Abstract
The mammalian genome contains numerous regions known as facultative heterochromatin, which contribute to transcriptional silencing during development and cell differentiation. We have analyzed the pattern of histone modifications associated with facultative heterochromatin within the mouse imprinted Snurf–Snrpn cluster, which is homologous to the human Prader-Willi syndrome genomic region. We show here that the maternally inherited Snurf–Snrpn 3-Mb region, which is silenced by a potent transcription repressive mechanism, is uniformly enriched in histone methylation marks usually found in constitutive heterochromatin, such as H4K20me3, H3K9me3, and H3K79me3. Strikingly, we found that trimethylated histone H3 at lysine 36 (H3K36me3), which was previously identified as a hallmark of actively transcribed regions, is deposited onto the silenced, maternally contributed 3-Mb imprinted region. We show that H3K36me3 deposition within this large heterochromatin domain does not correlate with transcription events, suggesting the existence of an alternative pathway for the deposition of this histone modification. In addition, we demonstrate that H3K36me3 is markedly enriched at the level of pericentromeric heterochromatin in mouse embryonic stem cells and fibroblasts. This result indicates that H3K36me3 is associated with both facultative and constitutive heterochromatin. Our data suggest that H3K36me3 function is not restricted to actively transcribed regions only and may contribute to the composition of heterochromatin, in combination with other histone modifications.
Modulation of the biochemical properties of nucleosomes through histone covalent modifications plays a crucial role in the regulation of transcription in mammals (Kouzarides 2007; Ruthenburg et al. 2007). These modifications can alter the structure of chromatin and stimulate the establishment of a chromatin state that is permissive to transcription, which is usually referred to as open chromatin, or euchromatin (Workman and Kingston 1998). On the other hand, specific enzymatic activities contribute to the formation of chromatin domains that are refractory to transcription, collectively called heterochromatin regions. A subset of these regions, which is present in all cell types and stable during the cell cycle, is referred to as constitutive heterochromatin. In contrast, other heterochromatin regions, established in a regulated manner during development and cell differentiation, are called facultative heterochromatin (Trojer and Reinberg 2007).
The profile of histone modifications on the mouse and human genomes has been analyzed in several cell types using chromatin immunoprecipitation followed by large-scale sequencing (Barski et al. 2007; Mikkelsen et al. 2007). These analyses revealed that histone modifications could be classified in two main groups that correlate with either active transcription and euchromatin, or gene repression and heterochromatin. Histone acetylation is primarily associated with gene activation. In contrast, the relationship between gene expression and histone lysine methylation is more complex as it depends on the position and state of methylation (mono-, di-, or trimethylation) (Martin and Zhang 2005). Studies using different model organisms have linked the methylation of histone H3 at both lysines 4 and 36 to transcriptionally active regions (Barski et al. 2007; Mikkelsen et al. 2007; Wang et al. 2008). While trimethylated histone H3 lysine 4 (H3K4me3) accumulates in promoter regions, trimethylated histone H3 at lysine 36 (H3K36me3) is deposited in the core and at the 3′ end of active genes (Bannister et al. 2005; Barski et al. 2007; Mikkelsen et al. 2007; Edmunds et al. 2008).
Several histone modifications have been linked to the formation of constitutive heterochromatin domains, including H3K9 trimethylation (H3K9me3) and trimethylated histone H4 at lysine 20 (H4K20me3) (Rea et al. 2000; Peters et al. 2001; Schotta et al. 2004). More recently, a third histone modification, H3K79me3, was shown to be involved in the formation of constitutive heterochromatin in mouse embryonic stem cells (Jones et al. 2008). Histone marks found in constitutive heterochromatin are often associated with facultative heterochromatin domains. For instance, H3K9me2/3 and H4K20me3 were found in subdomains of the inactive X chromosome in female somatic cells (Heard et al. 2001; Peters et al. 2002; Chadwick and Willard 2004). However, facultative heterochromatin formation is also controlled by mechanisms independent of those controlling constitutive heterochromatin. Indeed, facultative heterochromatin formation is often associated with histone H3 trimethylated at lysine 27 (H3K27me3), a mark that is absent in constitutive heterochromatin (Peters et al. 2003). H3K27me3, which is catalyzed in mammals by Polycomb repressive complex 2 (PRC2), contributes to the mechanisms of X-chromosome inactivation (Plath et al. 2003; Silva et al. 2003; Rougeulle et al. 2004). H3K27me3 was also shown to be involved in the control of genomic imprinting, which results in the exclusive or preferential expression of only one of the two parental alleles of specific genes (Lewis et al. 2004; Umlauf et al. 2004a). As in X-chromosome inactivation, facultative heterochromatin formation at imprinted loci involves additional mechanisms, such as DNA methylation and histone modifications like H3K9me3 and H4K20me3 (Wu et al. 2006; Delaval et al. 2007; Regha et al. 2007; Pannetier et al. 2008).
In this study, we analyzed in detail the pattern of histone modifications associated with facultative heterochromatin within a large imprinted domain of the mouse genome, called the Snurf–Snrpn cluster. In human, this highly conserved domain is referred to as the Prader-Willi syndrome (PWS) genomic interval (Nicholls and Knepper 2001). PWS is a complex neurogenetic disorder (Online Mendelian Inheritance in Man entry number 176270) with a population prevalence of 1 in 15,000. PWS is characterized by decreased fetal activity, muscular hypotonia, respiratory abnormalities, failure to thrive, short stature, obesity, mental retardation, and hypogonadism (Holm et al. 1993; Wharton and Bresnan 1989). PWS results from lack of paternal expression of one or several genes from the 15q11–q13 chromosomal region. This 3-Mb region includes several imprinted protein-coding genes, including MKRN3, MAGEL2, necdin (NDN), and SNRPN. A large number of non-coding RNAs are also transcribed from the paternal chromosome, including IPW and C/D-box snoRNA genes. The snoRNAs are organized in two large clusters, HBII-85 and HBII-52, that contain, respectively, 29 and 47 copies of snoRNAs. The mouse Snurf–Snrpn cluster (equivalent to the PWS genomic region in human) is located on chromosome 7 and contains all the aforementioned genes, which, as in human, are expressed from the paternally contributed chromosome. The mouse Snurf–Snrpn cluster is delimited on one side by the Peg12 gene (which is absent in human) and by Ube3a on the other side (Fig. 1A). While Peg12 belongs to the paternally expressed gene series, Ube3a has a more complex regulation: It is either maternally expressed or biallelically expressed, depending on the tissues examined. Ube3a is not involved in the PWS, but its deletion on the maternally contributed chromosome causes the Angelman syndrome (Nicholls and Knepper 2001). The expression of all PWS genes is dependent on the integrity of the Imprinting Center (IC), a cis-acting DNA element located in the 5′ region of Snrpn (Buiting et al. 1995; Ohta et al. 1999). In both human and mice, the PWS-IC is required for the transcription of genes located on the paternally contributed chromosome (Bielinska et al. 2000; Shemer et al. 2000; Bressler et al. 2001).
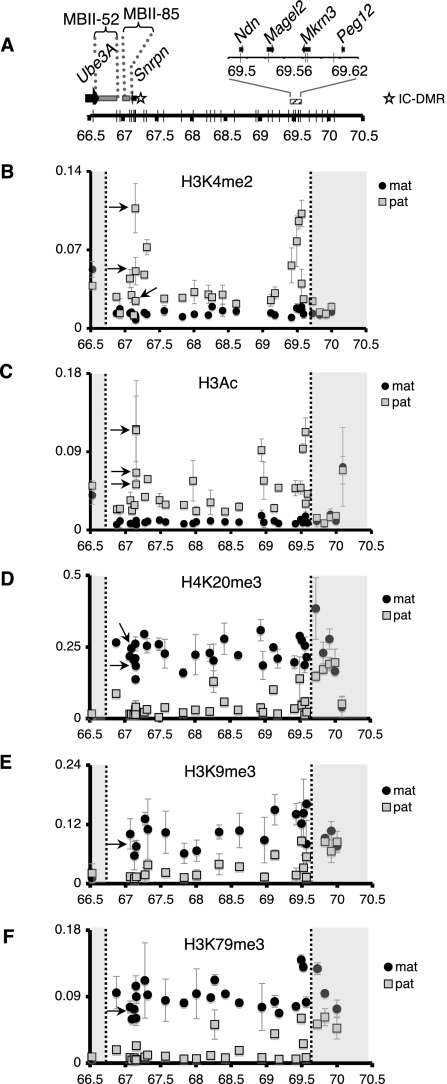
Figure 1.
