Abstract
The RUB1/NEDD-8 family of ubiquitin-related genes is widely represented among eukaryotes. Here we report that Cdc53p in Saccharomyces cerevisiae, a member of the Cullin family of proteins, is stably modified by the covalent attachment of a single Rub1p molecule. Two genes have been identified that are required for Rub1p conjugation to Cdc53p. The first gene, designated ENR2, encodes a protein with sequence similarity to the amino-terminal half of the ubiquitin-activating enzyme. By analogy with Aos1p, we infer that Enr2p functions in a bipartite Rub1p-activating enzyme. The second gene is SKP1, shown previously to be required for some ubiquitin–conjugation events. A deletion allele of ENR2 is lethal with temperature-sensitive alleles of cdc34 and enhances the phenotypes of cdc4, cdc53, and skp1, strongly implying that Rub1p conjugation to Cdc53p is required for optimal assembly or function of the E3 complex SCFCdc4. Consistent with this model, both enr2Δ and an allele of Cdc53p that is not Rub1p modified, render cells sensitive to alterations in the levels of Cdc4p, Cdc34p, and Cdc53p.
Keywords: Yeast, Cdc53p, Rub1p molecule, SCFCdc34, ubiquitin, auxin
The ubiquitin pathway catalyzes the post-translational modification of diverse proteins (for review, see Hochstrasser 1996; King et al. 1996). Ubiquitin is recruited initially from the pool of free ubiquitin by a ubiquitin-activating enzyme (E1 or Uba), passed to a ubiquitin-conjugating enzyme (E2 or Ubc), and transferred to the substrate protein often in concert with an ubiquitin–protein ligase (E3). In several instances the E3 function is performed by a complex composed of Cdc53, Skp1, and an F-box protein (SCF complex) (Feldman et al. 1997; Skowyra et al. 1997 ). The F-box protein contains a domain that binds Skp1p (the F-box) and confers substrate specificity to the SCF (Bai et al. 1996). E1 proteins are highly conserved throughout eukaryotes, with the E1 of humans and Saccharomyces cerevisiae displaying >50% amino acid identity. In yeast, E1 is encoded by the essential UBA1 gene and is believed to be the sole protein capable of activating free ubiquitin to initiate the ubiquitin–conjugation pathway (McGrath et al. 1991). However, sequence comparisons reveal several yeast genes displaying high degrees of sequence similarity to either the amino- or carboxy-terminal half of E1 (Dohmen et al. 1995; Hochstrasser 1996; Johnson et al. 1997; this study).
Proteins displaying sequence similarity to the amino-terminal half of E1 have been identified in many eukaryotic taxa. The first to be identified was the AXR1 protein of Arabidopsis thaliana (Leyser et al. 1993). Subsequently, members of this family have been found in humans (APP–BP1; Chow et al. 1996), Schizosaccharomyces pombe (Rad31p; Shayeghi et al. 1997), hamsters (SMC1; S. Handeli, pers. comm.), Candida albicans (CaAXR1; W. Jiang, unpubl.), and S. cerevisiae (Rhc31p/Aos1p/Enr1p and Enr2p; Shayeghi et al. 1996; Johnson et al. 1997; this study). Mutations in several of these genes suggest that members of the AXR1 family play roles in a variety of metabolic processes. The axr1 mutants of Arabidopsis are deficient in auxin response (Lincoln et al. 1990; Leyser et al. 1993; Timpte et al. 1995). A mutation in the SMC1 locus leads to the complex cell cycle arrest phenotype of the ts41 Chinese hamster cell line. In asynchronous cultures, the ts41 line arrests with one population of cells in G2, whereas a second population undergoes repeated, discrete S-phases without intervening mitoses (Handeli and Weintraub 1992). The human APP–BP1 was isolated as a binding partner of the amyloid precursor protein (APP) (Chow et al. 1996). The rad31 S. pombe mutants display sensitivity to ionizing and UV radiation, and may also be defective in a DNA damage cell cycle checkpoint (Shayegi et al. 1997).
A recent study suggests that the proteins related to the amino terminus of E1 are involved in the activation of ubiquitin-like proteins rather than of ubiquitin itself (Johnson et al. 1997). The ubiquitin-like proteins include the Smt3p/SUMO1/GMP1/PIC1/UBL1/sentrin family (Boddy et al. 1996; Matunis et al. 1996; Okura et al. 1996; Shen et al. 1996; Mahajan et al. 1997), the RUB1/NEDD-8 proteins (Hochstrasser 1996), the interferon-inducible ubiquitin-like dimer UCRP (Haas et al. 1987), and the baculoviral ubiquitin-like protein (Guarino 1990). Johnson et al. (1997) have shown that activation of yeast Smt3p requires the activity of two proteins, Aos1p and Uba2p. AOS1 belongs to the family of E1 amino-terminal relatives, whereas UBA2 has sequence similarity to the carboxy-terminal half of E1 (Dohman et al. 1995). The two proteins associate physically and together promote the formation of an Smt3p/Uba2p thiolester-linked complex, from which the activated Smt3p is passed to the E2 enzyme Ubc9p for subsequent conjugation to substrate proteins. No substrate proteins for Smt3p conjugation have been identified to date in yeast. However, in mammals a monomer of the SUMO-1 protein is conjugated to the Ran–GAP1 protein (Matunis et al. 1996; Mahajan et al. 1997). This conjugation is required for the proper targeting of Ran–GAP1 to the nuclear pore complex by promoting binding to Nup358p/RanBP2p (Mahajan et al. 1997).
By analogy with Aos1p (Johnson et al. 1997), AXR1, SMC1, and related proteins are likely to function as one component of a bipartite E1-like enzyme for the activation of ubiquitin-related proteins. Although the mode of activation may be similar between ubiquitin and its relatives, the consequences of post-translational modification may be different. Here we report that ENR2 (E1 amino terminus Related 2), a second yeast member of the AXR1 family, is required to conjugate Rub1p to Cdc53p, a protein that is essential for the G1- to S-phase transition in the cell cycle (Mathias et al. 1996).
In yeast, cell cycle progression is mediated by the activity of the cyclin-dependent kinase (CDK) Cdc28p. The activation state and specificity of Cdc28p are determined by cyclins and CDK inhibitors such as Far1p and Sic1p. During G1, Sic1p acts to inhibit CDK/cyclin B and prevent initiation of S-phase. The G1- to S-phase transition requires the degradation of Sic1p by the ubiquitin/proteosome pathway (Schwob et al. 1994). This process requires an E2 enzyme, Cdc34p (Goebl et al. 1988; Schwob et al. 1994; Deshaies et al. 1995), and an SCF complex containing the F-box protein Cdc4p.
Cdc53p and Skp1p form SCF complexes with a variety of F-boxes, including Grr1p and Met30p. SCFGrr1 is required for ubiquitination of the Cln cyclins, whereas SCFMet30 functions in sulfur metabolism (Thomas et al. 1995; M. Tyers, pers. comm.). The dynamics of the movement of Cdc53p and Skp1p between various complexes is largely unexplored. Here we report that Rub1p is conjugated to Cdc53p by a pathway that requires ENR2 and SKP1. Failure to modify Cdc53p because of either mutations in CDC53 or ENR2 renders cells sensitive to alterations in the abundance of Cdc34p and Cdc53p. Furthermore, cells respond to changes in the abundance of Cdc4p, Cdc34p, and Cdc53p by increasing the fraction of Cdc53p molecules that have Rub1p attached. We propose that Rub1p modification may be required for correct apportioning of Cdc53p among different SCF complexes or for optimal assembly of the SCF.
Results
ENR2 displays genetic interactions with CDC34 and SCFCdc4
ENR2 (YPL003W) was identified based on its sequence similarity to AXR1 (Fig. 1A). Deletions of the gene produced no obvious morphological phenotype, and enr2Δ cells grew at normal rates over a broad range of temperatures (data not shown). To determine whether ENR2 might play a role in ubiquitin-mediated processes in yeast, we constructed double mutants between enr2Δ and previously characterized mutations in genes encoding E1, E2, and E3 enzymes. First, we attempted to generate lines doubly mutant for enr2::LEU2 and cdc34-2. The two mutations displayed clear synthetic lethality. No viable double mutant segregants were recovered in >40 tetrads examined at either 30°C or 20°C. Analysis of the genotypes of viable segregants within each tetrad confirmed that each nonviable segregant was a double mutant (data not shown). Microscopic examination revealed that most enr2Δ, cdc34-2 spores germinated and underwent several rounds of division to form microcolonies before arresting. enr2Δ cdc34-2 cells have an appearance similar to cdc34-2 cells at near-restrictive temperatures (33°C), with multiple elongated and misshapen buds (Figure 2A–C).
Figure 1.

Alignment of ENR2 and related genes and of RUB1 and related genes. (A) Alignment of the predicted amino acid sequence of Enr2p with S. cerevisiae Aos1p, Arabidopsis AXR1, and human APP-BP1. Identical residues between three or more proteins are shaded. (B) Alignment of the predicted amino acid sequence of Rub1p with the NEDD-8 protein of mouse and the S. cerevisiae ubiquitin protein. Identical residues between ubiquitin and its homologs are shaded.
Figure 2.
