Abstract
We show the intracellular localization of the Escherichia coli replication origin (oriC) and chromosome terminus during the cell division cycle by FISH. In newborn cells, oriC is localized at the old-pole-proximal nucleoid border and the terminus at the new-pole-proximal nucleoid border. One copy of replicated oriC migrates rapidly to the opposite nucleoid border. These oriC copies are retained at both nucleoid borders, remaining at a constant distance from each cell pole. The terminus segment migrates from the nucleoid border to midcell and is retained there until the terminus is duplicated. The origin, terminus and other DNA regions show three migration patterns during active partitioning of daughter chromosomes.
Keywords: Cell division cycle, FISH, mitotic-like apparatus, oriC, positioning, segregation
Escherichia coli cells have a 4.6-Mb circular chromosome (Blattner et al. 1997) that is folded and organized in a compact form called the nucleoid. During the cell division cycle, replication of the chromosomal DNA initiates bidirectionally in a specific chromosome region, oriC. The two replication forks meet at the chromosome region opposite oriC. In this region, the progress of replication forks traveling clockwise and counter-clockwise is inhibited at specific DNA sequences, ter, in a direction-specific manner (Hill 1996). Thus, this region is replicated at the end of a round of chromosome replication and is called the replication terminus. The replicated chromosomal DNA molecules are physically separated by the processes of decatenation and/or recombination, and then each chromosome is partitioned with high fidelity into daughter cells (for review, see Hiraga 1992; Løbner-Olesen and Kuempel 1992; Rothfield 1994; Wake and Errington 1995). The partitioning mechanism is poorly understood in prokaryotic cells. Recently, bipolar localization of specific chromosome regions during the cell division cycle was reported, and this movement suggested that a mitotic-like apparatus was involved in chromosome partitioning in bacteria (Glaser et al. 1997; Lin et al. 1997; Mohl and Gober 1997; Webb et al. 1997; Wheeler and Shapiro 1997).
Donachie and Begg (1989) and Hiraga et al. (1990) observed that after inhibition of protein synthesis, replicated chromosomes do not separate from each other but remain together at midcell. When protein synthesis resumed, the chromosomes moved from midcell to cell quarter positions without cell elongation. This suggested that rapid movement of chromosomes was responsible for chromosome partitioning in bacteria (Hiraga 1990; Hiraga et al. 1990). Bipolar localization of the chromosome segment near oriC was visualized in Bacillus subtilis (Webb et al. 1997) and E. coli (Gordon et al. 1997). LacI-binding sequences were inserted into the chromosome near oriC, and the green fluorescent protein (GFP) fused to LacI was detected in living cells. Fluorescent foci of the GFP–LacI fusion protein bound to the chromosomal site near oriC showed that one copy of the replication origin region moved to the site of new pole formation, near the site of cell division, following replication at or near the cell poles. In addition, bipolar localization of the Spo0J protein was also observed by immunofluorescence microscopy or Spo0J–GFP in B. subtilis (Glaser et al. 1997; Lin et al. 1997). Spo0J binds to a DNA site in the origin-proximal third of the chromosome (Lin et al. 1997) and colocalizes with the oriC region (Lewis and Errington 1997). A spo0J mutant causes production of a significant proportion of anucleate cells and is blocked at the onset of sporulation (Ireton et al. 1994). These results suggested that Spo0J is involved in a mitotic-like apparatus responsible for the migration of the oriC region, and the binding site for Spo0J is a cis-acting site for partitioning, like a eukaryotic centromere. In Caulobacter crescentus, ParB binds to an AT-rich sequence, parS, downstream of the parAB operon, which is located within 80 kb of the replication origin (Mohl and Gober 1997). The ParA and ParB proteins are localized at cell poles in one stage of the cell cycle, and overproduction of ParA and ParB inhibits both cell division and chromosome partitioning. Thus, ParA and ParB may function as components of a mitotic-like apparatus, and the parS site is the only candidate for a centromere that has been sequenced in a bacterial chromosome.
Partition defective mutants that produce anucleate cells upon cell division were isolated in E. coli (Hiraga et al. 1989). The mukF, mukE, and mukB genes form an operon, and null mutants of each gene show a severe defect in chromosome partitioning and cell viability (Niki et al. 1991; Yamanaka et al. 1996). The mukB gene codes for a filamentous protein of 170 kD whose structure resembles that of motor proteins such as myosin or kinesin (Niki et al. 1992). MukB is presumably responsible for generating motive force during chromosome partitioning. In B. subtilis, the SpoIIIE protein that localizes at the center of the division septum promotes chromosome segregation into the prespore during sporulation (Wu and Errington 1994, 1997). The origin-proximal third of the chromosome is segregated into the prespore compartment by Spo0J, localized at the cell poles during sporulation (Glaser et al. 1997), and the rest of the chromosome is translocated to the prespore by SpoIIIE.
In addition to the bipolar behavior of the chromosome origin copies, the F plasmid also shows bipolar migration of replicated plasmid DNA molecules, which are actively partitioned into daughter cells on cell division (Gordon et al. 1997; Niki and Hiraga 1997). The F plasmid is partitioned to daughter cells with high fidelity owing to two trans-acting genes, sopA and sopB, and a cis-acting site, sopC (Ogura and Hiraga 1983). An F plasmid molecule having the sopABC partitioning system is localized at midcell in newborn cells. The replicated plasmid molecules migrate in opposite directions to cell positions one-fourth and three-fourths of the cell length and are tethered at these positions until the cell divides. Therefore, each daughter cell receives at least one F plasmid DNA molecule. P1 plasmid shows the same behavior during the cell division cycle (Gordon et al. 1997). Interestingly, Soj, in an operon with spo0J, and ParA in C. crescentus, belong to the ATPase family including the plasmid-encoded F-SopA and P1-ParA proteins, essential for plasmid partitioning. B. subtilis Spo0J and C. crescentus ParB are members of a family that includes F-SopB and P1-ParB. The basic mechanism of the mitotic-like apparatus for chromosome and plasmid migration may be common in various bacteria.
