Abstract
In metazoans, thousands of DNA replication origins (Oris) are activated at each cell cycle. Their genomic organization and their genetic nature remain elusive. Here, we characterized Oris by nascent strand (NS) purification and a genome-wide analysis in Drosophila and mouse cells. We show that in both species most CpG islands (CGI) contain Oris, although methylation is nearly absent in Drosophila, indicating that this epigenetic mark is not crucial for defining the activated origin. Initiation of DNA synthesis starts at the borders of CGI, resulting in a striking bimodal distribution of NS, suggestive of a dual initiation event. Oris contain a unique nucleotide skew around NS peaks, characterized by G/T and C/A overrepresentation at the 5′ and 3′ of Ori sites, respectively. Repeated GC-rich elements were detected, which are good predictors of Oris, suggesting that common sequence features are part of metazoan Oris. In the heterochromatic chromosome 4 of Drosophila, Oris correlated with HP1 binding sites. At the chromosome level, regions rich in Oris are early replicating, whereas Ori-poor regions are late replicating. The genome-wide analysis was coupled with a DNA combing analysis to unravel the organization of Oris. The results indicate that Oris are in a large excess, but their activation does not occur at random. They are organized in groups of site-specific but flexible origins that define replicons, where a single origin is activated in each replicon. This organization provides both site specificity and Ori firing flexibility in each replicon, allowing possible adaptation to environmental cues and cell fates.
In metazoans, DNA replication is initiated at thousands of chromosomal sites during each S phase. These DNA replication origins (Oris) should be activated only once at each cell cycle to avoid any amplification and maintain genome integrity. This is an important task in human or mouse cells, where 30,000 replication Oris are activated at each cell division. In prokaryotes as well as in bacterial and animal viruses, Oris are sequence-specific. In Saccharomyces cerevisiae, Oris are identified by specific DNA elements, called Autonomous Replication Sequences (ARS), which have a common AT-rich 11-bp ARS Consensus Sequence (ACS). However, sequence specificity identifies potential Oris but does not determine their selection. Indeed, of the 12,000 ACS present in S. cerevisiae genome only 400 (3.3%) are functional (Nieduszynski et al. 2006). In Schizosaccharomyces pombe, ARS were also identified, but they do not share a specific consensus sequence and are characterized by AT-rich islands (Segurado et al. 2003; Dai et al. 2005; Heichinger et al. 2006) and polydA/dT tracks.
In multicellular organisms, how Oris are defined remains elusive despite considerable efforts to unravel a replication origin code. Until recently, only a few Oris were identified in metazoans. They appear to have variable features, since they can be extremely site-specific, as the human lamin B2 Ori (Abdurashidova et al. 2000), or have a broad site specification like the DHFR Ori (Dijkwel and Hamlin 1995). No consensus motif with predictive value has been found yet and therefore it has been proposed that some unknown epigenetic features could identify metazoan Oris. In agreement, transcription and chromatin status were found to influence Ori specification at specific gene domains (Aladjem 2007; Mechali 2010).
Unraveling common features of metazoan Oris requires a large-scale identification procedure, the development of which was hampered by the lack of a genetic test, like the yeast ARS test, and by the fact that methods to map Oris were not always adapted to robust genome-scale analysis. The first genome-scale studies to localize Oris in human and mouse cells (Cadoret et al. 2008; Sequeira-Mendes et al. 2009) observed a correlation with unmethylated CpG islands (CGI) and some overlap with promoter regions (Delgado et al. 1998; Sequeira-Mendes et al. 2009). However, it was not clear whether CGI were a specific mark of Oris or of the associated promoters.
In order to identify new features of eukaryotic Oris, first we upgraded the method used to map nascent DNA strands (NS) at active Oris to a specificity and reproducibility compatible with genome-scale analysis. Then, we used this method in four cell systems: mouse embryonic stem cells (ES), mouse teratocarcinoma cells (P19), mouse embryonic fibroblasts (MEFs), and Drosophila cells (Kc). We characterized up to 2748 Oris on mouse chromosome 11 (P19 cells) and 6184 Oris in the Drosophila genome. The three mouse cell lines show common and also specific Oris, suggesting that Oris may contribute to cell identity specification. Ori-rich domains co-localize with the previously defined early replicating domains, whereas Ori-poor domains correspond to late replicating regions. Oris are also preferentially concentrated in transcription promoter regions in mouse cells. We found that Oris are strongly associated with CGI and exhibit a bimodal distribution that is suggestive of an asymmetric initiation event. Nucleotide sequence asymmetry is also present at the initiation site, and the analysis reveals specific GC-rich motifs in both mouse and Drosophila Oris at initiation sites. A strong correlation between HP1 binding sites and Oris was found at Drosophila chromosome 4, which is mainly constituted of heterochromatin. In combination with DNA combing data, our genome-scale results demonstrate that metazoan Oris are in large excess relative to their standard use, and computational simulations suggest that flexible Oris are clustered in groups that define the replicons, where activation of one Ori silences the others in the same group.
Results
Genome-scale mapping of DNA replication origins: General features
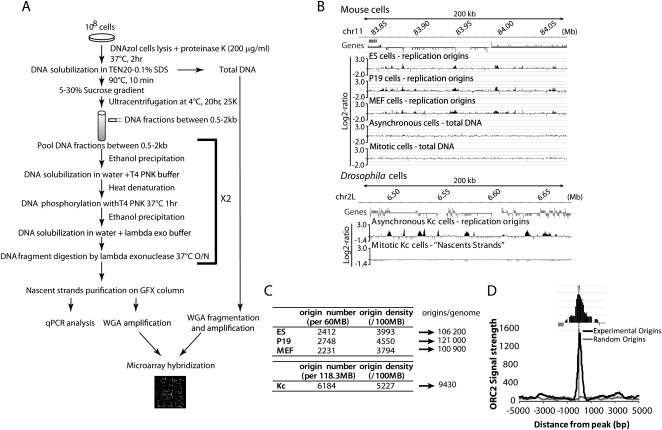
Ori mapping by purification of RNA-primed NS is a well-established procedure. NS purification is achieved through (1) size selection of RNA-primed NS at Oris and (2) specific enrichment using lambda exonuclease (Fig. 1A) as RNA-primed NS are resistant to this enzyme, while contaminating broken DNA is degraded efficiently. However, only a very small amount of NS (at most 20 ng per 108 cells; Cadoret et al. 2008) can be recovered and, consequently, the slightest contamination by broken DNA pieces would dramatically raise the background. In order to perform genome-scale mapping of Oris, we upgraded this procedure by improving NS purification (as detailed in Methods) and enrichment through two or three rounds of digestion with high specificity, custom-made exonuclease (Fermentas) to remove non-RNA-primed NS. The efficiency of this step is crucial, as explained in Methods. In addition, we used tiling microarrays (Nimblegen) in which oligonucleotides were spaced on average every 140 bp and which, therefore, gave a four- to sixfold more accurate resolution than in the previous genome-scale studies. Altogether, this improved method allowed us to detect a much larger number of consecutive positive oligonucleotides that scanned each Ori, and to score Oris with increased confidence.
Figure 1.