Distribution of histone modifications within the imprinted Snurf–Snrpn 3-Mb region. (A) Scheme of the genomic region analyzed by ChIP followed by allele-specific qPCR. The coordinates on chromosome 7 are indicated (bottom), as well as the location of the different transcription units. Arrows indicate the direction of transcription for protein-coding genes. The star indicates the location of the IC and its associated differentially methylated region (DMR). The vertical gray bars on the axis indicate the positions of 35 loci analyzed by allele-specific qPCR. (B–F) Distribution of H3K4me2 (B), H3Ac (C), H4K20me3 (D), H3K9me3 (E), and H3K79me3 (F) on the paternally and maternally contributed Snurf–Snrpn genomic regions. ChIP experiments were performed using brain chromatin prepared from fetuses conceived by crossing JF1/Ms males × C57BL/6 females. Data are expressed as relative abundance and are normalized to input. Values are the mean of two independent ChIP experiments ± standard deviation. Gray areas identify the flanking genomic domains located outside of the Snurf–Snrpn imprinted interval. Black arrows indicate positions in the Snurf–Snrpn IC. The paternally and maternally contributed Snurf–Snrpn genomic regions are indicated by gray squares and black circles, respectively.
Little is known about the epigenetic mechanisms that control the establishment and maintenance of the active (paternally contributed) or the silenced (maternally inherited) chromatin structure within the above-described 3-Mb imprinted interval. With this in mind, we performed a series of experiments by using a large-scale approach to better understand the epigenetic mechanisms that control the chromatin structure within the Snurf–Snrpn region. We used chromatin immunoprecipitation associated with an allele-specific qPCR strategy to map histone modifications within the imprinted cluster in mouse fetal brains. We found that the actively transcribed, paternally inherited 3-Mb interval is associated with histone acetylation and dimethylation of histone H3 at lysine 4. In contrast, we showed that silencing of the maternally contributed region is associated with repressive marks such as H4K20me3, H3K9me3, and H3K79me3, but not H3K27me3. Surprisingly, we found that H3K36me3, a mark classically associated with active transcription, is massively enriched on the transcriptionally silenced, maternally contributed, 3-Mb Snurf–Snrpn region. We further analyzed H3K36me3 distribution and showed that this histone mark is also associated with pericentromeric heterochromatin, indicating that H3K36me3 is associated with both facultative and constitutive heterochromatin. While H3K36me3 distribution within actively transcribed regions relies on the elongating RNAPII, we showed that H3K36me3 deposition within the silenced Snurf–Snrpn domain does not correlate with transcription events. Our findings strongly suggest that H3K36me3, besides marking an actively transcribed region, has an additional role in the composition of heterochromatin domains.
Results
To achieve a high-throughput allele-specific chromatin analysis within the complete imprinted Snurf–Snrpn domain, we designed an assay based on chromatin immunoprecipitation (ChIP) experiments associated with a quantitative PCR strategy that allows the distinction of the paternal and maternal chromosomes at defined positions. To discriminate the paternal and maternal genomes, we selected two related mouse strains, C57BL/6 and JF1/Ms, whose genomes can be distinguished by sequence polymorphisms. ChIP and Input DNA were amplified by qPCR using a set of 35 allele-specific primer pairs spanning the 3-Mb Snurf–Snrpn region. These primer pairs can discriminate the two parental alleles of all Snurf–Snrpn coding genes, four positions within the IC, and 22 randomly chosen, non-coding regions, located at ∼50-kb intervals (Fig. 1A; Supplemental Fig. S1).
The maternally and paternally inherited chromosomes are marked by distinct histone modification signatures within the 3-Mb Snurf–Snrpn region
We performed ChIP experiments using chromatin prepared from brains of embryonic day 17 (E17) F1 hybrid fetuses, obtained from C57BL/6 female × JF1/Ms male mouse crosses. We selected this developmental stage, which corresponds to the end of the neurogenic phase, because the expression level of Ndn is at its maximum in the brain at this stage. Despite this high level of expression, Ndn transcription occurs only from the paternally contributed chromosome, showing that the maternally inherited region is under the control of a potent transcription silencing mechanism (Gerard et al. 1999). We detected enrichment of dimethylated histone H3 at lysine 4 (H3K4me2) and acetylated histone H3 (H3Ac) on the active paternally contributed chromosome (Fig. 1B,C). We obtained a similar distribution for acetylated histone H4 (H4Ac) (Supplemental Fig. S2). The preferential enrichment of these histone marks on the paternally contributed region is valid at all positions tested, including the IC, which is required for the expression of paternally contributed genes within the Snurf–Snrpn interval. H3K4me2 and H3Ac are, however, more strongly enriched at the level of active transcription units such as Snrpn, Ndn, Mkrn3, and Magel2. The distribution of these histone marks was identical in the brain at postnatal day 1 (data not shown). H3K4me2 and H3Ac were found equally enriched on the maternally and paternally contributed chromosomes at the external limits of the Snurf–Snrpn imprinted region, downstream from Peg12 and at the Ube3a locus, respectively (Fig. 1B,C). This equal enrichment of active histone marks on both paternal and maternal alleles of Ube3a is likely due to a biallelic expression of this gene in most parts of the brain (Albrecht et al. 1997). In addition, in the regions of the brain in which imprinted expression of Ube3a occurs from the maternal chromosome, the paternally contributed chromosome expresses a large antisense Ube3a-ATS transcript, which overlaps with Ube3a (Landers et al. 2004) and thus might contribute to the presence of active histone marks on both parental alleles. In conclusion, the active, paternally inherited, Snurf–Snrpn 3-Mb region is associated with histone marks that were identified within transcribed regions of the genome in previous studies.
We next analyzed the distribution of histone marks usually associated with heterochromatin and gene silencing. The silenced, maternally contributed Snurf–Snrpn domain was enriched for trimethylated histone H4 at lysine 20 (H4K20me3) and trimethylated histone 3 at lysine 9 (H3K9me3) and at lysine 79 (H3K79me3) (Fig. 1D–F). Remarkably, these three histone marks were found, at all positions tested, five to 10 times more enriched on the maternally inherited Snurf–Snrpn domain than on the corresponding paternally inherited region. This enrichment for H3K9me3, H4K20me3, and H3K79me3 ends at the 5′ and 3′ limits of the 3-Mb Snurf–Snrpn imprinted region, at the level of Ube3a on one side, and at the region of the downstream Peg12 gene on the other (Fig. 1D–F). We performed similar ChIP experiments using brain chromatin from fetuses conceived by reciprocal parental crosses (i.e., C57BL/6 male × JF1/Ms female) and obtained the same distribution for all histone marks (H4K20me3 and H3K79me3 are shown in Supplemental Fig. S3). Hence, histone modifications segregate in opposite patterns that are dictated by the paternal or maternal origin of the imprinted genomic interval. H4K20me3 and H3K9me3 enrichment on the silent imprinted region is in accordance with the well-established role of these two modifications in heterochromatin and gene repression (Schotta et al. 2004). In contrast, we found that H3K27me3 was not enriched on the silenced, maternally contributed Snurf–Snrpn region, indicating that this histone modification is unlikely involved in gene silencing at this locus (Supplemental Fig. S4). We next analyzed the distribution of MacroH2A, a histone variant that is preferentially enriched on the inactive X chromosome and that could contribute to transcription silencing (Mietton et al. 2009). This analysis showed that MacroH2A was not enriched on the silenced imprinted interval (Supplemental Fig. S5). We also tested the distribution of H4K20me3, H3K9me3, and H3K79me3 histone marks in the postnatal (day 1) brain and found patterns very similar to those observed 3 d earlier, at fetal stage E17 (data not shown). In summary, the maternally inherited chromosome is enriched in H4K20me3, H3K9me3, and H3K79me3, which are hallmarks of constitutive heterochromatin.
