Abstract
Candida albicans and Candida dubliniensis are highly related pathogenic yeast species. However, C. albicans is far more prevalent in human infection and has been shown to be more pathogenic in a wide range of infection models. Comparison of the genomes of the two species has revealed that they are very similar although there are some significant differences, largely due to the expansion of virulence-related gene families (e.g., ALS and SAP) in C. albicans, and increased levels of pseudogenisation in C. dubliniensis. Comparative global gene expression analyses have also been used to investigate differences in the ability of the two species to tolerate environmental stress and to produce hyphae, two traits that are likely to play a role in the lower virulence of C. dubliniensis. Taken together, these data suggest that C. dubliniensis is in the process of undergoing reductive evolution and may have become adapted for growth in a specialized anatomic niche.
1. Introduction
Fungi are an important cause of human infection, and yeast species of the genus Candida are the most pathogenic fungi. While most Candida species are found in the environment, approximately a dozen or so are associated with colonization and infection of humans [1]. Candida species are common commensals of the oral cavity, intestinal tract and vagina, with newborns being colonized soon after birth. While these species are innocuous in most individuals, under certain circumstances they can opportunistically overgrow and cause a variety of diseases [2]. These diseases range from superficial infections of the vaginal and oral mucosae, to life-threatening systemic infections that can spread via the bloodstream to organs throughout the body. The risk factors for candidal vaginitis are poorly understood; however, other candidal infections are largely the result of host-related defects. These include depletion of CD4 T cells in HIV-infected individuals, which predisposes to oropharyngeal candidosis, or neutropenia and intestinal surgery, both of which are significant risk factors for systemic infection [1–3].
Candida albicans is widely recognized as being the most pathogenic yeast species and in the majority of epidemiological studies has been found to be the most common cause of superficial and systemic infections. Other species, such as Candida glabrata, Candida parapsilosis, and Candida tropicalis have also been associated with most forms of candidiasis and the relative distribution of each species can vary depending on geographic location, patient cohort, and previous exposure to antifungal drugs [2, 4]. In 1995, a new Candida species was identified in HIV-infected individuals with oropharyngeal candidosis in Dublin, Ireland [5]. This species, which was subsequently named Candida dubliniensis, is very closely related to C. albicans with which it shares many phenotypic properties, including the ability to produce hyphae and chlamydospores, traits previously specifically associated only with C. albicans [6–8]. Phylogenetic studies indicate that C. dubliniensis is the species that is most closely related to C. albicans, and it is often quite difficult to discriminate between the two species in clinical samples [9, 10]. Indeed it was only when DNA fingerprinting techniques were applied to the large-scale analysis of C. albicans populations in epidemiological studies that the first isolates of C. dubliniensis were originally identified [5]. Surprisingly, despite the close phylogenetic relationship of the two species epidemiological data show that C. albicans is far more prevalent than C. dubliniensis. In particular, in most analyses of systemic infection, C. albicans is found in >50% of cases, while if it is identified at all, C. dubliniensis has only been found in at most 2-3% of cases [11–13]. This apparent discrepancy between the ability of the two species to cause infection is also reflected in data obtained from comparative studies in a wide range of infection models (e.g., systemic and mucosal) which clearly show that C. albicans is significantly more pathogenic than C. dubliniensis [10, 14–18].
The identification of virulence-associated factors in Candida species is complicated by the fact that they are opportunistic pathogens that usually exist in harmony with the human host as part of the commensal flora and only cause infection when host deficiencies permit. Since it is by far the most pathogenic Candida species, C. albicans is the best-studied member of the genus in terms of pathogenesis. The most commonly cited C. albicans virulence factors include adhesins (e.g., Hwp1 [19] and the Als family [20]), extracellular enzymes (e.g., the secreted aspartyl proteinase (Sap) family [21] and phospholipases [22]), and most importantly of all, the ability to alternate between unicellular yeast and filamentous hyphal forms of growth [23]. Both morphological forms have been shown to be essential for virulence. Hyphae have been proposed to play a major role in adhesion, invasion, and biofilm formation while yeast cells are likely to be important for dissemination and initial colonization of host surfaces [24]. Comparative phenotypic analysis of C. albicans and C. dubliniensis has suggested that in vitro isolates of C. dubliniensis exhibit higher levels of proteinase activity, are more adherent to buccal epithelial cells, and undergo phenotypic switching at a higher rate than C. albicans [10, 25–27]. In addition, as described earlier, C. dubliniensis is the only Candida species, other than C. albicans that is able to produce hyphae [5, 6]. Given the close relationship between the two species and the fact that they are so alike phenotypically, at first glance, it is difficult to understand why there is such disparity in the capacity of C. albicans and C. dubliniensis to colonise and cause disease in humans. This short review appraises recent findings that help to clarify this conundrum and explain how C. albicans appears to have evolved to be a better commensal and opportunistic pathogen than C. dubliniensis.
