Abstract
Polyadenylation sites mark the ends of mRNA transcripts. Alternative polyadenylation (APA) may alter sequence elements and/or the coding capacity of transcripts, a mechanism that has been demonstrated to regulate gene expression and transcriptome diversity. To study the role of APA in transcriptome dynamics, we analyzed a large-scale data set of RNA “tags” that signify poly(A) sites and expression levels of mRNA. These tags were derived from a wide range of tissues and developmental stages that were mutated or exposed to environmental treatments, and generated using digital gene expression (DGE)–based protocols of the massively parallel signature sequencing (MPSS-DGE) and the Illumina sequencing-by-synthesis (SBS-DGE) sequencing platforms. The data offer a global view of APA and how it contributes to transcriptome dynamics. Upon analysis of these data, we found that ∼60% of Arabidopsis genes have multiple poly(A) sites. Likewise, ∼47% and 82% of rice genes use APA, supported by MPSS-DGE and SBS-DGE tags, respectively. In both species, ∼49%–66% of APA events were mapped upstream of annotated stop codons. Interestingly, 10% of the transcriptomes are made up of APA transcripts that are differentially distributed among developmental stages and in tissues responding to environmental stresses, providing an additional level of transcriptome dynamics. Examples of pollen-specific APA switching and salicylic acid treatment-specific APA clearly demonstrated such dynamics. The significance of these APAs is more evident in the 3034 genes that have conserved APA events between rice and Arabidopsis.
Messenger RNA (mRNA) polyadenylation is an essential post-transcriptional processing step in eukaryotic gene expression. Immediately following transcription, pre-mRNAs are capped, spliced, and cleaved at the 3′-untranslated region (3′ UTR) to generate new open ends allowing the addition of poly(A) tails (Zhao et al. 1999). A poly(A) tail at the 3′ end protects the mRNA from unregulated degradation, triggers export to the cytoplasm, and assists in recognition by the translational machinery (Zhao et al. 1999; Danckwardt et al. 2008). The location at which the pre-mRNA is cleaved, known as the poly(A) site, is heavily regulated by numerous protein factors called polyadenylation factors (Zhao et al. 1999; Hunt et al. 2008; Shi et al. 2009). Alternative polyadenylation (APA) is the use of two or more poly(A) sites that are more than 30 nt apart (Shen et al. 2008a; Xing and Li 2010). APA is a powerful pathway that increases the complexity of transcriptomes and proteomes because it leads to the production of two or more different proteins or non-functional variants from the same loci in the genome (Lutz 2008). Previous studies in mammals have shown that APA patterns are dynamically regulated across tissues and different stages of cells (Ji and Tian 2009; Ji et al. 2009). For example, in Caenorhabditis elegans, the average length of the 3′ UTR decreases progressively (Mangone et al. 2010). Analysis of protein expression levels of polyadenylation factors suggests that different APA patterns might result from cell-type-specific expression profiles of polyadenylation factors (Singh et al. 2009). The dynamic regulation patterns of APA add another layer to the regulation of transcriptomes, in addition to transcript initiation and alternative splicing.
Previous EST-based analyses showed that about a half of human and rice genes and up to one-third of algal protein-coding genes undergo APA (Zhang et al. 2005; Shen et al. 2008a,b), suggesting that APA has a global role in the regulation of gene expression. The cost and efficiency of EST-based data for APA studies, however, limit the number of poly(A) sites that can be found and prevent the study of APA in greater depth. Tiling microarray technology was also used to tally APA with an analog level of polyadenylation profile (Zheng et al. 2011). New, large-scale DNA sequencing technologies have made the study of the polyadenylated transcriptome easier. The digital gene expression (DGE) protocol sequenced by massively parallel signature sequencing (MPSS) or Illumina's GAII-based sequencing-by-synthesis (SBS) (Meyers et al. 2004c; Simon et al. 2009), called MPSS-DGE and SBS-DGE in this study, yield many millions of reads of 17 or more nucleotides, also called “signatures.” Each of these signatures is derived from a specific restriction enzyme (e.g., DpnII) in the 3′-most occurrence of a polyadenylated transcript. Thus, MPSS-DGE and SBS-DGE data can be useful in identifying the locations of poly(A) sites, in addition to tracking the frequency of the transcripts (Fig. 1; Meyers et al. 2004b). More than 36 million Arabidopsis and 46 million rice MPSS-DGE signatures from, respectively, 14 and 22 different libraries (tissues) have been made available to the public (Meyers et al. 2004a; Nobuta et al. 2007). These signatures were further mapped to the annotated genomes, resulting in more than 67,000 and 81,000 unique genome-matched signatures, each representing a unique set of poly(A) sites (Meyers et al. 2004a; Nobuta et al. 2007). The massive number of signatures provides an advantage over EST-based analysis, in which the extent of APA can be studied. Based on MPSS-DGE signatures of five libraries from Arabidopsis, a previous study estimates the extent of APA to be ∼25% (Meyers et al. 2004c). In recent years, less-expensive and higher-accuracy SBS-based methods have been used extensively in genomic and transcriptional studies (Irizarry et al. 2008; Li et al. 2009; Simon et al. 2009). However, a detailed analysis of plant APA using data generated by Illumina's SBS has yet to be generated.
