Abstract
The advent of new technologies for the imaging of living cells has made it possible to determine the properties of transcription, the kinetics of polymerase movement, the association of transcription factors, and the progression of the polymerase on the gene. We report here the current state of the field and the progress necessary to achieve a more complete understanding of the various steps in transcription. Our Consortium is dedicated to developing and implementing the technology to further this understanding.
INTRODUCTION
With the uncovering of the ever-growing fraction of the animal genome that is transcribed, transcription is more than ever the centerpiece of cell metabolism. Through biochemical analysis and genetics, many if not most of the proteins implicated in transcription have been identified. Decades of in vitro studies determined that the transcription process could be separated into three steps: preinitiation complex formation, initiation, and elongation. Each one of these steps may be subjected to regulation, accounting for the fine-tuning of gene expression. But biochemistry tells us only what is possible, not what actually happens, in the very specific milieu of a living cell. It has become feasible, using the new advances in microscopy, to interrogate the processes that make up transcription and break them down into their component parts. Accurate quantification is possible due to technology that has evolved over the years to detect and measure photons. The components of the transcription reaction can then be assigned rate constants describing their forward and reverse rates. As a result of these analyses, new models have arisen to fit these data. Among those are the observations that for some models the transcriptional complex can be transient, existing only for a few seconds, and that the entire process is inefficient, yet other factors can be stably associated for hours. Understanding the kinetic components that give rise to these disparate time constants will be an important function of the new technologies.
This review is dedicated to exploring the work that is contributing to the real-time analysis of transcription, an aspect that contributes data not addressed by chromatin immunoprecipitation, microarray studies, or other bulk assays that cannot resolve the events occurring in single cells. Many cell and tissue models are described including bacteria, polytene chromosomes, various reporter genes, and cell lines. Different approaches include fluorescence recovery after photobleaching (FRAP), fluorescence correlation spectroscopy (FCS), fluorescence resonance energy transfer (FRET), and multiphoton microscopy (MPM) (see sidebar, Methods Used to Analyze Transcription in Living Cells). This is a new field that is rapidly emerging, and these initial forays represent the beginning of a new territory in the area of gene expression research.
The review is organized by the model systems that have contributed to studies in live cell imaging: naturally occurring and artificial gene arrays, viral genes, steroid receptor responsive genes, and single copy endogenous genes. There are four sections: imaging gene arrays, imaging the nuclear organization of transcription, imaging single copy genes, and the analysis of imaging data.
IMAGING GENE ARRAYS
Some gene families or duplicated genes in vertebrate genomes have naturally regrouped into tandem arrayed genes. These genes therefore lie as neighbors on the same locus in the genome and are often under the control of the same transcriptional regulators. In addition, the fruit fly (Drosophila melanogaster) contains giant polytene chromosomes in certain larval cell types where the entire genome is multiplied. This spatial clustering of genes offers researchers many opportunities to study transcription, especially when using fluorescence microscopy, in which accumulation of fluorescence means a better signal-to-noise ratio. Another advantage of such arrays is that when studying a fluorescent molecule that interacts directly or indirectly with the arrays, the majority of the fluorescence will be due to interacting molecules (in contrast to freely diffusing molecules, which might be preponderant at a nonamplified locus). This increases the resolution on binding events that reflect catalytic activities. So far three cases of amplified genes have been used to study transcription as illustrated in Figure 1: polytene salivary gland chromosomes in Drosophila (95), the ribosomal DNA nucleolar clusters (36), and artificially developed mammalian gene arrays (68). These examples are covered more fully below.
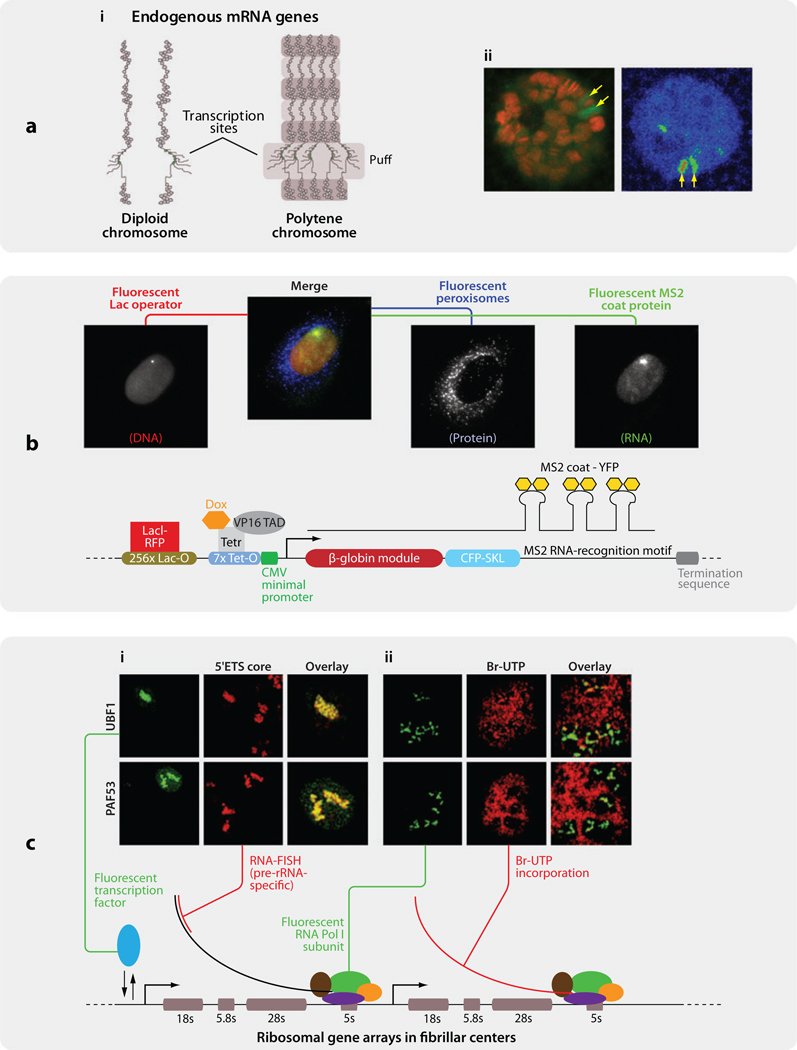
Figure 1.
Three models for visualizing transcription in vivo. (a) Drosophila polytene chromosome. (i) Diagram of diploid chromosome and polytene chromosome. Active transcription site of heat shock gene hsp70 loci at 87A and 87C exhibits “chromosome puffs” (yellow arrows). (ii) (Left) hsp70 loci are enriched with Pol II (red: Hoechst33342 stain; green: EGFP-Rpb3). (Reproduced with permission from Reference 95.) (Right) Upon heat shock, EGFP-Rpb3 exhibits a strong doublet at 87A and 87C loci that can be recognized in polytene nucleus (arrows). Image in pseudocolor. (b) An artificial reporter gene was stably inserted as multiple copies into a single locus in the genome of U2OS cells. The locus of integration was tracked with the use of a Lac-repressor-fused RFP. The MS2 stem loop structures on the RNA, which bind stably to MS2 coat proteins, fused with YFP, allowing the investigator to localize single molecules of mRNA in vivo (78).(c) Ribosomal RNA transcription occurring inside fibrillar centers in CMT3 cells. (i) PAF53, a Pol I subunit, and UBF1, a rDNA transcription factor, colocalize with nascent rRNA in fibrillar centers (green: PAF53-GFP or UBF1-GFP; red: fluorescent in situ hybridization probe directed to rRNA 5’ ETS core). (ii) GFP-tagged PAF53 and UBF1 accumulate in active rRNA transcription sites (green: PAF53-GFP or UBF1-GFP; red: Bromo-UTP stain) (Adapted with permission from Reference 24). Abbreviations: EGFP, enhanced green fluorescent protein; ETS, external transcribed spacer; Pol I (II), polymerase I (II)
Drosophila melanogaster Polytene Chromosomes
Drosophila polytene chromosomes are found in many larval cell types formed by endoreduplication during development, i.e., these cells undergo DNA replication without cell division. Polytene chromosomes from salivary gland cells contain approximately 1000 copies of DNA. Condensed and decondensed chromatin form unique band and interband structures that can be distinguished with a light microscope. The chromosome banding patterns were categorized and named by Bridges (13) and have been used as a marker for cytogenetic localization of individual genes along the polytene chromosomes (Figure 2a). Examining the unique cytogenetic pattern has allowed early genetic mapping such as gene deletion, gene duplication, chromosome translocation, and inversion. Furthermore, localizing protein factors on polytene chromosomes with antibody based immunostaining techniques has provided a means to study protein-DNA interaction in vivo.