Mitotic-like apparatuses thus seem feasible for chromosome and plasmid partitioning in bacteria. On the other hand, a gradual movement of bulk nucleoids in parallel with cell elongation has been described (van Helvoort and Woldringh 1994). The distance of nucleoid outer borders and cell poles is constant in living and fixed cells in all stages of the cell division cycle. Therefore, these authors concluded that the partitioning of daughter chromosomes occurs slowly and gradually throughout the cell division cycle and that E. coli does not have a mitotic-like apparatus for rapid chromosome movement during partitioning.
In contrast to the above conclusion, we show here statistical results by FISH indicating that one copy of the duplicated oriC region migrates rapidly along the nucleoid from one border to another. We also show a different dynamic migration pattern of the terminus region of the chromosome. By simultaneous use of DNA probes corresponding to different chromosomal regions and labeled with different fluorescent compounds, in situ hybridization revealed cell cycle-dependent nucleoid organization. We propose a model for the positioning of daughter chromosomes.
Results
Migration of the oriC DNA segment across the nucleoid
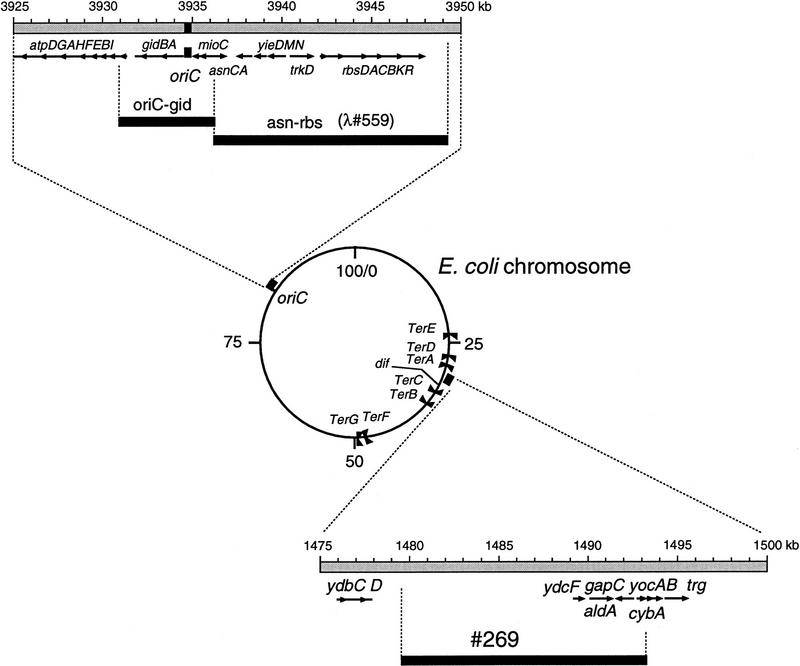
Bidirectional chromosome replication is initiated at oriC, which is a minimal 245-bp sequence (Oka et al. 1980) located at 84 min on the E. coli chromosome. To analyze the intracellular localization of oriC by FISH, we used two kinds of DNA segments as hybridization probes (Fig. 1). The oriC–gid DNA probe was prepared from a 6.3-kb DNA segment containing the oriC and gid genes. The asn–rbs DNA probe was prepared from about 13 kb of the oriC-flanking DNA segment containing the asn to rbs genes, but not oriC. There is an ∼2-kb gap between the minimal oriC region and the left border of the asn–rbs DNA segment. Cells of strain CSH50 growing exponentially in minimal glucose medium (doubling time 55 min) were fixed and hybridized with either probe (Fig. 2A,B).
Figure 1.
Map of the E. coli chromosome and location of the DNA probes used for FISH. The coordinates of the loci and of Kohara’s λ phage clones are given in kb according to EcoMap7 (Berlyn et al. 1996).
Figure 2.
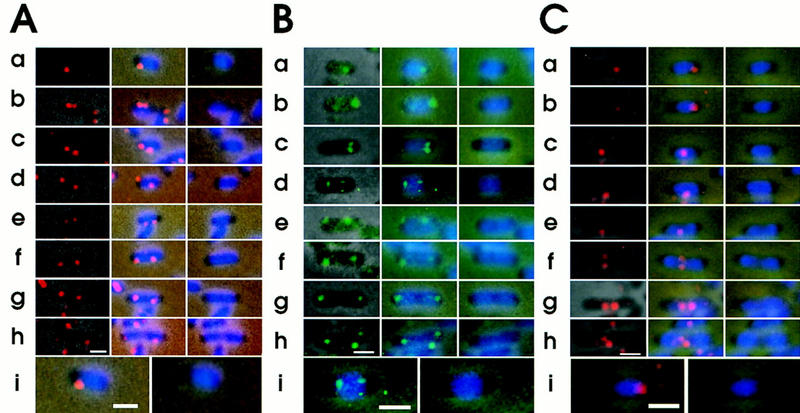
Intracellular localization of replication origin and terminus detected by FISH. Cells of strain CSH50, grown exponentially at 37°C in M9 minimal glucose medium, were fixed. The fixed cells were hybridized with the Cy3-labeled oriC–gid DNA probe (A), the fluorescein-labeled asn–rbs DNA probe (B), and the Cy3-labeled ter DNA probe (C). (Left columns) Combined images of the phase contrast micrograph and fluorescence micrograph for FISH; (middle columns) combined images of the phase contrast micrograph, fluorescence micrograph for FISH and fluorescence micrograph for DAPI; (right columns) combined images of the phase contrast micrograph, and fluorescence micrograph for DAPI. The cells from A row a, B row d, and C row a are shown at higher magnification. Bar, 1 μm.