Widespread trimethylation of H3K36 on the silenced, maternally contributed, Snurf–Snrpn domain
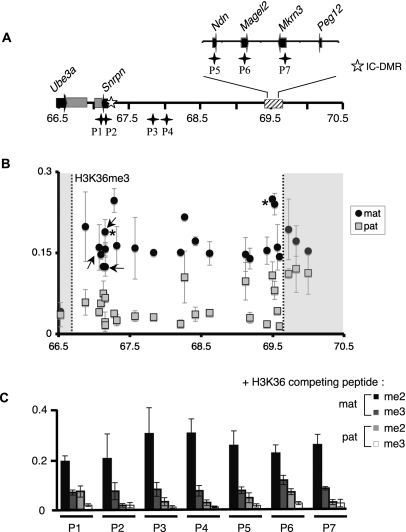
We next analyzed the distribution of trimethylated histone H3 at lysine 36 (H3K36me3) within the Snurf–Snrpn region. H3K36me3 has been mapped on the genome of several organisms, revealing that the deposition of this mark on chromatin is highly correlated with transcription elongation (Bannister et al. 2005; Vakoc et al. 2006; Barski et al. 2007; Guenther et al. 2007). We therefore expected H3K36me3 to be specifically enriched at the level of actively transcribed genes, which are only found on the paternally contributed chromosome within the Snurf–Snrpn interval. Strikingly, our ChIP analysis revealed a widespread H3K36me3 enrichment on the maternally inherited Snurf–Snrpn 3-Mb region (Fig. 2B). As for H4K20me3, H3K9me3, and H3K79me3 modifications, this maternal-specific enrichment for H3K36me3 was observed at all positions, including non-coding areas, known transcription units, as well as at the IC. This unexpected result suggests that H3K36me3 is associated with heterochromatin within the silenced Snurf–Snrpn region. As for the other histone modifications, H3K36me3 distribution on the Snurf–Snrpn domain in postnatal day 1 brain was highly similar to that observed at the late fetal stage E17 (data not shown). We verified the specificity of the antibody against trimethylated H3K36 by Western blotting (Supplemental Fig. S6). In addition, we performed ChIP experiments in the presence of competing peptides, which revealed that recognition of the H3K36me3 epitope is specifically blocked by an H3 peptide trimethylated at K36 but not by a peptide bearing the K36me2 modification (Fig. 2C). Hence, the antibody used in our study specifically recognizes the trimethylated form of H3K36. The distribution of H3K36me3 and the other histone marks on the maternally contributed Snurf–Snrpn interval is not restricted to brain tissue, as we found a similar allelic distribution in chromatin prepared from liver (Supplemental Fig. S7). The differential enrichment of histone marks on the paternal and maternal chromosomes is, however, less marked in liver chromatin than in the brain. In conclusion, the characterization of histone modifications within the 3-Mb Snurf–Snrpn genomic region revealed the existence of a facultative heterochromatin domain defined by the original association of the four histone modifications H4K20me3, H3K9me3, H3K79me3, and H3K36me3.
Figure 2.
H3K36me3 is broadly distributed within the silenced, maternally inherited Snurf–Snrpn region. (A) Representation of the 3-Mb Snurf–Snrpn region is shown as in Figure 1A. Black stars indicate the location of regions P1 to P7 that were analyzed in the competition assay. P1 and P2 primer sets are amplifying DNA fragments within the IC-DMR. (B) H3K36me3 distribution within the Snurf–Snrpn domain. ChIP experiments were performed as described in Figure 1. The statistical significance of the data was verified for two positions indicated by asterisks (P < 5.10−5; Student's t-test realized with eight independent experiments). (C) The specificity of H3K36me3 distribution within the Snurf–Snrpn region was confirmed by competition assay. Chromatin and antibodies against H3K36me3 were co-incubated with H3 peptides di- or trimethylated at lysine 36. qPCR assays were performed using seven discriminating primer pairs, which amplify loci located in or at the vicinity of coding genes (P1, P2, P5–P7), or in non-coding regions (P3 and P4) (see Supplemental Table S1). Values are mean ± SD of two independent ChIP experiments.
H3K36me3 is present on the paternally contributed chromosome at the level of transcribed regions
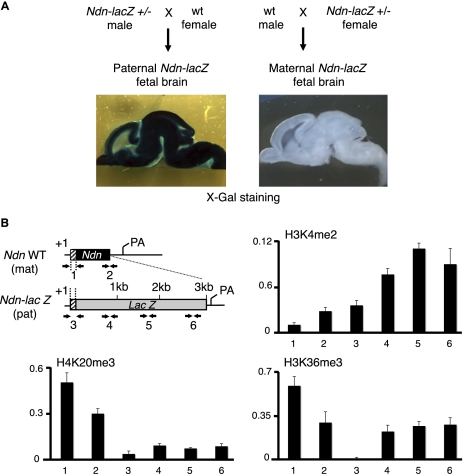
Several studies have associated H3K36me3 with actively transcribed regions in yeast and in metazoans (Krogan et al. 2003; Xiao et al. 2003; Bannister et al. 2005). More precisely, H3K36me3 was found to accumulate in the transcribed regions, as well as at the 3′ end of active genes in yeast and in mammals (Bannister et al. 2005; Barski et al. 2007; Mikkelsen et al. 2007; Edmunds et al. 2008). The finding that H3K36me3 is highly enriched on the silenced, maternally inherited Snurf–Snrpn cluster was thus at first glance in contradiction with previous published studies. We verified that our ChIP experiments could reveal a preferentially enrichment of this mark in the coding regions of several control genes expressed in the brain (Supplemental Fig. S8). Therefore, the preferential enrichment of H3K36me3 on the maternally contributed imprinted region did not necessarily mean that it was excluded from the paternally inherited region. Indeed, we observed a specific H3K36me3 enrichment at several positions on the paternally contributed chromosome, at the level of transcribed genes (Fig. 2B). To provide further evidence of H3K36me3 enrichment on specific regions of the paternal chromosome, we used a mouse line carrying an Ndn-lacZ fusion allele to monitor Ndn expression (Gerard et al. 1999). Maximal Ndn-lacZ expression from the paternal allele occurs in the brain during late gestation, and imprinting involves strict transcriptional silencing of the maternal allele (Fig. 3A). We prepared native chromatin from fetal brains carrying a paternal Ndn-lacZ fusion allele and a maternal wild-type Ndn allele. We performed ChIP experiments to detect histone modifications at the level of the Ndn promoter and in the body of the gene. Using appropriate pairs of oligonucleotides, we could distinguish the paternal and maternal Ndn alleles (Ndn-lacZ and wild-type Ndn, respectively) by qPCR (Fig. 3B). We confirmed the association of H3K4me2 with the actively transcribed paternally contributed allele, at the Ndn-lacZ promoter and in the body of the lacZ gene (Fig. 3B). In sharp contrast, H4K20me3 was exclusively detected on the silenced maternal allele. We found that H3K36me3 was present on both maternal Ndn and paternal Ndn-lacZ alleles, but with distinct deposition patterns. We detected high levels of H3K36me3 on the active paternally inherited Ndn-lacZ allele within the body and 3′ end of lacZ, but not at the promoter (Fig. 3B). This pattern is compatible with a deposition of H3K36me3 during transcription elongation. As expected from our large-scale approach, we detected H3K36me3 on the silenced, maternally contributed allele. In this case, however, H3K36me3 was present both at the promoter and gene body. In conclusion, H3K36me3 is deposited within actively transcribed regions on the paternally inherited Snurf–Snrpn region, whereas it is associated with heterochromatin on the silenced, maternally contributed 3-Mb region.
Figure 3.
H3K36me3 is distributed differentially on the maternally and paternally contributed Ndn alleles. (A) Brain sections prepared from E17 fetuses heterozygous for the Ndn-lacZ allele were stained with X-gal, showing robust transcription of the paternally contributed Ndn-lacZ allele and strict transcriptional silencing of the maternally contributed Ndn-lacZ allele. (B) Structure of the single exon Ndn gene, as well as the Ndn-lacZ allele in which lacZ is inserted as a fusion with the first 31 Ndn codons (the remaining of Ndn coding sequence is deleted). Mice were bred to obtain fetuses carrying the wild-type (wt) Ndn and Ndn-lacZ fusion alleles on the maternally and paternally contributed chromosomes, respectively. Arrows indicate the positions of the PCR primers used in the qPCR assay. Primer pairs 1 and 2 recognize the promoter and 3′ end of the silenced, maternally contributed Ndn allele, respectively. Primer pair 3 maps to the promoter of the actively transcribed, paternally contributed Ndn-lacZ allele, and primer pairs 4, 5, and 6 to the lacZ reporter gene body. (+1) The transcription initiation site; (pA) the mRNA polyadenylation signal. Enrichment of H3K36me3, H3K4me2, and H4K20me3 was determined by ChIP using chromatin prepared from E17 fetal brains carrying a paternally contributed (transcribed) Ndn-lacZ, and a maternally contributed (silenced) wild-type Ndn allele. For each ChIP assay, the antibody-bound fraction was amplified by qPCR with the indicated primer pairs. Histone modification relative abundance is normalized to input. Values are mean ± SD (n = 3 independent ChIP experiments).