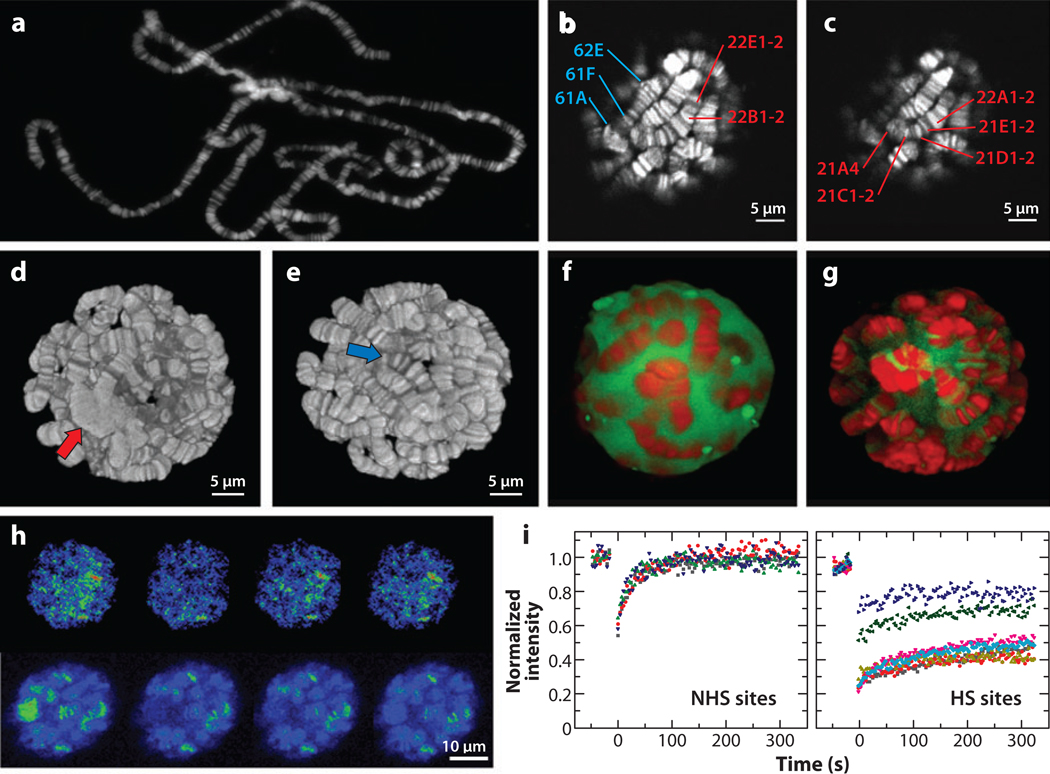
Figure 2.
Imaging transcription kinetics at Drosophila polytene chromosomes.(a) An image of Drosophila polytene chromosomes spread onto a glass slide and stained by Hoechst33342. (b, c) Optical sections of polytene chromosomes in a live salivary gland stained with Hoechst33342. The z-distance is 0.5 µm. Labels identify specific bands on chromosome arms 2L (red) and 3L (blue). (d, e) Three dimensional reconstructions of a polytene nucleus. Red and blue arrows indicate the centromeric region and telomere, respectively. Panels b–e are adapted with permission from Reference 95. (f, g) HSF expression and localization in salivary glands at NHS (f) and after HS (g). Shown are the three-dimensional reconstructions of the two-photon optical sections of polytene nuclei expressing HSF-EGFP (green) stained with Hoechst33342 (red). (h, i) FRAP of HSF-EGFP at endogenous gene loci in salivary glands. (h) Intensity images of a nucleus of a heat-shocked salivary gland in pseudocolor during FRAP. (i) FRAP curves at NHS and HS sites. (Adapted with permission from References 95 and 96). Abbreviations: EGFP, enhanced green fluorescent protein; FRAP, fluorescence recovery after photobleaching; HS, heat shock; HSF, heat shock factor; NHS, non-heat shock.
The naturally amplified chromatin template in Drosophila polytene chromosomes provides an opportunity to overcome the sensitivity limitation in visualizing transcription factors associated with endogenous gene loci in living cells. However, imaging transcription factors in living salivary gland tissues had been challenged by the thickness and optical properties of the tissue samples. Recently, Webb, Lis, and colleagues (94–96) have reported that MPM provides the experimental capability of resolving individual genetic loci (Figure 2b–e) and studying dynamic interactions of a green fluuorescent protein (GFP)-fused heat shock factor (HSF) or RNA polymerase II (Pol II) with active hsp70 gene loci in living tissues and in real time.
This is an elegant system for studying the dynamics of transcription regulation in vivo. The power of this approach lies in the combination of naturally amplified templates, Drosophila transgenic techniques, and MPM imaging, which provides optical sectioning deep within living tissues (99). The rapid and robust heat shock gene activation has allowed unambiguous localization of endogenous hsp70 loci and further assisted the visualization of the associated factors (Figure 2f,g). Furthermore, multicolor fluorescent proteins and mutant gene alleles can be introduced by simple genetic crosses. Some novel insights of transcription dynamics have arisen from the application of this method. For instance, the transient association (a few seconds) of a transcription activator with a gene promoter, the so called hit-and-run model (54), has been thought to be universal for all activators. However, in Drosophila polytene chromosomes, HSF is transiently associated with hsp83 gene loci (half life about 10 s) before heat shock but becomes stably associated with hsp70 gene loci (half-life > 5 min) during heat shock (Figure 2h,i) (95). Therefore, some transcription factors, such as HSF, are stably associated with their target genes and can provide a stable platform that supports multiple rounds of transcription. This may be a relatively common property of strong promoters, as suggested by other reports. (a) A stable association with DNA in vivo has been reported for yeast Gal4 activator (60); (b) higher transcription levels are correlated with a more stable association of Pol I subunits with rRNA genes (36). In addition, Pol II exhibits efficient recruitment to the locus and enters into elongation during early heat shock, and it is locally recycled during late heat shock (94). Furthermore, this MPM-based FRAP assay has been coupled with chromatin immunoprecipitation assays to study the dynamics of distinct populations of Pol II molecules: During early heat shock, FRAP was performed on hsp70 genes when the productively elongating forms of Pol II were eliminated by inhibiting P-TEFb kinase. This study shows that the remaining transcriptionally engaged/paused Pol II molecules near the transcription start site of the genes are stably associated with hsp70 genes (61). Although this experimental system will continue to provide unique insights into the dynamic mechanisms of gene regulation in Drosophila, it would be of interest to determine to what extent the transcription kinetics at micron-scale gene structures in polytene cells resembles the kinetics at nanoscale gene structures in diploid cells.
Natural Tandem Repeated Genes
The Misteli laboratory used the ribosomal genes’ organization into such arrays to study Pol I transcription. Their work offered the first attempt to measure kinetic rates for a RNA polymerase in living cells. They proposed an elongation rate of 5.7 kb per minute. They also observed that different subunits of the Pol I complex were loaded onto the genes with different kinetic rates, which can be interpreted as a pioneering subunit or subcomplex interacting with the DNA, serving as a docking platform for the other subunits (24). More recently, the same group analyzed the order of assembly of Pol I components and the regulation of this assembly during G1 /G0 and S phases. According to their findings, Pol I is assembled in an on-the-spot stepwise process that reflects transcriptional efficiency (36). This is in contrast to in vitro studies that show that a preassembled RNA polymerase holoenzyme can be recruited to a promoter site and efficiently transcribe an RNA molecule (22), or that there is no exchange between Pol I subunits in yeast (Pol I remains intact without subunit exchange through multiple rounds of transcription in Saccharomyces cerevisiae) (22, 75). Similar studies have yet to be done with Pol II, whose assembly might also be influenced by the promoter sequence of the studied gene and the various specific transcriptional regulators involved.
Another example of natural array is the CUP1 locus in the baker’s yeast S. cerevisiae (47). CUP1 exists as a small natural tandem array of 10 copies, transcribed by the Pol II. The transcriptional activator Ace1 binds to CUP1 promoter in the presence of copper and activates transcription. Fusion of three copies of GFP to Ace1 enabled visualization of the CUP1 array in live cells. The behavior of Ace1-GFP on the CUP1 promoter has been monitored by FRAP. A complete fluorescence recovery occurred within 2 min after photobleaching. It is similar but somewhat slower than that observed for other transcriptional activators. Furthermore, at longer timescales a slow cycling of Ace1-GFP binding to CUP1 can be detected. After computational analysis, Ace1 behavior is compatible with a model in which the slow cycle reflects the number of accessible binding sites at promoters and each accessible site can be bound by fast cycling molecules. It is suggested that the oscillation of histone occupancy at the locus accounts for cycling accessibility. At this promoter the fast cycle is responsible for transcription initiation and the slow cycle for adjusting the amount of mRNA synthesis. This simple natural model can be combined with powerful yeast genetics to explore further the implication of transcription factors and chromatin remodeling in the kinetics of transcription.
Gene arrays therefore offer a huge number of possibilities when it comes to studying transcription and related processes. A recent estimate suggested that such tandem gene arrays represent 14% of all genes in vertebrate genomes, although most are made of only two genes (62). That means that these gene arrays are available for investigators who wish to study transcription in a natural genomic context. However, these natural gene arrays offer low control over the fundamental mechanisms involved in transcription. A number of teams have therefore developed artificial gene arrays in which the reporter genes were tinkered with to study specific core mechanisms. These modified arrays represent a good compromise between natural conditions and control by the investigator.