Genome-scale mapping of replication origins by nascent strand (NS) chip. (A) NS isolation schematic. 0.5–2.5-kb NS were isolated from total genomic DNA by denaturation and sucrose gradient centrifugation. NS enriched by lambda exonuclease treatment were hybridized against total genomic DNA on high-density tiling arrays (see Supplementary Information). (B) Example of the distribution of replication origins in mouse (upper panel) and Drosophila cells (lower panel) along a 200-kb region. The log2-ratio between NS and total genomic DNA is shown. For genes, the position of the start site (high bar bordering the gene), exons (large gray boxes), and introns (thin gray boxes) are indicated (see Supplemental Fig. 2F). (C) Origin number and density per genome. (D) Immunoprecipitation of chromatin associated with ORC2 was carried out in P19 cells as described in Methods. Compilation of ORC2 signal strength data and correlation with the NS peaks is shown.
We used this procedure to obtain genome-scale maps of Oris in mouse ES, P19, and MEF cells as well as in Drosophila Kc cells in order to highlight conserved features between vertebrate and invertebrate Oris as well as to assess the impact of cell differentiation on the Oris repertoire. The full data set (obtained using up to four different biological replicates for each cell line) consisted of a continuous 60.4 Mbp fragment on mouse chromosome 11, which we considered representative of the mouse genome as it exhibits replication timing and transcription features that are comparable to those of the entire genome (Supplemental Fig. 1A), and of 118.3 Mbp of Drosophila genome. This allowed analyzing the overall distribution of Oris at a chromosome scale in comparison to the average 0.68 Mb regions of the previous genome-scale studies (Cadoret et al. 2008). NS maps showed enrichment at specific genomic locations with a high degree of reproducibility (see examples in Fig. 1B and Supplemental Fig. 1B–E). To control the rate of false positives, for each probe, the log2-ratio value was normalized and the P-value was computed by applying the false discovery rate (FDR) correction (Benjamin and Hochberg 1995). A probe was considered significant when the P-value was <5% (level of significance). With the FDR correction, potential Oris could be identified with high confidence. Moreover, as the minimum size of purified NS was 0.5 kb, Oris should be theoretically at least 1 kb (2 × 0.5 kb for the general case of a bidirectional Ori). Oris were thus defined as positive regions (log2-ratio > 0) of at least 1 kb containing significant probes (see details in Supplementary Information).
We further validated the Ori maps by quantitative PCR (qPCR) analysis of known Oris in the mouse Myc gene and Hoxb domain (Supplemental Fig. 1F,G) and Drosophila Histone gene locus (Supplemental Fig. 2A), as well as of randomly chosen putative Oris validated in this study (Supplemental Fig. 2B,C). Seventeen out of 18 validated Oris showed significant NS enrichment. Only background signal was observed when total DNA or “NS” from mitotic cells was used for hybridization (Fig. 1B; Supplemental Fig. 1B,D,G), or when NS were RNase-treated before exonuclease digestion (data not shown), confirming the specificity of the purification procedure. Using ChIP-chip analysis, we also confirmed the presence of ORC2, a key component of Oris, at the mouse Myc Oris (Fig. 1D), with a profile similar to the one observed in human cells (Ghosh et al. 2006). We also found that the ORC2 signal profile strongly correlated with the profiles of the NS peaks (Fig. 1D; Supplemental Fig. 3; see below).
By using exponentially growing cells we can potentially score all Oris activated during the entire S phase, and possible variations in Ori usage among cells would not prevent their detection. We identified 2412 Oris in ES, 2748 in P19, 2231 in MEF, and 6184 in Drosophila Kc cells (Fig. 1C). We noted that a large fraction of Oris (up to 44%) was common to the three mouse cell lines (Supplemental Fig. 4A). The Ori repertoires of ES and P19 cells, both pluripotent cells, were statistically more related to each other than to MEF Oris (see details in Supplementary Information and Supplemental Fig. 4). We also observed that Drosophila cells had a denser Ori repertoire. Finally, Ori sequences were significantly more conserved compared to non-Ori sequences in both Drosophila and mouse cells (P < 2 × 10−16), suggesting that important evolutionary conserved elements might be present in Oris (data not shown).
Overrepresentation of Oris at transcriptionally active promoters
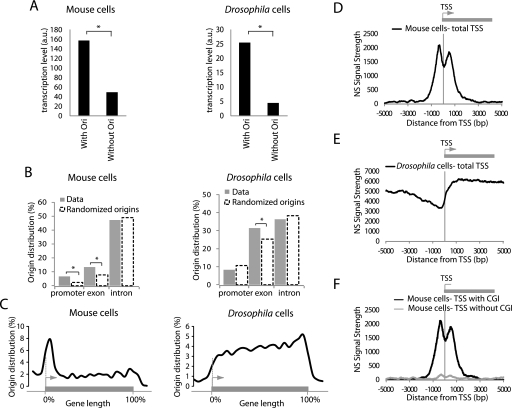
We next analyzed the association of Oris with genes. To determine if the observed correlation was significant, our data sets were compared with 1000 randomized Ori data sets, which contained the same number and length of Oris but randomly located, to evaluate association by chance. Oris were found in both inter-genic and genic regions (Supplemental Fig. 5A) with a significant preference for intragenic localization (P < 0.001). This association was not stochastic, as genes with Oris were significantly more actively transcribed than genes without Oris (Fig. 2A). Compared to randomization, Oris showed a significant association with promoters (in mouse cells) and exons (in mouse and Drosophila cells) (Fig. 2B; Supplemental Fig. 5B). Ori distribution relative to gene length was also analyzed. Gene lengths were normalized on a scale of 0 to 100 (corresponding to the start and end of genes). Mouse Oris were found all along the genes, although they were overrepresented at the start of genes (Fig. 2C). Conversely, Ori concentration at promoter regions was not observed in Drosophila.
Figure 2.
Replication origins in metazoans are linked to expressed genes. (A) Replication origins are significantly associated with transcribed genes (*: P < 0.001) in both mouse MEF and Drosophila Kc cells. (B) Association of replication origins with gene partitions in MEFs (left panel) and Drosophila Kc cells (right panel). Replication origins are found more frequently at gene promoters (mouse cells) and exonic sequences (mouse and Drosophila cells, *: P < 0.001). (C) Distribution of mouse replication origins along a gene. The position of each origin is allocated depending on the length of the gene adjusted to 100%. (D) Nascent strand signal strength at TSS in ES cells and (E) Drosophila Kc cells. The enrichment value is the log10 of the combined P-value associated with NS signal (see Supplementary Information). (F) NS signals in mouse ES cells are associated with CGI-positive TSS but not with CGI-negative TSS.
We then examined accurately the NS signal strength around transcription start sites (TSS) that had been aligned (see details in Supplementary Information). In mouse cells, we observed a striking bimodal distribution with two major NS peaks located on each side of TSS (Fig. 2D; Supplemental Fig. 6A,B). This pattern was lost when the location of TSS along the chromosome was randomized (data not shown), indicating that the bimodal distribution of NS signal strength around TSS was significant. The two putative replication initiation sites were separated by a shallow valley centered on the TSS, with the NS peaks located ∼600 bp upstream of and downstream from the TSS. A careful analysis showed four categories of TSS-linked Oris (Supplemental Fig. 7). Most of them (77%) exhibited the bimodal distribution. Other minor categories include unimodal TSS-linked Oris with the peak located upstream (11%), downstream (8%), or on the TSS (4%). We also noted that 67% of TSS with divergent transcription (Seila et al. 2008) contained a bimodal Ori, whereas the bimodal Ori-TSS association decreased to 35% when all TSS were considered. Conversely, in Drosophila cells, TSS were not enriched in Oris (Fig. 2E), and NS did not show a bimodal distribution but increased density within genes as opposed to promoter regions. This contrasting result led us to ask whether another element present at mammalian TSS, but not within TSS, was associated with Oris, both in Drosophila and mouse cells.