2. Comparative Genomic Analysis of C. albicans and C. dubliniensis
The C. albicans genome sequence was first published in 2004 [28], with improved annotation and analysis subsequently reported in 2005 [29] and 2007 [30]. In an early attempt to identify genomic differences that might serve to explain the disparity in the virulence of C. albicans and C. dubliniensis, Moran et al. cohybridized genomic DNA from each species to C. albicans whole genome microarrays to identify genes that are only present in C. albicans [31]. This relatively crude experiment suggested that there are 247 (approx. 4%) C. albicans genes that are either absent or highly (i.e., >60%) divergent in the C. dubliniensis genome. Interestingly, several genes strongly associated with C. albicans virulence are included in the list of absent/divergent genes. In 2009, in order to further investigate the genetic differences between the two species the Wellcome Trust Sanger Institute sequenced the entire C. dubliniensis genome [32]. Comparison of the two genome sequences revealed that, despite major karyotypic differences, the genomes of the two species are remarkably similar with 96.3% of genes exhibiting >80% identity, while 98% of genes are syntenic, thus, confirming the very close phylogenetic relationship and the relatively recent divergence of the two species (estimated to have occurred approx. 20 million years ago [33]). When transposable elements were discounted, comparison of the two genome sequences revealed that there are 29 C. dubliniensis-specific genes and 168 C. albicans-specific genes. The majority of the differences observed between the two species can be accounted for by the expansion of gene families in C. albicans, many of which have been previously associated with virulence. In particular, genes missing from the C. dubliniensis genome include those encoding hypha-specific virulence factors, such as the cell surface proteins Hyr1 and Als3 and two members of the secreted aspartyl proteinase family (i.e., Sap5 and Sap6), while the gene encoding the well-characterized epithelial adhesin Hwp1 is highly divergent [31, 32]. Hyr1 has been shown to confer resistance to neutrophil killing activity [34] and, along with Hwp1, has been shown recently to play an important role in oral mucosal biofilm formation [35]. Als3 has been shown to play an important role in adhesion to host cells and has been shown to have invasin-like [36] and iron-sequestering [37] activity, while the Saps are well-known virulence factors [21]. The biggest difference in gene family size between the two species is the TeLOmere-associated (TLO) family which is comprised of 14 genes in C. albicans, but only two genes in C. dubliniensis. Sequence comparisons suggest that the TLO genes encode transcriptional regulators, and preliminary analysis of the phenotype of C. dubliniensis Δtlo mutants suggests that these genes may play a role in the control of hypha formation [32]. In addition to these differences, a range of genes appear to be in the process of being lost by C. dubliniensis. There are 78 C. dubliniensis pseudogenes with intact positional orthologs in C. albicans, including genes identified as filamentous growth regulators (FGR) in haploinsufficiency studies [38]. These findings suggest that C. dubliniensis is undergoing a process of reductive evolution leading to the loss of genes that have been associated with C. albicans virulence. Interestingly, many of these genes are only expressed by the hyphal form of growth and are likely to play a prominent role in host-pathogen interaction.
