Abstract
Modification of microRNA sequences by the 3′ addition of nucleotides to generate so-called “isomiRs” adds to the complexity of miRNA function, with recent reports showing that 3′ modifications can influence miRNA stability and efficiency of target repression. Here, we show that the 3′ modification of miRNAs is a physiological and common post-transcriptional event that shows selectivity for specific miRNAs and is observed across species ranging from C. elegans to human. The modifications result predominantly from adenylation and uridylation and are seen across tissue types, disease states, and developmental stages. To quantitatively profile 3′ nucleotide additions, we developed and validated a novel assay based on NanoString Technologies' nCounter platform. For certain miRNAs, the frequency of modification was altered by processes such as cell differentiation, indicating that 3′ modification is a biologically regulated process. To investigate the mechanism of 3′ nucleotide additions, we used RNA interference to screen a panel of eight candidate miRNA nucleotidyl transferases for 3′ miRNA modification activity in human cells. Multiple enzymes, including MTPAP, PAPD4, PAPD5, ZCCHC6, ZCCHC11, and TUT1, were found to govern 3′ nucleotide addition to miRNAs in a miRNA-specific manner. Three of these enzymes–MTPAP, ZCCHC6, and TUT1–have not previously been known to modify miRNAs. Collectively, our results indicate that 3′ modification observed in next-generation small RNA sequencing data is a biologically relevant process, and identify enzymatic mechanisms that may lead to new approaches for modulating miRNA activity in vivo.
MicroRNAs (miRNAs) are small noncoding RNAs that play a regulatory role in gene function by binding to the 3′ untranslated region (UTR) of target mRNAs. Although the biogenesis and turnover of miRNAs has been the subject of much recent study (Chatterjee and Großhans 2009; Krol et al. 2010), mechanisms of post-transcriptional regulation of mature miRNAs are still not well understood. A few recent reports have suggested that post-transcriptional addition of nucleotides to the 3′ end of miRNAs is a mechanism for regulation of miRNA activity. Studies in plants and C. elegans have identified examples in which such modifications influence miRNA stability (Ramachandran and Chen 2008; Chatterjee and Großhans 2009; Lu et al. 2009; Ibrahim et al. 2010). In humans, effects on miRNA stability and on mRNA target repression have both been observed, dependent on the specific miRNA examined. For example, miR-122 was shown to be adenylated by the RNA nucleotidyl transferase PAPD4 (also known as GLD-2) in humans and mice, resulting in an increase in the stability of the miRNA (Katoh et al. 2009). In contrast, miR-26a was shown to be uridylated by the RNA nucleotidyl transferase ZCCHC11, which had no effect on miRNA stability, but rather reduced the ability of miR-26a to inhibit its mRNA target (Jones et al. 2009). Taken together, these studies indicate that understanding the prevalence and mechanisms of miRNA 3′ nucleotide additions will be important for understanding the post-transcriptional regulation of miRNA function, and could also inform strategies for therapeutic modulation of miRNA activity.
We and others have observed additions of nucleotides to the 3′ end of miRNAs on a global scale in animal miRNA high-throughput sequencing studies (Berezikov et al. 2006a,b; Cummins et al. 2006; Ruby et al. 2006; Landgraf et al. 2007; Bar et al. 2008; Wyman et al. 2009). For example, in our own high-throughput sequencing of small RNAs from human embryonic stem cells (hESC) and ovarian cancers (Bar et al. 2008; Wyman et al. 2009), we noticed substantial sequence variation at the 3′ end of the known miRNAs, frequently manifesting as nucleotide additions not matching the human genome sequence (i.e., 3′ nontemplated additions [3′ NTA]). However, it was not clear to what extent these were artifacts of cDNA library preparation as opposed to physiologic nucleotide additions. More recently, such miRNA variants resulting from 3′ NTA (so-called “isomiRs”) have been described in more detail in small RNA next-generation sequencing studies in humans, mice, and flies (Ruby et al. 2007; Wu et al. 2007; Kuchenbauer et al. 2008; Morin et al. 2008; Ahn et al. 2010; Burroughs et al. 2010; Chiang et al. 2010; Fehniger et al. 2010; Fernandez-Valverde et al. 2010; Guo and Lu 2010; Lee et al. 2010; Martí et al. 2010; Pantano et al. 2010; Berezikov et al. 2011). In one recent study (Burroughs et al. 2010) the enzyme PAPD4 was implicated as a miRNA nucleotidyl transferase, based on the finding that PAPD4 knockdown led to a decrease in the abundance of multiple miRNA variants bearing a nontemplated 3′ adenosine. However, not all miRNA variants were affected, and the extent of reduction was highly variable across miRNAs, suggesting that other enzymes may also be responsible for miRNA additions. Thus, the mechanisms of 3′ nucleotide additions on a global scale are still not well understood, even though the biological impact of such post-transcriptional modifications may be substantial.
In the present study, we systematically tested the extent to which miRNA 3′ NTA observed in sequencing data sets are physiologic additions versus artifacts of cDNA library preparation, and analyzed the additions on a global scale using multiple second-generation sequencing (i.e., 454 Life Sciences [Roche] and Illumina) data sets in human (Bar et al. 2008; Linsen et al. 2009; Pomerantz et al. 2009; Wyman et al. 2009), mouse (Baek et al. 2008), and C. elegans (Batista et al. 2008). To further investigate the 3′ end additions using an orthogonal platform ideally suited for quantitative analysis and high-throughput processing of samples, we adapted the nCounter Gene Expression Assay from NanoString Technologies (Geiss et al. 2008) to discriminate between and provide quantitative expression profiles of miRNA 3′ variants. The nCounter assay involves the hybridization of fluorescently labeled, bar-coded probes to the miRNAs of interest, which are then scanned and counted to quantify miRNA expression. We used the nCounter assay to profile 3′ end nucleotide additions during cell differentiation, which revealed that specific miRNAs undergo a change in 3′ NTA with differentiation, suggesting that 3′ NTA is a physiologically regulated process. We also conducted a systematic siRNA-based knockdown screen of RNA nucleotidyl transferases, using the nCounter assay as a readout, which identified multiple enzymes responsible for miRNA 3′ additions in human cells. Our results demonstrate the prevalence and mechanism of miRNA additions in humans, and introduce a new platform capable of quantitatively profiling miRNA 3′ NTA variants across a variety of biological samples.
Results
MicroRNA 3′ nontemplated nucleotide additions observed in next-generation sequencing data are a physiologic phenomenon
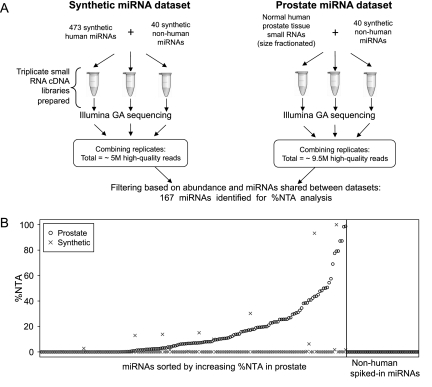
We sought to determine the extent to which nontemplated 3′ addition of nucleotides to miRNAs observed in next-generation small RNA sequencing data sets is a physiologic, in vivo phenomenon, as opposed to an artifact introduced during cDNA library preparation. We analyzed data from small RNA cDNA libraries prepared side-by-side and in triplicate from normal human prostate tissue RNA (Pomerantz et al. 2009) and from a pool of 473 chemically synthesized human miRNAs (Linsen et al. 2009) in order to compare 3′ NTA frequency between the physiologically derived versus chemically synthesized miRNAs (Fig. 1A). Each of the libraries also had 40 nonhuman chemically synthesized miRNAs spiked in (listed in the Supplemental Methods), with the expectation that if miRNA nucleotide addition were an artifact of cDNA library preparation, the spiked-in synthetic miRNAs would undergo additions in both sets of libraries. Sequencing on the Illumina Genome Analyzer platform produced 4,946,602 high-quality reads from the triplicate synthetic miRNA library and 9,417,361 high-quality reads from the triplicate prostate library, which were then analyzed for 3′ NTA using custom bioinformatics tools (details provided in the Supplemental Methods).