Replication origins are bimodal and enriched at CGI in mouse and Drosophila
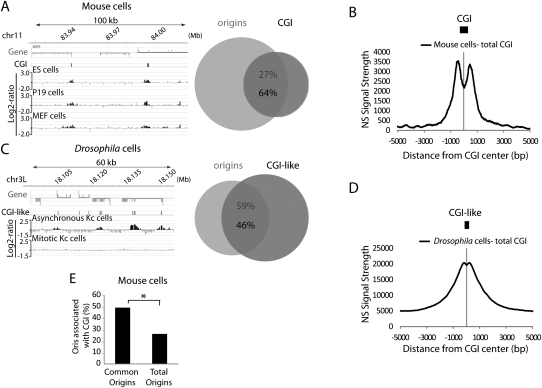
Mammalian promoters, particularly those of highly expressed genes, are CpG-rich while genes that are highly regulated during development are often CpG-poor or -free (Cross and Bird 1995). CpG-rich sequences, known as CGI, are usually defined as regions of at least 200 bp in length with 60% of CG and a ratio of CpG observed/CpG expected >0.6. We asked whether such elements could explain the bimodal NS signal strength around TSS, and we analyzed TSS with CGI (n = 820) and without CGI (n = 434) separately. In the three mouse cell lines, the NS bimodal signal strongly associated with CGI-positive TSS, but not with TSS that did not contain CGI. This result suggests that the overrepresentation of Oris at promoter regions was in reality a consequence of their association with CGI (Fig. 2F; Supplemental Fig. 6C,D). In addition, although CGI represented only 1.3% of the mouse genome, most of them (up to 73%) were strongly (P < 0.001) associated with Oris in the three mouse cell lines (Fig. 3A; Supplemental Fig. 6E,F), and the NS signal strength was bimodal around CGI as well (Fig. 3B; Supplemental Fig. 6E,F). Also, Oris that were common to the three mouse cell lines were significantly enriched in CGI (Fig. 3E).
Figure 3.
Association of replication origins with CGI in metazoans. Example of replication origins associated with CGI in (A) mouse ES cells and (C) Drosophila cells. The percentage of CGI/replication origin association is also shown. (B) NS signal strength around all CGI in mouse ES cells and (D) CGI-like regions in Drosophila Kc cells. The average size of CGI is shown in scale. (E) Common origins in mouse cells are strongly associated with CGI regions. The proportion of CGI-positive origins in the indicated groups of origins is shown.
In higher metazoans, CpG dinucleotides are subject to cytosine methylation, which results in their depletion from the genome over time during evolution (Cross and Bird 1995; Illingworth and Bird 2009). Although cytosine methylation is almost nonexistent in D. melanogaster, its genome contains regions with properties identical to those of mammalian CGI. We thus delimited ∼20,000 “CGI-like” regions that responded to the CGI definition and represented 5.9% of the Drosophila genome. Forty-six percent of these regions were significantly (P < 0.001) associated with 59% of Oris (Fig. 3C). Moreover, although the NS signal strength was not bimodal around Drosophila TSS (Fig. 2E), it was bimodal around the Drosophila CGI-like regions (Fig. 3D), like in mouse cells (Fig. 3B). The bimodal curve was less accentuated than in the mouse, possibly linked to the fact that the CGI-like elements found in Drosophila were smaller (346 bp vs. 606 bp in the mouse).
We conclude that CGI-related sequences are conserved determinants in a substantial part of mouse and Drosophila Oris, regardless of their genomic position. They do not need to be at promoter regions, or to rely on methylation, consistent with the presence of CGI-like sequences in exons in the Drosophila genome and the fact that CGI at mouse promoters are often demethylated. These results provide a novel possible function for CGI sequences in DNA replication that is conserved both in vertebrate and invertebrate species. Importantly, this role is, at least in Drosophila, independent of CpG methylation or of being localized close to a promoter region.
Oris are characterized by nucleotide asymmetry and CG-rich elements
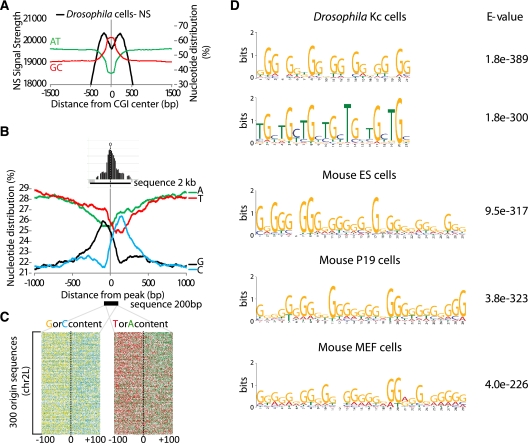
We further investigated the GC/AT nucleotide composition of these Oris. Replication initiation sites (defined by the NS peaks) were located outside the central CGI, in a region with an AT content that was found similar to that of the whole genome (Fig. 4A). We then asked whether the sequences flanking the NS peaks showed particular features. All Drosophila Ori sequences were aligned to their NS peaks (Fig. 4B) and the frequency of each nucleotide was calculated in a 1000-bp region upstream of and downstream from such peaks. This analysis revealed a clear nucleotide asymmetry with overrepresentation of T and G at the 5′ and of A and C at the 3′ of the NS peaks. This nucleotide bias could easily be visualized when individual Oris were aligned (Fig. 4C). Similar results were obtained in mouse cells although the skew was more marked for C and G (Supplemental Fig. 8). This asymmetry was not observed in randomized Oris (data not shown). We conclude that Drosophila and mouse initiation sites display a characteristic nucleotide asymmetry that is not observed at more upstream or downstream regions.
Figure 4.
Nucleotide skew and GC-rich elements at replication origins. (A) Origins were centered on Drosophila CGI-like regions. The mean AT and GC percentages of centered Oris are shown. Genome-scale NS signal strengths are represented by a black line. Note that the NS peaks (putative replication initiation sites) are not enclosed in the central CG-rich region. (B) Genome-scale nucleotide distribution of all Drosophila origins centered on the NS peak. Note the skew in nucleotide distribution with GT and AC enrichment at the 5′ and 3′ end of the origin peak, respectively. (C) Nucleotide distribution at and around the origin peak for origins in Drosophila Kc cells; 200 bp sequences of 300 replication origins were stacked and aligned around the NS peak. Four colors were used: green for A, red for T, yellow for G, and blue for C. The exact sequence can be read by enlarging the figure in Supplemental Data. A clear bias is observed for C or G, and A or T around the NS peak. (D) Motifs frequently found in Drosophila (top panel) and mouse (bottom panel) replication origins. The E-value is indicated (see Methods).