One or more apparent fluorescent foci were observed in more than 97% of cells using the oriC–gid probe. Cells with one, two, three, and four foci were 27%, 63%, 5.4%, and 4.6% of total cells having fluorescent foci, respectively. In short cells with a single focus, the focus was localized at a pole-proximal nucleoid border (Fig. 2A, row a). Two foci in short cells were either closely localized near one nucleoid border (Fig. 2A, row b) or separated from each other (Fig. 2A, rows c–e). In long cells, two foci were localized near the two borders of a nucleoid (Fig. 2A, rows f–h). Similar results were obtained with the asn–rbs DNA probe (Fig. 2B), indicating that one copy of the replicated oriC-flanking region, like oriC, also migrated to the opposite border of the nucleoid.
Localization of the oriC DNA segments at a constant distance from the nearest cell pole
To analyze statistically the intracellular distribution of the oriC DNA segment, we measured the distance from the center of each fluorescent focus to the nearest cell pole in 200 cells hybridized with the oriC–gid probe. In cells having a single fluorescent focus, the focus was mainly distributed around the one-fourth position of cell length (Fig. 3A,B). It should be noted that the focus of the oriC–gid probe was not localized at the extreme position of the cell pole or midcell. The average cell length of cells with a single focus was 1.45 ± 0.03 μm, suggesting that these cells were newborn or in an early stage of the cell division cycle (Fig. 3A; Table 1). The average distance between a focus and the nearest cell pole was 0.40 ± 0.02 μm.
Figure 3.
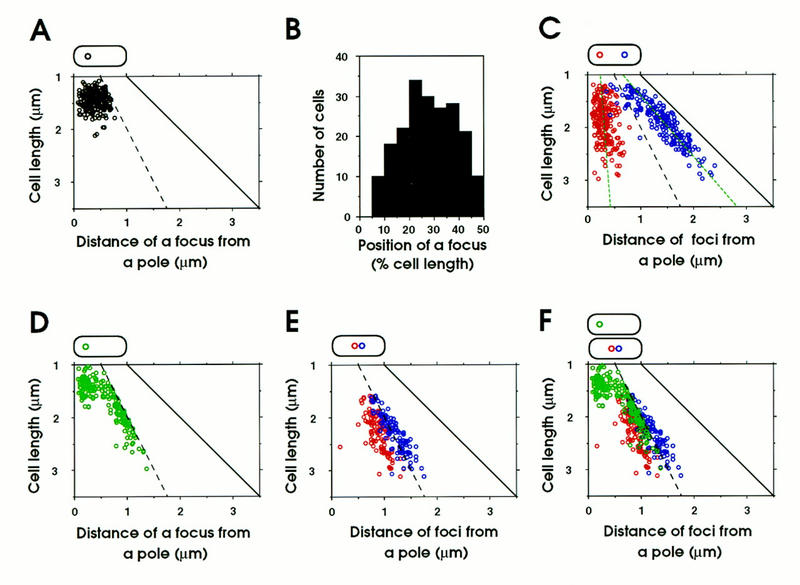
Analysis of intracellular distribution of replication origin and terminus in cells. Fixed cells were hybridized with a Cy3-labeled DNA probe. (A) Cells with a single fluorescent focus of oriC–gid. The distance between the center of the focus and the nearest pole was plotted versus cell length (n = 200). (B) Histogram of position of the oriC–gid focus in cells with a single focus. Position of the focus is given as a percent of cell length (n = 200). (C) Cells with two fluorescent ori–gid foci. The positions of the foci from a pole are plotted (red and a dark blue circles) versus cell length (n = 200). The green broken lines indicate regression lines. (D) Cells with one fluorescent focus of the terminus DNA segment (n = 200). (E) Cells with two fluorescent foci of the terminus DNA segment (n = 200). (F) Combination of D and E. The black broken line indicates midcell and the black solid line indicates the position of a pole.
Table 1.
Intracellular localization of fluorescent oriC foci
| Type of cellsa
|
Classification of cell length (μm)
|
Average cell length (μm) (L)
|
Average position of one-quarter (μm) (L) × 1/4
|
Distance of a focus from a pole (μm)
|
Number of sample cells
|
||
|---|---|---|---|---|---|---|---|
| left
|
right
|
average
|
|||||
| 1 | 1.0 ⩽ C.L. < 1.5 | 1.35 ± 0.02 | 0.34 | 0.39 ± 0.03 | 126 | ||
| 1.0 ⩽ C.L. < 2.0 | 1.63 ± 0.02 | 0.41 | 0.42 ± 0.04 | 72 | |||
| total cells | 1.45 ± 0.03 | 0.36 | 0.40 ± 0.02 | 198 | |||
| 2 | 1.0 ⩽ C.L. < 1.5 | 1.35 ± 0.03 | 0.34 | 0.28 ± 0.04 | 0.47 ± 0.06 | 0.38 ± 0.01 | 36 |
| 1.5 ⩽ C.L. < 2.0 | 1.75 ± 0.02 | 0.43 | 0.30 ± 0.03 | 0.47 ± 0.04 | 0.39 ± 0.03 | 86 | |
| 2.0 ⩽ C.L. < 2.5 | 2.23 ± 0.02 | 0.56 | 0.37 ± 0.04 | 0.55 ± 0.04 | 0.46 ± 0.03 | 65 | |
| 2.5 ⩽ C.L. < 3.0 | 2.71 ± 0.06 | 0.68 | 0.44 ± 0.10 | 0.60 ± 0.09 | 0.52 ± 0.07 | 13 | |
| total cells | 1.90 ± 0.06 | 0.48 | 0.33 ± 0.02 | 0.51 ± 0.03 | 0.42 ± 0.02 | 200 | |
Type 1 of cells had one fluorescent oriC focus; type 2 of cells had two fluorescent oriC foci.
In cells with two fluorescent foci, the foci were separated from each other and localized near the cell poles (Fig. 3C). Two foci were already separated from each other in short cells of length 1.3–1.5 μm, although there were a few cells with two closely located foci. The intracellular distribution of the oriC–gid foci indicated that oriC DNA segments were localized at an approximately constant distance from the nearest cell pole, regardless of cell length. The average distance between a focus and the nearest cell pole was 0.42 ± 0.02 μm (Table 1). The above results indicate that one of the duplicated oriC copies remained near a nucleoid border, whereas the other migrated rapidly toward the opposite nucleoid border, where it remained at a constant distance from the nearest cell pole throughout the cell division cycle, until these oriC copies were again replicated.