Figure 1.
%NTA observed in Illumina Genome Analyzer sequencing of a pool of chemically synthesized miRNAs or of prostate-tissue-derived miRNAs. (A) The schematic describes the workflow for the generation and sequencing of triplicate cDNA libraries corresponding to a pool of chemically synthesized miRNAs and normal human prostate tissue. (B, left) The %NTA for 167 shared miRNAs present at 10 or more reads in both the chemically synthesized miRNA and prostate sequencing data sets are plotted along the y-axis. Normal prostate tissue data are plotted as (○) and the synthetic miRNA data as (×). MicroRNAs are sorted along the x-axis with respect to increasing %NTA observed in prostate-tissue sequencing data. Only seven of the 167 miRNAs analyzed demonstrated greater %NTA in the synthetic miRNA pool compared with its prostate tissue %NTA. In most cases, these represented an additional nucleotide identical to the terminal nucleotide expected for the canonical miRNA sequence (data not shown), which is consistent with a typical type of error expected with solid-phase RNA oligonucleotide synthesis. (Right) The %NTA for 40 nonhuman (a mix of plant and C. elegans) spiked-in miRNAs is plotted for both normal prostate tissue and the synthetic miRNA sequencing data sets.
We compared the synthetic miRNA sequencing data to the prostate-tissue sequencing data, analyzing a set of 167 miRNAs that were shared between the two data sets and that passed filtering based on read abundance and inter-replicate variance criteria for these miRNAs (see Supplemental Methods for additional detail). For this set of 167 miRNAs, we calculated total matching reads (TMR) and percent nontemplated nucleotide additions (%NTA) for each miRNA in both data sets. TMR is the sum of: (1) all reads that exactly match the canonical miRNA sequence from miRBase (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008), plus (2) any matching reads that have nucleotide additions, regardless of whether those additions are templated (i.e., match the human genome sequence and miRNA precursor sequence) or nontemplated (i.e., do not match the human genome sequence and miRNA precursor sequence). %NTA is the fraction of TMR that has 3′ nucleotide additions that do not match the precursor sequence of the miRNA (hence, counting only nontemplated 3′ additions). We consider only nontemplated additions here because our interest is in unambiguous identification of post-transcriptional modifications to mature miRNAs.
Figure 1B depicts the %NTA for the 167 shared, abundant miRNAs sorted by increasing %NTA in the prostate sequencing data set. We observed a dramatic difference in the %NTA between the prostate tissue versus the chemically synthesized miRNA data sets, with the synthetic miRNAs only rarely demonstrating nucleotide additions, while over half of the prostate miRNAs examined had some extent of 3′ NTA. Notably, none of the 40 spiked-in synthetic nonhuman miRNAs showed any evidence of 3′ additions in either the prostate or chemically synthesized miRNA data sets (Fig. 1B). These data demonstrate that miRNA 3′ nontemplated nucleotide additions are predominantly a physiologic, in vivo process. In addition, we found no evidence of a relationship between miRNA abundance and the frequency of nontemplated nucleotide addition (Supplemental Fig. S1), also consistent with additions being a physiologic process rather than an artifact of library preparation.
Survey of miRNA 3′ NTA in multiple next-generation sequencing data sets representing diverse tissue types and species
To more broadly characterize miRNA 3′ NTA, we obtained and analyzed small RNA-sequencing data sets representing a diverse set of tissues, disease states, and developmental stages (Supplemental Table S1). These included cultures of normal human ovarian surface epithelial (HOSE) cells and tissue from three histological subtypes of ovarian cancer (Wyman et al. 2009); two prostate primary cell culture samples, one epithelial and one stromal; and undifferentiated and differentiated human embryonic stem cells (Bar et al. 2008). To assess the nature of miRNA 3′ NTA across diverse species, we also examined miRNA 3′ NTA in publicly available Illumina sequencing data sets from M. musculus (Baek et al. 2008) and C. elegans (Batista et al. 2008). For mouse, this consisted of 11 mouse cerebellum and medulloblastoma sequencing data sets representing diverse developmental stages, cell populations, and genotypes. For C. elegans, we examined 12 small RNA-sequencing data sets representing different developmental stages and genotypes. A complete list of all sequencing data sets and the data set identifiers used in this manuscript is given in Supplemental Table S1. Results of these analyses are presented below.
Nucleotide addition is miRNA specific
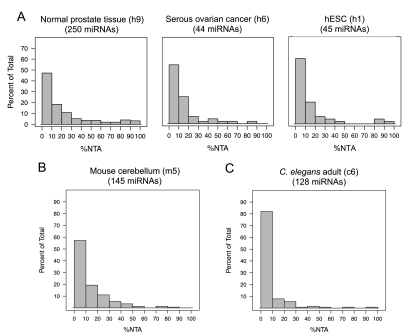
Analysis of miRNA-sequencing data sets demonstrated that although 3′ NTA occurs commonly, it is not universal and shows a strong predilection for specific miRNAs over others. In Figure 2A, histograms for three individual human data sets show that certain miRNAs have a very high frequency of additions (%NTA), while the majority of miRNAs are modified at a low frequency, if at all (histograms for six additional human data sets are shown in Supplemental Fig. S2A). A similar distribution of frequencies was observed when we analyzed 11 mouse cerebellum and medulloblastoma sequencing data sets representing several developmental stages and genotypes. The histogram for one of the 11 data sets analyzed is shown in Figure 2B and histograms for the other 10 are shown in Supplemental Fig. S2B. We also examined 12 C. elegans small RNA-sequencing data sets representing different developmental stages and genotypes. In C. elegans, the overall frequency of 3′ NTA observed was less than in human and mouse, but showed a similar distribution across sequencing data sets from seven developmental stages and multiple genotypes in which certain miRNAs show a high frequency of modification, whereas the majority of miRNAs are infrequently or never modified. A representative histogram of miRNA %NTA for the L1 stage of C. elegans larval development is shown in Figure 2C, with histograms for all 12 data sets shown in Supplemental Fig. S2C.
Figure 2.
3′ Nontemplated nucleotide additions are miRNA specific. (A) Histograms of %NTA observed in three representative human miRNA-sequencing data sets (normal prostate tissue [h9], serous ovarian cancer [h6], and human embryonic stem cells [h1]). MicroRNAs were required to have 10 or more reads in a given data set to be included, and the number of miRNAs that qualified is given in parentheses for each histogram. The y-axis scale is the same for all three human histograms. (B,C) Histograms of %NTA for representative data sets corresponding to mouse cerebellum or medulloblastoma (B) (m5 in Supplemental Table S1) and the L1 developmental stage of C. elegans (C) (c6 in Supplemental Table S1). MicroRNAs were required to have 10 or more reads in a given data set to be included. Data set identifiers are provided in parentheses for all histograms and refer to descriptions of these data sets provided in Supplemental Table S1.