Chromatin within the silenced Snurf–Snrpn region shares common properties with constitutive heterochromatin
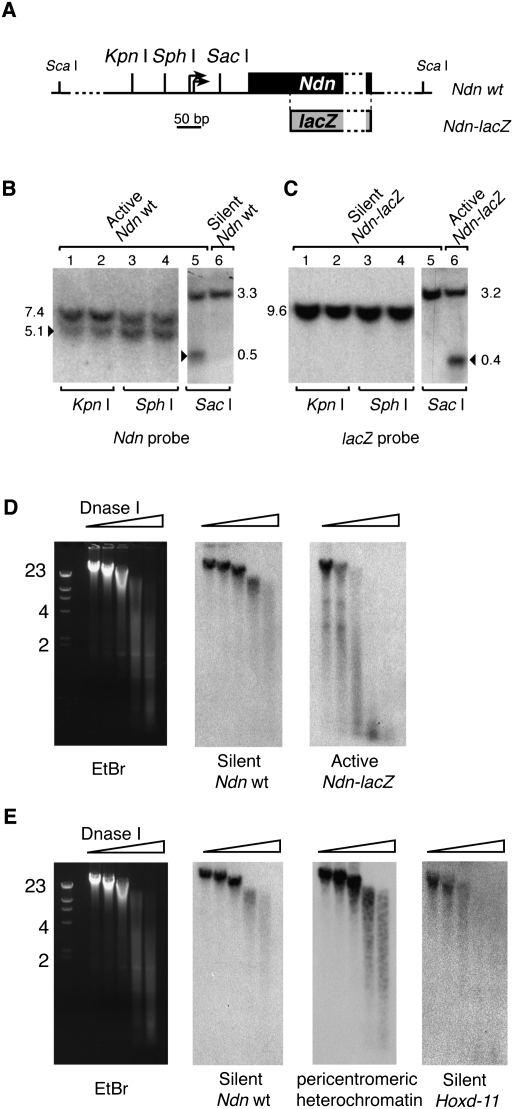
We investigated whether the combination of H4K20me3, H3K9me3, H3K79me3, and H3K36me3 could be associated with the establishment of a parental-specific chromatin structure. We first tested if chromatin at the maternally and paternally contributed Ndn promoters exhibited distinct accessibility to restriction enzymes. Nuclei derived from fetal brains heterozygous for the Ndn-lacZ allele were digested with restriction enzymes that cut at the level of the Ndn promoter (Fig. 4A). Our experiments showed that the silent maternal Ndn promoter was resistant to digestion by all restriction enzymes (Fig. 4B,C). In contrast, the paternal Ndn promoter was accessible to all tested restriction enzymes. In a second series of experiments, we tested the sensitivity of the paternal Ndn-lacZ and maternal wild-type Ndn alleles to DNase I, a nuclease suited to probe chromatin structure (Lu and Richardson 2004). Strikingly, maternal Ndn chromatin was highly resistant to DNase I digestion when compared to bulk genomic DNA (Fig. 4D), suggesting that the silenced Ndn allele is present in a chromatin conformation that occludes DNA access to this enzyme. Reciprocally, we found the active Ndn-lacZ allele more sensitive to DNase I digestion compared to bulk genomic DNA (Fig. 4D). The potent resistance to DNase I digestion at maternal Ndn was equivalent to that of pericentromeric heterochromatin (Fig. 4E), suggesting that silenced Ndn chromatin shares common properties with constitutive heterochromatin. To test the possibility that transcriptional silencing in fetal brain could be generally associated with a chromatin highly resistant to nucleases, we compared the DNase I digestion patterns of maternal Ndn and Hoxd11 genes, which are both transcriptionally silent in the fetal brain (Gerard et al. 1996). Hoxd11 was much more sensitive to DNase I digestion than silent Ndn (Fig. 4E). Thus, transcriptional silencing per se does not confer a chromatin structure resistant to nucleases. In summary, the presence of H4K20me3, H3K9me3, H3K79me3, and H3K36me3 at silent Ndn correlates with a chromatin structure that severely impairs DNA access to nucleases, whereas absence of these marks at the same locus correlates with an open chromatin structure, accessible to these enzymes.
Figure 4.
Transcriptional silencing at Ndn is associated with a chromatin structure that occludes DNA access to nucleases. (A) Ndn promoter organization with the position of restriction sites. The two transcription initiation sites are indicated with arrows. The position of lacZ in the fusion allele is shown. (B) Nuclei were prepared from E17 fetal brains and digested with KpnI, SphI, or SacI. DNA was prepared, cut with a second restriction enzyme (e.g., ScaI), and run on an agarose gel. DNA was then transferred to a nylon membrane, which was hybridized with a Ndn coding sequence [α-32P]-labeled probe. Ndn coding sequence is deleted in the Ndn-lacZ allele, and therefore this probe reveals only the wild-type Ndn allele. In all lanes, a single band indicates absence of digestion, whereas the presence of a second smaller DNA fragment (arrowhead) indicates digestion by the restriction enzyme at the Ndn promoter. In lanes 1 to 5, nuclei were prepared from fetal brains carrying a paternally contributed (active) wild-type allele and a maternally contributed (silenced) Ndn-lacZ fusion allele. In lane 6, nuclei were prepared from fetal brains carrying a paternally contributed Ndn-lacZ allele and a maternally contributed wild-type Ndn allele. (C) The membrane was then stripped and rehybridized with the lacZ probe. (D) Nuclei were prepared from E17 fetal brains carrying a maternally contributed (silenced) wild-type Ndn allele and a paternally contributed (active) Ndn-lacZ fusion allele, and digested with increasing amounts of DNase I. DNA was prepared, run on an agarose gel, and transferred to a nylon membrane that was hybridized with a first [α-32P]-labeled probe. After autoradiography, the membrane was stripped and rehybridized with a second probe. Ethidium bromide (EtBr) staining of DNA before transfer is shown on the left. The membrane was hybridized successively with Ndn exon probe, revealing the silenced wild-type Ndn allele, and lacZ probe, which reveals the active paternally contributed Ndn-lacZ allele. (E) The membrane was first hybridized with a probe against the silenced wild-type Ndn allele, then with a major satellite repeat probe, which reveals pericentromeric heterochromatin DNA, or a Hoxd11 probe.
H3K36me3 is detected within constitutive heterochromatin
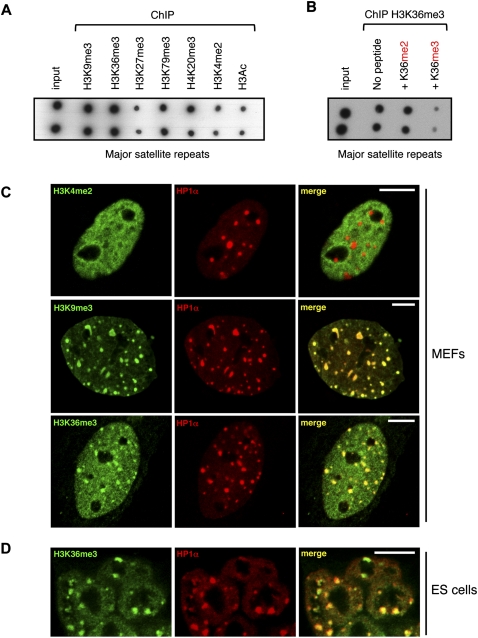
We next explored whether H3K36me3 could also be detected in constitutive heterochromatin, at the level of pericentromeric regions. Eluates of ChIP experiments were spotted onto a nylon membrane, which was then hybridized with a probe for major satellite repeats. We showed that pericentromeric regions were, indeed, associated with H3K9me3, H4K20me3, and H3K79me3, which are known marks of constitutive heterochromatin, but not with H3K27me3 or H3K4me2 (Fig. 5A). Strikingly, we found that pericentromeric regions were also enriched for the H3K36me3 modification. The specificity of the antibody against H3K36me3 was again verified by peptide competition assays (Fig. 5B). We also analyzed the distribution of H3K36me3 in the nucleus of mouse embryonic fibroblasts (MEFs) by immunofluorescence. Trimethylated H3K36 was found distributed throughout the nuclei, which is characteristic of an association with euchromatic regions (Fig. 5C). In addition, we detected H3K36me3 at local foci that colocalize with HP1α, a marker of pericentromeric regions (James et al. 1989). These foci are known as chromocenters, which results from the organization of pericentromeric regions in clusters (Guenatri et al. 2004). We compared the distribution of H3K36me3 in MEFs nuclei with H3K9me3 and H3K4me2 patterns (Fig. 5C). While H3K9me3 is strongly enriched at chromocenters and only weakly in the nucleoplasm, H3K4me2 is excluded from heterochromatic regions and present only in the nucleoplasm. H3K36me3, which is present both at chromocenters and in the nucleoplasm, combines these two patterns. Importantly, we also detected H3K36me3 associated with constitutive heterochromatin in mouse embryonic stem (ES) cells (Fig. 5D). The specificity of H3K36me3 association with pericentromeric heterochromatin was confirmed by peptide competition assays (Supplemental Fig. S9). Observation of mitotic figures revealed that H3K36me3 remains enriched at the level of pericentromeric regions of condensed chromosomes, and therefore that this heterochromatin marking is inherited across cell divisions (Supplemental Fig. S10). Altogether, our data show that H3K36me3 is associated with constitutive heterochromatin, in addition to its well-established association within actively transcribed genomic regions.