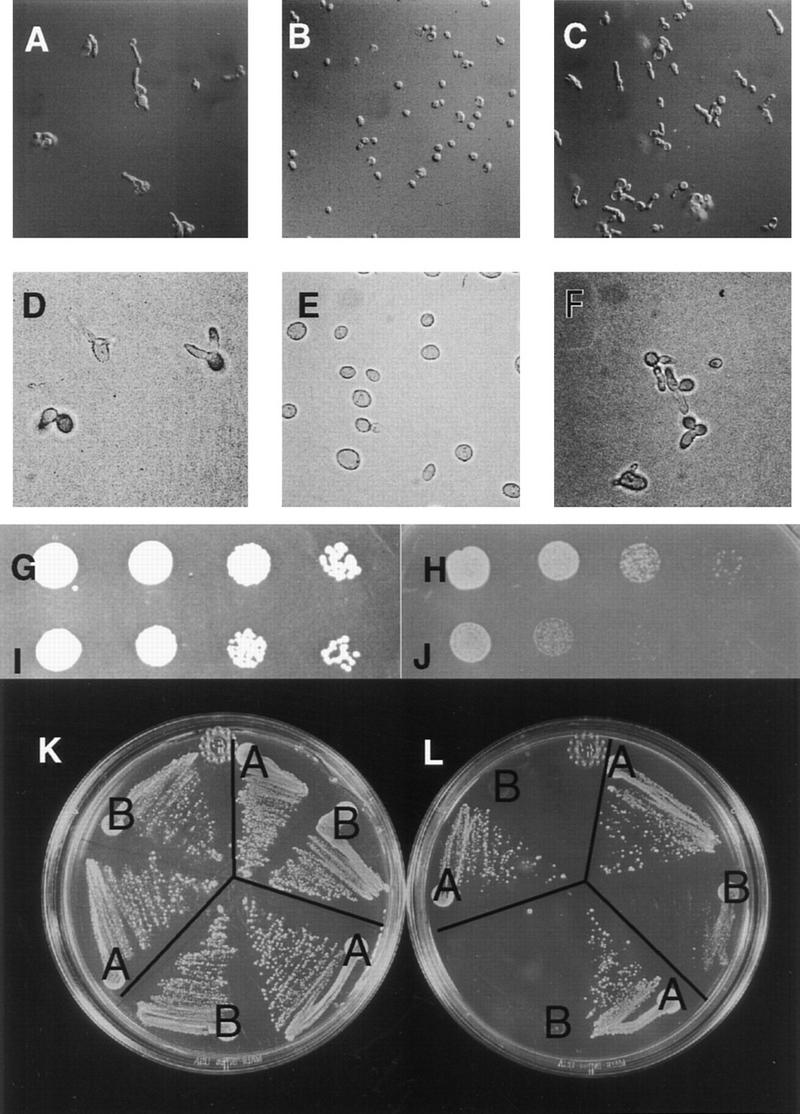
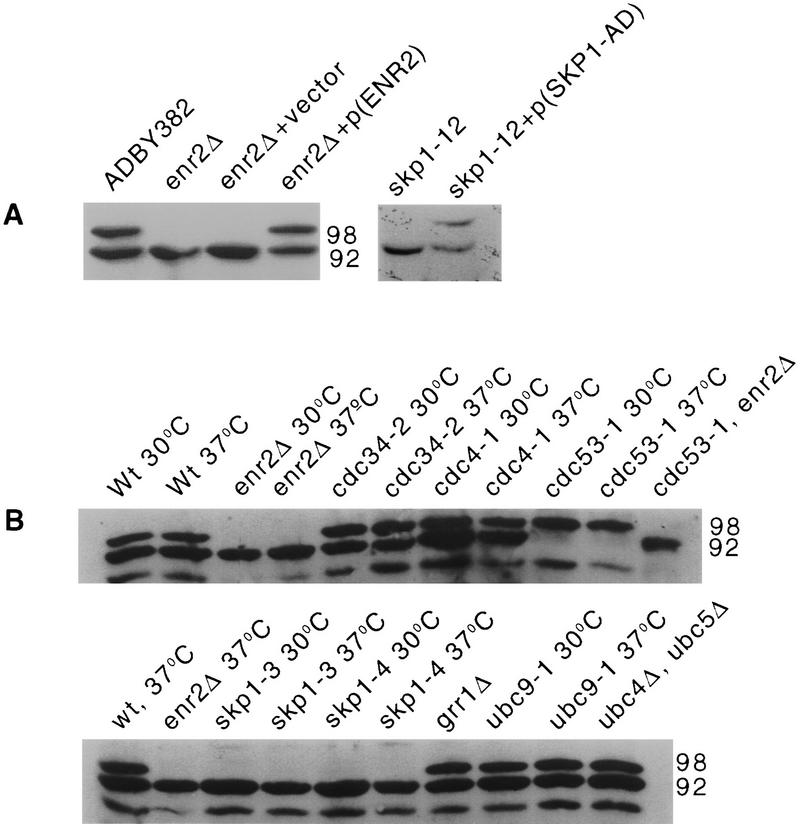
Genetic interactions of ENR2. (ROW 1) enr2Δ displays synthetic lethality with cdc34-2. (A) cdc34-2 enr2::LEU2 segregants, terminal phenotype at 20°C; (B) cdc34-2 ENR2 segregant at 20°C; (C) cdc34-2 ENR2 segregant at the nearly restrictive temperature 33°C. (ROW 2) enr2Δ enhances the temperature-sensitive phenotype of cdc4-1. (D) cdc4-1 enr2::LEU2 + empty vector at 25°C; (E) cdc4-1 enr2::LEU2 + p(ENR2, URA3, CEN4) at 25°C; (F) cdc4-1 enr2::LEU2 + p(ENR2, URA3, CEN4), terminal phenotype at 37°C. (ROW 3) enr2Δ enhances the ts phenotype of cdc53-1. (G) cdc53-1 enr2::LEU2 + p(ENR2, URA3, CEN4) at 30°C; (H) cdc53-1 enr2::LEU2 + p(ENR2, URA3, CEN4) at 33°C; (I) cdc53-1 enr2::LEU2 + empty vector at 30°C; (J) cdc53-1 enr2::LEU2 + empty vector at 33°C. Tenfold serial dilutions of individual transformants were grown on −Ura media with glucose as the carbon source for 2 days at the indicated temperatures. (ROW 4) enr2Δ enhances the temperature sensitivity of skp1-12. Three individual enr2::TRP1 skp1-12 segregants were transformed with either p(ENR2, URA3,CEN4) (streaks marked A) or with empty vector (streaks marked B), streaked on −Ura media with glucose as the carbon source, and incubated at either 30°C (K) or 33°C (L).
The Cdc34 protein is required for degradation of Sic1p during the G1- to S-phase transition in a process that also requires the E3 complex SCFCdc4. This complex contains Cdc53p, Skp1p, and Cdc4p. When the enr2Δ mutation was introduced into cdc4-1, cdc53-1, and skp1-12 strains, the result was enhancement of the temperature-sensitive mutant phenotype in each case (Fig. 2D–L). From these data, we conclude that ENR2 has a role in events that require Cdc34p SCFCdc4 activity. To determine whether ENR2 is also important for the function of other SCF complexes, we tested for genetic interactions with a strain deleted for GRR1, a component of SCFGrr1. No genetic interaction was observed in this case (data not shown).
ENR2 alters the physical state of Cdc53p
During the course of experiments to test for physical association between Enr2p and components of the SCFCdc4, we noticed that the physical state of Cdc53p varied depending on whether wild-type ENR2 was present in the strain. Cdc53p is normally found as two abundant isoforms of apparent molecular masses 92 kD and 98 kD (Mathias et al. 1996; Willems et al. 1996). In enr2Δ strains, only the 92-kD isoform is detectable, indicating that Enr2p is required for this modification. This was confirmed by transforming the strain with plasmid-borne ENR2 and restoring the larger form of Cdc53p (Fig. 3A).
Figure 3.
Formation of the 98-kD isoform of Cdc53p is dependent on the activity of ENR2 and SKP1. (A) Anti-Cdc53p Western blot of protein extracts from strains of the following genotypes: ENR2 (lane 1); enr2::LEU2 (lane 2); enr2::LEU2 + vector (lane 3); enr2::LEU2 + p(ENR2, URA3, CEN4) (lane 4); skp1-12 (lane 5); skp1-12 + p(SKP1–AD) (lane 6). All strains were grown to mid log phase at 30°C in selective SD liquid media as required. (B) Anti-Cdc53p Western blot of protein extracts from the indicated strains. The temperature-sensitive strains cdc34-2, cdc4-1, cdc53-1, skp1-3, skp1-4, and ubc9-1 were grown at either the permissive temperature of 30°C or shifted to the restrictive temperature of 37°C for 6 hr before preparation of protein extracts. These strains were checked microscopically for uniform arrest phenotype before extract preparation.