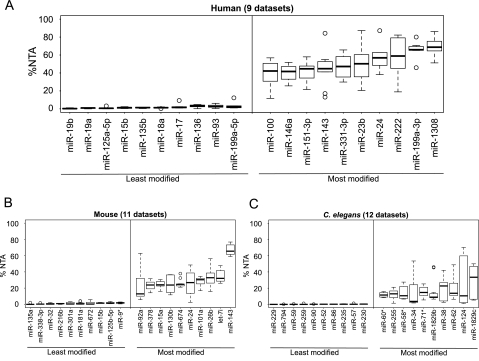
To determine whether there are some miRNAs that consistently show a high degree of 3′ NTA or consistently show an absence of it, we compared the %NTA of miRNAs across nine human sequencing data sets from different cell lines, tissues, and disease states (Supplemental Table S1, data sets h1–h9). A set of 73 miRNAs was detected with 40 or more reads in at least five of the nine data sets. Among the set of 73, we observed that certain miRNAs are consistently frequently modified across diverse tissue and cell types, while others are consistently rarely modified. A boxplot of %NTA for the 10 most and 10 least modified of these 73 miRNAs is shown in Figure 3A. The 10 miRNAs that are most often highly modified in our human-sequencing data sets are miR-100, miR-146a, miR-151-3p, miR-143, miR-331-3p, miR-23b, miR-24, miR-222, miR-199a-3p, miR-1308. Supplemental Table S2 gives the total matching reads (TMR) and %NTA for all miRNAs in each of these nine data sets.
Figure 3.
MicroRNA-specific 3′ nontemplated nucleotide addition is seen across small RNA sequencing data sets. (A) A boxplot of the miRNAs demonstrating the lowest %NTA (i.e., least modified) and highest %NTA (i.e., most modified) across nine human sequencing data sets (data sets h1–h9 in Supplemental Table S1). MicroRNAs were required to have at least 40 reads in five or more of the nine data sets. (B,C) Boxplots of the miRNAs demonstrating the lowest %NTA (i.e., least modified) and highest %NTA (i.e., most modified) across 11 mouse cerebellum or medulloblastoma data sets (B) (data sets m1–m11 in Supplemental Table S1) and 12 C. elegans sequencing data sets (C) (data sets c1–c12 in Supplemental Table S1). MicroRNAs were required to have at least 40 reads in five or more of the data sets for mouse and for C. elegans to be included in the analysis.
In mouse, there was a set of 81 miRNAs detected at 40 or more reads in at least five of the 11 sequencing data sets. The %NTA boxplot for the 10 most and 10 least modified of these 81 miRNAs is shown in Figure 3B. As observed in the human data sets, specific murine miRNAs were consistently highly modified across different biological samples. These miRNAs were only infrequently the mouse homologs of the most frequently modified human miRNAs, although this could be related to the expression of different sets of miRNAs between the mouse and human data sets analyzed. The TMR and %NTA for all miRNAs in each of the 11 mouse data sets is given in Supplemental Table S3. In C. elegans, we again saw that particular miRNAs were consistently among the most frequently modified miRNAs across developmental stages, as shown by the boxplot in Figure 3C. Supplemental Table S4 gives the TMR and %NTA for all miRNAs in each of the 12 C. elegans data sets.
3′ NTA is predominantly the result of adenylation and uridylation in human, mouse, and C. elegans
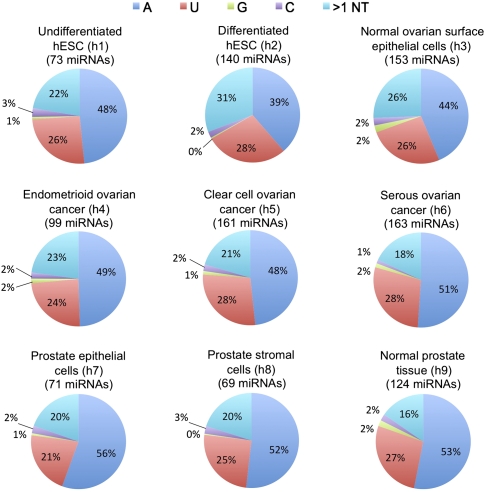
We next assessed specifically which nucleotides were added to generate 3′ NTA miRNA variants. The fractional contribution of each type of addition (A, U, G, C, and >1 nt) was calculated for each miRNA, and then averaged over all of the miRNAs detected at an abundance of at least 10 reads in each data set. Figure 4 shows the distribution of nucleotide additions across the nine human sequencing data sets. We found that mono-adenylation (blue) or mono-uridylation (red) were by far the most common type of 3′ NTA. To illustrate the distribution of additions on a per-miRNA basis, the complete list of nucleotide additions for abundant miRNAs (defined as 20 or more reads) in the normal prostate tissue sequencing data set is given in Supplemental Table S5. Throughout the nine human data sets examined, the distribution of added nucleotides was remarkably uniform, with mono-adenylation typically accounting for ∼50% of 3′ NTA and mono-uridylation for ∼25% of 3′ NTA. Chiang et al. (2010) reported finding uridine to be the most commonly added nucleotide calculated by summing all of the reads with added nucleotides for each of the four individual nucleotides for all of the miRNAs, and then calculated the percent from the sum. When the fractions of added nucleotides for the Chiang et al. (2010) manuscript are recalculated using our method described above, we found that adenosine is the most frequently added nucleotide for the mouse brain sequencing data sets of Chiang et al. (2010).
Figure 4.
Distribution of added nucleotides observed in nine human small RNA sequencing data sets. Distribution was calculated based on miRNAs with an abundance of at least 10 reads, and the number of miRNAs which qualified is given in parentheses for each pie chart. Fractional contribution of each nucleotide addition was calculated for each miRNA, and then averaged for all the miRNAs in each data set. Pie charts indicate what fraction of total additions each mononucleotide represented (on average) for each data set, and all multinucleotide additions were counted in the >1 category. Parenthetical data set identifiers refer to descriptions provided in Supplemental Table S1.
In mouse, we observed a similar distribution of predominant mono-adenylation and mono-uridylation (Supplemental Fig. S3A). In four of the mouse cerebellum sequencing data sets, however, an increased prevalence of mono-guanylation was observed (Supplemental Fig. S3A; data sets m3–m6 in Supplemental Table S1). These data sets were from specific mouse embryonic developmental stages (which were not represented in the human data sets) and were largely accounted for by three miRNAs (mmu-miR-16, mmu-miR-103, and mmu-miR-181a), which, although they were observed to be guanylated, had a low overall %NTA. Nonetheless, the results raise the possibility that although adenylation and uridylation are the most common modes of 3′ NTA in specific contexts and for specific miRNAs, additional types of 3′ NTA may be activated, perhaps with different biological consequences.
In small RNA-sequencing data sets corresponding to diverse developmental stages of C. elegans, we again observed mono-adenylation and mono-uridylation to be the most common forms of 3′ NTA (Supplemental Fig. S3B). Interestingly, however, the predominant form of modification in C. elegans was mono-uridylation (∼40%), followed by mono-adenylation (∼25%), compared with primarily mono-adenylation (∼40%–50%) and, secondly, mono-uridylation (∼25%) in human and mouse (Fig. 4; Supplemental Fig. S3A,B). This suggests that although we found miRNA 3′ nucleotide addition to be an evolutionarily conserved phenomenon, the degree to which specific mechanisms such as adenylation and uridylation are used for generating 3′ miRNA variants may differ depending upon the species.