One of the most prominent phenotypic differences between C. albicans and C. dubliniensis is their different capacity to tolerate environmental stress, with the former being far more tolerant of thermal, osmotic, and oxidative stress [5, 14, 39, 40]. Indeed, comparative growth at 45°C is commonly used as a simple diagnostic test to discriminate between the two species [41]. Comparative transcriptional profiling analysis revealed that although the two species express similar core stress responses, C. dubliniensis mounts a more robust response to thermal stress and a very poor transcriptional response to oxidative and osmotic stress [39]. Forward genetic screens using a C. albicans library to try and identify genes that might increase the tolerance of C. dubliniensis to environmental stress failed to identify any single gene that could complement oxidative and thermal sensitivity, suggesting that these are likely to be polygenic traits. However, the C. albicans ENA21 gene, which encodes a sodium efflux pump, was found to increase the salt tolerance of C. dubliniensis [39]. Since the C. dubliniensis ortholog of this gene appears to be functional but not upregulated in response to the presence of salt, it is likely that the differential salt stress susceptibility of the two species is due to differences in stress-related transcriptional regulatory pathways.
3. Comparative Analysis of Hypha Formation by C. albicans and C. dubliniensis
One of the most important and best-studied virulence factors of C. albicans is its ability to switch between yeast and filamentous growth forms (i.e., dimorphism), a trait also shared by C. dubliniensis [5]. However, although C. dubliniensis is capable of producing germ tubes and true hyphae, it does so far less efficiently than C. albicans, both in vivo and under a wide range of in vitro conditions [16, 42, 43]. Given the perceived importance of dimorphism in C. albicans virulence, we have previously suggested that the lower virulence of C. dubliniensis may, at least in part, be related to its relatively poor ability to switch between yeast and hyphal forms [16]. Evidence in support of this was obtained from murine systemic infection model studies [14, 15] and the neonatal orogastric infection model [16]. In the latter, stomach and kidney samples in infected animals contained only C. dubliniensis yeast cells, while C. albicans cells were found in both the yeast and hyphal forms [16].
We have used the RHE model of superficial infection [44] to compare the invasive potential of both species (Figure 1). In particular, in this model, C. albicans grows as both yeast and hyphae and invades the tissue causing major damage. In contrast, C. dubliniensis grows exclusively in the yeast form in this model, therefore, causing relatively limited tissue invasion and damage [16, 17]. In order to investigate why the two species differ so markedly in virulence in this model and in order to identify novel virulence-associated genes; Spiering et al. compared their global gene expression profiles during the early stages of RHE infection [17]. Both species showed similar expression profiles for ribosomal and general metabolic genes, however, unsurprisingly, C. albicans showed increased expression of hypha-specific virulence genes (e.g., ECE1, HWP1, HYR1, and ALS3) within 30 minutes of infection. In contrast, C. dubliniensis showed a far less robust transcriptional response and no expression of hypha-specific genes. In addition, several genes with unknown function were found to be specifically upregulated in C. albicans that are absent from or very divergent in the C. dubliniensis genome. One of these genes, named SFL2 due to its sequence similarity to the transcription factor-encoding gene SFL1, encodes a putative DNA-binding heat shock factor protein. When the gene was deleted in C. albicans, it resulted in the failure to produce hyphae under a wide range of growth conditions, including the RHE infection model. Interestingly, the Δsfl2 mutation had no effect on survival in the murine systemic infection model, although histological analysis revealed that the kidneys of infected mice were infected only with yeast cells, while the kidneys of mice infected with the wild-type parental strains contained both yeast and predominantly hyphal cells [17]. In a subsequent study, it has been shown that the Δsfl2 mutant exhibits reduced virulence in a mouse model of gastrointestinal infection, suggesting that Sfl2 is required for the penetration of the gut wall and subsequent dissemination throughout the body [45]. The C. dublinensis ortholog of SFL2 is only 50% identical and is not expressed under the same conditions as the C. albicans gene, therefore it is possible that the divergence of this gene and its apparent lack of expression may be partly responsible for its lower virulence.
Figure 1.