Then we addressed whether Cdc34p or components of SCFCdc4 acted in concert with ENR2 to modify Cdc53p. We found that both Cdc53p isoforms were present in cdc4-1 and cdc34-2 strains maintained at restrictive temperatures for 6 hr, but that only the 92-kD isoform was observed in skp1-3, skp1-4, and skp1-12 strains (Fig. 3B). The 98-kD form is restored in skp1-12 strains by transforming them with a SKP1–GAL4 activation domain fusion plasmid (Fig. 3A). As has been shown previously, only the 98-kD isoform is immunologically detectable in cdc53-1 backgrounds. The modification of Cdc53-1p, like that of the wild-type protein, is also dependent on ENR2 activity (Fig. 3B). Although Clnp stability has been shown to be enhanced in ubc9 (Seufert et al. 1995) and in grr1Δ backgrounds (Barral et al. 1995), Cdc53-1p remains modified in these strains (Fig. 3B). We conclude that the failure to modify Cdc53p in enr2Δ backgrounds is not simply a consequence of impaired G1/S transition, but instead that this modification requires the participation of ENR2 and SKP1.
Cdc53p is modified by addition of Rub1p and not ubiquitin
The 98-kD Cdc53p isoform cross-reacts with anti-ubiquitin antibodies in Cdc53p immunoprecipates, suggesting that the shift in mobility is caused by ubiquitination (Willems et al. 1996). However, there are several ubiquitin-related proteins encoded in the yeast genome that may cross-react with anti-ubiquitin antibodies. One of these is the product of the RUB1 gene (YDR139c) (see Fig. 1A). Rub1p is a member of a conserved family of ubiquitin-related proteins, which are also present in plants (Callis et al. 1995) and mammals (NEDD-8) (Kumar et al. 1993). Members of this family share ∼50% identity with ubiquitin. The RUB1 gene is dispensable under normal growth conditions in haploid cells, as deletion of the gene has no effect on growth rate or cell morphology (data not shown). To determine whether Rub1p is conjugated to Cdc53p, we examined Cdc53p in a rub1Δ background and in rub1Δ carrying GAL1::RUB1 on a plasmid. The results clearly show that Cdc53p modification is dependent on RUB1 (Fig. 4A). Overexpression of RUB1 from the GAL1 promoter leads to a dramatic increase in the ratio of Rub1–Cdc53p to Cdc53p, further supporting our hypothesis that Cdc53p is modified by Rub1p, and not by ubiquitin (Fig. 4A). This result also suggests that Rub1p is a limiting substrate for this modification when RUB1 is expressed from its normal chromosomal context.
Figure 4.
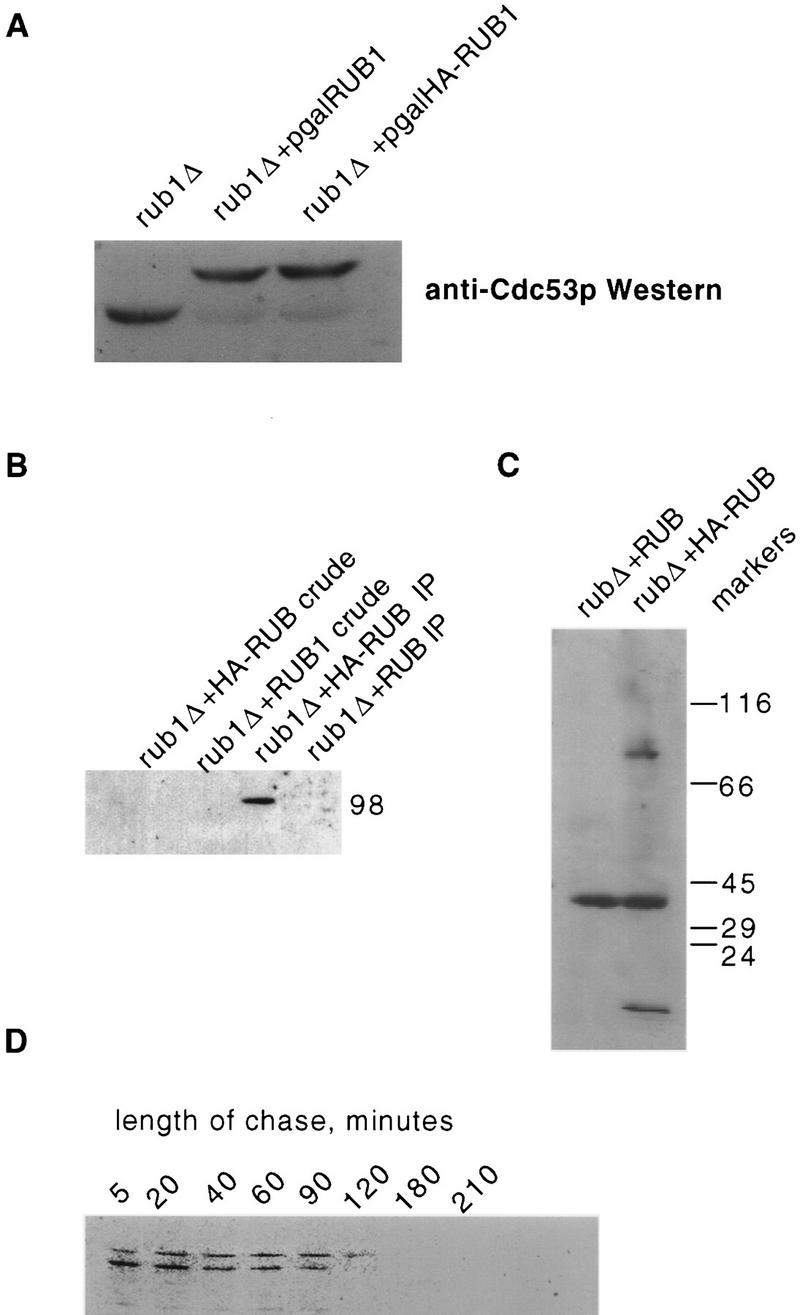
The 98-kD isoform of Cdc53p is formed through conjugation to Rub1p. (A) Anti-Cdc53p Western blot of soluble protein extracts from rub1::TRP1 + vector (lane 1); rub1::TRP1 + p(GAL1/10::RUB1, URA3, CEN) (lane 2), and rub1::TRP1 + p(GAL1/10::HA-RUB1, URA3, CEN) (lane 3). These strains were grown to mid-log phase in S-galactose −Ura before extract preparation. (B) Cdc53p was immunoprecipitated using anti-Cdc53p antibodies from a rub1::TRP1 strain expressing RUB1 encoding either native Rub1p or HA-tagged Rub1p, both under the control of the GAL1/10 promoter. These immunoprecipitates were then subjected to Western blot analysis using anti-HA mAb 12C5. rub1::TRP1 + p(GAL1/10::HA-RUB1) crude extract (lane 1); rub1::TRP1 + p(GAL1/10::RUB1) (lane 2); rub1::TRP1 + p(GAL1/10::HA-RUB1) immunoprecipitate (lane 3); rub1::TRP1 + p(GAL1/10::RUB1) immunoprecipitate (lane 4). (C) Anti-HA Western blots of total soluble protein from rub1::TRP1 strains expressing native or HA-tagged RUB1 demonstrating that a 98-kD band is the most prominant Rub1p conjugate. (D) Pulse-chase analysis demonstrates that both native Cdc53p and Rub1–Cdc53p are metabolically stable. A cdc53::HIS3 strain, kept viable by a CDC53–HA fusion, was given a 2-min pulse with [35S] methionine and cysteine in YPD media, washed twice in unsupplemented YPD, and then chased in YPD supplemented with unlabeled cysteine and methionine. Aliquots were taken at the indicated time points, and protein extracts were prepared and subjected to immunoprecipitation with anti-HA mAb 12C5. Immunoprecipitates were then subjected to SDS-PAGE and autoradiography. The half-life of the protein is ∼60 min.
To demonstrate more directly that Cdc53p is modified by Rub1p, we used a version of Rub1p that contains an HA epitope tag at its amino terminus, driven by the GAL1 promoter, and looked for the presence of the epitope tag in Cdc53p immunoprecipitates (Fig. 4B). The anti-Cdc53p antibody precipitates a single anti-HA cross-reactive protein of 98 kD from lysates containing GAL::HA–RUB1 plasmids. This protein is not detected in a control strain carrying untagged GAL::RUB1 (Fig. 4B) or in anti-Cdc34p immunoprecipitates of the GAL::HA–RUB strain (data not shown). Therefore, the 98-kD isoform is formed by conjugation of Rub1p to Cdc53p. In a separate experiment, a ∼98-kD protein is the only clearly visible species present in extracts from HA–RUB cells aside from unconjugated HA–Rub1p and a cross-reacting band also present in the control (Fig. 4C). A similar band is observed in lane 1 of Figure 4B. Thus, Cdc53p is probably the most abundant target for Rub1p conjugation. However, this does not preclude the existence of additional low abundance targets.
Because Cdc53p is the first known substrate for Rub1p modification, nothing is known about the fate of Rub1p-modified proteins. Ubiquitinated isoforms of short-lived proteins are often difficult to detect immunologically, therefore the abundance of Rub1–Cdc53p suggested that this modification was stable. We confirmed this directly through a pulse-chase analysis (Fig. 4D). A cdc53Δ strain carrying a carboxy-terminally HA-tagged version of Cdc53p was grown in YPD, pulsed with 35S-labeled methionine and cysteine for 2 min, washed twice in cold YPD, and then chased with YPD supplemented with cold cysteine and methionine. Both isoforms of Cdc53p are extremely stable and Rub1–Cdc53p persisted for at least 120 min. These results indicate that, at least in the case of Cdc53p, Rub1p modification is unlikely to serve as a tag for rapid degradation.