Development of a quantitative platform to profile relative abundance of miRNA variants resulting from 3′ nucleotide additions
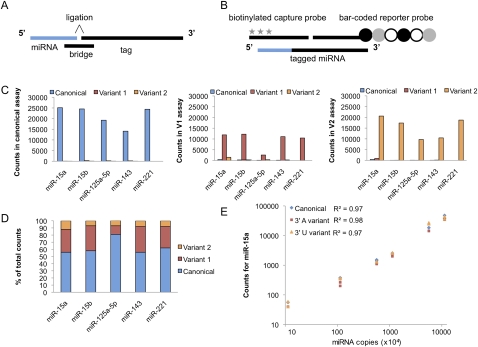
In order to profile 3′ nontemplated nucleotide additions to miRNAs in a more quantitative and high-throughput manner than is currently practical by next-generation sequencing, we developed a novel version of NanoString's nCounter miRNA Expression Assay. NanoString's hybridization-based technology uses color-coded, fluorescently labeled probe pairs to bar code selected target molecules, which are then scanned and counted (Geiss et al. 2008). The complexity of the bar codes, each containing one of four colors in each of six positions, allows a large diversity of target molecules present in the same sample to be individually distinguished during data collection. The NanoString miRNA assay uses an additional sample processing step to allow the detection of small RNAs (Fig. 5A). Preparation of the small RNA samples involves the ligation of a specific DNA tag onto the 3′ end of each mature miRNA; these tags are designed to normalize the melting temperatures of the targeted miRNA as well as to provide a unique identification for each miRNA species in the sample. Tagging is accomplished in a multiplexed ligation reaction using reverse-complement bridge oligonucleotides to direct the ligation of each miRNA to its designated tag. Following the ligation reaction, excess tags and bridges are removed, and the resulting material is hybridized with a panel of miRNA:tag-specific nCounter capture and bar-coded reporter probes (Fig. 5B). Following purification, each captured bar code is counted and tabulated in the nCounter assay (Geiss et al. 2008).
Figure 5.
Development of the nCounter miRNA assay to detect miRNA 3′ variants. (A) A schematic of the molecular components of the nCounter miRNA assay. The miRNA is shown in blue and DNA oligonucleotides in black. Each bridge oligonucleotide serves to template the 3′ end ligation of a particular miRNA species to a sequence-specific tag. Custom bridges were designed to discriminate between the 3′ end miRNA variants. (B) Following the removal of the excess tags and bridges, the tagged miRNAs are hybridized to specific capture and reporter probes attached to unique bar codes. The captured bar codes are individually resolved and counted in the NanoString nCounter assay system. The six-spot fluorescent bar code is represented by circles and the biotin capture moieties by stars. (C) Validation of the specificity of the nCounter assay. Three pools of synthetic RNA oligonucleotides, each containing canonical, variant 1, or variant 2 versions of five miRNAs were assayed. The graphs display the counts resulting when each of the three mixtures was individually assayed using the canonical (left), variant 1 (center), and variant 2 (right) bridge pools. (D) Validation of the accuracy of the nCounter assay. A mixture containing 60% canonical, 30% variant 1, and 10% variant 2 chemically synthesized miRNAs were assayed in each bridge pool. The relative abundance of each variant measured in the assay was then determined. (E) Validation of the linear range of the nCounter assays for miR-15a. Standard curves of synthetic oligonucleotides corresponding to the canonical, 3′ A (variant 1), or 3′ U (variant 2) form of miR-15a were assayed in their appropriate bridge pool. The graph displays the counts resulting from technical duplicates of standard curves ranging from 1 × 105 to 1 × 108 input copies of miRNA per reaction.
The standard nCounter miRNA assay detects only those miRNAs with canonical 3′ ends, as the tag ligation is a 3′ end-specific event. We modified the standard assay to be able to measure two 3′ variants each (arbitrarily designated variant 1 and variant 2). To measure the miRNA variants, we created pools of bridges that direct the tagging of the variant miRNA to the same tag as the canonical miRNA. Each sample is split in three and assayed separately with the canonical bridge pool, the variant 1 pool, and the variant 2 pool. By labeling each 3′ variant of a given miRNA species with the same tag in three parallel assays, we eliminate any potential tag-to-tag variation and ensure a direct comparison of the relative levels of each miRNA variant. A titration of six synthetic miRNA spike-ins is used in each assay as a control for assay-to-assay variation.
In initial pilot studies, we assayed a set of five synthetic miRNAs (miR-15a, miR-15b, miR-125a-5p, miR-143, and miR-221) and two variants for each miRNA. We validated the specificity of the platform by assaying three pools of synthetic RNA oligonucleotides representing canonical, variant 1, and variant 2 versions of our five pilot miRNAs (Fig. 5C). Each bridge pool (i.e., specific to canonical, variant 1, or variant 2) was used to assay the three mixtures of synthetic miRNA oligonucleotides (Fig. 5C). We found that each bridge pool distinguished the miRNA species of interest with high specificity, with minimal background detection of the other variants.
In order to assess accuracy, we also assayed mixtures of synthetic miRNAs containing 60% canonical miRNA, 30% variant 1, and 10% variant 2 for the set of five miRNAs. In four of the five cases, the ratios of canonical to variant miRNAs observed in the assay matched closely the known composition of the synthetic mixtures (Fig. 5D). Finally, to determine the linear range of the platform, we generated six-point standard curves for miR-15a and its two 3′ variants. Each standard curve was assayed in its respective bridge pool, with between 1 × 105 and 1 × 108 input copies of miRNA per reaction (Fig. 5E). We found that all three assays (canonical, variant 1, and variant 2) yielded a strong linear response to increases in concentration across our entire range of input values. These pilot studies demonstrate that the nCounter miRNA 3′ variant assay provides a quantitative platform for studying miRNA 3′ NTA.
Following our pilot experiments, we expanded the bridge pools to assay a set of 132 human miRNAs. The list of 132 miRNAs and their corresponding variants chosen for assay development is given in Supplemental Table S6. The miRNAs were chosen on the basis of showing an appreciable frequency of 3′ NTA in our next generation small RNA-sequencing data sets from human samples (h1–h9, Supplemental Table S1), as well as several miRNAs of special biological interest because of disease association or tissue specificity. The specific 3′ NTA variants for analysis were chosen largely on the basis of being the most common variants for specific miRNAs observed in the human small RNA sequencing data sets.