Figure 5.
H3K36me3 is detected within pericentromeric heterochromatin regions. (A) Dot-blot analysis of DNA prepared from fetal brain chromatin immunoprecipitated with antibodies against the indicated histone modifications. (B) The specificity of H3K36me3 deposition within pericentromeric heterochromatin was tested by competition assay. Chromatin and antibodies against H3K36me3 were co-incubated with H3 peptides di- or trimethylated at lysine 36. Eluates were analyzed by dot blot. (C) Immunofluorescence confocal microscopy analysis of H3K36me3 distribution in mouse embryonic fibloblasts (MEFs), in comparison with other histone modifications. H3K4me2, H3K9me3, or H3K36me3 distribution is shown in green, whereas the pericentromeric heterochromatin marker HP1α is revealed as a red signal. Green and red signals were merged in the right panel (yellow indicates a colocalization). (D) Immunofluorescence analysis of H3K36me3 distribution in undifferentiated mouse ES cells. Scale bar, 10 μm.
H3K36me3 deposition onto the silenced, maternally contributed Snurf–Snrpn cluster does not correlate with transcription events
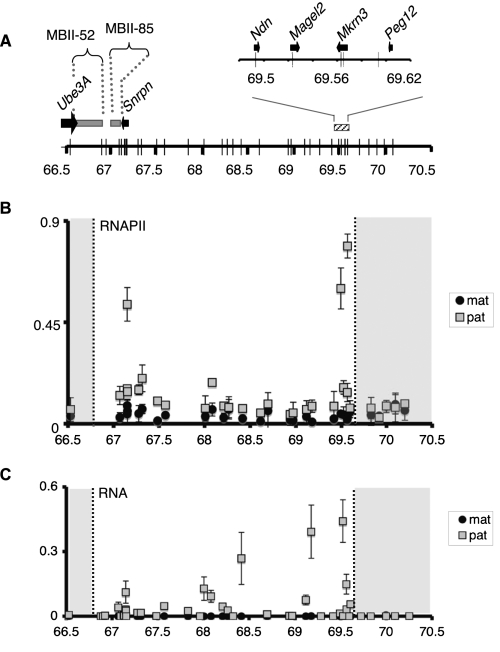
In yeast, Set2p is the sole enzyme that catalyzes the trimethylation of histone H3 at lysine 36 (Strahl et al. 2002). Set2p is recruited to transcriptionally active regions by interacting with the phosphorylated elongating form of RNA polymerase II (RNAPII) (Krogan et al. 2003; Li et al. 2003; Xiao et al. 2003). Setd2, one of several proteins homologous to Set2p, performs H3K36me3 deposition in mammals (Edmunds et al. 2008). As its yeast homolog, Setd2 interacts with the elongating RNAPII, which suggests that the association of H3K36me3 with transcription elongation is evolutionarily conserved (Sun et al. 2005). If trimethylation of H3K36 was catalyzed by Setd2 associated with RNAPII within the silenced Snurf–Snrpn region, we would expect to detect RNAPII on the maternally contributed imprinted domain. Indeed, whereas the 3-Mb maternally contributed Snurf–Snrpn region was characterized as transcriptionally silent, we cannot exclude that transcription events could arise within this silenced region. We explored this hypothesis by performing ChIP experiments using chromatin prepared from E17 fetal brains, with antibody against the C-terminal domain of RNAPII. We then analyzed RNAPII enrichment within the imprinted region using our set of primer pairs that discriminate the maternal and paternal regions at each locus. We focused our analysis on regions located outside of known, already characterized, transcription units. We observed a strong enrichment for RNAPII at three positions located exclusively on the paternally contributed Snurf–Snrpn cluster (Fig. 6B). In contrast, RNAPII could not be detected at any position on the maternally contributed Snurf–Snrpn domain. Hence, RNAPII is unlikely responsible for the accumulation of H3K36me3 on the silenced, maternally inherited 3-Mb interval. We further investigated RNA synthesis within the imprinted cluster using allele-specific amplification of RNAs. This experiment revealed that RNAs were transcribed from most loci within the Snurf–Snrpn interval, however, exclusively from the paternally inherited chromosome, which is in agreement with RNAPII distribution (Fig. 6C). Importantly, the experiment was also carried out using brain chromatin prepared from fetuses conceived by reciprocal mouse crosses, which confirmed that RNAs were transcribed exclusively from the paternally contributed Snurf–Snrpn 3-Mb domain (Supplemental Fig. S11). The facts that transcriptional events (and RNAPII) are observed only on the paternally contributed region and that H3K36me3 is mainly enriched on the maternally inherited domain suggest that the deposition of the H3K36me3 within the silenced 3-Mb Snurf–Snrpn interval occurs independently of RNAPII. We cannot, however, rule out the possibility that the transcription of short-life RNA species could contribute to the deposition of this mark within the repressed Snurf–Snrpn domain.
Figure 6.
H3K36me3 deposition onto the silenced, maternally contributed Snurf–Snrpn region is independent of transcription. (A) Representation of the 3-Mb Snurf–Snrpn region as in Figure 1A. (B) ChIP analysis of RNAPII binding to the Snurf–Snrpn region, using brain chromatin from fetuses conceived by mating JF1/Ms males with C57BL/6 females. Strong enrichment can be observed at positions 67,149,376; 69,493,157; and 69,565,295 that corresponds to the first intron of Snrpn and to Ndn and Mrkrn3 promoters, respectively. Values are mean ± SD of two independent ChIP experiments. (C) RT-qPCR analysis of RNAs extracted from the brains of E17 fetuses conceived by crossing JF1/Ms males with C57BL/6 females. Relative levels of paternally (gray squares) and maternally (black circles) expressed RNAs were quantified using the set of 35 allele-specific primer pairs and normalized to Hprt mRNA. This assay identifies Snrpn, Magel2, and Mkrn3 mRNAs, but not Ndn mRNA due to the absence of suitable polymorphisms inside the transcript. Values are mean ± SD (n = 3 independent experiments).
Several proteins that belong to the Setd2 family were recently involved in the deposition of H3K36me3 in mouse. In one study, Setd2 was identified as the major H3K36me3 HMT in MEFs (Edmunds et al. 2008). Another study identified Whsc1 as a second, potent H3K36me3 HMT (Nimura et al. 2009). We thus used a loss-of-function approach to test whether one of the proteins belonging to this family might be involved in H3K36me3 deposition within heterochromatin. shRNA vectors were designed against Setd2 and Whsc1, as well as against Whsc1l1 and Nsd1, that also show significant homology with Setd2. While we could efficiently down-regulate the expression of each targeted gene with several independent shRNA constructs (see Supplemental Methods), we did not detect a diminution of H3K36me3 deposition at the level of pericentromeric regions in ES and MEF cells (Supplemental Figs. S12, S13). Knockdown of Setd2 resulted, however, in a reduction of H3K36me3 fluorescence signal in the nucleoplasm of both cell types (Supplemental Figs. S12, S13), underlining a dominant role of this enzyme in this nuclear compartment. We next tested whether the down-regulation of Setd2 and Whsc1 might affect H3K36me3 accumulation within the Snurf–Snrpn region. As the mechanisms of genomic imprinting are severely perturbed in mouse ES cells (Dean et al. 1998; Humpherys et al. 2001), we performed these experiments using chromatin prepared from MEF cells at early passages. Knockdown of Setd2 but not that of Whsc1 resulted in a significant loss of H3K36me3 in the coding regions of several control genes (Supplemental Fig. S14). However, neither the knockdown of Setd2 nor that of Whsc1 led to an alteration of H3K36me3 levels within the Snurf–Snrpn region (Supplemental Fig. S14). These data suggest that H3K36me3 deposition at the level of the Snurf–Snrpn region, as well as in heterochromatin regions, might be directed by a new H3K36me3 HMT that remains to be identified. Alternatively, we cannot exclude a functional redundancy between Setd2 family members for H3K36me3 deposition onto heterochromatin.