We hypothesize that the primary biochemical defect in enr2 mutants is the failure to conjugate Rub1p to its normal targets, and that this defect is responsible for the observed genetic interactions between enr2Δ and the SCFCdc4. This was tested by constructing double mutants between rub1Δ and cdc34-2. Like the enr2Δ cdc34-2 mutants, rub1Δ cdc34-2 mutants show a pronounced cdc34-2 phenotype of multiple elongated buds at temperatures normally permissive for cdc34-2 (J.M. Laplaza and J. Callis, unpubl.). Similarly, rub1Δ strongly enhances the phenotype of cdc53-1 in double mutants (J.M. Laplaza and J. Callis, unpubl.). These results provide strong support for the view that absence of Cdc53–Rub1p in these genetic backgrounds compromises optimal function of SCFCdc4.
A conserved carboxy-terminal domain of Cdc53 is required for Rub1p conjugation
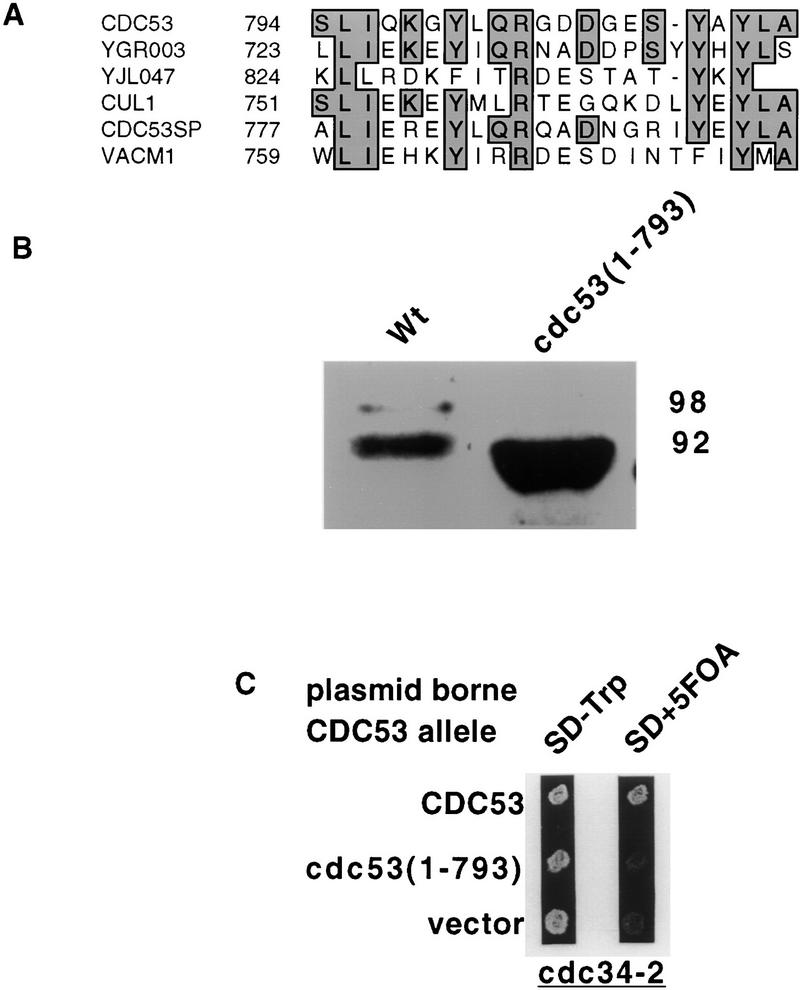
In a study to identify functional domains on Cdc53p we identified a region that was required for Rub1p modification. The most highly conserved region in the Cdc53p/Cullin family of proteins is located toward the carboxyl terminus of the protein (Fig. 5A). A plasmid that contained a cdc53 gene with a premature stop codon after residue 793 was transformed into a strain containing a temperature-sensitive cdc53-1 allele. This plasmid complemented the cdc53-1 allele, allowing growth of strain MGG10 at the nonpermissive temperature of 37°C (data not shown). To confirm that the cdc53(1–793) allele resulted in synthesis of a protein of the expected size, a lysate was made from NM53Δ cells containing cdc53(1–793) and full-length CDC53. Western blot analysis shown in Figure 5B indicates that Cdc53(1–793)p migrates faster than wild type and also migrate as a single band. Modified Cdc53(1–793)p was not observed even after overloading of the Cdc53(1–793)p lane. Thus, the terminal 22–amino acid residues of Cdc53p are required for its Rub1p modification mediated by Enr2p.
Figure 5.
The carboxy-terminal truncation mutant cdc53(1–793) does not become conjugated to Rub1p and shows synthetic lethality with cdc34-2. (A) Sequence alignment of the 21 carboxy-terminal amino acids deleted in cdc53(1–793) with the carboxyl terminus of other Cdc53/Cullin family members. YGR003 and YJ1047 are CDC53-related genes from yeast, and CUL1, CDC53SP, and VAC1 are CDC53-related genes from C. elegans, S. pombe, and rabbit, respectively. The carboxyl terminus is among the most highly conserved regions in this gene family. (B) Anti-Cdc53p Western blot of soluble protein from wild type and cdc53(1–793) strains. (C) Plasmid shuffle experiment to demonstrate that cdc53(1–793) displays synthetic lethality with cdc34-2. A cdc53::HIS3 cdc34-2 strain (NM53Δts34), which is kept viable by CDC53 on a URA3-marked plasmid, was transformed with TRP1 plasmids carrying either wild-type CDC53 or cdc53(1-793). Transformants were then patched on to 5-FOA medium to select for loss of the CDC53, URA3 plasmid.
cdc53(1–793) is synthetically lethal with a temperature-sensitive allele of cdc34
Previously, we have shown that cells unable to modify Cdc53p because of mutations in ENR2 or RUB1 are synthetically lethal with a cdc34-2 allele. To determine whether the truncation allele was also synthetic lethal with cdc34-2, a strain (NM53Δ34ts) (Table 1) was made that contains cdc34-2 and the cdc53 disruption, with the essential CDC53 activity provided by plasmid E3a (Table 2). This strain was transformed with plasmids containing either wild-type CDC53 or cdc53(1–793). The interaction between cdc34-2 and the different CDC53 alleles was assessed by the ability of each plasmid to allow colony formation after selection against E3a using 5-FOA medium (Fig. 5C). Cells that contained the plasmid bearing wild-type CDC53 were viable, as expected. However, cells that contained the plasmid expressing cdc53(1–793) were not viable. This confirms that cdc53(1–793) is synthetically lethal with cdc34-2.
Table 1.
Plasmid list
| Name
|
Features
|
Parent plasmid
|
Source
|
|---|---|---|---|
| pEApaCla | ApaI, ClaI genomic ENR2 region | pBluescript SK− | this study |
| penr2∷LEU2 | enr2∷LEU2 allele | pEApaCla | this study |
| penr2∷TRP1 | enr2∷TRP1 allele | pEApaCla | this study |
| pEUC | ClaI genomic ENR2 region, URA3, CEN6 | pRS316 and pEL2 | this study |
| BM3280 | Gala4 activation domain fusion with SKP1, LEU2, 2μ | pACT | Mark Johnston (Washington University, St. Louis, Mo) |
| MT839 | CDC53–HAX3, TRP1, CEN6 | pRS314 | Michael Tyers (University of Toronto, Canada) |
| E3a | CDC53, own promoter, URA3, CEN? | YEp24 | Mathias et al., (1995) |
| pCdc53 | CDC53, ADH promoter, TRP1, 2μ | pRS424 | this study |
| pCdc53(1-793) | cdc53(1–793), ADH promoter TRP1 | pRS424 | this study |
| pSJ4101 | CDC4, GAL10 promoter, LEU2 | pSJ101 | Liu et al. (1995) |
| YEp34-1 | CDC34, own promoter, URA3, 2μ | YEp24 | Liu et al. (1995) |
| pYL150 | CDC34, GAL10 promoter, LEU2 | pSJ101 | Liu et al. (1995) |
| pYLB2 | cdc34-2, GAL10 promoter, LEU2 | pSJ101 | Liu et al. (1995) |
| MT798 | CDC53, GAL10 promoter, LEU2 | YEp55 | Mike Tyers |
| p7127 | RUB1, GAL10 promoter, URA3 | pYES2 | this study |
| p7128 | RUB1, RUB1 promoter, URA3, ARS1 CEN4 | pSEYC102 | this study |
| p7147 | HA–RUB1, GAL10 promoter, URA3 | pYES2 | this study |
| pEL2 | ENR2, own promoter, LEU2, 2μ | YEp213 genomic library | this study |
| pMDM152 | GAL1/10 promoter, SIC1 LEU2 CEN4 | Michael Mendenhall (University of Kentucky, Lexington) | |
| pFHE52 | CLN2, MET3 promoter URA3 CEN4 | Duane Hall (University of Wisconsin, Madison) |
Table 2.