Migration of a terminus DNA segment from a nucleoid border to midcell
To detect the replication terminus of the E. coli chromosome by the FISH method, we used a fluorescent DNA probe, called the ter probe, which was prepared from the DNA segment located between the 1480- and 1493-kb positions of the chromosome map (Fig. 1). We detected one or two discrete fluorescent foci in CSH50 cells grown exponentially in minimal glucose medium (Fig. 2C). Statistical analysis revealed that cells with one or two foci were 74.7% and 25.3% of total hybridized cells, respectively; we did not observe cells with three or four foci. In short cells, the terminus DNA was localized near a nucleoid border (Fig. 2C, rows a and b) or at midcell (Fig. 2C, row c). The long cells have a single focus (Fig. 2C, rows d and e) or two foci located (Fig. 2C, rows f–h) at midcell. The statistical analysis clearly indicated that short cells (1.3–1.5 μm) have mainly a single focus (Fig. 3D). The average distance between a focus and the nearest cell pole was 0.32 ± 0.04 μm (Table 2). In cells longer than about 1.5 μm, a single focus and two foci were located in the midcell region (Fig. 3D,F; Table 2). A track of the migrating terminus DNA segment from the nucleoid border to midcell can be seen in Figure 3D. Thus, these statistical results suggest that the terminus DNA segment was initially localized near a new-pole-proximal nucleoid border and subsequently migrated toward midcell, where it was tethered until the segment was duplicated. The duplicated terminus DNA segments were gradually separated, and a septum was formed between them (Fig. 3E,F).
Table 2.
Intracellular localization of a single fluorescent focus of the terminus region
| Classification of cell length (μg)
|
Average cell length (μm) (L)
|
Average position of mid-cell (μm) (L) × 1/2
|
Distance of a focus from a pole (μm)
|
Number of sample cells
|
|---|---|---|---|---|
| 1.0 ⩽ C.L. <1.5 | 1.34 ± 0.03 | 0.67 | 0.32 ± 0.04 | 77 |
| 1.5 ⩽ C.L. <2.0 | 1.73 ± 0.02 | 0.87 | 0.66 ± 0.06 | 79 |
| 2.0 ⩽ C.L. <2.5 | 2.18 ± 0.02 | 1.09 | 0.99 ± 0.03 | 37 |
| 2.5 ⩽ C.L. <3.0 | 2.70 ± 0.13 | 1.35 | 1.15 ± 0.20 | 7 |
| Total cells | 1.69 ± 0.05 | 0.85 | 0.60 ± 0.05 | 200 |
Bipolarization of the nucleoid in newborn cells
To investigate the intracellular localization of the replication origin and terminus in single cells, we observed simultaneously both the oriC-flanking and terminus DNA segments in cells hybridized with these two probes labeled with different fluorescent compounds: the asn–rbs probe with a green fluorescent compound and the ter probe with a red fluorescent compound. Interestingly, the origin-flanking DNA segment was localized at one nucleoid border and the terminus DNA segment at the other in small cells (Fig. 4a). Thus, the nucleoid in newborn cells exhibits polarity, that is, bipolar organization, under growing conditions. In some small cells with two green foci at both nucleoid borders, a red focus was localized at midcell (Fig. 4c–f), suggesting that the terminus DNA segment migrated from the nucleoid border to midcell in an early stage of the cell division cycle. On the other hand, copies of the origin-flanking segment were separated from each other and localized at both nucleoid borders (Fig. 4c–f). The terminus segment was duplicated at midcell, and a septum was formed between these terminus segments (Fig. 4g,h). Therefore, after cell division, a newborn cell had a nucleoid in which the origin-flanking region was localized at the old-pole-proximal nucleoid border and the terminus was localized at the new-pole-proximal border.
Figure 4.
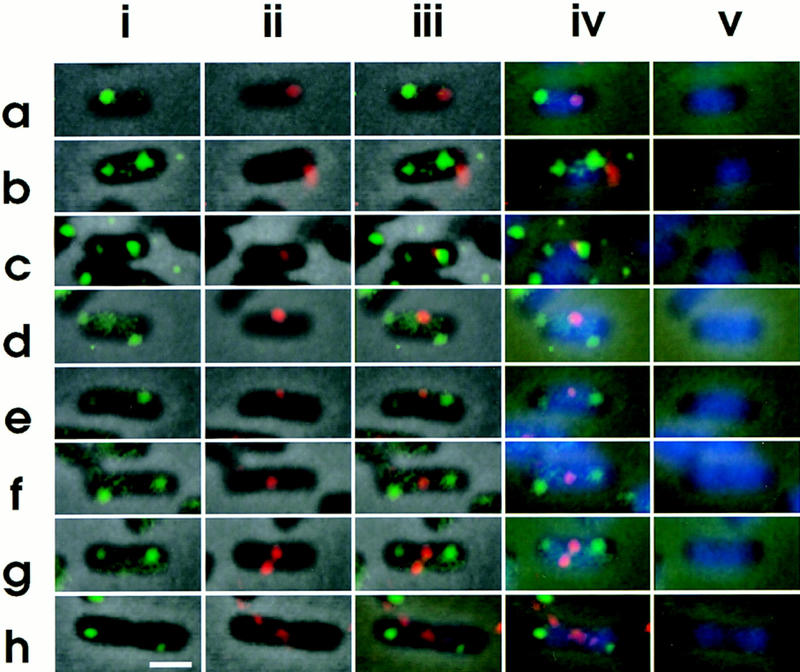
Intracellular localization of replication origin and terminus. Cells were hybridized simultaneously with the fluorescein-labeled asn–rbs DNA probe and the Cy3-labeled ter DNA probe. (i) asn–rbs; (ii) ter; (iii) asn–rbs and ter; (iv) asn–rbs, ter, and DAPI. (v) DAPI; Bar, 1 μm.