Although no consensus sequence has been associated with metazoan Oris (for review, see Mechali 2010), we investigated, using the MEME suite (http://meme.sdsc.edu/meme4_4_0), whether some enriched motifs could be identified in Oris. Due to its smaller genome, Drosophila sequences were first analyzed. We submitted 2-kb stretches of Drosophila DNA sequences centered on NS peaks using specific parameters (see Methods). The more frequent motifs recovered in different MEME runs using different batches of Drosophila or mouse Oris were GC-rich motifs with a repetitive nature. In Drosophila, two motifs (Fig. 4D) were associated with >60% Oris (data not shown). Interestingly, these motifs were often found in known Oris (data not shown). In mouse, G-rich motifs were also consistently recovered (Fig. 4D).
We further characterized Drosophila Oris using the R'MES program (https://mulcyber.toulouse.inra.fr/projects/rmes/), which investigates whether a motif is overrepresented in a set of sequences (Hoebeke and Schbath 2006). As R'MES is limited to 13 nucleotide motifs, we asked which among the 67,108,864 possible 13-mers occurred more frequently. (TGC)4T, its cyclic permutations, and complementary sequences were significantly overrepresented in agreement with the MEME analysis (see motif 2 in Fig. 4D). (TA)n sequences, which are very frequent in microsatellites, were not significantly associated with Oris (data not shown).
In summary, similar GC-rich motifs were found associated with metazoan Oris. Although it would be hazardous to conclude that there is a strong sequence specificity, these results altogether indicate that at least two general sequence features are associated with Oris: (1) a bias toward GC-rich elements and (2) a clear nucleotide asymmetry upstream of as well as downstream from these elements, at the position of NS synthesis.
In Drosophila heterochromatin, HP1 sites are associated with Oris
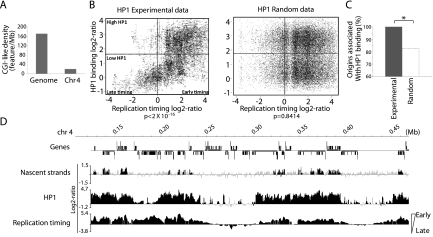
Drosophila chromosome 4 is unusual as it represents 1% of the genome and is organized mainly in heterochromatin that replicates early in S phase in Kc cells (Schwaiger et al. 2009). The density of CGI-like regions was eightfold lower than in other chromosomes (Fig. 5A), but this feature cannot explain the early replication timing of chromosome 4. In fission yeast, the HP1 ortholog Swi6 is involved in early S phase replication of heterochromatic pericentromeres and of the MAT locus (Hayashi et al. 2009). HP1 is believed not to be involved in Ori positioning, but rather in favoring Ori firing by recruiting DDK kinases (Hayashi et al. 2009). To evaluate a possible link between HP1 and replication in metazoans, Oris were correlated with reported HP1 binding sites (de Wit et al. 2007) and replication timing (Schwaiger et al. 2009) in Drosophila Kc cells. First, a strong positive correlation between early timing of replication and high HP1 binding was detected (P < 2 × 10−16, Fig. 5B, left panel). This association was lost when the HP1 probes were randomized (Fig. 5B, right panel). Moreover, 100% Oris were associated with HP1 sites (Fig. 5C,D), indicating that, in CGI-poor regions and in heterochromatin, HP1 binding sites may contribute to Ori recognition.
Figure 5.
Positive link between HP1 and origin firing/early replication in heterochromatic regions. (A) Density of CGI-like regions in the whole genome and on chromosome 4 in Drosophila. (B) Positive correlation between HP1 binding and early S phase replication timing in Drosophila chromosome 4. Scatter plots between experimental and randomized HP1 data sets and replication timing are shown. The P-value is indicated at the bottom of the panels. (C) Significant association between HP1 binding and origins on the entire Drosophila chromosome 4. (D) A 300-kb region of chromosome 4 showing the relationship between origin firing, HP1 binding and early replication timing.
DNA replication origins are organized in large, high-density domains
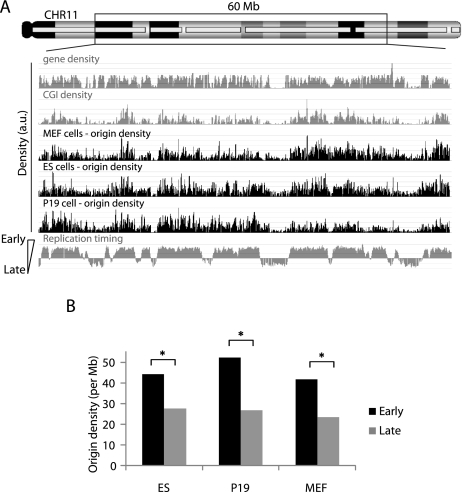
We then evaluated the higher order organization of Oris. In each cell line, Ori density along the chromosome was investigated (see details in Supplementary Information). In mouse cells, Ori density along chromosome 11 was not uniform, with areas of low Ori density separated by large high density areas (Fig. 6A). These regions were at similar positions in the three mouse cell lines (Fig. 6A). We also found that Ori density correlated well with the replication timing domains (Fig. 6A,B), which were previously identified by genome-scale analysis in mouse (Hiratani et al. 2008), suggesting that replication timing is controlled by Ori density. Drosophila Kc cells also showed Ori enrichment at early replication domains, albeit at a lower degree (data not shown).
Figure 6.
Early replication domains are characterized by high origin density. (A) Shown is the origin density in the three mouse cell lines calculated using a 100-kb sliding window along the chromosomal region. The computed gene and CGI densities are also illustrated. Origin density is positively correlated with early replication domains (*: P < 0.001). (B) Origin number is also positively correlated (*: P < 0.0001) with the early replication timing observed in mouse ES cells (Hiratani et al. 2008).
Hierarchic organization of metazoan Oris
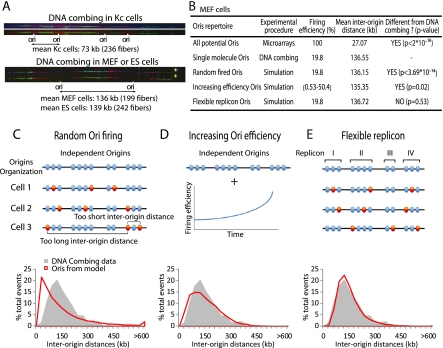
Genome-scale data score all the Oris that are activated in a given cell population and thus allow the identification of all potential sites which can serve as Oris, although they can vary from cell to cell within a given population. To study the actual Ori usage in Drosophila and mouse individual cells, DNA combing analysis was performed. Two consecutive pulses using different deoxynucleotide analogs allowed precise localization of Oris on single DNA molecules (Fig. 7A; Methods; Supplemental Methods). The size of the fibers analyzed ranged between 194 and 900 kb, and mouse cell lines presented similar inter-origin distances (136 kb in MEF and 139 kb in ES cells, Fig. 7A). This is in agreement with the mean inter-origin distance (137 kb) recently found in the human MRC-5 fibroblast cell line at the FRA3B locus (Letessier et al. 2011) where longer domains were analyzed. Conversely, a near twofold difference between the mouse (average 137.5 kb) and Drosophila (73 kb) inter-origin distances was observed (Fig. 7A).
Figure 7.
