRX1 Knockdown Cell Lines
NLRX1 knockdown cell lines (HEK293T) were challenged with influenza A/California/08/09 (H1N1) virus for 2 hrs at 37°C (MOI = 0.1). Following incubation, supernatant was replaced with fresh media and cells were incubated at 37°C. Cell free supernatants were harvested at select time points for viral titer. Cells were harvested and the RNA was extracted for rtPCR analysis.
Luciferase reporter assays
Detailed protocols for cell culture and luciferase assays have been previously published (Moore et al., 2008). Briefly, 293T cells were plated at 125,000/well in a 24 well plate. Cells were transfected using Fugene6 (Roche) with 150ng κB-luciferase reporter plasmid; 250ng myc-TRAF-3 or -6; 0, 100 or 250ng HA-NLRX1 and 250, 150 or 0ng of carrier pcDNA3.1 to bring the total to 900ng of DNA. To assess TNFα stimulation, cells were treated with 0, 5, 10, or 50ng of TNFα. After 24 hours, the cells were washed with PBS then lysed with 1x Reporter Lysis Buffer (Promega). ATP (gly-gly, KHPO4, MgSO4, EGTA, ATP, DTT) and luciferin (gly-gly, D-luciferin, DTT) buffers were added to the lysate and relative light units were measured on a luminometer.
Co-immunoprecipitation
Detailed protocols for cell culture, co-immunoprecipitation and antibodies used have been previously published (Moore et al., 2008). Briefly, 2×106 HEK293T cells were plated in a 10cm2 culture dish. Cells were transfected using Fugen6 (Roche) with 3ug HA-NLRX1 and 2ug of myc-TRAF-3 or myc-TRAF-6. After 24 hours, cells were washed with PBS, lysed in 1% TritonX-100. Lysates were centrifuged at 16,000rpm to remove nuclei. Myc-TRAFs were immunoprecipitated with myc-agarose beads overnight. The beads were washed three times and samples were run on NUPAGE Bis-Tris 4–12% gels. Blots were probed with HA-HRP antibody (Santa Cruz), myc-HRP (Santa Cruz) and GAPDH-HRP (Santa Cruz). Immunoblots for transfected NLRX1 were performed on immunoprecipitants of endogenous TRAF6. For endogenous immunoblots, cell extracts were pre-cleared with protein A/G agarose beads (Thermo Scientific) for 60min at 4°C and then incubated with 2μg of rabbit IgG, TRAF3, or TRAF6 primary antibodies overnight at 4°C. Protein A/G ultralink resin (Thermo Scientific) was then added to the samples and incubated for 2h at 4°C. Immunoprecipitated proteins were blotted for endogenous NLRX1 as described (Moore et al., 2008).
Statistical Analysis
Unless otherwise stated, all experiments were repeated a minimum of 3 times. All of the data are presented as the mean +/− the standard error of the mean (SEM). To assess complex data sets from both individual experiments and composite data, we utilized an Analysis Of Variance (ANOVA) followed by either the Tukey-Kramer HSD or Newman-Keuls Post Test for multiple comparisons. To assess single data points, we conducted a Student’s two-tailed t-test to determine significance. To assess survival, we generated survival curves using the product limit method of Kaplan and Meier. We utilized the log rank test to compare survival curves. A p-value < 0.05 was considered statistically significant for all data.
Supplementary Material
Supplemental Figure S1. Characterization of Nlrx1−/− mice. No overt morphological differences were observed between Nlrx1−/−, Nlrx1+/−, and wild type (WT) C57Bl/6 mice, up to 12 weeks of age and under specific pathogen free conditions. A. Nlrx1−/− mice produced normal litter sizes and mice were born within normal Mendelian and gender ratios. B. Whole spleens were collected from WT and Nlrx1−/− mice and the splenocyte composition was assessed by FACS. Splenic percentages of CD4+, CD8+, CD3+, CD19+ CD11c+, DX5/NK1.1+, and GR-1+ cells were similar between WT and Nlrx1−/− animals. C. Nlrx1 expression was assessed by RT-PCR, which revealed that Nlrx1 mRNA is widely expressed in a variety of mouse tissues. D. No significant differences were observed in Ifnβ transcription between WT and Nlrx1−/− MEFs under resting conditions. E–F. Basal and inducible levels of ROS production in macrophages and neutrophils from Nlrx1−/− and WT mice were assessed. No significant differences were detected following stimulation with various PAMPs (LPS, TNFα, polyI:C, Pam3Cys and fmlp). G. No significant differences were detected in IFN-β protein levels between WT or Nlrx1−/− plasmacytoid dendritic cells (pDCs) following challenges with R848, polyI:C, CpG A or CpG B. H. No significant differences were detected in Ifnβ transcript levels between the WT and Nlrx1−/− bone marrow derived macrophages in response challenges with either Legionella pneumophila or Listeria monocytogenes. I–J. No significant differences in IL-6 generation were detected in WT and Nlrx1−/− bone marrow derived macrophages stimulated with TNFα or Pam3CSK4. K. No significant differences were detected in pp65 levels between WT and Nlrx1−/− MEFS following stimulation with TNF-α. *p < 0.05;**p < 0.005. Related to Figures 1, 2 and 3.