Arrangement of four OriC DNA segments in a cell
According to Helmstetter et al. (1968), chromosome replication should be initiated just before cell division at a doubling time of slightly less than 60 min. We analyzed the number of oriC copies per individual cell in a growing culture (doubling time, 55 min) of strain CSH50 with a flow cytometer. We found that 73% of the cells had two oriC copies, whereas 37% had four oriC copies per cell. These results indicate that chromosome replication was initiated prior to cell division, resulting in four fluorescent oriC foci in the last stage. Indeed, as mentioned above, 4.6% of the cells had four fluorescent oriC foci and 5.4% had three. This suggests that the duplicated oriC DNA segments were separated from each other before cell division in a minority of cases, resulting in late stage cells with four oriC copies (Fig. 5). Two oriC copies localized at the two nucleoid borders (Fig. 5a) were duplicated at these positions (Fig. 5b,c). They were arranged along the long cell axis in cells with two nucleoids; the oriC copies were localized at the two borders of each nucleoid (Fig. 5d–f). In these cases, a copy of the replicated oriC segments migrated from a cell-pole-proximal border to a midcell-proximal border of a nucleoid. This observation is consistent with the migration of oriC DNA segments in living cells observed with the LacI–GFP system (Gordon et al. 1997).
Figure 5.
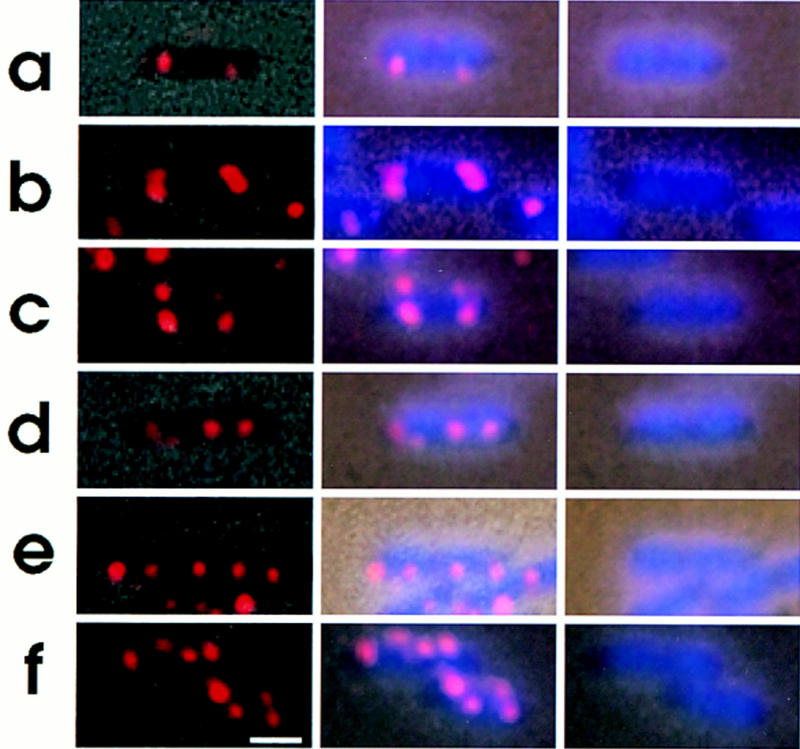
Intracellular localization of replication origin. Cells were grown, fixed and hybridized with the oriC–gid DNA probe as described in the legend to Fig. 2. (Left column) Combined images of the phase contrast micrograph and fluorescence micrograph of the same field; (middle column) combined images of fluorescence micrograph and phase contrast-DAPI micrograph; (right column) images of phase contrast-DAPI. Bar, 1 μm.
Discussion
In this paper we have shown the intracellular localization of specific chromosomal DNA segments during the cell division cycle (Fig. 6A). Our major observations and conclusion are the following: (1) In newborn cells, an oriC DNA segment is localized at an old-pole-proximal border of a nucleoid, but not at the cell pole; (2) one copy of the duplicated oriC DNA segments remains at the old-pole-proximal border, whereas the other copy migrates along the nucleoid to the opposite nucleoid border; (3) the distance between an oriC DNA segment and the nearest cell pole is constant throughout the cell division cycle; (4) the oriC-flanking region hybridized with the asn–rbs DNA probe shows a migration pattern similar to that of the oriC region; (5) the terminus DNA segment is localized at the new-pole-proximal nucleoid border in newborn cells; (6) the terminus DNA segment migrated from the nucleoid border to midcell in an early stage of the cell division cycle; (7) migration of the oriC and terminus DNA segments was not coupled to cell elongation; and (8) duplicated terminus DNA regions were separated from each other at the last stage of chromosome separation. These results indicate the existence of a mitotic-like apparatus. The distance between the nucleoid outer border and cell pole remains constant during all stages of the cell division cycle (van Helvoort and Woldringh 1994). The bulk replicated nucleoids thus seems to migrate gradually during cell elongation. However, the dynamic migration of the replication origin and terminus, which is not coupled to envelope elongation, is involved in the nucleoid movement for positioning of daughter chromosomes, as shown in Results.
Figure 6.