Discussion
We have characterized histone modifications within the Snurf–Snrpn region by performing ChIP experiments coupled with a parent-of-origin allelic qPCR assay. This large-scale approach provides, for the first time, a detailed characterization of this 3-Mb imprinted region. We analyzed histone modifications both in the vicinity of characterized coding genes and in non-coding regions, including the 2-Mb long gene desert that extends between Ndn and Snrpn.
We found pronounced allelic differences in histone modification distribution within the Snurf–Snrpn cluster. H3Ac and H3K4me2 were exclusively detected on the active, paternally contributed Snurf–Snrpn genomic interval. These marks were preferentially enriched in regions containing transcription units, which is reminiscent of the distribution of these histone modifications across the mouse and human genomes (Barski et al. 2007; Mikkelsen et al. 2007). The presence of these “active” histone modifications suggests that the paternally inherited Snurf–Snrpn region has a classical euchromatic structure. Our biochemical analysis, which showed that this region has an open chromatin structure, supports this idea.
We then focused our analysis on the mechanisms supporting the potent transcription repression that silences the maternally contributed Snurf–Snrpn genomic interval. We showed that the maternally contributed 3-Mb imprinted region is enriched in H4K20me3, H3K9me3, H3K79me3, and H3K36me3 histone marks. Several of these marks—H3K9me3, H4K20me3, and H3K79me3—have been previously associated with heterochromatin formation and gene repression, which suggests that they could participate in gene silencing within the Snurf–Snrpn interval (Schotta et al. 2004; Jones et al. 2008). We demonstrated that H3K27me3, which contributes to gene silencing at the imprinted Kcnq1 cluster, as well as to X-chromosome inactivation (Plath et al. 2003; Lewis et al. 2004; Rougeulle et al. 2004; Umlauf et al. 2004a), is not present on the silenced Snurf–Snrpn genomic interval, and thus is unlikely to be involved in transcription silencing in this imprinted region. We identified several H3K27me3-enriched loci on the paternally contributed Snurf–Snrpn interval, which might seem in contradiction with the active transcription status of this chromosomal domain (Supplemental Fig. S4). However, all active genes were negative for H3K27me3. Similarly, the IC did not show any H3K27me3 enrichment, except at the most distal position tested in this study, located 4.7 kb upstream of Snrpn exon 1. We also analyzed the incorporation of MacroH2A, an H2A histone variant that is enriched on the inactive X chromosome (Mietton et al. 2009), and showed that it was not enriched on the silenced, maternally inherited imprinted region. Altogether, these data demonstrate that the potent transcriptional silencing mechanism that restrict gene expression on the maternal Snurf–Snrpn cluster is divergent from the mechanism that controls X inactivation.
The involvement of H3K9me3 and H4K20me3 in the transcriptional silencing of imprinted genes was previously shown at the 0.5-Mb Igf2r cluster, as well as at the IC of other imprinted loci (Wu et al. 2006; Delaval et al. 2007; Regha et al. 2007; Pannetier et al. 2008). Interestingly, Barlow and colleagues showed that H3K9me3 and H4K20me3 were distributed in discrete foci interspaced with focal regions of active chromatin (Regha et al. 2007). Our analysis did not reveal the presence of active, interspaced, chromatin within the silenced 3-Mb Snurf–Snrpn region. We showed that all tested positions within the silenced 3-Mb region are enriched in H3K9me3 and H4K20me3, suggesting that this imprinted interval does not contain active chromatin subdomains on the maternal chromosome.
We used an in vivo mouse model system based on the insertion of a single-copy reporter gene at Ndn to investigate the chromatin structure at the silenced Snurf–Snrpn imprinted region. We show that the presence of H4K20me3, H3K9me3, H3K79me3, and H3K36me3 on the silenced Ndn gene correlates with a chromatin structure that occludes DNA access to several families of nucleases, whereas absence of the aforementioned marks at the paternal locus correlates with an accessible, open chromatin structure. A remarkable characteristic of this mechanism is its stability, since cells that reactivate the silenced Ndn allele in the fetal brain are detected with an extremely low frequency (Fig. 3A; data not shown). We found that the deposition of heterochromatin-associated histone marks within the Snurf–Snrpn imprinted region is higher in brain than in liver cells. A comparison of the amount of mRNA expressed from the genes of this genomic interval in liver and brain revealed that they are expressed at much higher levels in the brain (Supplemental Fig. S7). In the light of the fact that PWS is a developmental, neurological syndrome, we propose the following hypothesis: Imprinting in this genomic interval is likely required for the proper dosage of one or several genes in the brain, during late gestation. We propose that in the brain, a high level of heterochromatin-associated histone marks on the maternal chromosome contributes to the establishment of an efficient heterochromatin barrier that prevents the recruitment of the transcription factors that are active on the paternally contributed chromosome. As the genes of the Snurf–Snrpn region are expressed at low levels in the liver, the cells probably do not require the same efficient barrier to prevent their expression from the maternal chromosome. This lower selection pressure in the liver could explain why the mechanisms that deposit heterochromatin-associated marks are less efficient in these cells than in the brain.
Analysis of histone modifications at the Snurf–Snrpn cluster has revealed that H3K36me3 was mainly enriched on the silent maternal allele. It was an unexpected result because this histone modification is considered an “active” mark. Indeed, H3K36me3 was generally associated with transcriptionally active regions in yeast and mammals (Bannister et al. 2005; Vakoc et al. 2006; Barski et al. 2007; Guenther et al. 2007; Mikkelsen et al. 2007). More precisely, H3K36me3 was found to accumulate in the 3′ region of transcribed genes and to correlate with elongation of transcription. The link between H3K36me3 and transcription elongation is, however, not universal, since this modification was not detected in a subset of actively transcribed genes in Drosophila (Filion et al. 2010). In the present study, we show that H3K36me3 is associated with facultative heterochromatin within the imprinted Snurf–Snrpn cluster. We also provide evidence that H3K36me3 is enriched within pericentromeric regions, which shows that this mark is associated with constitutive heterochromatin.
How do we conciliate these two apparently contradictory H3K36me3 distributions, in transcribed regions and heterochromatin domains? In yeast, the Set2p HMTase associates with the phosphorylated C-terminal domain (CTD) of elongating RNAPII and catalyzes the trimethylation of H3K36 at the level of transcribed genes (Krogan et al. 2003; Li et al. 2003; Xiao et al. 2003). However, when Set2p is mistargeted to the promoter through artificial recruitment, it represses transcription (Strahl et al. 2002; Landry et al. 2003). Two studies demonstrated that Eaf3p, an Rpd3S subunit, binds H3K36me3-modified nucleosomes (Carrozza et al. 2005; Joshi and Struhl 2005). Once recruited, Rpd3S, which is a co-repressor protein complex, deacetylates histones within transcribed sequences. It was proposed that Rpd3S deacetylation could erase histone acetylation generated before or during the passage of elongating RNAPII. Interestingly, set2 and rpd3S mutants display aberrant intragenic transcription initiation events, which strongly suggests that a role of Rpd3S, and by extension H3K36me3, could be to prevent transcription initiation from intragenic cryptic promoters (Carrozza et al. 2005; Joshi and Struhl 2005). Thus, H3K36me3 could be involved immediately after a transcription event in the establishment of a chromatin state repressive to (intragenic) transcription. Similarly, in the fission yeast Schizosaccharomyces pombe, Set2 also associates with the coding regions of RNA polymerase II–transcribed genes, and specifically methylates H3-K36 (Morris et al. 2005). As in Saccharomyces cerevisiae, Set2 would recruit complex II, which resembles Rpd3S, and repress antisense transcription in coding regions (Nicolas et al. 2007).