Artificial Gene Arrays
The first purpose of an artificial array created by Tsukamoto et al. (89) was to study chromatin remodeling during transcriptional activation. By inserting this Tet-inducible reporter gene containing Lac operator sites into large arrays, they observed changes in the chromatin upon activation and verified that the fluorescent protein encoded was correctly expressed. This system was then improved by Janicki et al. (43), who inserted 24 repeats of the MS2 bacteriophage replicase translational operator, which allowed them to visualize the mRNA transcribed from the genes. This work has permitted a real-time parallel analysis of transcription and modifications in the chromatin, notably by following histone 3.3 depositions and HP1α depletion (43). The same system was then used to estimate various kinetic steps of transcription (21). Other studies on chromatin remodeling made use of a gene array first described in 2000 by McNally (54), made up of mouse mammary tumor virus long terminal repeats (MMTV-LTRs), which can be activated by glucocorticoid receptors (GRs) (Figure 3d) (45, 54, 58, 80). These studies have given great insight into the importance of various chromatin remodelers such as BRM and BRG1 during transcription. Finally, whether natural or artificial, gene arrays could be of great use in the study of many other nuclear processes, and the use of this powerful tool has only begun to reveal new and interesting observations.
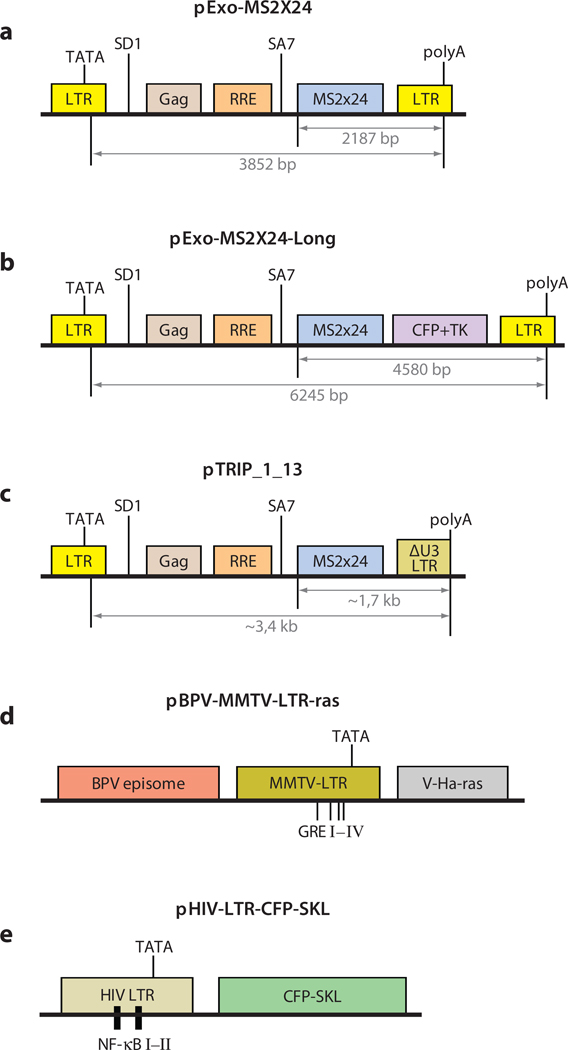
Figure 3.
Diagrams of vectors containing viral sequences used to produce tandem arrays for real-time live cell imaging.(a) Vector used by Molle and colleagues to determine the residency times of Tat and Cdk9 on the HIV-1-LTR-driven transgene (57). (b, c) Modifications of the vector presented in panel a used in Reference 8 to measure transcription elongation rate. (d) MMTV-LTR containing glucocorticoid receptor (GR) binding sites developed by McNally and colleagues was used to determine various kinetic parameters of gene activation by hormones (45, 54, 58, 80). Similarly, the vector presented in panel e has been used to study NF-κB interaction with its binding sites (9). The original vector designations and references in which they were first described are given. The kinetic parameters of transcription were obtained on tandem arrays in each of these vectors. Systems are discussed in the text. Diagrams are not drawn to scale. (For a compilation of other existing gene arrays, see Reference 68.) Abbreviations: BPV: bovine papilloma virus; Gag: retroviral gene coding for internal structural proteins; GRE: glucocorticoid-responsive element; NF-κB: nuclear factor κB; RRE: Rev-responsive element; SA7: splice acceptor 7; SD1: splicedonor 1; SKL: Ser-Lys-Leu peroxisome-targeting peptide; v-Ha-ras: Harvey viral ras.
One of the most surprising results concerning transcription obtained with gene arrays was that transcription in vivo is an inefficient process. The studies concerning Pol I by Dundr and colleagues (24) show that only about 10% of polymerases in the fibrillar centers are actually engaged in elongation. Similarly, Darzacq and colleagues (21) have shown that transcription in a Pol II array of genes was also inefficient, with only 1% of binding events resulting in the production of a complete mRNA. In the same study, they have determined the kinetics of different steps of transcription using FRAP experiments on an artificial gene array. They estimated that the elongation rate of Pol II is around 75 nucleotides per second, slightly slower than the 90 nucleotides per second published for Pol I (21, 24). These estimates are in accordance with previous findings of 50 and 100 nucleotides per second, respectively, using an independent method (76).
Viruses
Among viral species there exists a great variety of genomic structures and, consequently, mechanisms of replication and gene expression. Only a small part of all known viruses depends on host cell polymerases in both replication and transcription.
For retroviruses, such as HIV-1 and MMTV, replication is the process whereby genomesized RNA, which also functions as mRNA, is produced by host cell Pol II from the provirus integrated in the host genome. Thus, transcription is a means of their replication, as well as gene expression, and its tight regulation is important for the viral life cycle.
Some viruses use their own proteins to modify and redirect the activity of host cell’s transcriptional machinery. In the case of HIV-1, transcription is activated by the viral protein Tat, which recruits the elongation factor PTEFb [consisting of cyclin T and Cdk9, which phosphorylates the C-terminal domain of the large subunit of Pol II] to the nascent stembulge-loop leader RNA, TAR (trans-activation responsive) (Figure 4) (44). Recently, cell lines have been created harboring tandem arrays of a reporter that carries the elements required for HIV-1 RNA production (Figure 3a–c) (8, 57). In these cells the dynamics of the TAR:Tat:PTEFb complex components has been analyzed by FRAP at the transcription sites visualized by expressing a nuclear MS2 phage coat protein (MS2cp) fusion with a fluorescent protein. Comparison of Cdk9-GFP dynamics at sites activated by Tat or phorbol 12-myristate 13acetate/ionomycin showed that Cdk9 residency time at the HIV-1 transcription site was several times longer in the presence of Tat than in the absence of Tat (71 s and 11 s, respectively) and that it was similar to the residency time measured for Tat-GFP itself (55 s), suggesting that significant fractions of Tat and Cdk9 are present at the site as parts of the same complex, likely interacting with elongating Pol II (57). The transcription elongation rate measured by FRAP on MS2-GFP on the same HIV-1 tandem array and its variants with a longer transcribed region or without the U3 region in the 3’ LTR (which is required for efficient transcript 3’ -end formation) (Figure 3a–c) was estimated to be approximately 1.9 kb min−1 (8). The use of this HIV-MS2 tandem array also allowed the estimation of the Pol II residency time at the transcription site and its comparison to RNA production rates. The authors calculated a total polymerase residency time of 333 s, of which 114 s were attributed to elongation, 63 s to 3’-end processing and/or transcript release, and 156 s to polymerase remaining on the gene after RNA release (8).
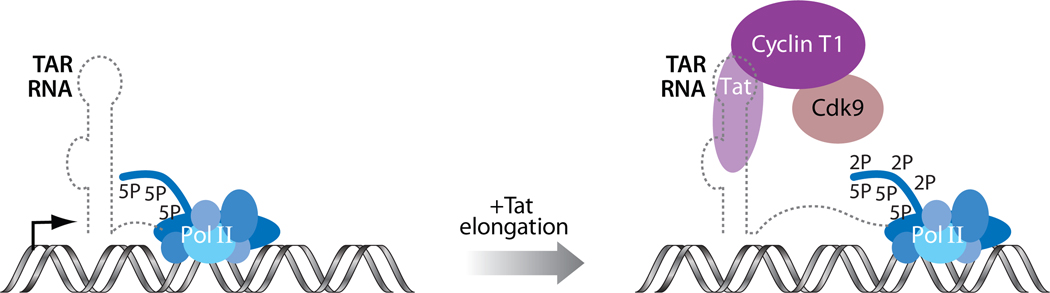
Figure 4.
The role of the TAR:Tat:P-TEFb complex in HIV transcription. RNA polymerase II (Pol II) is stalled soon after initiation of transcription of the HIV provirus. The viral protein Tat recruits host cellular P-TEFb to the nascent stem-bulge-loop leader RNA, TAR (trans-activation responsive). The elongation factor P-TEFb composed of the cyclin T and the kinase Cdk9 phosphorylates the C-terminal domain repeats of the large subunit of Pol II at the Ser2 positions to stimulate processive elongation.