Yeast strains
| Strain name
|
Relevant markers
|
Source
|
|---|---|---|
| ADBY382 | MATα, ade2, ade3, leu2, trp1, ura3 | Alan Bender (Indiana University) |
| DLYe382 | MATα, enr2∷LEU2, ade2, ade3, leu2, trp1, ura3 | this study |
| ADBY388 | MATa, ade2, ade3, leu2, lys3, ura3 | Alan Bender |
| BJ5405/BJ5407 | MATa/MATα, his3/his3, leu2/leu2, trp1/trp1, ura3/ura3 | Elizabeth Jones via Alan Bender |
| DLYe5405 | MATα, enr2∷TRP1, his3, leu2, trp1, ura3 | this study |
| MHY508 | MATα, ubc4∷HIS3, ubc5∷LEU2, his3Δ200, leu2,3-112, ura3-52, lys2-81, trp1-1 | Mark Hochstrasser (University of Chicago, IL) |
| MHY552 | MATα, ubc6∷HIS3, ubc7∷LEU2, his3Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1 | Mark Hochstrasser |
| Y0174 | ubc9∷TRP1, leu2∷ubc9 Pro-Ser∷LEU2, leu2-3,2-112, lys2-801, trp1-1, ura3-52 | Stephan Jentsch (Friedrich-MeischerLaboratorium der Max-Planck-Gesellschaft) |
| MHY612 | MATα, rad6∷LEU2, his3Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1 | Mark Hochstrasser |
| STX337-3D | MATα, cdc27-1, ade1, ade2, ade6, gal, his7, ura3, trp1, lys2, arg | Yeast Genetics Stock Center |
| STX92-1B | MATα, cdc16-1, lys2, tyr1, leu2, pet8, rad2, his2, his7, his6, ade2, gal1, mal | Yeast Genetics Stock Center |
| MGG12 | MATα, cdc53-1, trp1, his3-Δ200, ade2, ura3-52 | Mathias et al. (1996) |
| MGG314 | MATa cdc4-1, ade1, ade2, ura1, tyr1, lys2, his7 | Mark Goebl |
| MGG15 | cdc34-2, ura2-52, his3Δ200 | Mark Goebl |
| YPH 1161 | skp1-3 | Heiter and Connelly (1996) |
| YPH 1172 | skp1-4 | Heiter and Connelly (1996) |
| Y553 | MATα, skp1-11, can1-100, ade2-1, his3-11,-15, leu2-3,-112, trp1-1, ura3-1 | Bai et al. (1996) |
| Y555 | MATα, skp1-12, can1-100, ade2-1, his3-11,-15, leu2-3,-112, trp1-1, ura3-1 | Bai et al. (1996) |
| NMY53Δ | MATa, cdc53∷HIS3, his3, lys2, trp1, leu2, ura3, p(E3a) | segregant of MGG25, (this study) |
| YPH973 + CF | MATα, ctf13-30, ura3-52, lys2-801, lys2-801, ade2-101, ade2-101, his3Δ200, trp1-Δ1, TRP1, leu2Δ1, CFIII (URA3 SUP11 CEN3) | Phil Heiter via Weidong Jiang |
| JLY 250 | rub1∷TRP1, ade2-1, leu2-3,112, his3-11, trp1-1, ura3-1, GAL+ | Judy Callis (University of California, Davis) |
| ADBY388 | MATa, ade2, ade3, leu2, lys2, ura3 | Alan Bender |
| DLYe388 | MATa, enr2∷LEU2, ade2, ade3, leu2, lys2, ura3 | this study |
| YL10 | MATα, cdc34-2, his3, trp1, leu2, ura3 | Liu et al. (1995) |
| MGG10 | MATa, cdc53-1, ura3, trp1, ade2 | Mathias et al. (1996) |
| MGG25 | MATa/MATα, his3/his3, lys2/lys2, trp1/TRP1, leu2/leu2, ura3/URA3, ade2/ADE2, cdc53∷HIS3/CDC53 | Mathias et al. (1996) |
| NM53Δts34 | MATα, his3, trp1, leu2, ura3, cdc53∷HIS3, cdc34-2 + p[E3a] | this study |
Cells that contain cdc34-2 are characterized by a mutation in the catalytic domain of Cdc34p that renders the protein inactive at 37°C (Liu et al. 1995). In addition, the level of Cdc34p in cdc34-2 strains is substantially less than that found in CDC34 cells (Fig. 6A). Thus, it is possible that synthetic lethality observed between cdc34-2 and cdc53(1–793) is a consequence of the reduced levels of Cdc34p and not of the cdc34-2 mutation per se. We attempted to address this issue with the following experiment. In previous studies we have found that there is sufficient read-through of the GAL1/10 promoter under promoter-repressing conditions to permit the rescue of the cdc34-2 temperature-sensitive defect with a GAL::CDC34 gene (Fig. 6A). The rescue of the temperature-sensitive defect under these conditions occurs despite the fact that the abundance of immunodetectable Cdc34p in a cdc34-2 background is not increased by the presence of the plasmid (Fig. 6A, lane 1 vs. lane 2). On the basis of these observations, we reasoned that if the basis of the synthetic lethality between cdc53(1–793) and cdc34-2 was abnormally low Cdc34p levels, then this synthetic lethality should persist in the presence of GAL::CDC34 in promoter-repressing conditions, a situation where sufficient wild-type Cdc34p is produced to sustain growth at restrictive temperatures in cdc34-2 single mutants. If, on the other hand, the synthetic lethality resulted from an interaction between Cdc53(1–793)p and Cdc34-2p related to some property of the two mutant proteins aside from altered steady-state levels of Cdc34p, then the synthetic lethality should be alleviated. We transformed a cdc53Δ, cdc34-2 strain kept viable with CDC53 on a URA3 plasmid with one of three plasmids: wild-type CDC53 under the control of its own promoter on a TRP1 plasmid, cdc53(1–793) behind the ADH promoter on a TRP1 plasmid, or the TRP1 plasmid alone. These strains were all transformed with a third plasmid marked with LEU2, either CDC34 behind its own promoter, a GAL::CDC34 fusion, or a GAL::cdc34-2 gene. The strains were then tested for growth on S-dextrose + 5-FOA, and on S-galactose + 5-FOA. We found that this strain was unable to survive without the URA3-marked wild-type CDC53 plasmid in GAL promoter-repressing conditions, despite the fact that sufficient wild-type Cdc34p was produced in this strain to maintain viability at 37°C when no counterselection against wild-type CDC53 was applied (Fig. 6B, row 5, column 2 vs. column 4). This result indicates that under conditions where the level of Cdc34p is reduced, Cdc53(1–793)p is not able to support viability. It is possible that this failure is attributable to an effect of the Cdc53p truncation unrelated to Rub1p modification. However, given that cdc53(1–793) is capable of complete rescue of cdc53Δ in other contexts we favor the hypothesis that the inability to produce Rub1p–Cdc53p renders cells sensitive to decreased levels of Cdc34p. In support of this hypothesis, we observe a modest but consistent increase in the ratio of Rub1–Cdc53p to Cdc53p in cdc34-2 strains (Fig. 3B, wild-type lanes vs. cdc34-2 lanes).
Figure 6.