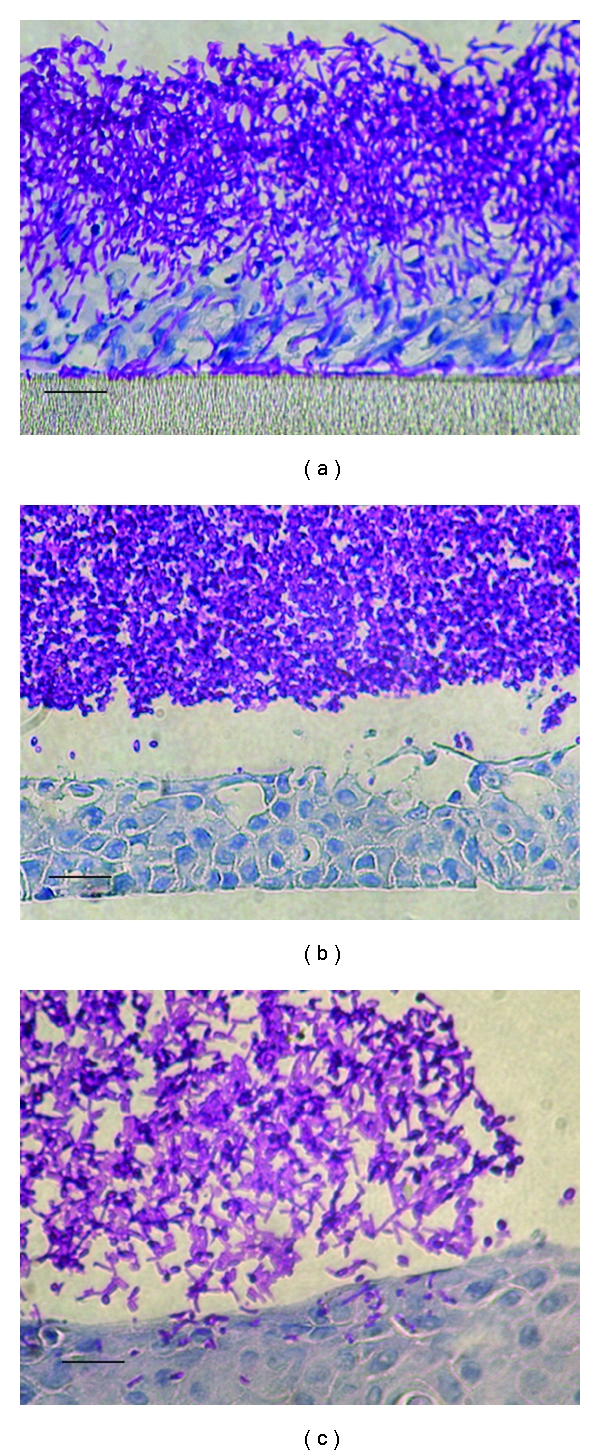
Photomicrograph of C. albicans SC5314 and C. dubliniensis CD36 infecting oral reconstituted human epithelial (RHE) tissue. (a) C. albicans originally grown in nutrient-rich YPD, note the presence of hyphae and extensive tissue invasion and damage; (b) C. dubliniensis originally grown in YPD, note the absence of hyphae and the limited level of invasion and tissue damage; (c) C. dubliniensis originally grown in Lee's medium, note the increased level of filamentation and invasion. Scale bars, approximately 25 μm.
Recent studies by our group have been directed towards investigating the molecular basis for differences in the signaling pathways responsible for filamentation in the two species. Comparative genome analysis suggests that orthologs of the known components of the major C. albicans morphogenetic pathways (e.g., Cph1-mediated MAPK and the Efg1-mediated Ras1-cAMP pathways) are highly conserved in C. dubliniensis, so the reduced capacity of C. dubliniensis to produce hyphae and express hypha-specific genes such as SFL2 is unlikely to be due to the absence of regulators involved in these pathways. Forced stimulation of the Ras1-cAMP pathway with a hyperactive RAS1G113V allele did not result in increased true hypha formation in C. dubliniensis, suggesting strong repression of the RAS1-cAMP pathway itself or downstream regulators [43]. One of the most important transcriptional regulators involved in the control of morphogenesis in C. albicans is Nrg1, which Staib and Morschhäuser. showed that it is differentially expressed by C. dubliniensis when grown on media such as Staib agar [46]. In C. albicans this protein targets the negative regulator Tup1 to specific sequences in the promoters of genes involved in hypha formation. NRG1 expression is rapidly downregulated in C. albicans cells incubated under hypha-inducing conditions, including when cells are phagocytosed by murine macrophages, which results in germination and escape from the phagocytes. However, in C. dubliniensis, NRG1 expression remains high under these conditions, preventing hypha formation and causing cells to remain in the yeast phase, which in the murine macrophage model results in failure to escape from the phagocytes and the death of the fungus [43]. Deletion of the NRG1 gene in C. dubliniensis resulted in an increase in the rate of hypha and particularly pseudohypha formation, which in turn led to increased survival when exposed to murine macrophages as well as increased virulence in the reconstituted human epithelial (RHE) cell model of oral candidosis. Surprisingly, the C. dubliniensis Δnrg1 mutant was no more virulent than its parent strain in the murine systemic infection model and formed mainly pseudohyphae in infected kidneys, suggesting an additional level of repression preventing true hypha formation in vivo [43].