Unlike P-TEFb, which stays at the transcription site induced by Tat for approximately 1 min (Figure 4), other transcription factors interacting with viral promoters interact transiently with the arrays. For example, mRFP tagged NF-κB proteins interact with a multicopy array of transgenes containing the HIV 5’ LTR for only a few seconds (Figure 3e) (9). FRAP of GFP-tagged GRs at the tandem array containing MMTV-LTR promoters showed even shorter residency times (Figure3d) (54, 80).
NUCLEAR ORGANIZATION OF TRANSCRIPTION
Transcription Factories
Pol II is a multisubunit enzyme responsible for the transcription of most eukaryotic genes. The composite holoenzyme generated by the association of Pol II with other large complexes involved in related functions such as capping, splicing, and polyadenylation ensures the efficient production of mature transcripts. The key element necessary for coupling transcription with all the maturation steps is the large subunit of Pol II and in particular its C-terminal domain, essential for tethering the different machineries and regulating them temporally. The discovery of this intricate network gave rise to the idea that a specialized molecular machine is assembled at the site of transcription, nucleating from the promoter of an active gene. Because several of these factories may cluster together to ensure high local concentrations and therefore efficient interactions with all the partners involved, it is important to understand how different transcription units are transcribed and how their identity, nuclear surroundings, and positions could affect their expression.
Over the past fifteen years Cook and collaborators have put forward the concept of a superstructure called transcription factories, an assemblage of transcription and RNA-processing enzymes containing multiple genes. Before the advent of new visualizing methodologies, the transcription sites in mammalian cells were marked by elongation of nascent RNA in the presence of [3 H]uridine, [32 P]uridine, or BrUTP, and subsequent observation at the fluorescent or electron microscope (39, 41,42, 66). With these techniques they were able to see multiple nuclear foci sensitive to α amanitin and containing splicing components (41). Those foci remained visible also after nucleolytic removal of most of the chromatin, highlighting the presence of an underlining structure responsible for the clustering of transcription units in which transcripts are both synthesized and processed (39). Moreover, these results are consistent with polymerases confined by the nucleoskeleton into factories and transcription occurring as templates slide past attached polymerases (40). Quantitative analysis (42) also showed that a typical factory contains approximately 30 engaged polymerases. Because two-thirds or more transcription units are associated with one polymerase at any time, each factory could contain at least 20 different transcription units.
A recent work on transcription factories (27) increased the resolution obtained with the electron microscope by coupling this technique with electron spectroscopic imaging. Electron spectroscopic imaging is a high-resolution and potent ultrastructural method that can be used to map atomic distribution in unstained preparations. Combining immunolabeling of the newly synthesized BrU-RNA with the distribution maps of nitrogen (N) and phosphorus (P) enabled specific atomic signature marking, allowing these nucleoplasmic sites to be identified. Template and nascent RNAs were attached to the surface of enormous protein-rich structures 87 nm in diameter and with a mass of 10 MDa. These structures appear porous, large enough to contain all the different protein complexes required for the complete maturation of the transcript. This finding suggested the idea that the polymerase was anchored, probably at the surface of the core, and that the DNA diffuses or loops to come in contact with a specific factory. Eskiw et al. (27) suggest that only a minority of all the machinery in the site is active, but that the high local concentrations will guarantee robust and efficient processivity.
Other questions concern how many transcription factories exist in a cell and how they should be classified. The first level of organization is the division of the three polymerases into different factories (67, 93). A more complicated issue is determining the influence of the genes’ characteristics (promoter or presence of introns) on their arrangement within the nucleus. Using replicating minichromosomes from Cos7 cells analyzed by FISH (fluorescent in situ hybridization) and 3C (chromosome conformation capture), Xu & Cook (93) examined whether the factories were specialized and the importance of the genes’ distinctive characteristics. Their results confirmed that plasmids were concentrated in transcribing foci and that those being copied by different polymerases were not transcribed by the same foci. Moreover, units transcribed by Pol II, with different promoters (CMV and U2) or with the same promoter but with or without an intron in the coding sequence, are seen in nonoverlapping foci.
Even more intriguing are the results from live cells where the fluorescent tagging system of Pol II large subunit has been exploited (87). The GFP-tagged version was stably expressed in a Chinese hamster ovary cell line bearing a temperature-sensitive Pol II mutant, tsTM4, and it was observed that the fluorescent version was functional and normally assembled in the complex and rescued the phenotype. Because each factory contains only a few polymerases, it would be difficult to image those foci in live cells; however, significant results could be obtained by the study of polymerase kinetics in the nucleus of living cells. FRAP and FLIP (fluorescence loss in photobleaching) experiments in the nucleus gave important information on fractions of the enzyme in different states (48). They revealed two kinetic components in the Pol II population: A fast mobile component showed that 75% of the molecules were diffusing freely and the immobile component showed that 25% of the molecules were transiently immobile with a t1/2 of 20 min. This latter fraction was likely the active one, since incubation with DRB (5,6 dichloro-1- β -d-ribofuranosyl-benzimidazole), a potent inhibitor of elongation, eliminated it. Their model of the transcription cycle supports the idea that the enzyme spends most of the time diffusing and exchanging between the nucleoplasm and a promoter or a transcription factory. Once bound, a third of the time is mainly dedicated to elongation. With the improved use of this photobleaching technique, a more detailed analysis also resolved a third component resistant to DRB but sensitive to heat shock, representing the bound but not yet engaged fraction (37).
Whether these factories exist in most cells is a question that needs to be addressed with more sensitive technologies. Given that resolution problems pervade the experiments concerning testing of this concept, it will fall to the more quantitative, high-resolution methods to determine whether there simply exist gene-rich regional concentrations of transcription, or whether the factories are truly higher-order structures.
Gene Positioning
The influence of the position of a gene with respect to the nuclear periphery on transcriptional competence has been extensively studied in recent years. Historically, the nuclear periphery has been seen as a nuclear substructure enriched in heterochromatin and thereby an area of transcriptional repression. However, data from yeast showing that active genes are often found in the nuclear periphery and in association with the nuclear pore complex led to a series of studies investigating the influence of nuclear positioning on transcriptional competence (3, 17).
In an early study, Cabal et al. (14) showed that the yeast GAL1 gene changed its position from a mostly internal position to a preferential location at the nuclear periphery when the gene was activated, supporting the idea that the nuclear periphery harbors active genes. To do this, they used a fluorescently labeled GAL1 locus in living cells by inserting an array of 112 TetO operators downstream of the GAL1 gene, which upon coexpression of GFP-tagged TetR turns fluorescent (14). Nuclear positioning and movement of a locus were then followed using 4D live cell microscopy. Importantly, they found that the movement of the locus was not fully constrained but restricted to a 2D sliding movement at the nuclear envelope and was suggested to act as a gating mechanism to allow efficient mRNA processing and export.
Genes in yeast have been analyzed using this technique and were shown to move to the nuclear periphery upon activation (2, 12, 14, 23, 88). The requirements for the translocations, however, were often gene specific. In addition to components of the nuclear pore complex, promoters or elements in the 3’ UTR, the SAGA complex of transcription factors, and components of the mRNA export machinery are involved (2, 12, 14, 23, 74, 88). Similarly, gene movement to the periphery has been suggested to occur before transcription starts for some genes, but it has also been suggested to occur as a result of transcription for other genes (12, 14, 23, 74, 88). It still remains to be shown if general principles exist that mediate the perinuclear localization of active genes in yeast and what fraction of genes use this mechanism to regulate their expression. Peripheral localization has also been suggested to mediate epigenetic memory over many generations (11).
These data from yeast led to the question whether such a mechanism may exist in higher eukaryotes. In yeast and in higher eukaryotes, chromatin loci in general are not statically positioned within the nucleus. In yeast as well as in higher eukaryotes, chromatin during interphase is mobile but mostly constrained within a radius of approximately 0.5–1 µm. That is less than 1% of the volume of a typical 10-µm spherical mammalian nucleus but half of the diameter of a yeast nucleus (52). In yeast, if a locus is located at the nuclear periphery, diffusion and the accessibility of binding sites at the nuclear periphery might be sufficient to allow tethering, as most genes likely encounter the nuclear periphery at least occasionally. In higher eukaryotes, however, if such events existed, it might require a more active movement, as a locus would have to move several microns to attach to the nuclear periphery or to nuclear pores. Peripheral heterochromatin is often interrupted at nuclear pores, indicating the presence of euchromatin in the vicinity of nuclear pores and making it possible that, like in yeast, active genes might get tethered to nuclear pore complexes to stimulate expression.