Replication origins are organized in a functional hierarchical manner along the chromosome. (A) DNA combing analysis performed in Drosophila Kc (top panel) and mouse (bottom panel) cells after two consecutive labeling pulses of IdU and CldU. (B) Summary of the experimental and simulated inter-origin distance distributions for MEF cells. For the “Increasing Ori efficiency” model, the values for the firing efficiency represent the initial and final origin firing efficiency during simulations. (C) “Random Ori firing” model. In this model, origins are completely independent and are activated randomly (red circles). Very short and long inter-origin distances are observed. (D) In the “Increasing Ori efficiency” model, origins are completely independent and activated randomly, but with increasing firing efficiency throughout S phase progression. (E) “Flexible Replicon” model. In this model, origins are linked within functional units where activation of one origin silences the others in the same group. The bottom panels present the computer-simulated results for each model. The gray profile is the distribution of inter-origin distances obtained by DNA combing of MEF cells. The red line represents the simulated distribution of inter-origin distances according to each model. The “Flexible Replicon” model is the only model to yield a simulated distribution of inter-origin distances that is statistically indistinguishable from the DNA combing data.
The pattern of Ori usage was further investigated by combining our genome-scale Ori data (from Kc, ES, and MEF cells) and the inter-origin distances obtained by DNA combing. For simplicity, only the MEF replication dynamics will be explained in more detail. If all mapped Oris were activated in all cells (100% firing efficiency) the resulting, very short, inter-origin distance distribution would be significantly different from the distribution observed using DNA combing (Fig. 7B). Indeed, the comparison of genome-scale and DNA combing data suggests that, in MEFs, one of every five Oris on average was activated in a given DNA molecule (19.8% firing efficiency, Fig. 7B). Similar values were obtained for ES and Kc cells. Our results are consistent with the notion that metazoan Oris are redundant, and that only a small proportion of them is effectively used at each cell cycle.
We then assessed how different models of metazoan DNA replication could explain our data. We first considered the “Random Ori firing” model, in which Oris are randomly activated at a density based on the DNA combing experiments. The mean inter-origin distance of fired Oris was then identical to the value obtained by DNA combing (Fig. 7B); however, the simulated inter-origin distance distribution (Fig. 7C in red; Supplemental Fig. 9A,B) was different from the experimental distribution obtained in combing experiments (Fig. 7C in gray; Supplemental Fig. 9A,B). Specifically, the “Random Ori firing” model led to populations of short and long inter-origin distances that were not observed in the DNA combing experiments. The presence of a group of large inter-origin distances is in agreement with the random gap problem (i.e., random firing leads to large gaps of unreplicated DNA that will persist at the end of S phase; Laskey 1985; Hyrien et al. 2003).
Then, we evaluated the “Increasing Ori efficiency” model (Rhind 2006). This model is based on the idea that Ori firing efficiency is not constant but actually increases during S phase (Fig. 7D). This increase in efficiency ensures Ori firing in late S phase in order to fill the remaining stretches of unreplicated DNA. To implement this model, fork speed was analyzed by DNA combing: Replication forks in mouse cells were about twice faster than in Drosophila cells (1.77 vs. 0.81 kb/min, Supplemental Fig. 10A). Using these experimental values, Ori activation was simulated with increasing firing efficiency during S phase progression (Supplemental Fig. 10B). The “Increasing Ori efficiency” model solved the random-gap problem, as indicated by the disappearance of the population of large inter-origin distances observed in the “Random Ori” model (Fig. 7D). However, the simulated inter-origin distribution remained significantly different from the one observed by DNA combing (Fig. 7B,D; Supplemental Fig. 9). The simulated inter-origin distance distribution was wider and contained a much larger short-distance population than the one derived from the DNA combing data.
We then considered a “Flexible Replicon” model in which adjacent Oris are functionally linked together over a defined distance that delineates a replicon, providing a multiple firing choice. In each replicon/group of Oris, one Ori is randomly activated and silences the others (Fig. 7E). Oris were classified using hierarchical cluster analysis (see Supplementary Information) to generate a dendrogram of Ori repartition along the chromosome. Clusters were obtained by cutting the dendrogram at the height that gave the strongest correlation with DNA combing data (Supplemental Fig. 10C–E). The simulated inter-origin distances were very similar to the DNA combing values in these conditions (Fig. 7B,E; Supplemental Fig. 9). This analysis suggests that replicons are on average 56 kb in length, contain 4.3 Oris, and the inter-replicon distance is 117 kb in MEF cells (Supplemental Fig. 10F). The “Flexible Replicon” model was also applicable to ES and Kc cells (Supplemental Figs. 9, 10F). The model is thus robust and can accommodate changes in Ori density and firing efficiency.
Overall, these findings suggest that Oris are in large excess in metazoans and have a flexible use. However, Ori firing flexibility is not stochastic in the whole genome, but only inside each replicon. Metazoan replicons appear constituted of groups of potential and flexible adjacent Oris where activation of one Ori suppresses the surrounding Oris. The inter-origin distance is therefore the average distance between activated Oris in each group of flexible Oris.
Discussion
This high resolution, genome-scale analysis of Oris allowed the identification of 13,575 Oris in four cell lines from two different metazoan species and the discovery of common organization and sequence features. The combined analysis of genome-scale and DNA combing data suggests that metazoan Oris are organized in replicons in which Ori flexibility is an essential feature.
Metazoan origins are bimodal and are enriched at actively transcribed genes and transcription start sites
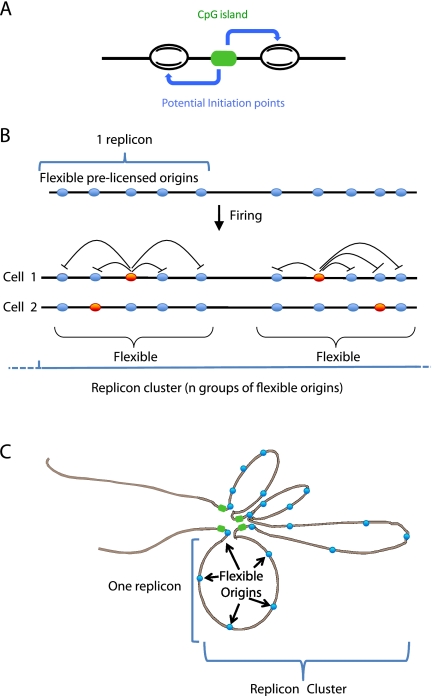
In metazoans, replication timing and transcriptional activity are connected. Early activated Oris are in actively transcribed genes, whereas late replication is associated with poorly transcribed regions. We show here a strong correspondence between Ori density and timing of replication. Ori-rich regions are in early replicating domains; conversely, Ori-poor regions correspond to late replicating domains. In mouse, but not in Drosophila, we found a significant enrichment of Oris at promoter regions, particularly at TSS, where most Oris have a bimodal structure, with two peaks of NS bordering the TSS regions. We then show that this bi-modal structure is mainly linked to CGI elements often found at promoter regions. We postulate that the bimodal nature of Oris is due to initiation with two opposite start sites for the leading strand synthesis, separated by ∼1 kb of sequence that might contain the Ori genetic determinants. The fusion of these two replication bubbles would rapidly lead to a single bubble at Oris (Fig. 8A). Such mechanism is similar to the asymmetric bidirectional model of replication proposed for the human DBF4 Ori (Romero and Lee 2008) and is reminiscent of initiation at the E. coli Ori (Fang et al. 1999), where the DNA helicase proceeds for at least 100 nt before priming DNA synthesis.