Recent investigations by O'Connor et al. have suggested that repression of filamentation in C. dubliniensis is mediated by nutrients. In order to improve our understanding of how environmental signals trigger these pathways O'Connor et al. investigated the effects of nutrient availability on the rate of hypha formation [42]. One of the most common incubation conditions for inducing hypha formation in C. albicans is incubation in the nutrient-rich medium YPD supplemented with 10% (vol/vol) fetal calf serum at 37°C. Under these conditions >80% of C. albicans cells produced germ tubes/filaments within two hours, in contrast only ~20% of C. dubliniensis cells were observed to produce hyphae under the same induction conditions. However, when C. dubliniensis cells were incubated in water supplemented with 10% (vol/vol) fetal calf serum (WS) at 37°C, the level of hypha producing cells increased to 90% (i.e., similar to the level of hypha formation by C. albicans), suggesting that a nutrient-rich environment, in particular the presence of complex mixtures of peptides, suppressed hypha formation in this species. This was confirmed when the addition of peptone and peptone and glucose was found to significantly reduce the levels of hyphae, suggesting that nutrient starvation is a prerequisite for hypha formation by C. dubliniensis [42]. These morphological changes were coupled with changes in the expression of genes encoding key transcriptional regulators, such as NRG1 and UME6, which were significantly altered in WS, with the former downregulated by 70% and the latter upregulated 30-fold. Overexpression of the UME6 gene (which encodes a protein required for hyphal extension), using a doxycycline-inducible promoter led to C. dubliniensis cells being able to produce true hyphae, even in nutrient-rich media such as YPD. Similarly, preculture of C. dubliniensis cells in nutrient poor media, such as Lee's medium, pH 4.5, prior to the induction of hyphae in YPDS also resulted in a transient ability of C. dubliniensis cells to produce hyphae which increased the ability of these cells to adhere to and invade epithelial tissue in the RHE model (see Figure 1) and increased survival in the murine macrophage infection model [42].
These data suggest that factors controlling UME6 expression in C. dubliniensis are repressed by the presence of nutrients, and unlike C. albicans, this repression cannot be lifted by a shift to alkaline pH, which occurs when serum is added to the medium. UME6 is likely to be regulated by Efg1 and Eed1 and is therefore under the control of the Ras1-cAMP pathway. Few studies have investigated how nutrients regulate this pathway in C. albicans or C. dubliniensis. Preliminary investigations in our laboratory have shown that rapamycin, an inhibitor of the nutrient sensing kinase Tor1 (a kinase that plays a central role in the control of responses to nutrient availability [47]), can stimulate transient hypha formation in C. dubliniensis in nutrient-rich YPD serum [48]. This derepression of hypha formation in the presence of nutrients is concomitant with a reduction of NRG1 and an increase in UME6 expression. These data suggest that differences in Tor1 activity may play a role in the differential ability of C. albicans and C. dubliniensis to form hyphae. The molecular basis for the difference in the activity of Tor1 in the two species is currently under investigation.