Recent live cell studies suggested that repositioning of genes from or to the nuclear periphery might have some influence on gene expression in higher eukaryotes, but that it might not be a major factor mediating gene expression. Imaging the naturally amplified Drosophila polytene nuclei in living salivary gland tissues by MPM did not reveal a preferred localization of the loci upon transcription induction. The genes could be found in the nuclear interior as well as at the nuclear periphery (94). Consistently, a GFP-tagged locus tethered to the nuclear periphery by a lamin B1 fusion maintained its transcriptional competence, indicating that sole peripheral or internal/central nuclear positioning does not influence transcription (51). However, another study suggested that expression of a subset of genes can reversibly be suppressed when tethered to the periphery, whereas many genes are not affected (30). Using DNA FISH, Reddy et al. (70) showed that genes can be silenced when targeted to the inner nuclear membrane. Together these results suggest that the nuclear periphery is not incompatible with active transcription but that it is not a primary determinant of whether genes are active. Different cis and trans-acting factors are likely to determine whether peripherally localized genes in higher eukaryotes can be transcribed. However, chromatin movements in higher eukaryotes seem to actively play a role in regulating gene expression. Chromatin can frequently exhibit long-range movements of >2 µm during the cell cycle (90). Migration of an interphase chromosome site from the nuclear periphery to the interior has been observed 1–2 h after targeting a transcriptional activator to this site, showing a contrary localization to that in yeast (19). More surprisingly this movement was perturbed in specific actin and myosin I mutants, suggesting some kind of motor-driven movement. Similarly, actin dependent intranuclear repositioning occurs with the U2 snRNA gene locus (25). If and how motor proteins mediate such long-distance chromatin movements still remain to be determined.
IMAGING ENDOGENOUS GENES Steroid Receptors
Perhaps the most well-studied transcription factors of endogenous genes in living cells are nuclear receptor (NR) regulated. These ligand activated transcription factors constitute the nuclear hormone receptor superfamily and are involved in regulating a vast array of eukaryotic genes. NR transcription is initiated by agonist binding to the receptor, forming either a homodimer or heterodimer complex. The corepressors (histone deacetylases, NR specific corepressors) associated with the dimer are then replaced by coactivators such as histone acetylases (SRC/p160 family or CBP/p300) and histone methylases (CARM-1, PRMT-1). In addition, ATP-coupled chromatin remodeling complexes (SWI/SNF) are recruited. Eventually, the basal transcription machinery is assembled, followed by the initiation of Pol II. After initiation, transcription can be influenced by NR factors such as vitamin D receptor interacting protein and thyroid-associated protein (38). Thus, NR transcription is an excellent model system for observing the cooperative interactions among enhancers, repressors, transcription factors, and basal transcription components (63). The view that has emerged from live cell studies utilizing fluorescence techniques such as FRAP, FRET, and FCS is that these NR complexes are highly dynamic: Individual species have dwell times on the order of seconds to minutes. However, these same complexes can result in cycles of transcriptional progression that can last hours or days (56). NR-regulated transcription is therefore dynamically responsive to changes in agonist concentration and also capable of long-term changes of gene expression.
Live cell studies of NR-regulated transcription can be divided into those that study nuclear dynamics in general and those that focus on a particular locus. The first approach provides information about multiple possible transcription sites within the nucleus in addition to nonspecific interactions. The second approach has the benefit of providing specific information about interactions and dynamics at an active transcription site but usually requires modification of the locus—either multimerization of an endogenous gene (54) or creation of an artificial locus (85). The first example of this approach, which has been used by a number of investigators since its inception, was a large tandem array of a mouse mammary tumor virus/Harvey viral ras (MMTV/v-Haras) reporter, which contains about 200 copies of the LTR and thus includes 800 to 1200 binding sites for the GR (54). This same array has been used for FRAP studies of the GR (6, 45, 55, 83), the androgen receptor (AR) (50), and the progesterone receptor (PR) (69). For each of those receptors, an agonist dependent decrease in receptor mobility (increase in t1/2) was observed [GR, t1/2 : 1–1.6 s (55); AR, t1/2 : 0.2–3.6 s (50); PR, t1/2 : 0.6–3.7 s; (69)]. A similar agonist-dependent decrease in mobility was also observed for general nuclear bleaching of the estrogen receptor [ER, t1/2 :0.8–5.9 s (85)]. These observations demonstrate that the recovery time reflects the interaction of the NR with the locus in a specific fashion. In fact, Schaaf & Cidlowski (73) demonstrated that higher-affinity ligands result in slower recovery times, and Kino et al. (49) directly showed a positive correlation between FRAP t1/2 times and transcriptional activity, with higher transcriptional activity corresponding to longer effective recovery times.
In contrast, other receptors do not show an agonist-dependent increase in t1/2 for general nuclear recovery. The retinoic acid receptor (RAR), the thyroid hormone receptor (TR), the peroxisome proliferator-activated receptor (PPAR), and the retinoid X receptor (RXR) all have the same recovery time with or without ligand [RAR, t1/2 : 1.9–2.3 s; TR, t1/2 : 1.8–1.8 s (53); PPAR, t1/2 : 0.13–0.15 s; RXR, t1/2 : 0.2–0.25 s (29)]. In the case of PPAR, this lack of measurable difference may reflect some constitutive activity of the receptor (29).
In all FRAP experiments, the recovery dynamics will reflect both specific and nonspecific interactions. In the case of transient transfections, in which an excess of receptor may be present, nonspecific interactions are likely to be a significant contribution to the dynamics for both locus-specific recovery and general nuclear recovery. The recovery curve is likely a convolution of more than one kinetic process. In computational models of AR dynamics, the recovery was separated into two distinct kinetic components: a fast component (due to diffusion or transient binding) of 1–5 s and a slow component of 60 s (28, 50).
This slow component presumably represents a longer-lived interaction in the vicinity of the gene such as with chromatin or nuclear matrix (28, 49, 55, 69, 73, 83), although the nature of this interaction is not clear and may vary between receptors.
In addition to receptor dynamics, several studies have addressed the kinetic behavior of coactivators involved in NR-regulated transcription. Becker et al. (6) observed the receptor coactivator GRIP1 (glucocorticoid receptor interacting protein 1) at the active MMTV array and measured a recovery time that was indiscernible from the GR t1/2 (5 s), suggesting that the binding and release of these proteins may be coupled. CBP and SRC-1 (ER coactivators) have t1/2 times of 4 s and 8 s, respectively (85); BRM and BRG1, subunits of the SWI/SNF chromatin remodeling complex, have t1/2 times of 2 s and 4 s, respectively (45).
Taken together, the remarkable aspect of these data is that these recovery times are all less than or equal to 11 s (Table 1). Consider, for example, a typical NR transcription complex: NR t1/2 = 5 s, SRC1 t1/2 = 8 s, CBP t1/2 = 4 s (85), BRM t1/2 = 2 s, BRG1 t1/2 = 4 s (45), and GRIP1 t1/2 = 5 s (6). The only molecular species that has a dwell time on the order of minutes is the elongating polymerase (t1/2 5 min) (6). How might these transient interactions lead to transcriptional cycles that are observed in the timescale of hours? One idea that has been proposed is that of a transcriptional ratchet, in which permanent changes—methylation, acetylation, phosphorylation—accumulate at a transcription site as a result of the transient interactions described above (56). There are several suggestive directions about how such long-lived interactions might occur. SRC1 recovery becomes progressively slower at longer times after stimulation of ER with estradiol (t1/2 = 8.0–30.2 s) (85); chromatin decondensation seems to depend on polymerase elongation (59). Live cell experiments that follow the change in dynamics over an induction period are likely to be informative as well.
Table 1.