Figure 8.
Origins, replicons, and replicon clusters. (A) The presence of a CpG island or CGI-like region allows the positioning of two potential initiation sites upstream of and downstream from the region. (B) Replicons are organized as functional units containing several potential DNA replication origins. Activation of one origin within a replicon silences the others. The origin choice within each replicon can occur either stochastically or be dictated by specific cell fates. Replicon clusters include several consecutive replicons which are activated simultaneously (Berezney et al. 2000). (C) Representation of replicons as chromatin loops where activation of one origin silences the other origins contained in the same replicon.
We also observed that 67% of divergent TSS have an Ori (data not shown). Divergent transcription at TSS in mouse ES cells is associated with CpG-rich promoters, where antisense and sense short RNAs of 16–30 nt are synthesized upstream of and downstream from the TSS at two sites separated by 400–500 bp (Seila et al. 2008). The distance between the 3′ ends of these transcripts is close to the mean distance we observed between the two NS peaks. It could be asked whether such short transcripts might be used for initiation of DNA replication, like in E. coli (Baker and Kornberg 1988; Skarstad et al. 1990), or Epstein-Barr Virus (Rennekamp and Lieberman 2011), where initiation of DNA replication is facilitated by transcription by RNA polymerases.
Metazoan origins exhibit common sequences features
We found a link between CGI and Oris in all cell types analyzed, as reported for a subset of human and mouse Oris (Delgado et al. 1998; Cadoret et al. 2008; Gomez and Antequera 2008; Sequeira-Mendes et al. 2009). CGI are essential elements for transcriptional control and imprinting in mammals and are regulated by DNA methylation. However, CGI-like regions are also present in Drosophila and they were significantly associated with Oris, which showed a bimodal NS distribution at these sites, like in mammals. Their association with Oris in Drosophila is intriguing as methylation is rather poor in this species. These data suggest that CGI, or some elements embedded in these regions, had a primary role in DNA replication and they further evolved to be used in transcriptional control. As the number of CGI elements in the genome is much lower than the number of potential Oris, other Ori classes must be present. However, CGI elements appear to be an important class of determinants of Ori localization, which explain why Oris are enriched at promoter regions without being obligatorily linked to transcription.
Two independent bioinformatics approaches showed that the Oris described in this study contain conserved features, which, although not as strict as the S. cervisiae ACS, reveal some bias toward GC-rich elements. NS enrichment peaks are not localized at the CpG-rich domain itself, but on its sides, suggesting that this domain might be a binding site for factors controlling NS synthesis upstream and downstream (Fig. 8A). The NS synthesis sites are not GC-rich but are characterized by more AT-rich sequences, in agreement with an easier opening of DNA (Fig. 4A).
Another feature is the strong nucleotide skew we observed at NS positions, with a general bias for GT at the 5′ side and for CA at the 3′ side in both Drosophila and mouse Oris. Interestingly, bacterial Oris have a similar nucleotide skew (Lobry 1996), whereas S. cerevisiae ARS have only an (A/T) skew (Breier et al. 2004). We observed nucleotide asymmetry only around initiation sites and not in more upstream or downstream regions, another strong indication that these are true Oris. Nucleotide skew might be therefore a universal Ori property, possibly involved in the structure of DNA at Oris. It has been suggested that the nucleotide skew was a consequence of the mutational bias associated with DNA replication (Touchon et al. 2005).
Positive association of active Oris with HP1 in Drosophila chromosome 4
Heterochromatic DNA is generally believed to constitute late replicating domains. However, accumulating evidences indicate that a subset of heterochromatin DNA replicates early in S phase (Hayashi et al. 2009) and this is the case for Drosophila chromosome 4 (Schwaiger et al. 2009). We detected a positive relationship between HP1 binding and early replication, as recently reported (Schwaiger et al. 2010). Moreover, we found a strong correlation between HP1 binding sites and Oris at this chromosome. This is in agreement with the interaction of HP1 with the Origin Recognition Complex (ORC) in higher eukaryotes (Pak et al. 1997). HP1 binding sites could help Ori recognition in compact heterochromatin regions. The fission yeast HP1 homolog Swi6 is required for early replication of heterochromatic regions (Hayashi et al. 2009). Swi6 stimulates Ori firing by recruiting DDK and facilitating pre–Initiation Complex formation. It would be interesting to investigate whether HP1 also stimulates Ori usage through DDK recruitment in higher eukaryotes. Although chromosome 4 represents only 1% of the Drosophila genome, this finding indicates that DNA replication could be facilitated by other means at specific chromatin domains, and strengthens the role of HP1 in Ori localization.
Replicons are groups of flexible origins
Our data show that, in metazoans, DNA replication firing appears at first as a relatively inefficient system as reported in yeast (Friedman et al. 1997; Dai et al. 2005; Heichinger et al. 2006). On average, there are four- to fivefold more potential Oris than used. Therefore, a replicon cannot be considered as the distance between two Oris. Rather, our data suggests that a replicon is a group of several adjacent and flexible potential Oris, in which only one Ori is activated per cell and per cell cycle, and the others are silenced (Fig. 8B). The possibility to use several Oris in each replicon would increase their firing probability. In other words, flexibility is not stochastic in the whole genome but only inside each replicon. Such Ori flexibility and abundance in each replicon might be needed to respond to variations in growth conditions, problems encountered by the replication fork and to ensure complete duplication. For instance, when the concentration of nucleotides is decreased, new Oris are activated in the hamster Gna13 domain (Anglana et al. 2003; Ge et al. 2007). This model is also in agreement with the conservation of replication foci (clusters of replicons) in subsequent cell cycles, since flexibility will be mainly inside replicons. It is also in agreement with the notion of initiation zone used for the DHFR domain, where multiple Oris can be found at close intervals (Dijkwel and Hamlin 1995). If some sites are deleted, others, close-by, become activated (Mesner et al. 2003). Several potential Oris per replicon might allow choosing the more suitable Ori to be activated in a given chromatin context, which could vary according to the transcriptional status or cell identity. The proposed “Jesuit Model: Many are called, few are chosen” (DePamphilis 1993) appears therefore to apply to the Flexible Replicon model.
Pioneer former work (for review, see Berezney et al. 2000) as well as our DNA combing experiments indicate that Oris are often synchronously activated in clusters which can form replication foci. We thus propose that a replication cluster includes consecutive groups of adjacent flexible Oris (each group constituting a replicon) that are activated synchronously (Fig. 8C). The selection of a given Ori within each replicon might depend on the cell fate or the organization of the chromatin domain. The Ori interference mechanism has been described in yeast (Brewer and Fangman 1993; Lebofsky et al. 2006), where firing at one Ori inhibits close-by Oris and this phenomenon could lead to the 100–120-kbp average size of the replicon. Activation of one Ori might promote looping out of the replicon resulting in the silencing of the other potential Oris (Fig. 8B).
DNA replication origins: A barcode defining cell fate and cell identity?