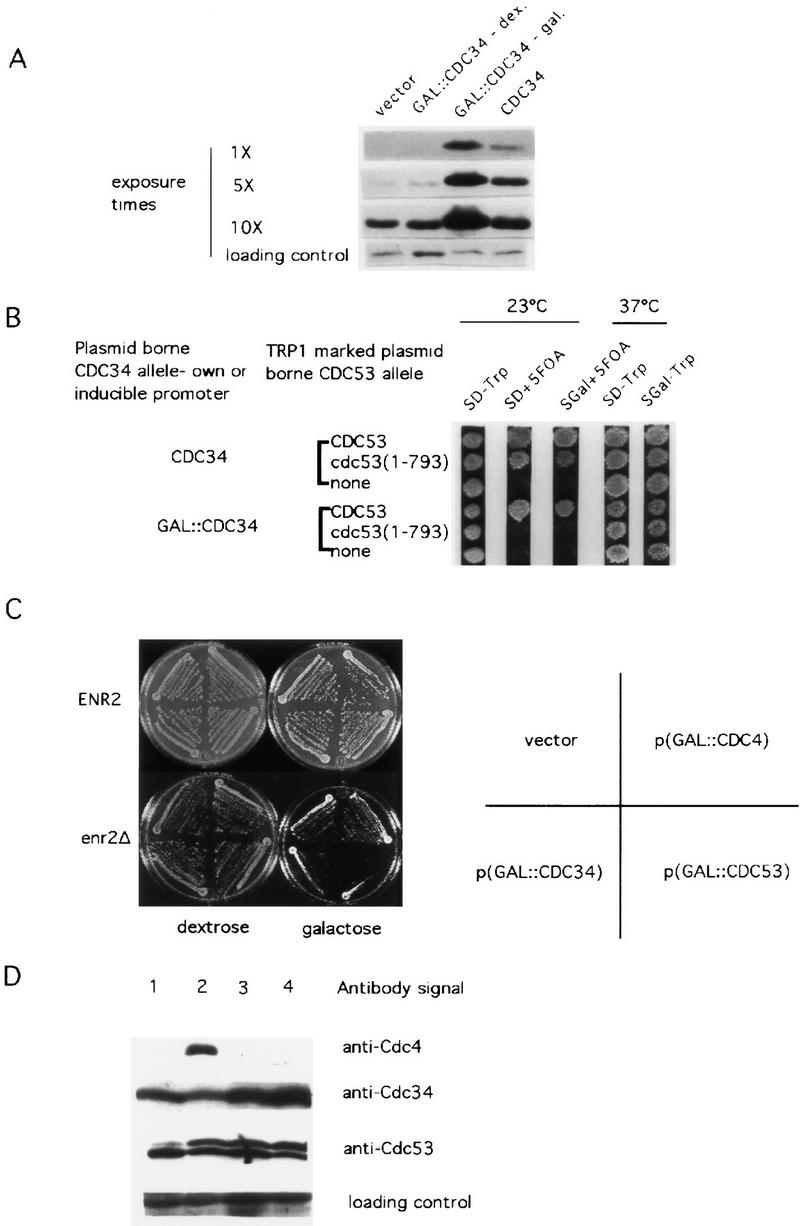
The absence of Rub1p conjugation to Cdc53p renders cells sensitive to altered levels of Cdc34p and Cdc53p. (A) Anti-Cdc34p Western blot demonstrating the relative abundance of Cdc34p in NMY53Δts34 strains transformed with GAL:: CDC34 in promoter-repressing conditions (lane 2), with GAL::CDC34 in promoter-inducing conditions (lane 3), and with CDC34 under the control of its own promoter (lane 4). Three exposure times of the same Western blot are shown. An anti-Cdc34p cross-reacting band serves as a loading control. (B) Both increased and decreased abundance of Cdc34p is lethal to cdc53(1–793) strains. A cdc53::HIS3, cdc34-2 strain (NMY53Δts34) is kept viable by CDC53 on a URA3-marked plasmid. This strain was transformed with either vector, CDC53, or cdc53(1–793) on a TRP1-marked plasmid. A third plasmid marked with LEU2 was then introduced into these resulting strains. The LEU2-marked plasmids contained either CDC34 behind its own promoter, CDC34 driven by the GAL1/10 promoter, or the cdc34–2 allele driven by the GAL1/10 promoter. The resulting strains were then tested for their ability to survive on 5-FOA medium in promoter-inducing or repressing conditions, and to survive at high temperature. (C) enr2Δ strains are sensitive to overexpression of CDC34 and CDC53. enr2::TRP1 and ENR2 strains were transformed with vector, GAL::CDC4, GAL::CDC34, or GAL::CDC53, and grown on either S-dextrose or S-galactose plates. (D) Overexpressing CDC4 and CDC34 in wild-type cells leads to an increase in the relative abundance of the Rub1–Cdc53p isoform. ADBY382 was transformed with either GAL::CDC4, CDC34, or GAL::CDC34, and lysates were prepared after growth of the cells in S-galactose medium. ADBY382 transformed with vector (lane 1); GAL::CDC4 (lane 2); CDC34 from its own promoter (lane 3); GAL::CDC34 (lane 4). Westen blots were probed with anti-Cdc4, anti-Cdc34, or anti-Cdc53 antibodies as indicated. An anti-Cdc4p cross-reacting band served as a loading control.
The effect of increasing the level of SCF components and Cdc34p in backgrounds incapable of Rub1p conjugation to Cdc53p was evaluated by expressing CDC4, CDC34, or CDC53 in enr2Δ and cdc53(1–793) strains. Overexpression of CDC34 (Fig. 6B), and CDC4 (not shown) was severely inhibitory to cdc53(1–793) strains, and overexpression of CDC34 and CDC53, but not CDC4, was found to be inhibitory to enr2Δ cells (Fig. 6C). It is unclear why a discrepancy exists in the behavior of enr2Δ cells and cdc53(1–793) with regard to the sensitivity of the two mutants to overexpression of CDC4. We suspect that the sensitivity of cdc53(1–793) cells to high levels of Cdc4p either represents a consequence of the mutation unrelated to the failure to become modified by Rub1p, or that the slightly elevated Cdc53p levels in the cells when cdc53(1–793) is expressed under the control of the ADH promoter, combined with the elevated levels of Cdc4p, are lethal.
To determine whether alterations in the levels of SCF components affect the amount of modified Cdc53p, a standard laboratory strain, ADBY382, was transformed with plasmids carrying GAL/10-driven CDC4, or CDC34, and the ratio of Rub–Cdc53p to Cdc53p was compared against a vector-only control. Western blotting against Cdc34p and Cdc4p confirmed that steady-state protein levels were increased under inducing conditions (Fig. 6D). Overexpressing either Cdc4p, Cdc34p, or Cdc53p resulted in a dramatic increase in steady-state ratios of Rub1–Cdc53p to Cdc53p (Fig. 6D).
Discussion
Our results identify Cdc53p as the first known target for conjugation to the RUB1/NEDD-8 family of ubiquitin-related proteins. Unlike most ubiquitinated proteins, a single Rub1p molecule is conjugated to Cdc53p and the modification does not target the protein for rapid degradation. We have identified two genes whose activities are required for this conjugation event, ENR2 and SKP1. The biochemical function of Enr2p is suggested by its similarity to Aos1p. The activation of a second yeast ubiquitin-related protein, Smt3p, requires the cooperative action of two proteins—one related to the amino terminus of E1 (Aos1p) and the other related to the carboxyl terminus of E1 (Uba2p) (Johnson et al. 1997). By analogy, the activation of Rub1p is likely to occur through the interaction of Enr2p with an unidentified protein similar to Uba2p.
The role of Skp1p in Rub1p conjugation is less clear. We cannot rule out the possibility that the absence of Rub1–Cdc53p in skp1 backgrounds is a secondary effect of the skp1 lesions, although several pieces of data argue against this interpretation. First, mutations in other genes whose functions are implicated in the G1–S transition do not affect the formation of Rub1–Cdc53p, including cdc4, cdc34, and grr1. The absence of Rub1–Cdc53p in skp1 backgrounds is therefore clearly not simply a consequence of arrest or delay at the G1–S boundary. Furthermore, in three of the four skp1 alleles examined, Cdc53p is not modified at both permissive and restrictive temperatures. Second, we observe a strong enhancement of both the skp1-11 and skp1-12 phenotype in enr2Δ double mutants, strongly suggesting the involvement of ENR2 and SKP1 in common functions. Therefore, we favor the hypothesis that SKP1 functions directly in Rub1p conjugation. One possibility is that Skp1p is part of an E3 complex that is required for Rub1p conjugation.
The Rub1p pathway is dispensible in S. cerevisiae cells growing under standard laboratory conditions. Both rub1Δ and enr2Δ cells grow at normal rates over a wide range of temperatures. Furthermore, the behavior of both strains was similar to the wild type when exposed to a variety of stress conditions including acute heat shock, UV irradiation, as well as plating on medium containing cadmium or canavanine (data not shown). enr2Δ cells are not sensitive to overexpression of two targets of SCF complexes, Sic1p and Cln2p, whereas the mutant strains cdc34, cdc4, cdc53, and skp1 show pronounced sensitivity to overexpression of these genes (Schwob et al. 1994; Bai et al. 1996; Willems et al. 1996). Direct measurement of Sic1p protein stability in an enr2Δ background also failed to detect a difference between enr2Δ and wild-type strains (D. Lammer and M. Estelle, unpubl.).
In contrast we found that the Rub1p pathway is critical when the function of the SCF is compromised by mutations in CDC34, CDC4, CDC53, or SKP1. This affect is probably attributable to an inability to modify Cdc53p, as cdc53(1–739) also displays synthetic lethality with cdc34-2. In addition, both enr2Δ and cdc53(1–793) are sensitive to overexpression of CDC34 and CDC53. Because both overexpression and underexpression lead to an increase in the relative amount of Rub1–Cdc53p, this sensitivity may be caused by the inability to conjugate Rub1p to Cdc53p. One possibility is that modification of Cdc53p affects SCF assembly. To date, three such complexes have been described: SCFCdc4p, SCFGrr1p, and SCFMet30p (Feldman et al. 1997; Li and Johnston 1997; Skowyra et al. 1997; M. Tyers, pers. comm.). In animals, modification of RanGAP1 by the ubiquitin-related protein SUMO-1 is required for binding of RanGAP1 to Nup358/RanBP2 (Matunis et al. 1996; Mahajan et al. 1997). Similarly, modification of Cdc53p may affect interactions with its various binding partners.
Although the precise physiological role of Rub1p conjugation is unclear, our results indicate that the modification is relevant under certain circumstances. It is also important to note that mutations in genes related to ENR2 result in dramatic defects in cell growth. In Chinese hamsters, the smc1 mutation is responsible for cell cycle defects in the ts41 cell line (Handeli and Weintraub 1992). The SMC1 protein is nearly identical to APP-BP1 (S. Handeli, pers. comm.) and displays a two-hybrid interaction with the mammalian Rub1p homolog NEDD-8, suggesting a role for SMC1 in an analogous NEDD-8 conjugation system. In the plant A. thaliana, mutations in the AXR1 gene result in a defect in auxin-stimulated cell division and cell elongation. Recent results indicate that AXR1 functions in activation of Arabidopsis RUB proteins (J.C. del Pozo, J. Callis, and M. Estelle, unpubl.).
We have identified the carboxy-terminal 21 amino acids of CDC53 as being necessary for conjugation to Rub1p. This sequence is not sufficient for Rub1p modification, analogous to the destruction box for ubiquitination, because it does not result in Rub1p modification of heterologous proteins when placed in cis (N. Mathias and M. Goebl, unpubl.). However, the carboxyl terminus may be a secondary signal for Rub1p modification, analogous to the requirement for phosphorylation of many proteins before their recognition by the ubiquitination system (Lanker et al. 1996; Verma et al. 1997). Another possibility is that the carboxy-terminal region is the actual site of Rub1p conjugation.