FRAP experiments summarized
| Receptor | Cofactor/muta tion |
Nuclear/array | Ligand | Agonist = 1; partial antagonist = 2; antagonist = 3 |
t1/2 (s) | Immobile fractionb |
Reference |
|---|---|---|---|---|---|---|---|
| AR | MMTV array | None | 0.2 | (50) | |||
| AR | MMTV array | R1881 | 1 | 3.6 | |||
| AR | MMTV array | DHT | 1 | 5.3 | |||
| AR | MMTV array | TST | 1 | 5 | |||
| AR | MMTV array | Bicalutamide | 3 | 0.5 | |||
| AR | MMTV array | OHF | 3 | 0.5 | |||
| AR | MMTV array | CPA | 2 | 1.1 | |||
| AR | MMTV array | RU486 | 2 | 4.3 | |||
| AR | Nuclear | R1881 | 1 | <5.90a | (28) | ||
| ER | Nuclear | None | 0.8 | (86) | |||
| ER | Nuclear | Estradiol | 1 | 5.9 | |||
| ER | Nuclear | 4-HT | 2 | 5.3 | |||
| ER | Nuclear | ICI | 3 | - | |||
| ER | SRC | Nuclear | Estradiol | 1 | 9.8 | ||
| ER | Artifcial array—LacO |
Estradiol | 1 | - | |||
| ER | SRC | Artifcial array—LacO |
None | 2.1 | (85) | ||
| ER | SRC | Artifcial array—LacO |
Estradiol | 1 | 8 | ||
| ER | CBP | Artifcial array—LacO |
Estradiol | 1 | 4.2 | ||
| ER | Nuclear | None | 1.6 | 0.14 | |||
| ER | Nuclear | Estradiol | 1 | 5.8 | 0.44 | (53) | |
| GR | MMTV array | Dexamethasone | 1 | 5 | (6) | ||
| GR | GRIP (p160 family) |
MMTV array | Dexamethasone | 1 | 5 | ||
| GR | RNAPII | MMTV array | Dexamethasone | 1 | 300 | ||
| GR | MMTV array | RU486 | 3 | <5 | (84) | ||
| GR | MMTV array | Corticosterone | 3 | <5 | |||
| GR | Nuclear | None | 1 | (73) | |||
| GR | Nuclear | Dexamethasone | 1 | 1.9 | |||
| GR | Nuclear | Triamcinolone | 1 | 1.87 | |||
| GR | Nuclear | Corticosterone | 1 | 1.13 | |||
| GR | Nuclear | RU486 | 3 | 1.43 | |||
| GR | Nuclear | ZK98299 | 3 | 1.07 | |||
| GR | Nuclear | Dexamethasone | 1 | 0.43–>1.11 | (49) | ||
| GR | MMTV array | Dexamethasone | 1 | 1.6 | (55) | ||
| GR | 407C–GR mutant |
MMTV array | Dexamethasone | 1 | 1.34 | ||
| GR | N525-GR mutant |
MMTV array | None | 1 | 0.69 | ||
| GR | MMTV array | RU486 | 3 | 0.82 | |||
| GR | MMTV array | Dex-Mes | 2 | 1.4 | |||
| GR | BRG1 | MMTV array | Dexamethasone | 1 | 3.9 | (45) | |
| GR | BRM | MMTV array | Dexamethasone | 1 | 1.95 | ||
| RAR | Nuclear | None | 1.9 | 0.18 | (53) | ||
| RAR | Nuclear | Retinoic acid | 1 | 2.3 | 0.18 | ||
| TR | Nuclear | None | 1.8 | 0.12 | (53) | ||
| TR | Nuclear | Triiodithyronine | 1 | 1.8 | 0.14 | ||
| PR | MMTV array | None | 0.6 | (69) | |||
| PR | MMTV array | R5020 | 1 | 3.7 | |||
| PR | MMTV array | RU486 | 3 | 11 | |||
| PR | MMTV array | ZK98299 | 3 | 1.8 | |||
| PPAR α | Nuclear | None | 0.13 | 0.017 | (29) | ||
| PPAR α | Nuclear | wy14643 | 1 | 0.15 | 0.024 | ||
| PPAR α | Nuclear | None | 0.1 | 0.014 | |||
| PPAR α | Nuclear | L-165041 | 1 | 0.16 | 0.03 | ||
| PPAR α | Nuclear | None | 0.1 | 0.05 | |||
| PPAR α | Nuclear | Rosiglitazone | 1 | 0.12 | 0.033 | ||
| RXR α | Nuclear | None | 0.2 | 0.019 | |||
| RXR α | Nuclear | 9-cis retinoic acid |
1 | 0.25 | 0.064 |
Two-component fit.
Immobile fraction is only reported in a subset of studies.
Abbreviations: AR, androgen receptor; Dex-Mes, dexamethasone mesylate; ER, estrogen receptor; GR, glucocorticoid receptor; GRIP, glucocorticoid receptor interacting protein; MMTV, mouse mammary tumor virus; PPAR α, peroxisome proliferator-activated receptor α; PR, progesterone receptor; RAR, retinoic acid receptor; RNAPII, RNA polymerase II; RXR α, retinoid X receptor α; TR, thyroid hormone receptor.
Imaging a Single Gene
Imaging the transcription of a single gene is potentially a powerful approach because it obviates the averaging inherent in gene array studies. This way, the behavior of individual transcription units can be quantified and their variability assessed. However, this has been difficult to achieve because of technical challenges: specifically detecting the desired locus and then observing the small numbers of factors involved in transcribing a single gene.
When a major challenge must be overcome, the tool of choice in vivo is fluorescence microscopy. Although a single fluorescent protein molecule can be detected when immobilized on a surface, it is difficult to resolve in the context of a living cell, where it undergoes fast diffusion or transport and where the fluorescent background can be high. So far, only a few experiments have managed to provide direct observation of gene expression at the single gene level.
A series of recent experiments demonstrated that it is possible to detect single protein products resulting from the expression of a single gene in live bacteria (15, 18, 97). From the distribution of proteins synthesized over time, it is then possible to test different models of transcription. In the first experiment (15), the reporter was a β-galactosidase protein, which produces a fluorescent product upon hydrolysis of a synthetic substrate. Hydrolysis of a large number of substrate molecules by a single enzyme provides the signal amplification necessary to observe a single protein. By observing discrete values in the rate of hydrolysis, the authors could indeed resolve single protein numbers. In subsequent experiments, the reporter was a fluorescent protein fused to a membrane protein (18, 97). When bound to the membrane, the protein is slowly diffusing and it is therefore possible to accumulate enough fluorescence to resolve a single protein.
These experiments studied reporter genes under the control of the Lac repressor. In this classic system, two operator sequences on the DNA can be bound by a tetramer repressor. Upon full induction, the repressor unbinds the DNA and the cell fully expresses the lac genes downstream. In the absence of inducer, protein is produced in infrequent bursts (0.5–1 per cell cycle) in which a few (2–4) proteins are produced. The distribution of the number of proteins produced per burst is consistent with a model in which a burst results from the transcription of a single mRNA molecule, finally yielding a few proteins. In the regime of moderate inducer concentration, both low-expressing cells (0–10 proteins) and high-expressing ones (hundreds of proteins) are observed. This bimodal distribution results from the presence of frequent, small bursts (similar to the noninduced state) and infrequent, large bursts of protein production. The authors proposed a model in which the small bursts consist of a partial dissociation of the repressor from one of the operator sequences; one RNA molecule is transcribed typically before the repressor binds back the operator sequence. In contrast, the large bursts correspond to full dissociation of the repressor from both operator sequences. In this case, many mRNA molecules are transcribed before another repressor binds the DNA, leading to the production of a large number of proteins.
A similar detection approach was used to study transcription factor dynamics in Escherichia coli at the single molecule level (27). Lac repressor (Lac I) molecules fused to a fluorescent protein could be detected when bound to their promoter sequence by imaging for long periods of time (1 s) to average out the background of freely diffusing molecules. This made it possible to measure the kinetics of association of Lac I to its promoter in vivo. The authors also used short light excitation pulses to characterize the diffusion of the repressor as well as its nonspecific binding to DNA. From these results emerged a picture of Lac I dynamics: Searching for its target sequence, the protein spends 87% of its time in short events (<5 ms), where it is nonspecifically bound to DNA and undergoes 1D diffusion while it scans the DNA. These short events are separated by periods where the repressor diffuses in three dimensions between different DNA segments.
It is also possible to directly visualize mRNA molecules using a technique that exploits the high affinity between RNA stem loops and the bacteriophage MS2 coat protein (7). By introducing repeats of the stem loop coding sequence in the desired gene, and expressing the MS2 coat protein fused to a fluorescent protein, one can detect single mRNA molecules. This technique has been used in numerous studies to characterize single mRNA motion and localization in E. coli (34), yeast (5, 7), mammalian cells (33, 72, 77), and Drosophila oocytes (31, 91, 98).
Golding et al. (35) used this technique to study transcription in E. coli by utilizing an inducible reporter gene under the control of the Plac/ara promoter. By measuring the distribution of mRNA molecules per cell, the authors tested two models for transcription. The simplest model, in which transcription events were randomly initiated according to a Poisson process (with a constant probability per unit time), could not fit the data; a more elaborate model, in which the gene can switch between an “off” state (no transcription takes place) and an “on” state (transcription is randomly initiated) successfully described the data. The gene stays “on” for typically 6 min, during which it produces approximately two transcripts. In contrast, the “off ” state lasts much longer (37 min), which results in a burst-like transcription behavior, even in full-induction conditions.
Bursts of transcription have also been observed on dscA, an endogenous developmental gene in the social amoeba Dictyostelium discoideum (20). Although the authors could not detect single mRNA particles, they could resolve the sites of transcription because of the high fluorescence accumulated by the multiple nascent mRNAs. As differentiation occurred, they could observe the dscA gene switch between the “on” and “off” states, which displayed similar lifetimes (5.2 and 5.8 min, respectively), in contrast with the E. coli result. Over the course of development, variation was only observed in the fraction of the population expressing the gene, but the lifetime of the “on” and “off” states remained constant. In addition, the authors observed transcriptional memory, as a gene was more likely to enter the “on” state if it had been transcribing before than when it was undergoing de novo transcription.
In spite of their quantitative differences, both studies could be modeled the same way, using a simple two-state system. The nature of the event(s) that dictates the transition between the “on” and “off” states has yet to be discovered, but it could consist of DNA conformational change and/or chromatin remodeling, binding (or release) of an activator (or repressor), or transcription pausing/ reinitiation.
These studies demonstrate the potential of single-molecule techniques in studying transcription in vivo and open avenues for future research. Upcoming directions could involve expanding these observations to different systems and/or trying to combine imaging of different factors at a given locus.
ANALYSIS OF KINETICS Mobility of Transcription Factors
Transcription factor mobility represents the process of a genome-wide search for specific target sites. Since the discovery of the GFP, more precise observations of nuclear protein mobility have been enabled. In vivo techniques identify populations of transcription factors on a real-time timescale, breaking down many assumptions previously held about this topic.