We propose that in the Snurf–Snrpn interval, H3K36me3 could participate in gene silencing by preventing transcription initiation events in heterochromatin. We wondered whether the deposition of H3K36me3 within the silenced Snurf–Snrpn region was linked to transcription elongation or to a different mechanism. Setd2, which is the best characterized H3K36 HMTase in mammals (Edmunds et al. 2008), is recruited to its genomic target regions by elongating RNAPII. We analyzed RNAPII distribution within the Snurf–Snrpn interval and showed that we could detect the enzyme exclusively on the active, paternally inherited chromosome. Moreover, we found that transcripts have also an exclusive paternal origin. Our data are in agreement with a recent large-scale study that analyzed the allelic expression of several imprinted clusters in the mouse brain and reported the presence of numerous transcripts, exclusively expressed from the paternal allele, within the interval between Snrpn and Ndn (Gregg et al. 2010). As H3K36me3 was enriched on the silenced, maternally contributed region, our data suggest that its deposition does not depend on RNAPII and ongoing transcription.
It is now clear that cross talk exists among histone modifications and that both chromatin structure and gene expression are controlled by a complex interplay between them. It is tempting to speculate that in heterochromatic regions, H3K36me3 acts in concert with the previously characterized H3K9me3 and H4K20me3 repressive histone marks to control and maintain gene repression. Supporting this hypothesis, Grewal and colleagues showed that, in fission yeast, mutations in Set2, which deposits H3K36me3, and Clr4, the HMTase responsible for the methylation of H3K9, resulted in cumulative defects in heterochromatin silencing, suggesting that the two enzymes act in parallel pathways to promote heterochromatin formation (Chen et al. 2008). Our results suggest that H3K36me3, in addition to H3K9me3, might also contribute to heterochromatin formation in mammalian cells. The recent finding that H3K36me3 stimulates the recruitment of DNA-methyltransferase DNMT3A onto nucleosomes is compatible with our hypothesis (Dhayalan et al. 2010). Interestingly, this last study showed that the interaction between DNMT3A and H3K36me3 stimulates the methylase activity of DNMT3A in vitro, suggesting a cross talk between H3K36me3 and DNA methylation. H3K36me3 could thus contribute to the establishment of a transcriptionally unfavorable chromatin conformation by recruiting (and perhaps stimulating) repressive complexes.
In conclusion, this study demonstrated that trimethylated histone H3 at lysine 36 is specifically enriched in facultative and constitutive heterochromatin in mouse, and that this histone modification correlates with gene silencing within these heterochromatin domains. We propose that H3K36me3 would contribute to the composition of heterochromatin, in association with other DNA and histone modifications.
Methods
Mouse strains
The mouse strains used were C57BL/6 (Mus musculus domesticus), JF1/Ms (Mus musculus molossinus), and (C57BL/6 X JF1/Ms) F1 hybrids. The Ndntm2Stw mutant mouse line that carries the Ndn-lacZ fusion allele was described previously (Gerard et al. 1999). We crossed these mice with (C57BL/6N × CBA) F1 mice to obtain E17 fetuses heterozygous for the Ndn-lacZ allele.
Chromatin immunoprecipitation
We carried out immunoprecipitation on E17 and P1 brain or P1 liver chromatin. We prepared native chromatin fragments of two to six nucleosomes in length, as described previously (Umlauf et al. 2004b). Chromatin immunoprecipitation was carried out as follows: 30 μg of chromatin was incubated overnight at 4°C with 5–10 μg of commercial antibodies or 2 μL of H3K27me2/3 antibodies in ChIP buffer (50 mM Tris at pH 7.5, 5 mM EDTA, 50 mM NaCl, 0.1 mM PMSF, 1× protease inhibitor cocktail). Antibody references and batches are indicated in Supplemental Table S3.
Immunoprecipitated chromatin was recovered by rotating with Protein G Sepharose for 3 h, and beads were washed sequentially at room temperature with ChIP buffer, washing buffer A (75 mM NaCl, 20 mM Tris-HCl at pH 8.0, 10 mM EDTA), washing buffer B (same as buffer A with 125 mM NaCl), and washing buffer C (buffer A with 175 mM NaCl). Bound chromatin was eluted at room temperature in Elution buffer (50 mM Tris-HCl at pH 7.5, 50 mM NaCl, 5 mM EDTA, 1% SDS). DNA was purified by phenol–chloroform extraction and precipitated by isopropanol. Pellets were washed with 70% ethanol and resuspended in 10 mM Tris (pH 8.0). Each assay was performed two or three times on independent chromatin preparations to control sample variation. For competition assays, 30 μg of chromatin was incubated overnight with 10 μg of H3K36me3 antibodies and 1 μg/mL H3K36me2 or H3K36me3 competing peptides (Abcam).
Real-time PCR analysis after chromatin immunoprecipitation
To measure allele-specific chromatin differences in the Snurf–Snrpn 3-Mb region, we developed a quantitative real-time PCR assay based on 35 allele-specific primer sets that allow the discrimination of maternal and paternal alleles within the region spanning positions 66,527,171 to 70,251,665 on chromosome 7 (NCBI build 37). Details on primer design are available in the Supplemental Methods. The 35 primer sets can discriminate the two parental alleles of all Snurf–Snrpn coding genes, four positions within the imprinting center, and a series of randomly chosen non-coding regions (Fig. 1A). Except for RNAPII ChIP experiments, we performed a pre-amplification step with the Thermoprime Taq polymerase (Abgene), using primers flanking the allelic differences (these primers amplify the JF1/Ms and C57BL/6 genomes with the same efficiency). This step improves mismatch detection (Shively et al. 2003). Pre-amplification was performed as follows: 1 cycle for 4 min at 94°, 15 cycles for 20 sec at 94°C, for 30 sec at 56°C, and for 1 min at 72°C, and 1 cycle for 10 min at 72°C. The resulting 500–700-bp amplified products (IP or IN) were diluted at 1/25e and used as templates for allele-specific qPCR assays. Allelic qPCR reactions were performed in two separate wells using a common primer and either the C57BL/6-specific primer or the JF1/Ms-specific primer. All reactions were carried out in a total volume of 10 μL, in 384-well plates. Liquid handling of the 384-well plates was performed with a Baseplate robotic workstation (The Automation Partnership). The composition of the quantitative PCR assay included 2.5 μL of DNA (pre-amplified immunoprecipitated DNA or corresponding pre-amplified input), 0.5 mM forward and reverse primers, and 1× SYBR Green PCR Master Mix (Applied Biosystems). The amplifications were performed as follows: 2 min at 95°C, 40 cycles for 15 sec at 95°C and for 60 sec at 60°C in the ABI/Prism 7900HT real-time PCR machine (Applied Biosystems). The real-time fluorescence data were analyzed with the Sequence Detection System 2.3 (Applied Biosystems). Each pre-amplification and qPCR reaction was performed in triplicate. Values were considered valid and included in the charts only when the two allele-specific amplifications passed the following quality controls: (1) Amplification reactions were checked for the presence of a single specific peak in the melting curve. (2) We considered only CTs from qPCR triplicates with an SD <0.5. To standardize between experiments, we calculated the relative abundance by dividing 2−CT IP by 2−CT IN, both values first being normalized for dilution factors. qPCR analysis of control regions after H3K36me3 chromatin immunoprecipitation is described in the Supplemental Methods.
Quantitative RT-PCR
Quantitative RT-PCR protocols (including the allele-specific assay) and primers used are described in the Supplemental Methods.
shRNA vectors
shRNA vectors that target Setd2, Nsd1, Whsc1, and Whsc1l1 mRNAs were constructed using the pHYPER shRNA vector, as described (Berlivet et al. 2010). Sequences encoding the shRNAs, which were designed using the DSIR software (http://biodev.extra.cea.fr/DSIR/DSIR.html), are listed in Supplemental Table S6. For all targeted genes, a strong decrease of mRNA levels was observed for two to four shRNA vectors (see the Supplemental Methods).