4. Conclusions
Candidal pathogenicity involves the complex interplay of a wide range of virulence-associated factors. Comparative analysis of C. albicans and C. dubliniensis genomic and transcriptomic data has revealed that the reasons for the differences in the capacity of these two species to cause disease are also complex and are not due to a simple defect in C. dubliniensis. Instead these studies have revealed genetic differences in the two species, which, at least in part, may explain the differences in their capacity to tolerate stress and to filament. It is clear that the C. dubliniensis genome is missing important virulence genes (e.g., ALS3 and HYR1), is in the process of losing others (e.g., the FGR genes), has failed to expand certain gene families (e.g., the SAP and TLO families), and has undergone some degree of transcriptional rewiring (e.g., the Tor pathway and Sfl2). All of these differences suggest that the main discrepancy between these two closely related species relates to differences in hypha formation and the expression of hypha-specific products (summarized in Table 1). We propose that C. dubliniensis is in the process of undergoing reductive evolution, whereby its genetic repertoire is diminishing in comparison with C. albicans and their common ancestor. One of the main phenotypic manifestations of this is the narrowing of environmental conditions permissive for hypha formation, perhaps as a result of specialization for survival in a specific (as yet unidentified) anatomic niche where hyphae are not required for colonization or growth. By further investigating the molecular basis for the differences between C. albicans and C. dubliniensis, we hope to improve our understanding of candidal virulence, in particular the relative contribution of hyphae and hypha-specific proteins to the pathogenesis of candidal infections.
Table 1.
Comparison of C. albicans and C. dubliniensis.
| C. albicans | C. dubliniensis | References | |
|---|---|---|---|
| Growth and morphology | |||
| Growth at ≥42°C | Yes | No | [39, 41] |
| Growth in high salt media | Yes | No | [39, 40] |
| Hypha formation in YPD + serum | Yes | Poor | [16, 42] |
| Hypha formation in water + serum | Yes | Yes | [42] |
| RHE infection model | yeasts and hyphae | yeasts only | [17] |
|
| |||
| Genome | |||
| Chromosome number | 8 | 9–11 chromosome-sized fragments | [7] |
| No. of species-specific genes | 168 | 29 | [32] |
| ALS3 | Present | Absent | [32] |
| HYR1 | Present | Absent | [32] |
| SAP4, 5 and 6 | All three genes | One gene | [32] |
| HWP1 | Present | Divergent | [32] |
| TLO family | 14 genes | 2 genes | [32] |
Acknowledgments
The authors would like to acknowledge support from the Dublin Dental University Hospital, the Health Research Board (Grant HRA/2009/3), and Science Foundation Ireland (Grant SFI 11 RFP.1/GEN/3042).
References
- 1.Calderone RA, editor. Candida and Candidiasis. Washington, DC, USA: ASM Press; 2002. [Google Scholar]
- 2.Ruhnke M. Epidemiology of Candida albicans infections and role of non-Candida albicans yeasts. Current Drug Targets. 2006;7(4):495–504. doi: 10.2174/138945006776359421. [DOI] [PubMed] [Google Scholar]
- 3.Coleman DC, Bennett DE, Sullivan DJ, et al. Oral Candida in HIV infection and AIDS: new perspectives/new approaches. Critical Reviews in Microbiology. 1993;19(2):61–82. doi: 10.3109/10408419309113523. [DOI] [PubMed] [Google Scholar]
- 4.Krcmery V, Barnes AJ. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. Journal of Hospital Infection. 2002;50(4):243–260. doi: 10.1053/jhin.2001.1151. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141(7):1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. Journal of Clinical Microbiology. 1998;36(2):329–334. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan DJ, Moran GP, Coleman DC. Candida dubliniensis: ten years on. FEMS Microbiology Letters. 2005;253(1):9–17. doi: 10.1016/j.femsle.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Citiulo F, Moran GP, Coleman DC, Sullivan DJ. Purification and germination of Candida albicans and Candida dubliniensis chlamydospores cultured in liquid media. FEMS Yeast Research. 2009;9(7):1051–1060. doi: 10.1111/j.1567-1364.2009.00533.x. [DOI] [PubMed] [Google Scholar]
- 9.McManus BA, Coleman DC, Moran G, et al. Multilocus sequence typing reveals that the population structure of Candida dubliniensis is significantly less divergent than that of Candida albicans . Journal of Clinical Microbiology. 2008;46(2):652–664. doi: 10.1128/JCM.01574-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilfillan GD, Sullivan DJ, Haynes K, Parkinson T, Coleman DC, Gow NAR. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology. 1998;144(4):829–838. doi: 10.1099/00221287-144-4-829. [DOI] [PubMed] [Google Scholar]