Dynamics
The use of fluorescent proteins made it possible to conduct experiments on the transcription factors of yeast (47), bacteria (26), and mammalian cells (54), enlightening many aspects of the dynamic behavior of transcription factors, from Brownian motion to anomalous diffusion to cyclic binding (dynamic equilibria) at binding sites to dynamic complex formation. Transcription factors are generally impaired in their diffusion throughout the nucleus by unspecific interactions with other nuclear components (64). Furthermore, a tagged nuclear protein might exhibit more than one apparent diffusion constant due to complex formation. Most models of FRAP have been applied to homogeneously and globally distributed binding sites, easily approximated by diffusion.
The pioneering work on the construction of localized cluster binding sites in the genome (54, 89) made it possible to address specific binding of transcription factors in the nucleus. Sprague et al. (80) used such a system to prove that recoveries resulting from bleaching of the tandem array area could not fit a model accounting for only diffusion. The new model involves “on” and “off” rates of the transcription factor’s binding to the promoter array and provides information on the binding dynamics of the system. A similar construct has been used by the Singer laboratory (21) to analyze transcriptional mechanisms by bleaching the Pol II and nascent RNAs on a tandem array. This particular system was demonstrated to be kinetically independent of the availability of the freely diffusing components, therefore making it possible to disregard the diffusing component (80) and to use first-order differential equations to model the reactions.
The work of Natoli’s laboratory (9) on NF-κB promoter binding microdynamics showed that stable bindings were actually states of dynamic equilibrium between promoter-bound and nucleoplasmic dimers (Figure 3e). In a subsequent study, Karpova et al. (47) showed that the yeast transcription factor Ace1p fit an accessibility model in which the slow cycle of binding reflects the number of accessible binding sites at promoters and each accessible site can be bound by fast-cycling molecules.
Complex formation of transcription factors on their promoters is likewise of a highly dynamic nature: The factor and its partners do not associate with and are not released from target promoters as a single and stable complex (9, 36, 71). Bosisio et al. (9) specified that NF-κB residence time on specific sites defined a stochastic window during which general transcription factors and possibly additional activators must collide with the same regulatory region for transcription to occur. Furthermore, Gorski et al. (36) successfully proved the role of complex formation in regulating transcription. Remodeling factors also play a critical role in the regulation of gene expression and in governing the dynamics of transcription factors (46, 47).
Modeling
FRAP analysis quantifies and sets a kinetic model characterized by parameters of diffusion coefficients, chemical rates, and residence times translatable into differential equations. Taking into account only diffusion, a convenient way of displaying fluorescence recovery curves is the form defining the mobile fraction:
Under assumptions of a Gaussian intensity profile laser, this gives the closed form solution of the normalized fluorescence recovery:
where Po designates the total laser power, K the bleaching parameter, Co the initial fluorophore concentration, and A the attenuation factor of beam during the observation of the FRAP recovery. q is the product of all quantum efficiencies of light absorption, emission, and detection. Furthermore, parameter v and the characteristic diffusion time τ D are given by υ = (1 + 2t/τ D)−1 and τ D = w2/4D, respectively, where t is time and w is the radius of the laser beam at e−2 intensity/height. r(υ) is given by the gamma function (4). P(2K | υ) is the probability distribution tabulated in Reference 1.
However, this might be only an approximation when it comes to systems in which specific binding cannot be ignored. A complete solution to FRAP reaction-diffusion equations has been proposed (81) in which various special cases of FRAP with binding (diffusion/binding dominant) can be covered by a set of differential equations including the diffusion terms given above, as well as the chemical kinetics of binding.
Whether a dynamic system is diffusion limited depends on the magnitude of two parameters: diffusion time and association rate. The relative magnitude of these two parameters reflects potential interplay between diffusion and binding and thus determines whether a recovery is diffusion coupled or uncoupled (79). A simple method of testing whether a system is diffusion limiting is to vary the spot size of the bleach: If the recovery is dependent on the spot size, the system is diffusion limiting and must be included in the analysis (83).
Apart from diffusion modeling with differential equations, Rino et al. (71) successfully portrayed modeling of splicing protein kinetics in the nucleus with a method involving kon and koff rates in a Monte Carlo simulation. Other types of modeling might explain dynamic behaviors (for more details, see Reference 65).
CONCLUSION
Controlled manipulation of the biological system by using drugs has proved to be a useful tool to perturb biological mechanisms in order to obtain deeper understanding of the mechanisms involved (32, 21). Other biological manipulation stems from the construction of binding defective mutants of the transcription factors under analysis (46, 82). Photoactiveable and photoswitchable fluorophores have a particular advantage, however, in that they can be used with single-cell, single-molecule sensitivity, and they produce photons, which are easily measured, quantified, and converted to the dynamic behavior of transcription (92). Further, hyper-resolution techniques provide a tool to produce single-molecule dynamic measurements at the subdiffraction level. Using these approaches, it will be possible to answer crucial questions about how transcription factor dynamics regulate gene expression, how transcription factors sort the right genes, and how they search for their targets. Short residence times, stochastic formation of complexes, anomalous diffusion with continuous assembly, and disassembly of the transcription factors is only the beginning of a complex story about the dynamic behavior of transcription factors.
THE FUTURE
The current conclusions regarding transcription dynamics are based mainly on synthetic genes and cell lines that give us some insight into the processes involved in gene expression. However, the next important step is to apply the technology to minimally perturbed systems, endogenous genes, and primary cells or tissues. To achieve this we will need more sensitive systems capable of processing weak signals. Additionally, high-speed imaging will be required to separate transient and rapid events from the diffusional rates occurring in the background. Brighter fluorochromes with lower photobleaching or novel labeling systems capable of multiplexing will also be required. Finally, the digitization of the data will allow for the type of mining that is common with microarray databases, but required now are algorithms capable of extracting data from large image sets, particularly those that contain 4D information (a time series in three dimensions). The field therefore will assemble expertise from engineers, computer scientists, chemists, physicists, and biophysicists. As these explorations evolve, they will lead to leaps in understanding the biological basis of gene expression.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Gerry Rubin and Kevin Moses for the support of this Consortium and John Lis and Watt Webb for reading portions of this manuscript and giving permission for use of published figures. Support for personnel is from the HHMI, the NIH, and the CNRS. The authors thank Shailesh Shenoy for his help in preparing the manuscript.
Abbreviations Used
- FCS
fluorescence correlation spectroscopy
- FISH
fluorescent in situ hybridization
- FRAP
fluorescence recovery after photobleaching
- FRET
fluorescence resonance energy transfer
- GFP
green fluorescent protein
- GR
glucocorticoid receptor
- HSF
heat shock factor
- LTR
long terminal repeat
- MMTV
mouse mammary tumor virus
- MPM
multiphoton microscopy
- NR
nuclear receptor
- Pol I (II)
RNA polymerase I (II)
- PR
progesterone receptor
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abramowitz M, Stegun IA. Handbook of Mathematical Functions. New York: Dover; 1965. [Google Scholar]
- 2.Abruzzi KC, Belostotsky DA, Chekanova JA, Dower K, Rosbash M. 3’ -end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation. EMBO J. 2006;25:4253–4262. doi: 10.1038/sj.emboj.7601305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 4.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 6.Becker M, Baumann C, John S, Walker DA, Vigneron M, et al. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–1194. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 8.Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, et al. The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. A hyperdynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee P-C, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bridges CB. Salivary chromosome maps. J. Hered. 1935;26:60–64. [Google Scholar]
- 14.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, et al. SAGA interacting factors confine subdiffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 15.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 16.Carter DR, Eskiw C, Cook PR. Transcription factories. Biochem. Soc. Trans. 2008;36:585–589. doi: 10.1042/BST0360585. [DOI] [PubMed] [Google Scholar]
- 17.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 18. Choi PJ, Cai L, Frieda K, Xie XS. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science. 2008;322:442–446. doi: 10.1126/science.1161427. The number of proteins expressed from a gene under the control of LacO is linked to dissociation of the repressor.
- 19.Chuang C-H, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 20. Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr. Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. This work demonstrates the stochastic behavior of an endogenous, developmentally regulated gene.
- 21. Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, et al. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. This work demonstrates the power of combining live cell imaging of transcription with mathematical modeling to determine rate constants for each of the various components of the process.
- 22.Darzacq X, Singer RH. The dynamic range of transcription. Mol. Cell. 2008;30:545–546. doi: 10.1016/j.molcel.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, et al. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. This is the first work to measure polymerase kinetics.
- 25.Dundr M, Ospina JK, Sung M-H, John S, Upender M, et al. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. Specific binding, diffusion tracking, and colocalization of the transcription factor to its DNA binding domains are measured.
- 27.Eskiw CH, Rapp A, Carter DR, Cook PR. RNA polymerase II activity is located on the surface of protein-rich transcription factories. J. Cell Sci. 2008;121:1999–2007. doi: 10.1242/jcs.027250. [DOI] [PubMed] [Google Scholar]
- 28.Farla P, Hersmus R, Geverts B, Mari PO, Nigg AL, et al. The androgen receptor ligand-binding domain stabilizes DNA binding in living cells. J. Struct. Biol. 2004;147:50–61. doi: 10.1016/j.jsb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Feige JN, Gelman L, Tudor C, Engelborghs Y, Wahli W, Desvergne B. Fluorescence imaging reveals the nuclear behavior of peroxisome proliferator-activated receptor/retinoid X receptor heterodimers in the absence and presence of ligand. J. Biol. Chem. 2005;280:17880–17890. doi: 10.1074/jbc.M500786200. [DOI] [PubMed] [Google Scholar]
- 30.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 32.Fromaget M, Cook PR. Photobleaching reveals complex effects of inhibitors on transcribing RNA polymerase II in living cells. Exp. Cell Res. 2007;313:3026–3033. doi: 10.1016/j.yexcr.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 33.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proc. Natl. Acad. Sci. USA. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. A real-time quantitative study of the expression of a single gene in bacteria demonstrating burstlike transcription.