Our data show that, although flexible, Oris are at specific positions that appear to be mostly conserved among different cells. Pluripotent cells (ES or P19) have slightly more Oris than differentiated cells (MEF), but the size of the replicon (Fig. 7) and the length of S phase (data not shown) are similar. Pluripotent ES cells may have fewer constraints than differentiated cells thus allowing an extended Ori choice. This is in agreement with the changes in Ori choice observed during differentiation in Xenopus (Hyrien et al. 1995), Physarum (Maric et al. 2003), Sciara fly development (Lunyak et al. 2002), human B cell development (Norio et al. 2005), and in the chicken Globi locus (Dazy et al. 2006). In contrast, Drosophila or Xenopus early embryos have no transcriptional constraints and can use all the potential Oris to accelerate S phase. Indeed, in early embryos, Oris are activated at very close intervals, every 10–20 kb in Xenopus (Hyrien and Méchali 1993; Walter and Newport 1997; Lemaitre et al. 2005) and every 8–12 kb in Drosophila (Blumenthal et al. 1974). These values are close to the use of every Ori. Since here we show that potential mouse and Drosophila Oris are at conserved specific sites in the same species, one could ask whether the Oris activated in Xenopus and Drosophila early embryos are as random as previously thought. A maximum usage of specific sites rather than random Ori usage might regulate embryonic chromosome replication. The Ori position in the genome might therefore define a barcode that organizes chromosomal replication patterns, in which the choice and usage of each bar (Ori position) are defined according to cell growth and fate.
Methods
Cells and cell culture
MEFs derived from 13.5-d mouse embryos were cultured as previously described (Hiratani et al. 2008) and used at passage 4 or 5. P19 cells were cultured as previously described (Gregoire et al. 2006). The ES cell line CGR8 was cultured in standard ES cell medium. Drosophila Kc cells were cultured in Schneider's medium (Invitrogen) supplemented with 10% insect cell culture-tested FBS (Sigma). When necessary, mouse cells were synchronized in prometaphase with 100 ng/mL nocodazole for 12 h. Kc cells were synchronized in prometaphase by incubation with 4 mM thymidine overnight, released in fresh medium for 4 h, and incubation with 1 μg/mL nocodazole overnight.
Nascent strand (NS) preparation
DNA purification
Dividing cells (2.5–5 × 108) were washed in PBS, harvested, and lysed in 15 mL DNAzol (Invitrogen) at room temperature (RT) for 5 min. Samples were digested with 200 μg/mL proteinase K at 37°C for 2 h. We found that combining the proteinase K treatment with DNAzol significantly improved the yield of NS. Insoluble material was discarded by centrifugation at 3000g at 4°C for 15 min and genomic DNA was precipitated with 15 mL of 100% ethanol at RT for 5 min. DNA was transferred to a new tube and washed with 5 mL of 70% ethanol at RT for 5 min and air-dried. DNA was resuspended in 2 mL TEN20 (10 mM Tris-Cl pH 7.9, 2 mM EDTA, 20 mM NaCl, 0.1% SDS, 1000 U RNasin) at 70°C, boiled for 10–15 min and chilled on ice.
NS purification by sucrose gradient
One milliliter of denatured genomic DNA was loaded onto a 30-mL neutral 5 to 30% sucrose gradient prepared in TEN300 (10 mM Tris pH 7.9, 2 mM EDTA, 300 mM NaCl) and centrifuged in a Beckman SW28 rotor at 24,000 RPM, 4°C, for 20 h. One-milliliter fractions were withdrawn from the top of the gradient using a wide-bore pipette tip. Fifty microliters of each fraction was run with appropriate size markers on a 2% alkaline agarose gel at 40–50 volts, overnight at 4°C. The gel was neutralized with 1× TBE and stained with Gel Red (Interchim). Fractions corresponding to 0.5–2.5 kb were pooled and precipitated with 2.5 volumes ethanol at −80°C for 15 min. Pellets were washed with 1 mL of 70% ethanol and suspended in 20 μL of water with 100 U RNasin (NEB).
Lambda exonuclease treatment
After addition of 2 μL of T4 polynucleotide kinase (PNK) 10× buffer (NEB), fractions were boiled for 5 min and chilled on ice. Phosphorylation with T4 PNK was performed in 1× PNK buffer containing 0.2 U/μL PNK in a volume of 100 μL at 37°C for 1 h. After heat inactivation at 75°C for 15 min, DNA was precipitated with 2.5 volumes ethanol–0.3 M sodium acetate (Na-acetate) at −80°C for 15 min. Pellets were washed with 1 mL of 70% ethanol and suspended in 50 μL of water with 100 U RNasin. Digestion with 100 U lambda exonuclease was in exo buffer (67 mM Glycine-KOH pH 9.4, 2.5 mM MgCl2, 50 μg/mL BSA) in 100-μL total volume at 37°C overnight. We found that the quality of the lambda exonuclease is crucial and deserves to be always tested before use. For the experiments described here, we used a custom-made preparation by Fermentas (20 U/μL). NS were extracted once with phenol/chloroform/isoamylalcohol and once with chloroform/isoamylalcohol, then precipitated with 2.5 volumes ethanol–0.3 M Na-acetate at −80°C for 15 min. Pellets were washed with 1 mL of 70% ethanol and suspended in 20 μL of water. NS were subjected to one or two further cycles of T4 PNK phosphorylation and exonuclease digestion. We observed that the second round of exonuclease treatment significantly improves the NS preparation. Aliquots of digested and undigested DNA were run on a 2% agarose gel to confirm the efficiency of the exonuclease treatment. Finally, NS were purified using the CyScribe GFX Purification Kit (GE Healthcare) and eluted in 60 μL of water.
NS amplification and Chip data analysis
Ten microliters of purified NS were amplified using the WGAII kit (Sigma), omitting the first step of fragmentation. Amplification products were purified with NucleoSpin columns (Machrey Nagel). Proper unbiased amplification was monitored by qRT-PCR. Hybridization, washing and scanning of microarrays were done by Nimblegen Service Laboratory. Details about the Nimblegen microarrays used and the data analysis are available as Supplemental Data.
Quantitative real-time PCR analysis
qRT-PCR analysis of NS samples was performed using the SYBR Green PCR master mix (Roche) in a Lightcycler 480 real-time PCR thermocycler (Roche). For relative quantification, dilutions of total genomic DNA were used to construct the standard curves. One microliter of NS or genomic DNA was used per reaction and all experiments were done in triplicate.
DNA combing
The complete procedure is detailed in Supplemental Data. Briefly, asynchronous cell populations were labeled with 40 mM IdU for 20 min followed by a second 20-min pulse with 40 mM CldU. After staining of proteinase K-treated DNA plugs with YOYO-1 (Molecular Probes) and digestion with agarase (New England Biolabs), DNA fibers were combed on silanized cover slips (Michalet et al. 1997). Immunodetection was done with mouse anti-BrdU (Becton Dickinson) and rat anti-BrdU (Sera Lab) antibodies and DNA was stained with the anti-ssDNA antibody (Chemicon). Image acquisition was performed with a fully motorized Leica DM6000B microscope equipped with a CoolSNAP HQ CDD camera and controlled by MetaMorph (Roper Scientific). Inter-origin distances and fork speed were measured manually using the MetaMorph software.