Supplemental Figure S2. No difference in IL-1β or TNF-α was detected in lungs harvested from Nlrx1−/− or wild type mice. A–B. Lungs were harvested at specific time points and homogenized to assess local cytokine levels. No significant differences in IL-1β and TNF-α levels were detected between the Nlrx1−/− and WT mice over the course of the infection. All studies utilized 5–7 animals per group. *p < 0.05. Related to Figure 4.
Supplemental Figure S3. IL-6 receptor antagonism did not alleviate airway epithelial cell denuding in the Nlrx1−/− mice. A. Mice treated with the IL-6 receptor antagonist Actemra (αIL-6) demonstrated greater decreases in body temperature following influenza infection. B. Actemra reduced airway inflammation in both WT (not significant) and Nlrx1−/− mice. C–D. WT mice that were treated with Actemra demonstrated a significant increase in airway epithelial cell denuding compared to untreated WT mice. However, no significant differences were observed between Actemra treated and untreated Nlrx1−/− animals. All studies utilized 7 animals per group. *p < 0.05;**p < 0.005. Related to Figure 4.
Supplemental Figure S4. Influenza virus infection results in the up-regulation of a variety of genes associated with the in vivo type I interferon response. A. Schematic illustrating the experimental design for expression profiling. Three mice from each genotype and condition were challenged intratracheally (i.t.) with 6×104 PFU/ml of influenza virus A/PR/8/34. The total lung RNA from each mouse was pooled and profiled for gene expression. B. Table of genes that were overexpressed in lungs isolated from mock treated Nlrx1−/− mice compared to mock treated WT mice. Only 3 genes were identified as being upregulated in the mock treated Nlrx1−/− mice. C. Table of genes that were overexpressed in WT mice compared to mock treated WT mice. Influenza virus significantly up-regulates a diverse assortment of genes associated with the type I interferon response in WT mice. Related to Figure 5.
Supplemental Figure S5. Inoculation with the influenza NS1mut39 virus results in increased histopathology in WT mice. WT and Nlrx1−/− mice were challenged with 6X104 PFU/ml of either a WT influenza A/PR/8/34 virus (PR8 control virus) or the recombinant influenza NS1mut39 virus. A. Survival was assessed daily for 15 dpi. No significant differences in mortality were observed between the WT and Nlrx1−/− mice. B. Lungs were harvested 3 dpi and homogenized to assess influenza viral titers, which were determined by standard plaque assay. The NS1mut39 virus showed a small, but significant, decrease in viral load, compared to the WT control virus in both sets of animals. C. Lungs were harvested 3 dpi and fixed by inflation and immersion in buffered PFA. Sections through the main bronchiole of the left lobe revealed increased airway epithelial cell denuding and small airway obstruction throughout the lungs of mice that were challenged with the NS1mut39 virus. No significant differences in lung histopathology were observed between Nlrx1−/− mice treated with WT virus and the NS1mut virus. D. Simplified schematic illustrating the synergistic effect of the viral NS1 protein and host NLRX1 protein on the IFN-I response. Related to Figure 5.
HIGHLIGHTS.
Nlrx1−/− mice were generated and characterized.
NLRX1 attenuates IFN induction by preventing the interaction between RIG-I and MAVS.
NLRX1 functions as a negative regulator of IFN-I and IL-6 during influenza infection.
NLRX1 attenuates inflammation by intersecting the TRAF6 pathway to affect NF-κB.
Acknowledgments
The authors would like to thank Dr. Wendy S. Barclay (Imperial College London) for providing the influenza A/PR/8/34mut39 virus. We would also like to thank Dr. Jacqueline M. Katz and Nancy J. Cox of the Influenza Division for their support. This work is supported by: U54-AI057157 (SERCEB); U19-AI067798, U19-AI077437 (J.P.Y. Ting), F32-AI-082895-01; T32-AR007416 and T32-CA009156 (I.C. Allen).