The frequency of 3′ NTA to specific miRNAs changes with differentiation of human embryonic stem cells, suggesting that post-transcriptional nucleotide addition is a physiologically regulated process in humans
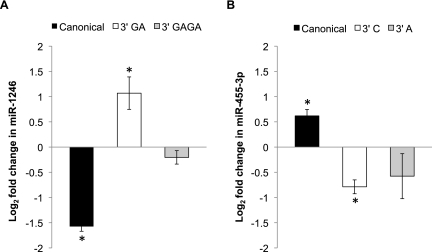
To investigate whether miRNA 3′ additions are a physiologically regulated process, we determined whether the frequency of miRNA 3′ NTA varies with a change in biological state, using differentiation of embryonic stem cells as a model system. We used the NanoString nCounter assay to profile 3′ NTA in undifferentiated H1 human embryonic stem cells versus spontaneously differentiated cells derived from the same cell line. To ensure that we restricted our analysis to miRNAs and miRNA variants for which robust measurements were available, we filtered the data to consider only the miRNAs for which at least 50 counts were reliably obtained in at least two out of three bridge pools (i.e., canonical, variant 1, or variant 2) in both of the data sets (i.e., the undifferentiated and differentiated hESC data sets). This comprised 10 miRNAs and their corresponding variants (given in Supplemental Table S7). We found that the fractional abundance of five miRNA variants changed significantly after differentiation (P < 0.05 in at least one variant assay for a given miRNA) (Supplemental Table S7). Depending on the specific miRNA, cell differentiation could be associated with either an increase or a decrease in variant abundance. The two most marked examples were miR-1246 (differentiation was associated with a significant increase in the fractional abundance of one of the variants and decrease in that of the canonical sequence) (Fig. 6A) and miR-455-3p (differentiation was associated with a decrease in fractional abundance of both the variants and an increase in that of the canonical sequence) (Fig. 6B). These results suggest that nontemplated nucleotide additions to different miRNAs could be mediated by unique mechanisms. More importantly, our results demonstrate that miRNA 3′ additions in human cells are dynamic and can vary in response to biological stimuli, indicating that 3′ NTA is a physiologically regulated process.
Figure 6.
MicroRNA additions are altered in response to differentiation. The NanoString nCounter assay was used to profile differential expression of miRNA 3′ variants in undifferentiated versus differentiated H1 human embryonic stem cells. MicroRNAs were filtered by requiring at least 50 counts in at least two of the three assays (canonical, V1, or V2) in both the undifferentiated and differentiated hESC data sets. The plotted miRNAs are the two showing the greatest change in the fractional abundance of 3′ variants following differentiation. Error bars represent ±SD of the fold change across two biological replicates, and asterisks indicate P-values <0.05. Supplemental Table S7 provides data corresponding to all 10 miRNAs that met filtering criteria, including three additional miRNAs showing significant changes in relative variant abundance. (A) The graph shows the fold change in the canonical and 3′ variant forms of miR-1246. For miR-1246, differentiation is associated with a significant increase in the fractional abundance of the 3′ GA variant, and a corresponding decrease in the canonical sequence. (B) For miR-455-3p, differentiation is associated with a significant decrease in the abundance of the 3′ C variant and a corresponding increase in the canonical miRNA fractional abundance.
Identification of nucleotidyl transferases responsible for miRNA 3′ NTA
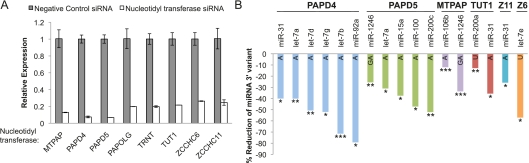
Previous studies have suggested that members of the RNA nucleotidyl transferase family, including PAPD4, PAPD5, and ZCCHC11, can mediate miRNA 3′ additions in humans (Jones et al. 2009; Katoh et al. 2009; Burroughs et al. 2010). However, RNA nucleotidyl transferases comprise a much larger family of enzymes, and the existing data on these three enzymes do not fully account for the widespread 3′ NTA of miRNAs observed on a genomic scale. We hypothesized that additional nucleotidyl transferases are involved in mediating miRNA 3′ nontemplated nucleotide additions, and that even for PAPD4, PAPD5, and ZCCHC11, additional miRNA substrates may exist beyond ones that have been identified to date. To understand the mechanism of 3′ NTA to a range of different miRNAs, we used RNA interference to individually suppress expression of eight different nucleotidyl transferases in HCT-116 colon-cancer cells, followed by quantitative analysis of canonical and variant miRNAs using the nCounter miRNA variant analysis platform developed above. These eight candidate miRNA 3′ modifying enzymes represent all but four of the 12 known RNA nucleotidyl transferases in humans (Martin and Keller 2007). These specific enzymes were chosen based on a high likelihood of having specific effects (e.g., poly(A) polymerase was excluded because of expected broad effects on polyadenylation), evidence for robust expression in HCT-116 cells (Supplemental Table S8), and availability of effective reagents for RNA interference. As a negative control, we performed RNA interference directed against cyclophilin B (a commonly used negative control gene in siRNA experiments), which is not expected to affect miRNA nucleotide addition.
We confirmed the success of knockdown with quantitative reverse transcription–PCR (qRT–PCR) (Fig. 7A), which demonstrated at least 75% reduction in expression of each enzyme compared with cells transfected with negative control siRNA targeting cyclophilin B (also known as peptidylprolyl isomerase B). We also used qRT–PCR to assess the specificity of the nucleotidyl transferase knockdowns by measuring all eight nucleotidyl transferases in each knockdown experiment (Supplemental Fig. S4). Overall, we found the knockdown of each enzyme to be highly specific (i.e., knockdown of a given nucleotidyl transferase was not associated with a concomitant decrease in other nucleotidyl transferases; Supplemental Fig. S4). Interestingly, in a few cases knockdown of one enzyme was associated with an increase in expression of one or more other nucleotidyl transferase(s) (e.g., knocking down PAPD4 led to an increase in PAPD5, while TUT1 and MTPAP knockdown led to an increase in PAPOLG), raising the possibility that compensation of function by one enzyme for another may occur in some cases. This is worth mention because it suggests that our results may underestimate the full effect of knockdown of individual nucleotidyl transferases on 3′ NTA, because of such cases of potential compensatory increase in expression of other enzymes.
Figure 7.
MicroRNA 3′ nontemplated nucleotide additions are regulated by multiple nucleotidyl transferase enzymes. (A) qRT–PCR demonstrates the successful suppression of eight different nucleotidyl transferases in HCT-116 cells transfected, with siRNAs individually targeting each enzyme. Bars represent mean expression ±SD of biological replicates relative to cells transfected with siRNAs against the cyclophilin B negative control. Normalization was performed using qRT–PCR quantification of the endogenous control gene GUSB to control for variations in RNA input. (B) Bar graphs for each enzyme show percent reduction in miRNA 3′ variants following suppression of each of the six nucleotidyl transferases indicated above the x-axis. MicroRNAs shown represent variants that (1) were significantly reduced (FDR < 10%) in the cells treated with siRNAs targeting the enzyme of interest versus those treated with the negative control, siCyclophilin, and (2) showed at least a 5% absolute decrease in fractional abundance of the variant miRNA. The letter(s) within each bar for a given miRNA identifies which 3′ variant displays significant reduction. Z11 refers to the enzyme ZCCHC11 and Z6 to ZCCHC6. (*) miRNAs meeting criteria of FDR < 10%; (**) FDR < 5%; (***) FDR < 2%. The false discovery rate (FDR) is determined using the method of Benjamini and Hochberg. The change in fractional abundance and associated FDR for all canonical and 3′ miRNA variants retained after filtering for abundance is given in Supplemental Table S9.