Cdc53p is a member of a protein family called the Cullins, after the CUL1 locus of Caenorhabditis elegans. Loss of function cul1 mutations cause increased cell proliferation in a number of embryonic lineages in the worm, suggesting a tumor-suppressing function for this class of proteins in multicellular eukaryotes (Kipreos et al. 1996). One human homolog, Hs-CUL-2, has been shown to associate with the von Hippel–Lindau tumor-suppresser gene products, elongin B and elongin C (Pause et al. 1997). Furthermore, elongin C is a Skp1-like protein, and elongin B is a ubiquitin homolog. The elongin B protein contains a glycine comparable to the COOH glycine of ubiquitin and Rub1p suggesting the possibility that elongin B may modify HsCul2. (Aso et al. 1995; Krumm and Groudine 1995). One of the most highly conserved regions within the CDC53/CUL1 family is the carboxy-terminal domain that we have shown is required for Rub1p modification. At present, we do not know whether any plant or animal Cullins are RUB modified. If they are, it is possible that the modification has a similar function in these systems as in yeast.
The discovery of distinct biochemical pathways for the activation of the yeast ubiquitin-related proteins Smt3p and Rub1p raises many interesting questions regarding the metabolism of these proteins. On the basis of amino acid sequence, 13 yeast genes are suspected to encode ubiquitin-conjugating enzymes. For some of these, biochemical activity with ubiquitin has not been reported leaving open the possibility that they function in RUB1 or SMT3 pathways instead. In a recent study, yeast Ubc9p was shown to function as an E2 for Smt3p (Johnson and Blobel 1997). Similarly, at least 16 yeast genes encode potential deubiquitinating enzymes (Hochstrasser 1996). Some proportion of these may also be specific to Rub1p or Smt3p conjugates. Finally, additional studies will be required to determine whether any Rub1p-conjugated proteins are substrates for the proteasome.
The finding that Rub1p and ubiquitin are immunologically cross-reactive will necessitate the reevaluation of many reports of ubiquitin modification. Perhaps most important to reexamine will be those cases where, like the 98-kD isoform of Cdc53p, the modified form of the protein appears to be metabolically stable. For example, many neurodegenerative diseases, including Alzheimer’s disease, are associated with the accumulation of ubiquitin cross-reactive material (Mayer et al. 1996). Currently, we have no direct biochemical evidence for alternative targets of Rub1p conjugation. However, we have found that the kinetochore protein mutation ctf13–30 is suppressed by high level expression of ENR2 (D. Lammer and M. Estelle, unpubl.). This result suggests that Rub1p modification may also have a role in some aspect of kinetochore function.
Materials and methods
Media and yeast manipulations, yeast strains, and plasmids
Standard media and methods were used for growth and genetic manipulation of yeast (Ausubel et al. 1987). Yeast strains and plasmids used in this study are listed in Tables 1 and 2. Some yeast transformations were performed using a Frozen E-Z Kit (Zymo Research) according to the manufacturer’s protocol. All physiological comparisons between double mutant and single mutant genotypes were performed by selecting double mutant segregants and transforming these with either vector or the wild-type copy of one of the mutant genes on a plasmid vector, and multiple segregants were analyzed for each.
Plasmid construction
Plasmid and genomic DNA were prepared using standard methods, and used to generate PCR products as described previously (Liu et al. 1995). Full-length CDC53 was cloned into pGEM7 (Promega Corp.) as follows. A SphI and HindIII restriction digest of pYcDE53-1 yields a fragment containing the ADHI promoter and a portion of CDC53 encoding the first 752 residues of Cdc53p. This fragment was ligated into pGEM7 that had been digested with the same enzymes to create pNM53Δ. A PCR fragment containing a portion of CDC53 that encodes residues 753–815 was digested with HindIII and SacI and ligated into pNM53Δ digested with the same enzymes to generate pNM53FL. The PCR product was generated using primers that annealed at the 5′ side of the HindIII site in the CDC53 gene (5′-ATACGATAGCGAATTAGGAAACAAACGCTTGACGGAAG-3′) and annealed at the 3′ end of the gene (5′-AAAGAGCTCGAATTCAATCACACACAACGAGAACGATC-3′). An ApaI–SacI fragment from pNM53FL that contains full-length CDC53 downstream of the ADH1 promoter was ligated into pRS424 restricted with the same enzymes to create pCdc53. A premature stop codon was engineered into CDC53 as follows. A PCR fragment containing two successive stop codons after residue 793 was digested with XbaI and SacI and ligated into pNM53FL digested with the same enzymes to create pNM(1–793). The PCR product was generated using a primer that anneals toward the 5′ end of the gene (5′-AAAGGGTTCTGAAAGTTTCCCCGACGACATAC-3′) with 5′-GGGGAGCTCTTATTATTCCAAGAAAATCTGCCTTTCTG-3′, which anneals toward the 3′end of the CDC53 gene. ApaI–SacI fragments generated from this plasmid were cloned into pRS424 to create pCdc53(1–793). p(ENR2 Apa Cla) was constructed by digesting PCR-amplified genomic DNA from the ENR2 region with ApaI and ClaI and ligating this fragment into pBluescript SK− digested with the same enzymes. The PCR fragment was amplified from genomic yeast DNA with the following primers: 5′-TGCTTGCCGGAATATCATCT-3′ and 5′-CACCTCCTCCAAAAAAGGCCATT-3′. The insert of this plasmid was used to screen a genomic library cloned into the YEp318 vector; one isolate that contained the complete ENR2 locus and ∼3 kb of upstream sequence was designated pEL2. A ClaI fragment from pEL2 containing ENR2 and its complete promoter was subcloned into ClaI-digested pSJ316 to generate pEUC. p(enr2::LEU2) was constucted by digesting p(ENR2 Apa Cla) with BglII and SalI and replacing this fragment with a BamHI–SalI fragment of pJJ252 containing the LEU2 locus. p(enr2:: TRP1) was constructed in an identical fashion, except the BglII–SalI fragment was replaced by a BamHI–SalI fragment from pJJ248 containing the TRP1 locus.
The S. cerevisiae genomic clone c9302 (ATCC 70929) was used as template for amplification of the coding region of S. cerevisiae RUB1 using the 5′ primer 5′-CGCGGATCCTATGATTGTTAAAGTGAAGACACTGAC and the 3′ primer 5′-CTCGGATCCGAATTCCTCGAGTCAACCACCTCTTAGTGTTAATACCAAG. The PCR fragment was ligated subsequently into pYES2 (Invitrogen, Inc.) and pYES2–HA using the BamHI and XhoI sites, and the sequence was verified by dideoxy sequencing. pYES2–HA was constructed by ligating a double-stranded oligonucleotide that encodes for the HA epitope into the HindIII and BamHI restriction sites of pYES2 (M. West, unpubl.).
An NdeI–NcoI fragment from S. cerevisiae c9302 containing the RUB1-coding regions, containing 458 bp and 262 bp of the 5′ and 3′ region, respectively, was cloned into a Bluescript plasmid (Stratagene) with the polylinker modified to contain an NcoI and NdeI site. The fragment was then moved as a BamHI–SalI fragment into the BamHI–SalI sites of pSEYC102, replacing the lacZ-coding region.
Strain construction
Yeast strains are listed in Table 1. DLYe382 and DLYe388 are segregants from an ADBY382/ADBY388 diploid transformed with plasmid p(enr2::LEU2). The plasmid was digested with ApaI and BamHI to liberate the insert before transformation. The replacement of ENR2 by the enr2::LEU2 allele was confirmed by Southern blot analysis. DLYe5405 and DLYe5407 are segregents from a BJ5405/BJ5407 diploid transformed with plasmid p(enr2::TRP1). The plasmid was digested with ApaI and BamHI to liberate the insert before transformation. The replacement of ENR2 by the enr2::TRP1 allele was confirmed by Southern blot analysis.
The enr2::LEU2, cdc34-2 strains shown in Figure 1 are segregents from the following crosses. DLYe388 was crossed to MGG15 and a MATa, cdc34-2, ade2, ade3, leu2, lys2, ura3, segregent from this cross was backcrossed to DLYe382; the strains shown in Figure 1 are representative segregents of the indicated genotypes from this second cross.
The enr2::LEU2, cdc4-1 strain shown in Figure 1 is a segregant from a cross between MGG314 and DLYe382. Double mutant segregents were transformed with vector or p(EUC) to make comparisons between cdc4-1 and cdc4-1, enr2Δ. The cdc4–1 strain in Figure 1 was a single mutant segregent from this cross. Several other double mutant segregents were transformed with the two plasmids and showed similar results.
The enr2::LEU2, cdc53-1 strain shown in Figure 1 is a segregant from a cross between MGG12 and DLYe388. The double mutant segregents were transformed with vector or with p(EUC) to make comparisons between cdc53-1 and cdc53-1, enr2Δ. Several other double mutant segregents were transformed with the two plasmids and showed similar results.
The enr2::TRP1, skp1-12 strains shown in Figure 1 are segregants from a cross between Y555 and DLYe5405. Double mutant segregants were transformed with vector or with p(EUC) to make comparisons between skp1-12 and skp1-12, enr2Δ.