- 11.Kibbler CC, Seaton S, Barnes RA, et al. Management and outcome of bloodstream infections due to Candida species in England and Wales. Journal of Hospital Infection. 2003;54(1):18–24. doi: 10.1016/s0195-6701(03)00085-9. [DOI] [PubMed] [Google Scholar]
- 12.Odds FC, Hanson MF, Davidson AD, et al. One year prospective survey of Candida bloodstream infections in Scotland. Journal of Medical Microbiology. 2007;56(8):1066–1075. doi: 10.1099/jmm.0.47239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clinical Microbiology Reviews. 2007;20(1):133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilela MMS, Kamei K, Sano A, et al. Pathogenicity and virulence of Candida dubliniensis: comparison with C. albicans . Medical Mycology. 2002;40(3):249–257. doi: 10.1080/mmy.40.3.249.257. [DOI] [PubMed] [Google Scholar]
- 15.Ásmundsdóttir LR, Erlendsdóttir H, Agnarsson BA, Gottfredsson M. The importance of strain variation in virulence of Candida dubliniensis and Candida albicans: results of a blinded histopathological study of invasive candidiasis. Clinical Microbiology and Infection. 2009;15(6):576–585. doi: 10.1111/j.1469-0691.2009.02840.x. [DOI] [PubMed] [Google Scholar]
- 16.Stokes C, Moran GP, Spiering MJ, Cole GT, Coleman DC, Sullivan DJ. Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans . Fungal Genetics and Biology. 2007;44(9):920–931. doi: 10.1016/j.fgb.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Spiering MJ, Moran GP, Chauvel M, et al. Comparative transcript profiling of Candida albicans and Candida dubliniensis identifies SFl2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryotic Cell. 2010;9(2):251–265. doi: 10.1128/EC.00291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koga-Ito CY, Komiyama EY, De Paiva Martins CA, et al. Experimental systemic virulence of oral Candida dubliniensis isolates in comparison with Candida albicans,Candida tropicalis and Candida krusei . doi: 10.1111/j.1439-0507.2010.01899.x. Mycoses. In press. [DOI] [PubMed] [Google Scholar]
- 19.Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283(5407):1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 20.Hoyer LL. The ALS gene family of Candida albicans . Trends in Microbiology. 2001;9(4):176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- 21.Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiology and Molecular Biology Reviews. 2003;67(3):400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niewerth M, Korting HC. Phospholipases of Candida albicans . Mycoses. 2001;44(9-10):361–367. doi: 10.1046/j.1439-0507.2001.00685.x. [DOI] [PubMed] [Google Scholar]
- 23.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans . Trends in Microbiology. 2004;12(7):317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nature Reviews Microbiology. 2010;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannula J, Saarela M, Alaluusua S, Slots J, Asikainen S. Phenotypic and genotypic characterization of oral yeasts from Finland and the United States. Oral Microbiology and Immunology. 1997;12(6):358–365. doi: 10.1111/j.1399-302x.1997.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 26.De Repentigny L, Aumont F, Bernard K, Belhumeur P. Characterization of binding of Candida albicans to small intestinal mucin and its role in adherence to mucosal epithelial cells. Infection and Immunity. 2000;68(6):3172–3179. doi: 10.1128/iai.68.6.3172-3179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCullough M, Ross B, Reade P. Characterization of genetically distinct subgroup of Candida albicans strains isolated from oral cavities of patients infected with human immunodeficiency virus. Journal of Clinical Microbiology. 1995;33(3):696–700. doi: 10.1128/jcm.33.3.696-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones T, Federspiel NA, Chibana H, et al. The diploid genome sequence of Candida albicans . Proceedings of the National Academy of Sciences of the United States of America. 2004;101(19):7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braun BR, van het Hoog M, d’Enfert C, et al. A human-curated annotation of the Candida albicans genome. PLoS Genetics. 2005;1(1):36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van het Hoog M, Rast TJ, Martchenko M, et al. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biology. 2007;8(4, article no. R52) doi: 10.1186/gb-2007-8-4-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran G, Stokes C, Thewes S, Hube B, Coleman DC, Sullivan D. Comparative genomics using Candida albicans DNA microarrays reveals absence and divergence of virulence-associated genes in Candida dubliniensis . Microbiology. 2004;150(10):3363–3382. doi: 10.1099/mic.0.27221-0. [DOI] [PubMed] [Google Scholar]