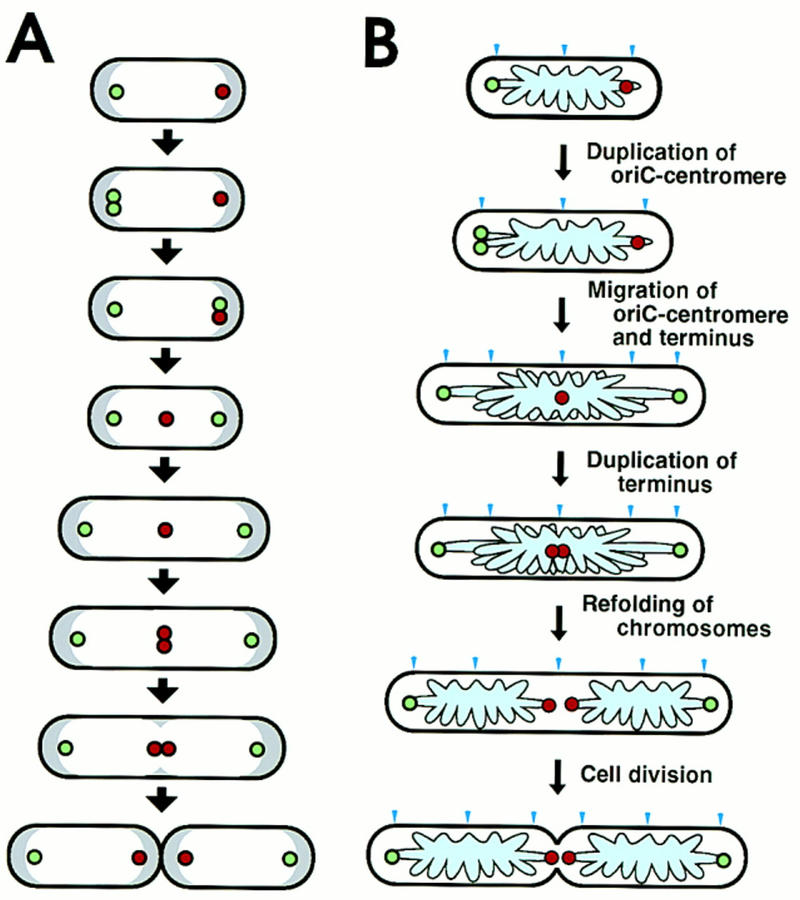
Schema of intracellular localization of replication origin and terminus during cell division cycle and a model of chromosome partioning in E. coli. (A) In cells growing with a 60-min doubling time, a replicated origin DNA segment (green circle) on the chromosome is localized near a nucleoid border in newborn cells. One copy of the replicated origin segment remains at the same position, whereas the other migrates to the opposite nucleoid border, where the replication terminus (red circle) is localized. Subsequently, the replication terminus migrates to midcell. The distance between an origin DNA segment and the nearest cell pole is constant during the cell division cycle, despite cell elongation. One round of chromosome replication is completed, and the terminus segment is duplicated at midcell. A septum is formed at midcell, resulting in newborn cells that have a replication terminus localized at the nucleoid border near a pole newly created by cell division. On the other hand, the replication origin is localized at the other nucleoid border, near the old pole. White and gray regions indicate nucleoid and cytosol spaces, respectively. (B) A model of partitioning of daughter chromosomes. A putative cis-acting sequence for migration, oriC–centromere, is located in a loop containing oriC. The specific migration patterns of oriC–centromere (green circle) and terminus (red circle) are shown in cells during the cell division cycle. The pale blue region and arrow head indicate the folded chromosome and the cell division site, respectively.
E. coli chromosomal DNA is folded into about 50 loops per chromosome (Pettijohn and Hecht 1974). The oriC and terminus DNA segments may be located in loops of the folded chromosome rather than in the core structure. The migration of oriC DNA segment in an early stage of the cell division cycle is important for chromosome partitioning as described in a model (Fig. 6B). However, the oriC DNA sequence itself presumably does not have a function for the migration of oriC DNA segments along the nucleoid, since oriC plasmids are unstably maintained, although oriC plasmids carrying the sopABC segment of F plasmid are stably maintained (Ogura and Hiraga 1983; Hiraga 1992). A putative cis-acting specific DNA sequence(s) responsible for oriC migration may be located in the oriC-containing loop. After migration, the cis-acting sequence(s) may be attached to a peripheral site of the cell membrane, with the result that oriC is localized at a constant distance from the nearest cell pole. In C. cresentus, a cis-acting parS site for chromosome partitioning, located ∼80 kb from the replication origin, may act as a centromere (Mohl and Gober 1997). In B. subtilis, the binding sites of Spo0J, which is responsible for chromosome partitioning, are localized in the origin–proximal third of the chromosome. It is likely that bacterial cells have a cis-acting site(s) like a eukaryotic centromere near the replication origin. A DNA loop containing the terminus region may have another cis-acting DNA sequence(s) that ensures the specific migration pattern of the terminus region.
There are differences between the number of oriC copies per cell and the number of fluorescent oriC–gid foci. The results indicate that the oriC segment is duplicated before cell division under our growth conditions, and these oriC copies are located close to one another in newborn cells and in early stages, so that they are observed as a single fluorescent focus in 27% of the cells. The duplicated oriC copies seem to be close to each other for approximately the first 15 min of the 55 min doubling time, enough for duplication of the origin-proximal third of the chromosome. For the dynamic migration of oriC and its flanking regions, the putative centromere region(s) must be duplicated, specific proteins must bind to the duplicated regions, and an unidentified mitotic-like apparatus must be activated.
A growing cell has three types of sites related to cell division. The first is the next cell division site at midcell. The second is future cell division sites at positions one-fourth and three-fourths of the cell length. The third is the old cell division sites at both cell poles. The old cell division sites have potential for septum formation, but this is suppressed by the minCD gene products (de Boer et al. 1989). It is thought that the specialized structures that organize cell division, called periseptal annuli, exist at these sites (de Boer et al. 1990). Cell quarter sites are important for the sopABC partitioning system of mini-F plasmid. Mini-F plasmid DNA is localized at midcell in newborn cells. After replication, duplicated molecules migrate in opposite directions from midcell to the cell quarter positions and are tethered there until the cell divides (Niki and Hiraga 1997). The DNA-protein complexes of sopC DNA, SopA and SopB (Hirano et al. 1998) may play an important role for migration and tethering at the cell quarter positions. Mini-P1 plasmid molecules migrate similarly from midcell to cell quarter positions (Gordon et al. 1997). Furthermore, using immunofluorescence microscopy, we have found that SeqA fluorescent foci are duplicated and migrate from midcell to the cell quarters in a cell cycle-dependent manner (Hiraga et al. 1998). It is thought that SeqA is involved in replication initiation by the sequestration of oriC and/or modulation of the cellular level of DNA activity (von Feiesleben et al. 1994; Lu et al. 1994; Slater et al. 1995; Garwood and Kohiyama 1996). The SeqA foci were not formed at oriC but in another chromosomal region. The intracellular localization of SeqA-associated DNA–protein complexes was abnormal in mukB null mutants. The SeqA-associated DNA–protein complex may play an important role, direct or indirect, in the migration and tethering of bulk daughter chromosomes at cell quarter sites (Hiraga et al. 1998). Putative centromere sites in a chromosomal loop containing oriC are presumably tethered at the old cell division site in newborn cells. The cell division sites must be closely related to chromosome and plasmid migration and partitioning.