Transfection of MEF and ES cells
The shRNA plasmid vectors were introduced into MEF and ES cells by electroporation, using a Lonza (Amaxa) electroporation apparatus, as well as ES cells and MEF-specific electroporation kits (Lonza VPH-1001 and VPD-1004 nucleofector kits, respectively). For ES cells: 4 × 106 cells were electroporated with 20 μg of shRNA plasmid vector, following the manufacturer's instructions. Cells were seeded in plates with feeder cells and incubated for 24 h in D15 medium (DMEM with 15% serum, supplemented with LIF and antibiotics). For immunofluorescence analysis, ES cells were seeded on coverslips with feeder cells, in 24-well plates. Cells were then selected with 2 μg/mL puromycin for 72 h. For MEF cells: 2 × 106 cells were electroporated with 10 μg of shRNA plasmid vector according to the manufacturer's instructions. Cells were seeded and incubated for 24 h in D10 medium (DMEM with 10% serum, supplemented with antibiotics). Cells were then selected with 1.2 μg/mL puromycin for 72 h before being collected for the ChIP experiments.
Fetal brain nuclei preparation and nuclease digestion
Reagents, protocols, and hybridization probes for Southern blot are described in the Supplemental Methods.
Histology
E17 brains were dissected, fixed in PBS with 4% paraformaldehyde (PFA) for 1 h, and included in PBS with albumin (30%, w/v), gelatin (0.5%, w/v) and glutaraldehyde (1%, v/v). The brains were cut into 300-μm sections using a Leica vibratom, fixed for 30 min in 4% PFA, and stained overnight with X-gal.
Immunofluorescence
MEFs were derived from E13 embryos and analyzed at passages 3 or 4. Immunostaining was performed essentially as previously described (Galvani et al. 2008). Briefly, cells grown on coverslips were washed with PBS, then fixed with formaldehyde 1.6% (Methanol-free formaldehyde; Polysciences Inc., #18814) and permeabilized with 0.2% Triton X-100 in PBS. After blocking with 50 mg/mL bovine serum albumin with 0.1% Tween 20 in PBS, cells were incubated with primary antibodies overnight at 4°C (H3K36me3 antibody, abcam ab9050, 1 μg/mL; H3K9me3 antibody, abcam ab8898, 1 μg/mL; H3K4me2 antibody, abcam ab32356, dilution 1/1000, HP1α antibody, Euromedex 2HP-1H5-As, dilution 1/1000). Washes between primary and secondary antibodies were in 0.1% Tween 20 and 0.05% TX-100 in PBS, and washes after incubation with the secondary antibody were in 0.1% Tween 20. Coverslips were mounted on glass slides with ProLong Gold (Invitrogen) antifade reagent and sealed with nail varnish. Image acquisition was carried out on an LSM510 confocal microscope (Zeiss), in confocal mode (pinhole size set at 1 Airy unit).
Acknowledgments
We thank F. Ochsenbein and C. Francastel for discussions, and S. Jounier and the Gene Identification team at CNG for technical help. We thank D. Reinberg for providing the H3K27me2/3 antibody, R. Feil for the JF1/Ms mouse strain, and N. Gilbert for probes. M.G.'s team is supported by the Association pour la Recherche sur le Cancer (ARC). S.D. acknowledges la Ligue Nationale Contre le Cancer for support (Equipe Labelisée).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.118091.110.
References
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL 1997. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet 17: 75–78 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T 2005. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Berlivet S, Houlard M, Gerard M 2010. Loss-of-function studies in mouse embryonic stem cells using the pHYPER shRNA plasmid vector. Methods Mol Biol 650: 85–100 [DOI] [PubMed] [Google Scholar]
- Bielinska B, Blaydes SM, Buiting K, Yang T, Krajewska-Walasek M, Horsthemke B, Brannan CI 2000. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat Genet 25: 74–78 [DOI] [PubMed] [Google Scholar]
- Bressler J, Tsai TF, Wu MY, Tsai SF, Ramirez MA, Armstrong D, Beaudet AL 2001. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat Genet 28: 232–240 [DOI] [PubMed] [Google Scholar]
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B 1995. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet 9: 395–400 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF 2004. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci 101: 17450–17455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI 2008. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses JJ, Reik W, Feil R 1998. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 125: 2273–2282 [DOI] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R 2007. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J 26: 720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A 2010. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285: 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL 2008. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27: 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani A, Courbeyrette R, Agez M, Ochsenbein F, Mann C, Thuret JY 2008. In vivo study of the nucleosome assembly functions of ASF1 histone chaperones in human cells. Mol Cell Biol 28: 3672–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard M, Chen JY, Gronemeyer H, Chambon P, Duboule D, Zakany J 1996. In vivo targeted mutagenesis of a regulatory element required for positioning the Hoxd-11 and Hoxd-10 expression boundaries. Genes Dev 10: 2326–2334 [DOI] [PubMed] [Google Scholar]
- Gerard M, Hernandez L, Wevrick R, Stewart CL 1999. Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet 23: 199–202 [DOI] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C 2010. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science 329: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 166: 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL 2001. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107: 727–738 [DOI] [PubMed] [Google Scholar]
- Holm VA, Cassidy SB, Butler MG, Hanchett JM, Greenswag LR, Whitman BY, Greenberg F 1993. Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics 91: 398–402 [PMC free article] [PubMed] [Google Scholar]
- Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM III, Biniszkiewicz D, Yanagimachi R, Jaenisch R 2001. Epigenetic instability in ES cells and cloned mice. Science 293: 95–97 [DOI] [PubMed] [Google Scholar]
- James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol 50: 170–180 [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, et al. 2008. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4: e1000190 doi: 10.1371/journal.pgen.1000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers M, Bancescu DL, Le Meur E, Rougeulle C, Glatt-Deeley H, Brannan C, Muscatelli F, Lalande M 2004. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res 32: 3480–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sutton A, Hesman T, Min J, Xu RM, Johnston M, Sternglanz R 2003. Set2-catalyzed methylation of histone H3 represses basal expression of GAL4 in Saccharomyces cerevisiae. Mol Cell Biol 23: 5972–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 36: 1291–1295 [DOI] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR III, Workman JL 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278: 8897–8903 [DOI] [PubMed] [Google Scholar]
- Lu Q, Richardson B 2004. Methods for analyzing the role of DNA methylation and chromatin structure in regulating T lymphocyte gene expression. Biol Proced Online 6: 189–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y 2005. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- Mietton F, Sengupta AK, Molla A, Picchi G, Barral S, Heliot L, Grange T, Wutz A, Dimitrov S 2009. Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol 29: 150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Shibata Y, Noma K, Tsukamoto Y, Warren E, Temple B, Grewal SI, Strahl BD 2005. Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot Cell 4: 1446–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL 2001. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2: 153–175 [DOI] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI 2007. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol 14: 372–380 [DOI] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y 2009. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature 460: 287–291 [DOI] [PubMed] [Google Scholar]
- Ohta T, Gray TA, Rogan PK, Buiting K, Gabriel JM, Saitoh S, Muralidhar B, Bilienska B, Krajewska-Walasek M, Driscoll DJ, et al. 1999. Imprinting-mutation mechanisms in Prader-Willi syndrome. Am J Hum Genet 64: 397–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannetier M, Julien E, Schotta G, Tardat M, Sardet C, Jenuwein T, Feil R 2008. PR-SET7 and SUV4-20H regulate H4 lysine-20 methylation at imprinting control regions in the mouse. EMBO Rep 9: 998–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O'Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet 30: 77–80 [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Regha K, Sloane MA, Huang R, Pauler FM, Warczok KE, Melikant B, Radolf M, Martens JH, Schotta G, Jenuwein T, et al. 2007. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell 27: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P, Heard E 2004. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol 24: 5475–5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer R, Hershko AY, Perk J, Mostoslavsky R, Tsuberi B, Cedar H, Buiting K, Razin A 2000. The imprinting box of the Prader-Willi/Angelman syndrome domain. Nat Genet 26: 440–443 [DOI] [PubMed] [Google Scholar]
- Shively L, Chang L, LeBon JM, Liu Q, Riggs AD, Singer-Sam J 2003. Real-time PCR assay for quantitative mismatch detection. BioTechniques 34: 498–504 [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4: 481–495 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z 2005. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 280: 35261–35271 [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D 2007. Facultative heterochromatin: Is there a distinctive molecular signature? Mol Cell 28: 1–13 [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R 2004a. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296–1300 [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Feil R 2004b. Site-specific analysis of histone methylation and acetylation. Methods Mol Biol 287: 99–120 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA 2006. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 26: 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RH, Bresnan MJ 1989. Neonatal respiratory depression and delay in diagnosis in Prader-Willi syndrome. Dev Med Child Neurol 31: 231–236 [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67: 545–579 [DOI] [PubMed] [Google Scholar]
- Wu MY, Tsai TF, Beaudet AL 2006. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev 20: 2859–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 17: 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]