Footnotes
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency or Centers for Disease Control and Prevention
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdul-Sater AA, Said-Sadier N, Lam VM, Singh B, Pettengill MA, Soares F, Tattoli I, Lipinski S, Girardin SE, Rosenstiel P, et al. Enhancement of reactive oxygenspecies production and chlamydial infection by the mitochondrial Nod like family member NLRX1. J Biol Chem. 2010;285:41637–41645. doi: 10.1074/jbc.M110.137885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Roberts PC, Kipperman T, Bhalla KN, Compans RW, Archer DR, Barber GN. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/Caspase-8 death signaling pathway. J Virol. 2000;74:1513–1523. doi: 10.1128/jvi.74.3.1513-1523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185:1681–1691. doi: 10.4049/jimmunol.0903900. [DOI] [PubMed] [Google Scholar]
- Buchweitz JP, Harkema JR, Kaminski NE. Time-dependent airway epithelial and inflammatory cell responses induced by influenza virus A/PR/8/34 in C57BL/6 mice. Toxicol Pathol. 2007;35:424–435. doi: 10.1080/01926230701302558. [DOI] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Chen LM, Zeng H, Gomez JA, Plowden J, Fujita T, Katz JM, Donis RO, Sambhara S. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol. 2007;36:263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- Hayman A, Comely S, Lackenby A, Hartgroves LC, Goodbourn S, McCauley JW, Barclay WS. NS1 proteins of avian influenza A viruses can act as antagonists of the human alpha/beta interferon response. J Virol. 2007;81:2318–2327. doi: 10.1128/JVI.01856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Song T, Wei C, Ni C, Zheng Z, Xu Q, Ma H, Li L, Zhang Y, He X, et al. Negative regulation of MAVS-mediated innate immune response by PSMA7. J Immunol. 2009;183:4241–4248. doi: 10.4049/jimmunol.0901646. [DOI] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenzel S, Till A, Winkler M, Hasler R, Lipinski S, Jung S, Grotzinger J, Fickenscher H, Schreiber S, Rosenstiel P. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- Le Goffic R, Pothlichet J, Vitour D, Fujita T, Meurs E, Chignard M, Si-Tahar M. Cutting Edge: Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol. 2007;178:3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- Lechner J, Malloth N, Seppi T, Beer B, Jennings P, Pfaller W. IFN-alpha induces barrier destabilization and apoptosis in renal proximal tubular epithelium. Am J Physiol Cell Physiol. 2008;294:C153–160. doi: 10.1152/ajpcell.00120.2007. [DOI] [PubMed] [Google Scholar]
- Lich JD, Williams KL, Moore CB, Arthur JC, Davis BK, Taxman DJ, Ting JP. Monarch-1 suppresses non-canonical NF-kappaB activation and p52 dependent chemokine expression in monocytes. J Immunol. 2007;178:1256–1260. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Neerincx A, Lautz K, Menning M, Kremmer E, Zigrino P, Hosel M, Buning H, Schwarzenbacher R, Kufer TA. A role for the human nucleotide binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J Biol Chem. 2010;285:26223–26232. doi: 10.1074/jbc.M110.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′ phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, Arnoult D, Girardin SE. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PG, Dash P, Aldridge JR, Jr, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- Williams KL, Lich JD, Duncan JA, Reed W, Rallabhandi P, Moore C, Kurtz S, Coffield VM, Accavitti-Loper MA, Su L, et al. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem. 2005;280:39914–39924. doi: 10.1074/jbc.M502820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaojun X, Wang HY, Cui J, Zhu L, Matsueda S, Wang Q, Yang X, Hong J, Songyang Z, Chen ZJ, Wang R. NLRX1 negatively regulates NF-kB signaling and immune responses by targeting TRAF6 and IKK. Immunity. 2010 doi: 10.1016/j.immuni.2011.02.022. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Xiao N, Liu F, Ren H, Gu J. Inhibition of RIG-I and MDA5-dependent antiviral response by gC1qR at mitochondria. Proc Natl Acad Sci U S A. 2009;106:1530–1535. doi: 10.1073/pnas.0811029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Characterization of Nlrx1−/− mice. No overt morphological differences were observed between Nlrx1−/−, Nlrx1+/−, and wild type (WT) C57Bl/6 mice, up to 12 weeks of age and under specific pathogen free conditions. A. Nlrx1−/− mice produced normal litter sizes and mice were born within normal Mendelian and gender ratios. B. Whole spleens were collected from WT and Nlrx1−/− mice and the splenocyte composition was assessed by FACS. Splenic