- 35.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 36. Gorski SA, Snyder SK, John S, Grummt I, Misteli T. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol. Cell. 2008;30:486–497. doi: 10.1016/j.molcel.2008.04.021. This work demonstrates the power of combining live cell imaging of transcription with mathematical modeling to determine rate constants for each of the various components of the process.
- 37.Hieda M, Winstanley H, Maini P, Iborra FJ, Cook PR. Different populations of RNA polymerase II in living mammalian cells. Chromosome Res. 2005;13:135–144. doi: 10.1007/s10577-005-7720-1. [DOI] [PubMed] [Google Scholar]
- 38.Hinojos CA, Sharp ZD, Mancini MA. Molecular dynamics and nuclear receptor function. Trends Endocrinol. Metab. 2005;16:12–18. doi: 10.1016/j.tem.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J. Cell Sci. 1996;109(Pt. 6):1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 40.Iborra FJ, Pombo A, McManus J, Jackson DA, Cook PR. The topology of transcription by immobilized polymerases. Exp. Cell Res. 1996;229:167–173. doi: 10.1006/excr.1996.0355. [DOI] [PubMed] [Google Scholar]
- 41.Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson DA, Iborra FJ, Manders EM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeang KT, Xiao H, Rich EA. Multifaceted activities of the HIV-1 transactivator of transcription. Tat. J. Biol. Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 45.Johnson TA, Elbi C, Parekh BS, Hager GL, John S. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol. Biol. Cell. 2008;19:3308–3322. doi: 10.1091/mbc.E08-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karpova TS, Chen TY, Sprague BL, McNally JG. Dynamic interactions of a transcription factor with DNA are accelerated by a chromatin remodeller. EMBO Rep. 2004;5:1064–1070. doi: 10.1038/sj.embor.7400281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, et al. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. This work demonstrates the rapid turnover of transcription factors at the promoter.
- 48.Kimura H, Sugaya K, Cook PR. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kino T, Liou SH, Charmandari E, Chrousos GP. Glucocorticoid receptor mutants demonstrate increased motility inside the nucleus of living cells: time of fluorescence recovery after photobleaching (FRAP) is an integrated measure of receptor function. Mol. Med. 2004;10:80–88. doi: 10.2119/2005-00026.Kino. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klokk TI, Kurys P, Elbi C, Nagaich AK, Hendarwanto A, et al. Ligand-specific dynamics of the androgen receptor at its response element in living cells. Mol. Cell Biol. 2007;27:1823–1843. doi: 10.1128/MCB.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 53.Maruvada P, Baumann CT, Hager GL, Yen PM. Dynamic shuttling and intranuclear mobility of nuclear hormone receptors. J. Biol. Chem. 2003;278:12425–12432. doi: 10.1074/jbc.M202752200. [DOI] [PubMed] [Google Scholar]
- 54. McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. This seminal paper was the first to show the dynamic behavior of transcription factors at an active gene locus in a living cell.
- 55.Meijsing SH, Elbi C, Luecke HF, Hager GL, Yamamoto KR. The ligand binding domain controls glucocorticoid receptor dynamics independent of ligand release. Mol. Cell Biol. 2007;27:2442–2451. doi: 10.1128/MCB.01570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metivier R, Reid G, Gannon F. Transcription in four dimensions: nuclear receptor-directed initiation of gene expression. EMBO Rep. 2006;7:161–167. doi: 10.1038/sj.embor.7400626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molle D, Maiuri P, Boireau S, Bertrand E, Knezevich A, et al. A real-time view of the TAR:Tat:PTEFb complex at HIV-1 transcription sites. Retrovirology. 2007;4:36. doi: 10.1186/1742-4690-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller WG, Rieder D, Karpova TS, John S, Trajanoski Z, McNally JG. Organization of chromatin and histone modifications at a transcription site. J. Cell Biol. 2007;177:957–967. doi: 10.1083/jcb.200703157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller WG, Walker D, Hager GL, McNally JG. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 2001;154:33–48. doi: 10.1083/jcb.200011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nalley K, Johnston SA, Kodadek T. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature. 2006;442:1054–1057. doi: 10.1038/nature05067. [DOI] [PubMed] [Google Scholar]
- 61.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan D, Zhang L. Tandemly arrayed genes in vertebrate genomes. Comp. Funct. Genomics. 2008:545269. doi: 10.1155/2008/545269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 64.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 65.Phair RD, Misteli T. Kinetic modelling approaches to in vivo imaging. Nat. Rev. Mol. Cell Biol. 2001;2:898–907. doi: 10.1038/35103000. [DOI] [PubMed] [Google Scholar]
- 66.Pombo A, Cook PR. The localization of sites containing nascent RNA and splicing factors. Exp. Cell Res. 1996;229:201–203. doi: 10.1006/excr.1996.0360. [DOI] [PubMed] [Google Scholar]
- 67.Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rafalska-Metcalf IU, Janicki SM. Show and tell: visualizing gene expression in living cells. J. Cell Sci. 2007;120:2301–2307. doi: 10.1242/jcs.008664. [DOI] [PubMed] [Google Scholar]
- 69.Rayasam GV, Elbi C, Walker DA, Wolford R, Fletcher TM, et al. Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol. Cell Biol. 2005;25:2406–2418. doi: 10.1128/MCB.25.6.2406-2418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 71.Rino J, Carvalho T, Braga J, Desterro JM, Luhrmann R, Carmo-Fonseca M. A stochastic view of spliceosome assembly and recycling in the nucleus. PLoS Comput. Biol. 2007;3:2019–2031. doi: 10.1371/journal.pcbi.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rook MS, Lu M, Kosik KS. CaMKIIalpha 3’ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J. Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schaaf MJ, Cidlowski JA. Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity. Mol. Cell Biol. 2003;23:1922–1934. doi: 10.1128/MCB.23.6.1922-1934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleoporepromoter interaction of genes in yeast. Mol. Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 75.Schneider DA, Nomura M. RNA polymerase I remains intact without subunit exchange through multiple rounds of transcription in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2004;101:15112–15117. doi: 10.1073/pnas.0406746101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sehgal PB, Derman E, Molloy GR, Tamm I, Darnell JE. 5,6-Dichloro-1-Beta-Dribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous RNA chains in HeLa cells. Science. 1976;194:431–433. doi: 10.1126/science.982026. [DOI] [PubMed] [Google Scholar]
- 77.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shav-Tal Y, Singer RH, Darzacq X. Imaging gene expression in single living cells. Nat. Rev. Mol. Cell Biol. 2004;5:855–861. doi: 10.1038/nrm1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 80.Sprague BL, Muller F, Pego RL, Bungay PM, Stavreva DA, McNally JG. Analysis of binding at a single spatially localized cluster of binding sites by fluorescence recovery after photobleaching. Biophys. J. 2006;91:1169–1191. doi: 10.1529/biophysj.105.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sprouse RO, Shcherbakova I, Cheng H, Jamison E, Brenowitz M, Auble DT. Function and structural organization of Mot1 bound to a natural target promoter. J. Biol. Chem. 2008;283:24935–24948. doi: 10.1074/jbc.M803749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stavreva DA, McNally JG. Fluorescence recovery after photobleaching (FRAP) methods for visualizing protein dynamics in living mammalian cell nuclei. Methods Enzymol. 2004;375:443–455. doi: 10.1016/s0076-6879(03)75027-7. [DOI] [PubMed] [Google Scholar]
- 84.Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stenoien DL, Nye AC, Mancini MG, Patel K, Dutertre M, et al. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol. Cell Biol. 2001;21:4404–4412. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, et al. FRAP reveals that mobility of oestrogen receptor-alpha is ligand and proteasome-dependent. Nat. Cell Biol. 2001;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- 87.Sugaya K, Vigneron M, Cook PR. Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J. Cell Sci. 2000;113(Pt. 15):2679–2683. doi: 10.1242/jcs.113.15.2679. [DOI] [PubMed] [Google Scholar]
- 88.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 89.Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat. Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- 90.Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 2003;160:685–697. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev. Cell. 2006;11:251–262. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 92.Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annu. Rev. Biophys. 2008;37:417–444. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 93.Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J. Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol. Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 95. Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. This work first describes the method of MPM imaging Drosophila polytene chromosomes to study transcription factor kinetics at endogenous gene loci.
- 96.Yao J, Zobeck KL, Lis JT, Webb WW. Imaging transcription dynamics at endogenous genes in living Drosophila tissues. Methods. 2008;45:233–241. doi: 10.1016/j.ymeth.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 98.Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, et al. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.