We used the NanoString platform to monitor changes in miRNA 3′ NTA fractional abundance as a result of the suppression of each RNA nucleotidyl transferase. MicroRNAs were filtered by retaining only those that had a count of at least 50 for the canonical in all four of the negative control (i.e., siCyclophilin) knockdown lanes. We calculated the percentage of each variant miRNA as a fraction of the total abundance of the miRNA (i.e., counts obtained for canonical plus variant 1 counts plus variant 2 counts), which ensured that any decrease in a given variant observed with knockdown of specific nucleotidyl transferases would reflect a change in propensity for nucleotide addition, rather than being due to an overall decrease in all forms of the miRNA as a result of decreased transcription, for example. The relative percentages of each canonical and variant species found in the knockdown cells were then compared with negative control cells treated with siRNAs targeting cyclophilin B. A t-test conducted across groups representing biological replicates was performed, followed by an adjustment for multiple hypothesis testing based on the false discovery rate (FDR) method of Benjamini and Hochberg (Benjamini and Hochberg 1995; van der Laan et al. 2004) to identify nucleotidyl transferase knockdowns associated with a significant reduction (FDR < 10%) of specific miRNA 3′ NTA variants. The most compelling cases were considered to be those knockdowns in which a statistically significant decrease in at least one 3′ NTA variant was observed along with a concomitant statistically significant increase in the fractional abundance of the canonical miRNA sequence.
We found that suppression of seven out of eight nucleotidyl transferases led to a significant reduction in at least one miRNA 3′ variant (FDR < 10%). In addition to PAPD4, PAPD5, and ZCCHC11, which have previously been described as affecting miRNA additions (Jones et al. 2009; Katoh et al. 2009; Burroughs et al. 2010), we found significant reductions in miRNA variants after suppression of MTPAP, PAPOLG, TUT1, and ZCCHC11 (Supplemental Table S9). To highlight nucleotidyl transferases showing the most substantial effect on 3′ NTA of specific miRNAs, we selected cases in which effects of enzyme knockdown on specific miRNA variants were both statistically significant (FDR < 10%) and had at least a 5% reduction in the fractional abundance of the variant (Fig. 7B). Loss of PAPD4 resulted in the most dramatic changes in miRNAs with 3′ A additions: six miRNAs with a 3′ A addition showed significant decreases, with some miRNA variants such as miR-92a and let-7b showing >70% reduction (Fig. 7B). Suppression of PAPD5 led to significant reductions in four miRNA 3′ variants which all featured a 3′ A, and one miRNA variant, miR-1246, with a dinucleotide 3′ GA (Fig. 7B). The variant miR-1246 with a 3′ GA also showed a significant reduction with the MTPAP knockdown. While both PAPD4 and PAPD5 suppression predominantly affected variants with a 3′ A, knockdown of two enzymes (TUT1 and ZCCHC6) was associated with a decrease in 3′ uridylation. Suppression of TUT1 led to a significant reduction in the variant of miR-200a with a 3′ U, and loss of ZCCHC6 decreased expression of let-7e with a 3′ U (Fig. 7B), which suggests that different enzymes may show specificity for certain nucleotide additions. Taken together, our results suggest multiple enzymes affect the post-transcriptional modification of miRNAs by 3′ NTA.
To validate the accuracy of the assays in Figure 7B, we tested the ability of our adapted nCounter miRNA variant profiling assay to distinguish between the canonical and variant forms of the miRNAs from Figure 7B. We profiled pools of chemically synthesized versions of the canonical and variant miRNAs of interest and, again, the assay demonstrated excellent discrimination between miRNA 3′ end nucleotide variants (Supplemental Fig. S5). We also tested the linear range of these miRNA assays by creating pools of synthetic miRNAs (containing the canonical or variant version of each miRNA) and generating 6-point standard curves assaying from 1 × 105 to 1 × 108 input copies of each miRNA or miRNA 3′ variant per reaction. We assayed each dilution point of the standard curves in the appropriate bridge pool, and found that all assays showed a strong, linear response to changes in concentration (Supplemental Fig. S6).
Discussion
Our study provides confirmation on a broad scale that miRNA 3′ nontemplated nucleotide additions that have been observed in small RNA next-generation sequencing data are a physiologic and biologically regulated phenomenon in human cells, complementing and extending work presented in other reports (Kuchenbauer et al. 2008; Morin et al. 2008; Burroughs et al. 2010; Chiang et al. 2010; Fehniger et al. 2010; Fernandez-Valverde et al. 2010; Lee et al. 2010; Martí et al. 2010; Berezikov et al. 2011). We find that across multiple species, these modifications are miRNA specific and that adenylation and uridylation are the primary modes of 3′ NTA, which is consistent with the previously described specificities of known RNA nucleotidyl transferase enzymes in their modification of other RNA substrates (Martin and Keller 2007). We attempted to identify specific sequence features in miRNAs that might govern modification. However, we did not find any motifs or sequence characteristics that showed significant association with modifications (data not shown). It is of interest, however, that the degree to which adenylation versus uridylation is used for miRNA modification varies across species, with uridylation predominating among C. elegans miRNAs, whereas adenylation is more prominent in mouse and human. Although the basis for this difference is not yet known, we speculate that cross-species differences in expression patterns of nucleotidyl transferases and their miRNA substrates may underlie this observation.
The development of a novel, bridged ligation-based assay permitted us to quantitatively profile 3′ NTA variants (along with their corresponding canonical miRNA species) following knockdown of a panel of candidate miRNA nucleotidyl transferases in order to investigate the mechanism of 3′ NTA. Prior studies had primarily implicated two nucleotidyl transferases, PAPD4 and ZCCHC11, and to a lesser extent, PAPD5, in miRNA 3′ NTA (Katoh et al. 2009; Burroughs et al. 2010). Strikingly, we found that knockdown of seven of the eight RNA nucleotidyl transferases we examined (including PAPD4, ZCCHC11, and PAPD5) was associated with a significant reduction in 3′ NTA of one or more miRNA variants. Individual enzymes tended to show specificity for particular miRNAs, and the involvement of multiple additional enzymes points toward much more complexity in mechanisms of miRNA 3′ NTA than has previously been appreciated. One caveat of our analysis is that we have examined only the steady-state levels of 3′ NTA. Thus, the mechanisms by which each enzyme modulates the observed miRNA 3′ NTA frequency—whether it be via altered kinetics of 3′ NTA or effects on miRNA degradation—remain to be determined. Remarkably, MTPAP is known to localize to the mitochondria and to polyadenylate the 3′ ends of mitochondrial transcripts (Tomecki et al. 2004), and thus it was surprising to find that suppression of this heretofore mitochondria-specific enzyme affected miRNA additions. TUT1, on the other hand, was found to regulate both 3′ A and 3′ U additions, which supports previous reports that describe TUT1 both as a uridyltransferase and as a nuclear poly(A) polymerase (Trippe et al. 1998; Mellman et al. 2008). Additionally, we identified the first known role for ZCCHC6, an enzyme with unknown substrates that is homologous to the miRNA-modifying enzyme ZCCHC11 (Rissland et al. 2007).