Motif search
Enriched motifs in Oris were identified using the MEME bioinformatics suite (http://meme.sdsc.edu/meme4_4_0/cgi-bin/meme.cgi). The settings were: zero or one occurrence, motif length between 6 (minimum) and 50 pb (maximum). A fifth order Markov model was generated as a background distribution model to take into account repetitive sequences. From 5% of all Oris, 2 kb of DNA sequences centered on the NS peak were randomly selected. Independent analyses were performed which showed that the results were not dependent on the Oris sample. As an additional negative control, randomly selected genomic sequences were also analyzed. For each motif, an E-value was computed. E-values are commonly used for assigning significance to the optimal reported motifs. When the E-value is high, the confidence in the motif prediction is low, whereas low E-values are significant. Genomic frequencies of motifs were generated with the help of the FIMO server (http://meme.sdsc.edu/meme4_4_0/cgi-bin/fimo.cgi). Occurrences having P-value < 1 × 10−5 were used in this study. Overrepresented motifs were searched with R'MES (Hoebeke and Schbath 2006). The motif length was set to the maximum (13 nt); as background distribution, we used for Drosophila Oris a Markov model of the sixth order, in which the expected number of occurrences of each motif was estimated using the compound Poisson distribution.
Data access
The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE29183.
Acknowledgments
We thank E. Andermarcher for critical reading of this manuscript, Benjamin Leblanc and Aubin Thomas for their proficient help with bioinformatic analyses, Giacomo Cavalli for providing Drosophila Kc cells, as well as our laboratory members for helpful suggestions. We acknowledge the Montpellier DNA Combing Facility for providing silanized surfaces. C.C. was supported by the “Fondation pour la Recherche Médicale” (FRM) and P.C. was supported by the FRM and the “Association pour la Recherche contre le Cancer” (ARC). The research leading to these results has received funding from the European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013 Grant Agreement no. 233339). This work was also supported by the “Agence Nationale de la Recherche” (ANR) (ANR-08-BLAN-0092-0), the ARC and the “Ligue Nationale Contre le Cancer” (LNCC).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.121830.111.
References
- Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287: 2023–2026 [DOI] [PubMed] [Google Scholar]
- Aladjem MI 2007. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet 8: 588–600 [DOI] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M 2003. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 [DOI] [PubMed] [Google Scholar]
- Baker TA, Kornberg A 1988. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: An RNA-DNA hybrid near oriC. Cell 55: 113–123 [DOI] [PubMed] [Google Scholar]
- Benjamin Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B 57: 289–300 [Google Scholar]
- Berezney R, Dubey DD, Huberman JA 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol 38: 205–223 [DOI] [PubMed] [Google Scholar]
- Breier AM, Chatterji S, Cozzarelli NR 2004. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol 5: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL 1993. Initiation at closely spaced replication origins in a yeast chromosome. Science 262: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Bird AP 1995. CpG islands and genes. Curr Opin Genet Dev 5: 309–314 [DOI] [PubMed] [Google Scholar]
- Dai J, Chuang RY, Kelly TJ 2005. DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci 102: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazy S, Gandrillon O, Hyrien O, Prioleau MN 2006. Broadening of DNA replication origin usage during metazoan cell differentiation. EMBO Rep 7: 806–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B 2007. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet 3: e38 doi: 10.1371/journal.pgen.0030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S, Gomez M, Bird A, Antequera F 1998. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J 17: 2426–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML 1993. Eukaryotic DNA replication: Anatomy of an origin. Annu Rev Biochem 62: 29–63 [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Hamlin JL 1995. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol Cell Biol 15: 3023–3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Davey MJ, O'Donnell M 1999. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell 4: 541–553 [DOI] [PubMed] [Google Scholar]
- Friedman KL, Brewer BJ, Fangman WL 1997. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells 2: 667–678 [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ 2007. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Kemp M, Liu G, Ritzi M, Schepers A, Leffak M 2006. Differential binding of replication proteins across the human c-myc replicator. Mol Cell Biol 26: 5270–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M, Antequera F 2008. Overreplication of short DNA regions during S phase in human cells. Genes Dev 22: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire D, Brodolin K, Mechali M 2006. HoxB domain induction silences DNA replication origins in the locus and specifies a single origin at its boundary. EMBO Rep 7: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H 2009. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol 11: 357–362 [DOI] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P 2006. Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245 doi: 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebeke M, Schbath S 2006. R'MES: Finding Exceptional Motifs, version 3. User Guide. http://genome.jouy.inra.fr/ssb/rmes
- Hyrien O, Méchali M 1993. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J 12: 4511–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270: 994–997 [DOI] [PubMed] [Google Scholar]
- Hyrien O, Marheineke K, Goldar A 2003. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays 25: 116–125 [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP 2009. CpG islands—‘a rough guide'. FEBS Lett 583: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Laskey RA 1985. Chromosome replication in early development of Xenopus laevis. J Embryol Exp Morphol Suppl89: 285–296 [PubMed] [Google Scholar]
- Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A 2006. DNA replication origin interference increases the spacing between initiation events in human cells. Mol Biol Cell 17: 5337–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M 2005. Mitotic remodeling of the replicon and chromosome structure. Cell 123: 1–15 [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M 2011. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123 [DOI] [PubMed] [Google Scholar]
- Lobry JR 1996. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol 13: 660–665 [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Ezrokhi M, Smith HS, Gerbi SA 2002. Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol Cell Biol 22: 8426–8437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric C, Benard M, Pierron G 2003. Developmentally regulated usage of Physarum DNA replication origins. EMBO Rep 4: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M 2010. Eukaryotic DNA replication origins: Many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738 [DOI] [PubMed] [Google Scholar]
- Mesner LD, Li X, Dijkwel PA, Hamlin JL 2003. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol Cell Biol 23: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. 1997. Dynamic molecular combing: Stretching the whole human genome for high-resolution studies. Science 277: 1518–1523 [DOI] [PubMed] [Google Scholar]
- Nieduszynski CA, Knox Y, Donaldson AD 2006. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev 20: 1874–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL 2005. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell 20: 575–587 [DOI] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91: 311–323 [DOI] [PubMed] [Google Scholar]
- Rennekamp AJ, Lieberman PM 2011. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J Virol 85: 2837–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N 2006. DNA replication timing: Random thoughts about origin firing. Nat Cell Biol 8: 1313–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero J, Lee H 2008. Asymmetric bidirectional replication at the human DBF4 origin. Nat Struct Mol Biol 15: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schubeler D 2009. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev 23: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Kohler H, Oakeley EJ, Stadler MB, Schubeler D 2010. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res 20: 771–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F 2003. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep 4: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446 doi: 10.1371/journal.pgen.1000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K, Baker TA, Kornberg A 1990. Strand separation required for initiation of replication at the chromosomal origin of E. coli is facilitated by a distant RNA–DNA hybrid. EMBO J 9: 2341–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Nicolay S, Audit B, Brodie of Brodie EB, d'Aubenton-Carafa Y, Arneodo A, Thermes C 2005. Replication-associated strand asymmetries in mammalian genomes: Toward detection of replication origins. Proc Natl Acad Sci 102: 9836–9841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport JW 1997. Regulation of replicon size in Xenopus egg extracts. Science 275: 993–995 [DOI] [PubMed] [Google Scholar]