percentages of CD4+, CD8+, CD3+, CD19+ CD11c+, DX5/NK1.1+, and GR-1+ cells were similar between WT and Nlrx1−/− animals. C. Nlrx1 expression was assessed by RT-PCR, which revealed that Nlrx1 mRNA is widely expressed in a variety of mouse tissues. D. No significant differences were observed in Ifnβ transcription between WT and Nlrx1−/− MEFs under resting conditions. E–F. Basal and inducible levels of ROS production in macrophages and neutrophils from Nlrx1−/− and WT mice were assessed. No significant differences were detected following stimulation with various PAMPs (LPS, TNFα, polyI:C, Pam3Cys and fmlp). G. No significant differences were detected in IFN-β protein levels between WT or Nlrx1−/− plasmacytoid dendritic cells (pDCs) following challenges with R848, polyI:C, CpG A or CpG B. H. No significant differences were detected in Ifnβ transcript levels between the WT and Nlrx1−/− bone marrow derived macrophages in response challenges with either Legionella pneumophila or Listeria monocytogenes. I–J. No significant differences in IL-6 generation were detected in WT and Nlrx1−/− bone marrow derived macrophages stimulated with TNFα or Pam3CSK4. K. No significant differences were detected in pp65 levels between WT and Nlrx1−/− MEFS following stimulation with TNF-α. *p < 0.05;**p < 0.005. Related to Figures 1, 2 and 3.
Supplemental Figure S2. No difference in IL-1β or TNF-α was detected in lungs harvested from Nlrx1−/− or wild type mice. A–B. Lungs were harvested at specific time points and homogenized to assess local cytokine levels. No significant differences in IL-1β and TNF-α levels were detected between the Nlrx1−/− and WT mice over the course of the infection. All studies utilized 5–7 animals per group. *p < 0.05. Related to Figure 4.
Supplemental Figure S3. IL-6 receptor antagonism did not alleviate airway epithelial cell denuding in the Nlrx1−/− mice. A. Mice treated with the IL-6 receptor antagonist Actemra (αIL-6) demonstrated greater decreases in body temperature following influenza infection. B. Actemra reduced airway inflammation in both WT (not significant) and Nlrx1−/− mice. C–D. WT mice that were treated with Actemra demonstrated a significant increase in airway epithelial cell denuding compared to untreated WT mice. However, no significant differences were observed between Actemra treated and untreated Nlrx1−/− animals. All studies utilized 7 animals per group. *p < 0.05;**p < 0.005. Related to Figure 4.
Supplemental Figure S4. Influenza virus infection results in the up-regulation of a variety of genes associated with the in vivo type I interferon response. A. Schematic illustrating the experimental design for expression profiling. Three mice from each genotype and condition were challenged intratracheally (i.t.) with 6×104 PFU/ml of influenza virus A/PR/8/34. The total lung RNA from each mouse was pooled and profiled for gene expression. B. Table of genes that were overexpressed in lungs isolated from mock treated Nlrx1−/− mice compared to mock treated WT mice. Only 3 genes were identified as being upregulated in the mock treated Nlrx1−/− mice. C. Table of genes that were overexpressed in WT mice compared to mock treated WT mice. Influenza virus significantly up-regulates a diverse assortment of genes associated with the type I interferon response in WT mice. Related to Figure 5.
Supplemental Figure S5. Inoculation with the influenza NS1mut39 virus results in increased histopathology in WT mice. WT and Nlrx1−/− mice were challenged with 6X104 PFU/ml of either a WT influenza A/PR/8/34 virus (PR8 control virus) or the recombinant influenza NS1mut39 virus. A. Survival was assessed daily for 15 dpi. No significant differences in mortality were observed between the WT and Nlrx1−/− mice. B. Lungs were harvested 3 dpi and homogenized to assess influenza viral titers, which were determined by standard plaque assay. The NS1mut39 virus showed a small, but significant, decrease in viral load, compared to the WT control virus in both sets of animals. C. Lungs were harvested 3 dpi and fixed by inflation and immersion in buffered PFA. Sections through the main bronchiole of the left lobe revealed increased airway epithelial cell denuding and small airway obstruction throughout the lungs of mice that were challenged with the NS1mut39 virus. No significant differences in lung histopathology were observed between Nlrx1−/− mice treated with WT virus and the NS1mut virus. D. Simplified schematic illustrating the synergistic effect of the viral NS1 protein and host NLRX1 protein on the IFN-I response. Related to Figure 5.