Our study identified four new miRNA nucleotidyl transferases (MTPAP, TUT1, ZCCHC6, and PAPOLG) that have significant effects on 3′ NTA to specific miRNAs, supporting the notion that 3′ NTA represents a multifaceted layer of post-transcriptional regulation for miRNAs by an ensemble of miRNA-modifying enzymes. Our study likely represents a minimal estimate of the enzymes and miRNA substrates involved, as there was evidence in several cases of a compensatory increase in the expression of other nucleotidyl transferases after knockdown of a given enzyme, which could have obscured some of the effects on 3′ NTA. Additionally, some of the enzymes that did show an effect may be false negatives as a result of incomplete suppression of protein levels in siRNA experiments. Importantly, the finding that 3′ NTA is a regulated and miRNA-specific process mediated by distinct enzymes raises the possibility that inhibiting particular RNA nucleotidyl transferases could provide a new strategy for modulating miRNA activity for therapeutic as well as research purposes. In this vein, the enzymes we have identified may be worth investigating as targets for small molecules to potentially modify miRNA function via inhibiting (or potentially enhancing) 3′ NTA. Nucleotidyl transferases may also show differential expression in different disease states. In a brief survey of Oncomine (Rhodes et al. 2007), for example, we found ZCCHC6 to be in the top 1% of genes overexpressed in prostate carcinoma versus normal tissue (Tomlins et al. 2007). Thus, we speculate that differential expression of nucleotidyl transferases could impact miRNA 3′ NTA in different biological states. In the future, quantification of 3′ NTA variants in samples from varied biological sources using the NanoString assay developed here and/or other technologies will facilitate understanding of the potential biological effects of these enzymes.
Quantification of 3′ NTA variants distinct from canonical miRNAs in biological samples poses a problem for current technologies. Although next-generation sequencing can certainly identify 3′ variants, it is not currently an assay for which parameters such as quantitative precision, quantitative accuracy, linear range of quantification, or absolute quantification are well established. Additionally, the throughput and turnaround time for analysis of large numbers of biological or clinical samples is still limited in practice. Although quantitative reverse transcription–PCR (qRT–PCR) approaches are routinely used for miRNA quantification, currently available qRT–PCR platforms are not equipped to measure and discriminate between variants with high accuracy (Wu et al. 2007; Lee et al. 2010). The NanoString assay we have introduced here is an important advance for enabling further studies in this area, as it permits quantitative analysis of miRNAs and variants on a platform that has sufficient throughput and precision to be routinely used for analysis of large numbers of biological samples.
Although 3′ NTA can clearly have functional effects on animal miRNAs by increasing miRNA stability in the case of hsa-miR-122 (Katoh et al. 2009), or affecting the efficacy of mRNA target repression in the case of hsa-miR-26a (Jones et al. 2009), investigation of the biological functions of miRNA 3′ NTA is still at a nascent stage. Future studies will be required to determine whether 3′ NTA affects the expression of miRNA targets or the stability of miRNAs on a global scale. A recent study has noted an increase in uridylated miR-223 during miRNA decay (Baccarini et al. 2011). Although our analysis has evaluated only the steady-state levels of miRNA additions, we speculate that 3′ NTA may serve as either a signal for or a consequence of targeting of miRNAs for degradation. The functional effects of 3′ NTA on miRNAs may be varied and miRNA specific, making such modifications a potentially versatile mechanism for generating functional diversity and complexity in an otherwise relatively limited miRNA transcriptome. Our study has identified a multitude of new miRNA variants along with the nucleotidyl transferases that may mediate their genesis. These findings, along with our introduction of a quantitative platform for miRNA variant analysis, should enable further studies of the biological roles of miRNA 3′ NTA.
Methods
Preparation of samples and library construction
Preparation and sequencing of the cDNA libraries corresponding to the synthetic miRNA pool and normal prostate tissue small RNAs in this study have already been described (Linsen et al. 2009; Pomerantz et al. 2009). Briefly, 473 human and 40 nonhuman miRNAs (four A. thaliana, 26 C. elegans, and 10 O. sativa) were synthesized and pooled at equal concentrations of 2.14 nM to generate a synthetic miRNA library (Linsen et al. 2009). The 40 nonhuman miRNAs at varying concentrations (50 nM–190.7 fM) were 5′ phosphorylated by T4 polynucleotide kinase (Invitrogen) and spiked with radiolabeled 18–24-nt markers, and gel purified. The resulting synthetic pools were spiked into normal prostate RNA derived from two individuals, and the 18–24-nt fraction was gel purified. Normal prostate RNA was from histologically confirmed normal prostate tissue obtained with informed consent under IRB supervision at the time of radical prostatectomy for clinically localized prostate cancer. Linker ligation and amplification of both the synthetic miRNA pool and prostate RNA samples were performed as described in Mitchell et al. (2008). Small RNA cDNA libraries were sequenced on the Genome Analyzer (Illumina) according to the manufacturer's instructions. Small RNA cDNA libraries for 454 sequencing from cultured prostate epithelial and stromal cells were generated as described previously (Bar et al. 2008). RNA from prostate epithelial cell and prostate stromal cell cultures was a generous gift from B. Knudsen. Small RNA cDNA libraries were sequenced on the 454 platform according to the manufacturer's instructions.
Processing of the sequencing data
Processing of the sequencing data was performed using a set of custom bioinformatics scripts (detailed in the Supplemental Methods). Briefly, sequence reads were pruned of any relevant linker sequences and then compared with canonical miRNA sequences from miRBase (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008). Any reads that exactly matched the canonical miRNA sequence (with or without additional nucleotides) contributed to the total matching reads (TMR) count, and any with additional 3′ nucleotides that did not match the precursor sequence contributed to the nontemplated addition (NTA) count.
NanoString nCounter miRNA assay
Total RNA or synthetic miRNA pools (IDT; 30 pmol per oligonucleotide) were used as input for nCounter miRNA sample preparation reactions. All sample preparation was performed according to the manufacturer's instructions, with the following exception: for assays examining end-variant expression, nCounter Human miRNA Tag Reagent (which contains a mixture of tags and bridges) (see Fig. 5) was replaced with a custom reagent containing the same nCounter Human miRNA tags mixed with a novel pool of bridge oligos, each designed to template the ligation of a particular 3′ end variant to a specific tag (3′ end variants are given in Supplemental Table S6). Two such pools were generated; only one variant for each miRNA was present in each pool, and the variants of a given miRNA species were always ligated to the same tag. Following ligation, sample preparation reactions were purified and diluted according to the manufacturer's instructions. Hybridization reactions were performed according to the manufacturer's instructions with 5 μL of the fivefold diluted sample preparation reaction. All hybridization reactions were incubated at 65°C for a minimum of 18 h. Hybridized probes were purified and counted on the nCounter Prep Station and Digital Analyzer (NanoString) following the manufacturer's instructions. For each assay, a high-density scan (600 fields of view) was performed.
NanoString nCounter miRNA data analysis
For platform validation using synthetic oligonucleotides, NanoString nCounter miRNA raw data was normalized for lane-to-lane variation with a dilution series of six spike-in positive controls. The sum of the six positive controls for a given lane was divided by the average sum across lanes to yield a normalization factor, which was then multiplied by the raw counts in each lane to give normalized values. The normalized counts for the synthetic oligonucleotide mixtures (canonical or variant) in each of the three bridge pools were used to determine the specificity of the assays. To assess accuracy, the relative abundance of the canonical, V1, and V2 counts for each miRNA in a mixture of five canonical miRNAs and their 3′ variants (containing 60% canonical, 30% variant 1, 10% variant 2) was determined. To assess the linear range of the assays, pools of the canonical, variant 1, or variant 2 RNA oligonucleotides were generated and diluted to yield three six-point standard curves. We assayed the ability of the three bridge pools to detect miRNA inputs ranging from 105 to 108 copies per reaction.