In summary, we present a model of chromosome partitioning in E. coli (Fig. 6B) based on three different migration patterns of chromosomal regions during the cell division cycle; (1) migration along a nucleoid of a putative centromere located near oriC; and (2) migration of a cis-acting sequence(s) in the terminus region from a nucleoid border to midcell. The migration along a nucleoid of one copy of duplicated DNA regions containing a putative centromere and oriC determines the direction of positioning of daughter chromosomes. Subsequently, the terminus region migrates to midcell, that is, between the two oriC regions. Newly synthesized DNA strands accumulate in cells during the progression of replication forks. After the termination of chromosomal replication, SeqA-associated DNA–protein complexes migrate to the cell quarter sites and presumably act as refolding centers of the daughter chromosomes, resulting in two separated nucleoids (Hiraga et al. 1998). Duplicated terminus regions are pulled in opposite directions and finally are separated into two nucleoids during migration and/or refolding of bulk daughter chromosomes at the cell quarter positions.
To resolve dimeric daughter chromosomes to monomers before cell division, the terminus region has a hyperrecombination site for the XerCE-dependent site-specific recombination and a terminal recombination region (TRZ) (Hill 1996). Interestingly, both dif, which is the XerCE recognition site, and TRZ function only when they are ocated at the proper position in the chromosomal terminus (Louarn et al. 1994; Cornet et al. 1996; Kuempel et al. 1996). The dif locus might be positioned in a TRZ loop of the folded chromosome, and TRZ regions are finally separated in the process of chromosome partitioning (Louarn et al. 1994). The dif resolution specificity might be dependent on the chromosomal location that is finally pulled apart (Baker 1991). Our observation of the migration pattern of the terminus is consistent with this model of TRZ and dif.
Materials and methods
Bacteria, plasmids, and culture conditions
An E. coli K-12 derivative, CSH50 [ara (Δlac–pro) strA thi] (Miller 1972), was used. Cells were cultivated at 37°C in M9 medium (Miller 1972) supplemented with glucose (0.5%), proline (50 μg/ml), and thiamine (2 μg/ml) for FISH. Under growing conditions, the doubling time of CSH50 cells was 55 min.
Fluorescent probes for in situ hybridization
To detect the replication origin DNA segment of the E. coli chromosome in fixed cells by in situ hybridization, we used the oriC–gid probe DNA segment containing mioC, oriC, gidAB, and a part of atpI, which was amplified from CSH50 chromosomal DNA by PCR. The primer sequences for the oriC–gid DNA segment were 5′-TCAGGATAAATACGCCCAGTGCCATAGACAGCG-3′ and 5′-AGCTCTGACGGGGTGCTCCAGTCATCATAATCC-3′. We also used Kohara’s λ phage clones; no. 559 for the origin-flanking region and no. 269 for the terminus DNA segment (Kohara et al. 1987). The cloned chromosomal DNA segments of these λ clones were amplified by PCR according to Oshima et al. (1996). The PCR products (1 μg) were labeled with Cy3–dCTP (Amersham) or Fluorescein-11–dUTP (Amersham) by use of a random-primed DNA labeling kit (Boehringer). After removing nonincorporated substrates by ethanol precipitation, the labeled probe DNA was resolved in hybridization buffer (50% formamide, 2× SSC, 100 μg/ml salmon sperm DNA). The probe DNA in the hybridization mixture was sonicated for 15 sec and denatured by heating to 80°C for 10 min before hybridization.
FISH and image analysis
To fix cells, an equal volume of fixation solution [methanol:acetic acid (3 : 1)] was added directly to a bacterial culture growing exponentially in M9 glucose medium at 37°C. The FISH was carried out according to the procedure described previously (Niki and Hiraga 1997). All images were recorded with a cooled CCD camera, C5810-01 (Hamamatsu, Japan), by use of a phase contrast and fluorescence microscopy system (Nikon). The images were transferred directly to a Power Macintosh and processed by use of Adobe Photoshop 4.0-J software. The image was printed on a Pictrography 3000 (Fuji).
Measuring number of oriC copies in individual cells by flow cytometry
The number of oriC copies in individual cells was determined by flow cytometory (Bio-Rad) according to the method described previously (Løbner-Olesen et al. 1989).
Acknowledgments
We thank R. D’Ari for critically reading the manuscript and comments. This work was supported by Grant-in-Aid for Scientific Research on Priority Areas and a grant for International Scientific Research Programs for Joint Research from the Ministry of Education, Science, Sports, and Culture of Japan, and by a grant from the Human Frontier Science Program (contact no. RG-386/95M).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL niki@gpo.kumamoto-u.ac.jp; FAX 96-371-2408.