NM53Δ was constructed as follows. A heterozygous cdc53 disruption strain MGG25 cdc53::HIS3/CDC53 (Table 1; Mathias et al. 1996) was transformed with plasmid E3a, which expresses wild-type CDC53, marked by the URA3 gene. Transformants were sporulated and asci dissected on YPD medium. Ura+ His+ colonies were identified, one of which was named NM53Δ. Strain NM53Δts34 is a meiotic product of a cross between NM53Δ and YL10 and contains the cdc34-2 temperature-sensitive allele and the cdc53 disruption.
A PCR-based technique was used for the construction of the rub1 deletion strain. Primers were constructed with 40 bp identical to the RUB1 −10 to +30 or to the RUB1 +307 to +346 3′ untranslated region followed by 18 or 21 bp, respectively, of sequence outside the auxotrophic markers common to all pRS vectors. The TRP1 auxotrophic marker was amplified using the pRS314GU plasmid as template. The PCR fragment was purified from agar using GeneClean (BIO 101, Inc.); strain W303-1B was transformed using a modified Li-PEG procedure, and Trp+ colonies were selected. Total DNA was purified from Trp+ transformants. Using primers containing sequences outside the replaced region, the RUB1 locus was amplified from DNA of individual TRP+ colonies to distinguish between insertion of the PCR fragment at the RUB1 locus and a TRP1+ revertant.
Western blot analysis
Western blots were performed using two different protocols depending on the experiment. Extracts were prepared as described in Liu et al. (1995) and proteins were separated by SDS-PAGE. In some cases, proteins were transferred onto polyvinylidene fluoride membranes with 10 mm 3-[cyclohexylamino]-1-propane sulfonic acid (pH 11). In other cases transfer was performed according to Ausubel et al. (1987). Generation and affinity purification of anti-Cdc4, anti-Cdc34, and anti-Cdc53 antibodies have been described previously (Goebl et al. 1994; Mathias et al. 1996). Anti-HA monoclonal antibody 12CA5 was purchased from Boehringer Mannheim. Secondary antibodies were peroxidase conjugated goat anti-rabbit or goat anti-mouse (Sigma and Amersham). Detection was by the enhanced chemiluminesence method as described by the manufacturer (Amersham).
Cdc53p pulse-chase and immunoprecipitation
NMY53Δ carrying pMT389 (CDC53–HA), was grown in YPD until early log phase, centrifuged, and resuspended in 2 ml of YPD supplemented with 150 μCi of Amersham in vivo labeling Redimix of [35S]methionine and cysteine. Cells were pulsed for 2 min, then washed with two changes of cold YPD before being resuspended in 100 ml of YPD supplemented with unlabeled methionine and cysteine for outgrowth. Aliquots (10 ml) were taken at time points between 5 and 210 min, washed in water, and then frozen at −80°C before protein extraction. Protein extracts were prepared by the glass bead disruption method (Ausabel et al. 1987) in a buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mm EDTA, 5 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μm sodium metabisulfate, and PMF. The protein extracts were preadsorbed with 50 μl of a protein A–agarose slurry (Boehringer Mannheim) for 3 hr at 4°C. Five micrograms of monoclonal anti-HA antibodies (Boehringer Mannheim) were added to the supernatant and were incubated for 1 hr at 4°C, after which 50 μl of protein A–agarose slurry was added for an additional 3 hr. The beads were washed twice in extraction buffer for 20 min and three times in 50 m Tris-HCl (pH 7.5), 500 mm NaCl, 01% NP-40. The beads were then boiled for 5 min in 1× SDS-PAGE loading buffer, and extracts were run on a 10% polyacrylamide gel, dried on Whatman paper, and subjected to autoradiography.
HA–Rub1–Cdc53p immunoprecipitation
Immunoprecipitations were performed on extracts of rub1Δ cells carrying plasmids with HA–RUB1 or RUB1 behind a GAL1/10 promoter after 6 hr of growth in liquid SM–Ura galactose media. Protein extraction and immunoprecipitations were performed as above, except that the primary antibody was affinity-purified anti-Cdc53p and immunoprecipitates were used for Western blotting with the monoclonal anti-HA antibody 12CA5 (Boehringer Mannheim).
Acknowledgments
We thank Jürgen Dohmen, Mark Johnson, Michael Tyers, Phil Heiter, Mark Hochstrasser, Michael Mendenhall, Alan Bender, Steven Elledge, Bruce Futcher, Stephan Jentsch, Ben Hall, and Lee Hartwell for generously supplying strains and plasmids, Erica Johnson for providing information in advance of publication, and the yeast and Arabidopsis groups of Indiana University, especially Alan Bender, Jose Bonner, Tom Donahue, and the Bender laboratory for helpful discussions and technical advise during the course of this work. We also thank Mark Hochstrasser and Andrew Murray (D.L. and M.E.) and Dan Finley for helpful discussions (J.C.) of our preliminary results and Alan Bender for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health (NIH) (GM43644) to M.E., a National Science Foundation grant (93-06759), and a National Science Foundation Presidential Young Investigator Award (NSF 91-58453) to J.C. and an NIH grant (GM45460) to M.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mestelle@bio.indiana.edu; FAX (812) 855-6705.
References
- Aso T, Weliky-Conaway J, Conaway RC. The RNA polymerase II elongation complex. FASEB J. 1995;9:1419–1428. doi: 10.1096/fasebj.9.14.7589983. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Brooklyn, NY: Greene Publishing Associates; 1987. [Google Scholar]
- Bai C, Sen P, Hofmann K, Mei L, Goebl M, Harper JW, Elledge SJ. Skp1p connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes & Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- Callis J, Carpenter T, Sun C-W, Vierstra RD. Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics. 1995;139:921–939. doi: 10.1093/genetics/139.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow N, Koernberg JR, Chen XN, Neve RL. APP-BP1, a novel protein that binds to the carboxyl-terminal region of the amyloid precursor protein. J Biol Chem. 1996;271:11339–11346. doi: 10.1074/jbc.271.19.11339. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential gene encoding a homologue of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Goebl MG, Jochem J, McGrath JP, Varshavsky A, Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- Guarino LA. Identification of a viral gene encoding a ubiquitin-like protein. Proc Natl Acad Sci. 1990;87:409–413. doi: 10.1073/pnas.87.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AL, Ahrens P, Bright P, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- Handeli S, Weintraub H. The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M, and G1. Cell. 1992;71:599–611. doi: 10.1016/0092-8674(92)90594-3. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul1-1, a C. elegans CDC53 homologue, is a negative cell cycle regulator required for exit from the cell cycle. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Krumm A, Groudine M. Tumor suppression and transcriptional elongation: The dire consequences of changing partners. Science. 1995;269:1400–1401. doi: 10.1126/science.7660121. [DOI] [PubMed] [Google Scholar]
- Kumar S, Yoshida Y, Noda M. Cloning a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun. 1993;195:393–399. doi: 10.1006/bbrc.1993.2056. [DOI] [PubMed] [Google Scholar]
- Lanker S, Valdivieso MH, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Li FN, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis pathway through Skp1: Coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Steussy CN, Goebl MG. Intragenic suppression among CDC34 mutations defines a class of ubiquitin-conjugating catalytic domains. Mol Cell Biol. 1995;15:5635–5644. doi: 10.1128/mcb.15.10.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mathias N, Johnson SL, Winey M, Adams A, Goetsch L, Pringle JR, Byers B, Goebl MG. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer RJ, Tipler C, Arnold J, Laszlo L, Al-Khedhairy A, Lowe J, Landon M. Endosome-lysosomes, ubiquitin and neurodegeneration. Adv Exp Med Biol. 1996;389:261–269. doi: 10.1007/978-1-4613-0335-0_33. [DOI] [PubMed] [Google Scholar]
- McGrath JP, Jentsch S, Varshavshy A. UBA1: An essential yeast gene encoding ubiquitin activating enzyme. EMBO J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei CF, Chang HM, Yeh ET. Protection against Fas/Apo-1 and tumor necrosis factor mediated cell death by a novel protein, sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- Pause A, Lee S, Worrell RA, Chen DYT, Burgess WH, Linehan WM, Klausner RD. The von Hippel-Lindau tumor suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Bohm TB, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40/SIC1 controls the G1 to S phase transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Shayeghi M, Doe CL, Tavassoli M, Watts FZ. Characterization of Schizosaccharomyces pombe RAD31, a UBA-related gene required for DNA damage tolerance. Nucleic Acid Res. 1997;25:1162–1169. doi: 10.1093/nar/25.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Pariington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. Association of UBE21 with RAD52, UBL1, p53 and RAD51 proteins in a yeast two-hybrid system. Genomics. 1996;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Thomas D, Kuras L, Barbey R, Cherest H, Blaiseau PL, Surdin-Kerjan Y. Met30p, a yeast transcriptional inhibitor that responds to S-adenosylmethionine, is an essential protein with WD repeats. Mol Cell Biol. 1995;15:6526–6534. doi: 10.1128/mcb.15.12.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 1995;8:561–569. doi: 10.1046/j.1365-313x.1995.8040561.x. [DOI] [PubMed] [Google Scholar]
- Verma R, Renny Feldman RM, Deshaies RJ. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/Cdk activities. Mol Biol Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]