- 32.Jackson AP, Gamble JA, Yeomans T, et al. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans . Genome Research. 2009;19(12):2231–2244. doi: 10.1101/gr.097501.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Molecular Genetics and Genomics. 2007;278(4):455–465. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- 34.Luo G, Ibrahim AS, Spellberg B, Nobile CJ, Mitchell AP, Fu Y. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. Journal of Infectious Diseases. 2010;201(11):1718–1728. doi: 10.1086/652407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dwivedi P, Thompson A, Xie Z, et al. Role of Bcr1-activated genes Hwp1 and Hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0016218. Article ID e16218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phan QT, Myers CL, Fu Y, et al. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biology. 2007;5(3, article e64) doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almeida RS, Brunke S, Albrecht A, et al. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathogens. 2008;4(11) doi: 10.1371/journal.ppat.1000217. Article ID e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhl MA, Biery M, Craig N, Johnson AD. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C.albicans . EMBO Journal. 2003;22(11):2668–2678. doi: 10.1093/emboj/cdg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enjalbert B, Moran GP, Vaughan C, et al. Genome-wide gene expression profiling and a forward genetic screen show that differential expression of the sodium ion transporter Ena21 contributes to the differential tolerance of Candida albicans and Candida dubliniensis to osmotic stress. Molecular Microbiology. 2009;72(1):216–228. doi: 10.1111/j.1365-2958.2009.06640.x. [DOI] [PubMed] [Google Scholar]
- 40.Alves SH, Milan EP, De Laet Sant’Ana P, Oliveira LO, Santurio JM, Lopes Colombo A. Hypertonic sabouraud broth as a simple and powerful test for Candida dubliniensis screening. Diagnostic Microbiology and Infectious Disease. 2002;43(1):85–86. doi: 10.1016/s0732-8893(02)00368-1. [DOI] [PubMed] [Google Scholar]
- 41.Pinjon E, Sullivan D, Salkin I, Shanley D, Coleman D. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans . Journal of Clinical Microbiology. 1998;36(7):2093–2095. doi: 10.1128/jcm.36.7.2093-2095.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Connor L, Caplice N, Coleman DC, Sullivan DJ, Moran GP. Differential filamentation of Candida albicans and Candida dubliniensis is governed by nutrient regulation of UME6 expression. Eukaryotic Cell. 2010;9(9):1383–1397. doi: 10.1128/EC.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moran GP, MacCallum DM, Spiering MJ, Coleman DC, Sullivan DJ. Differential regulation of the transcriptional repressor NRG1 accounts for altered host-cell interactions in Candida albicans and Candida dubliniensis . Molecular Microbiology. 2007;66(4):915–929. doi: 10.1111/j.1365-2958.2007.05965.x. [DOI] [PubMed] [Google Scholar]
- 44.Schaller M, Zakikhany K, Naglik JR, Weindl G, Hube B. Models of oral and vaginal candidiasis based on in vitro reconstituted human epithelia. Nature Protocols. 2007;1(6):2767–2773. doi: 10.1038/nprot.2006.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song W, Wang H, Chen J. Candida albicans Sfl2, a temperature-induced transcriptional regulator, is required for virulence in a murine gastrointestinal infection model. FEMS Yeast Research. 2011;11(2):209–222. doi: 10.1111/j.1567-1364.2010.00710.x. [DOI] [PubMed] [Google Scholar]
- 46.Staib P, Morschhäuser J. Differential expression of the NRG1 repressor controls species-specific regulation of chlamydospore development in Candida albicans and Candida dubliniensis . Molecular Microbiology. 2005;55(2):637–652. doi: 10.1111/j.1365-2958.2004.04414.x. [DOI] [PubMed] [Google Scholar]
- 47.Rohde JR, Bastidas R, Puria R, Cardenas ME. Nutritional control via Tor signaling in Saccharomyces cerevisiae. Current Opinion in Microbiology. 2008;11(2):153–160. doi: 10.1016/j.mib.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan DJ, Moran GP. Differential virulence of Candida albicans and C. dubliniensis a role for tor1 kinase? Virulence. 2011;2(1):77–81. doi: 10.4161/viru.2.1.15002. [DOI] [PubMed] [Google Scholar]