References
- Baker T. … and then there were two. Nature. 1991;352:794–795. doi: 10.1038/353794a0. [DOI] [PubMed] [Google Scholar]
- Berlyn MKB, Low B, Rudd KE. Linkage map of E. coli K-12. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger ED, editors. Escherichia coli and Salmonella typhimurium: Cellular and molecular biology. Washington, DC.: American Society for Microbiology; 1996. pp. 1715–1902. [Google Scholar]
- Blattner FR, Plunkett III G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Cornet F, Louarn J, Patte J, Louarn JM. Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes & Dev. 1996;10:1152–1161. doi: 10.1101/gad.10.9.1152. [DOI] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- de Boer PAJ, Cook WR, Rothfield LI. Bacterial cell division. Annu Rev Genet. 1990;24:249–274. doi: 10.1146/annurev.ge.24.120190.001341. [DOI] [PubMed] [Google Scholar]
- Donachie WD, Begg KJ. Chromosome partition in Escherichia coli requires postreplication protein synthesis. J Bacteriol. 1989;171:5405–5409. doi: 10.1128/jb.171.10.5405-5409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood J, Kohiyama M. A novel cytoplasmic hemimethylated oriC binding activity. J Biol Chem. 1996;271:7404–7411. doi: 10.1074/jbc.271.13.7404. [DOI] [PubMed] [Google Scholar]
- Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes & Dev. 1997;9:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight A, Losick R, Murray AW, Wright A. Chromosome and low copy plasmid segregation in E. coli: Visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- Helmstetter CE, Cooper S, Pierucci O, Revelas L. The bacterial life sequence. Cold Spring Harbor Symp Quant Biol. 1968;33:809–822. doi: 10.1101/sqb.1968.033.01.093. [DOI] [PubMed] [Google Scholar]
- Hill TM. Features of the chromosomal terminus region. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger ED, editors. Escherichia coli and Salmonella typhimurium: Cellular and molecular biology. Washington, DC.: American Society for Microbiology; 1996. pp. 1602–1614. [Google Scholar]
- Hiraga S. Partitioning of nucleoids. Res Microbiol. 1990;141:50–56. doi: 10.1016/0923-2508(90)90097-a. [DOI] [PubMed] [Google Scholar]
- ————— Chromosome and plasmid partition in Escherichia coli. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffé A. Chromosome partitioning in Escherichia coli: Novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Ogura T, Niki H, Ichinose C, Mori H. Positioning of replicated chromosomes in Escherichia coli. J Bacteriol. 1990;172:31–39. doi: 10.1128/jb.172.1.31-39.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Ichinose C, Niki H, Yamazoe M. Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA-protein complexes in E. coli. Mol Cell. 1998;1:381–387. doi: 10.1016/s1097-2765(00)80038-6. [DOI] [PubMed] [Google Scholar]
- Hirano M, Mori H, Onogi T, Yamazoe M, Niki H, Ogura T, Hiraga S. Autoregulation of the partition genes of the mini-F plasmid and the intracellular localization of their products in Escherichia coli. Mol Gen & Genet. 1998;257:392–403. doi: 10.1007/s004380050663. [DOI] [PubMed] [Google Scholar]
- Ireton K, Gunther NWI, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: Application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kuempel P, Hogaard A, Nielsen M, Nagappan O, Tecklenburg M. Use of a transposon (Tndif) to obtain suppressing and nonsuppressing insertions of the dif resolvase site of Escherichia coli. Genes & Dev. 1996;10:1162–1171. doi: 10.1101/gad.10.9.1162. [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Errington J. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol Microbiol. 1997;25:945–954. doi: 10.1111/j.1365-2958.1997.mmi530.x. [DOI] [PubMed] [Google Scholar]
- Lin DC-H, Levin PA, Grossman AD. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A, Kuempel PL. Chromosome partitioning in Escherichia coli. J Bacteriol. 1992;174:7883–7889. doi: 10.1128/jb.174.24.7883-7889.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A, Skarstad K, Hansen, M.K. von FG, Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989;57:881–889. doi: 10.1016/0092-8674(89)90802-7. [DOI] [PubMed] [Google Scholar]
- Louarn J, Cornet F, Francois V, Patte J, Louarn JM. Hyperrecombination in the terminus region of the Escherichia coli chromosome: Possible relation to nucleoid organization. J Bacteriol. 1994;176:7524–7531. doi: 10.1128/jb.176.24.7524-7531.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Campbell JL, Boye E, Kleckner N. SeqA: A negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mohl DA, Gober JW. Cell cycle–dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- Niki H, Hiraga S. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- Niki H, Jaffé A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E.coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 1992;11:5101–5109. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Hiraga S. Partition mechanism of F plasmid: Two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983;32:351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- Oka A, Sugimoto K, Takanami M, Hirota Y. Replication origin of the Escherichia coli K-12 chromosome: The size and structure of the minimum DNA segment carrying the information for autonomous replication. Mol & Gen Genet. 1980;178:9–20. doi: 10.1007/BF00267207. [DOI] [PubMed] [Google Scholar]
- Oshima T, Aiba H, Baba T, Fujita K, Hayashi K, Honjo A, Ikemoto K, Inada T, Itoh T, Kajihara M, et al. A 718-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 12.7-28.0 min region on the linkage map. DNA Res. 1996;3:137–155. doi: 10.1093/dnares/3.3.137. [DOI] [PubMed] [Google Scholar]
- Pettijohn DE, Hecht R. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harbor Symp Quant Biol. 1974;38:31–41. doi: 10.1101/sqb.1974.038.01.006. [DOI] [PubMed] [Google Scholar]
- Rothfield LI. Bacterial chromosome segregation. Cell. 1994;77:963–966. doi: 10.1016/0092-8674(94)90435-9. [DOI] [PubMed] [Google Scholar]
- Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- van Helvoort JMLM, Woldringh CL. Nucleoid partitioning in Escherichia coli during steady-state growth and upon recovery from chloramphenicol treatment. Mol Microbiol. 1994;13:577–583. doi: 10.1111/j.1365-2958.1994.tb00452.x. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U, Rasmussen KV, Schaechter M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol. 1994;14:763–772. doi: 10.1111/j.1365-2958.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- Wake RG, Errington J. Chromosome partitioning in bacteria. Annu Rev Genet. 1995;29:41–67. doi: 10.1146/annurev.ge.29.120195.000353. [DOI] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC-H, Grossman AD, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L. Bacterial chromosome segregation: Is there a mitotic apparatus? Cell. 1997;88:577–579. doi: 10.1016/s0092-8674(00)81898-x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- ————— Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol & Gen Genet. 1996;250:241–251. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]