For the hESC and the nucleotidyl transferase knockdown data sets, NanoString nCounter miRNA raw data was normalized for lane-to-lane variation with a dilution series of six spike-in positive controls using the vsn R package (Huber et al. 2002). Using vsn, each lane of nCounter data is calibrated by an affine transformation, and then the data is transformed by a variance-stabilizing transformation. Once normalized, the data were filtered (described in detail for each data set below), and then relative abundances of the canonical, V1, and V2 counts for each miRNA were calculated, representing the fraction of the total abundance that each of the three sequences contributed. Results from assays corresponding to four miRNAs were filtered out universally because of intrinsically high background (miR-16, miR-192, and miR-196a), or, in one case, because the assay design precluded distinguishing between two miRNA family members (miR-20a assay).
For the H1 hESC data, we had two biological replicates each for canonical, V1, and V2 for the undifferentiated hESC and the differentiated hESC. Using the averaged two lanes, the data were filtered to require a miRNA to have at least 50 counts in two of the three bridge pools (canonical, V1, or V2) for both the undifferentiated and the differentiated hESC sample. Ten miRNAs met these criteria. Differential addition of nucleotides was calculated based on differences in fractional abundance of the variants. P-values were calculated for each miRNA between the undifferentiated and differentiated samples using the biological replicates.
For the nucleotidyl transferase knockdown data, we had four biological replicates of our negative control (siCyclophilin) and two replicates of the nucleotidyl transferases knockdowns for each of the canonical, V1, and V2 lanes assayed. The data were filtered based on counts in the four siCyclophilin samples. If a miRNA had at least 50 counts in two of three assays (canonical, variant 1, or variant 2) for all four samples, it was retained. The counts were then converted to relative percentages of the canonical, variant 1, and variant 2 sequences. Relative percentages were compared between each of the knockdown samples (averaged across two lanes) and the four siCyclophilin lanes (averaged across four lanes). P-values were calculated for each miRNA comparing canonical, V1, and V2 results from the negative control siCyclophilin samples to the nucleotidyl transferase knockdown samples. The false discovery rate (FDR) was determined by the Benjamini-Hochberg method using the multtest R package (Benjamini and Hochberg 1995; van der Laan et al. 2004).
RNA for stem cell differentiation experiment
RNA used was from undifferentiated H1 human embryonic stem cells or from plates of H1 cells allowed to spontaneously differentiate (i.e., in a nondirected manner) for 7 or 9 d after the removal of fibroblast growth factor. The RNAs corresponded to aliquots from our earlier study, in which the culture of the cells and RNA isolation was performed (Bar et al. 2008). A total of 100 ng of RNA was used as input into the sample preparation reaction for the NanoString nCounter assay. Two biological replicates of undifferentiated and differentiated cells were run.
Cell culture, siRNA transfection, RNA isolation, and qRT–PCR
HCT-116 cells were maintained in McCoy's media (GIBCO) with 10% FBS (Atlanta Biologicals Inc). Transfections were performed with Lipofectamine RNAiMax (Invitrogen) and 30 nM of ON-TARGETplus SMARTpool siRNAs (Dharmacon) directed against each enzyme of interest or the negative control gene, cyclophilin B. Duplicate transfections for each enzyme knockdown, and quadruplicate transfections of siCyclophilin were performed. RNA was isolated with the miRNeasy RNA isolation kit (QIAGEN) 72 h post-transfection. cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Expression analysis via qRT–PCR was performed with Taqman gene expression assays (Applied Biosystems), using expression of GUSB to normalize for variations in RNA input. A total of 400 ng of RNA was used as input into the sample prep reaction for the NanoString nCounter assay.
Data access
The data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE26970.
Acknowledgments
This work was supported by Chromosome Metabolism Training Grant 5 T32 CA09657-16 (S.K.W.), a Rosetta Inpharmatics Fellowship in Molecular Profiling (S.K.W.), an American Cancer Society/Canary Foundation Postdoctoral Fellowship (S.K.W.), a Public Health Service National Research Service Award (T32 GM07270) from the National Institute of General Medical Sciences (E.C.K.), a Jaconnette L. Tietze Young Scientist Award (M.T.), a Prostate Cancer Foundation Creativity Award (M.T), a National Institutes of Health DK-085714 Transformative R01 Grant (M.T.), and a Damon Runyon-Rachleff Innovation Award (M.T.). We thank the Fred Hutchinson Genomics Shared Resource for sequencing services; Merav Bar and Beatrice Knudsen for providing RNA samples; and Jason Arroyo, Ingrid Ruf, and Lynn Amon for helpful advice and discussions.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.118059.110.
References
- Ahn HW, Morin RD, Zhao H, Harris RA, Coarfa C, Chen ZJ, Milosavljevic A, Marra MA, Rajkovic A 2010. MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Mol Hum Reprod 16: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarini A, Chauhan H, Gardner TJ, Jayaprakash AD, Sachidanandam R, Brown BD 2011. Kinetic analysis reveals the fate of a microrna following target regulation in mammalian cells. Curr Biol 21: 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. 2008. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26: 2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH 2006a. Diversity of microRNAs in human and chimpanzee brain. Nat Genet 38: 1375–1377 [DOI] [PubMed] [Google Scholar]
- Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, et al. 2006b. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res 16: 1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. 2011. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res 21: 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 20: 1398–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Großhans H 2009. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461: 546–549 [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. 2006. The colorectal microRNAome. Proc Natl Acad Sci 103: 3687–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehniger TA, Wylie T, Germino E, Leong JW, Magrini VJ, Koul S, Keppel CR, Schneider SE, Koboldt DC, Sullivan RP, et al. 2010. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res 20: 1590–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Taft RJ, Mattick JS 2010. Dynamic isomiR regulation in Drosophila development. RNA 16: 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26: 317–325 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S 2004. The microRNA Registry. Nucleic Acids Res 32: D109–D111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lu Z 2010. Global expression analysis of miRNA gene cluster and family based on isomiRs from deep sequencing data. Comput Biol Chem 34: 165–171 [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18: S96–S104 [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H 2010. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci 107: 3906–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP 2009. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol 11: 597–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, et al. 2008. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res 18: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K 2010. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 16: 2170–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL 2009. Adenylation of plant miRNAs. Nucleic Acids Res 37: 1878–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí E, Pantano L, Bañez-Coronel M, Llorens F, Miñones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X 2010. A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res 38: 7219–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W 2007. RNA-specific ribonucleotidyl transferases. RNA 13: 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA 2008. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451: 1013–1017 [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci 105: 10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano L, Estivill X, Marti E 2010. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res 38: e34 doi: 10.1093/nar/gkb1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Beckwith CA, Regan MM, Wyman SK, Petrovics G, Chen Y, Hawksworth DJ, Schumacher FR, Mucci L, Penney KL, et al. 2009. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res 69: 5568–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X 2008. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321: 1490–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. 2007. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9: 166–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ 2007. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol 27: 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP 2004. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res 32: 6001–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. 2007. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 39: 41–51 [DOI] [PubMed] [Google Scholar]
- Trippe R, Sandrock B, Benecke BJ 1998. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res 26: 3119–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan MJ, Dudoit S, Pollard KS 2004. Augmentation procedures for control of the generalized family-wise error rate and tail probabilities for the proportion of false positives. Stat Appl Genet Mol Biol 3: doi: 10.2202/1544-6115.1042 [DOI] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N 2007. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE 2: e1020 doi: 10.1371/journal.pone.0001020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O'Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS, Tewari M 2009. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE 4: e5311 doi: 10.1371/journal.pone.0005311 [DOI] [PMC free article] [PubMed] [Google Scholar]