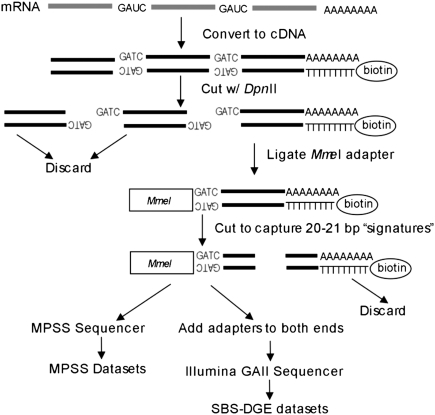
Figure 1.
Sample preparations steps for MPSS-DGE and SBS-DGE sequencing. mRNAs were isolated and reverse-transcribed into cDNA, with a biotin tag attached to an oligo-d(T) primer. This was digested with the DpnII enzyme, and only the cDNA fragment closest to poly(A) tail is retained for the next step. An MmeI adapter was added to the 5′ end of cDNA fragments, and the resulting fragments were digested with MmeI, which cuts 20–21 bp downstream from the recognition site, to generate 20–21-bp short fragments called signatures. All signatures were then sequenced by MPSS (Meyers et al. 2004b) or SBS using an Illumina GAII sequencer (for SBS-DGE) (Venu et al. 2011) after adding sequencing adapters on both ends.
Among deep (or next-generation) transcriptome sequencing technologies, the most widely used protocol is RNA-seq, in which cDNAs are fragmented to generate libraries of random length. For the purpose of tallying the 3′-most segments of the cDNAs for poly(A) site analysis, however, RNA-seq is not very efficient because relatively few sequences with a poly(A) track can be found (Mangone et al. 2010). The restriction-enzyme-anchored tags such as the DGE method used here have important advantages over RNA-seq methods. For example, the DGE data preserve the approximate location of the poly(A) site for a given transcript, while RNA-seq does not. Taking advantage of this unique characteristic of DGE-type transcript data, we analyzed the extent and the conservation of APA in Arabidopsis and rice, two diverse (dicot and monocot) plant lineages, using MPSS-DGE and SBS-DGE data. We also studied tissue-specific usage of APA events and identified several candidate genes that account for differences between tissues. These analyses offer valuable information for studying alternative transcript processing and the potential role of these alternates in gene expression as well.
Results
Distribution and verification of MPSS-DGE and SBS-DGE signatures among Arabidopsis and rice genes
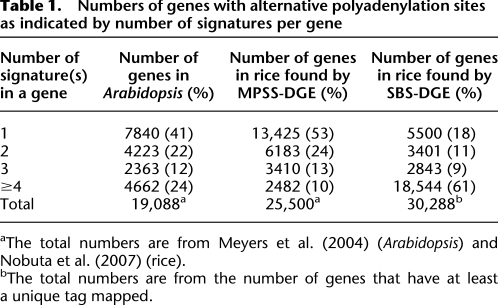
Previously in Arabidopsis, a total of 67,735 MPSS-DGE signatures of 17 bp were sequenced and matched to 19,088 annotated genes (Meyers et al. 2004c). Analysis based on five different plant organs identified more than 4000 genes (that account for 26.1% of total genes found in these five libraries) that have more than one signature matching to the sense strand, suggesting multiple 3′ ends. Since these data were published in 2004, 12 additional libraries were made available, including four inflorescences of different mutants, two stress-treated leaves, germinating seedlings, and four additional organs (Meyers et al. 2004b). We have now analyzed all 17 libraries and found that 11,248 Arabidopsis genes have multiple signatures, which is ∼60% of the 19,088 genes detected by MPSS-DGE among these libraries. To avoid unreliable signatures or counting signatures multiple times, each signature we studied had only one genomic location and was also expressed at significant levels (>3 TPM, transcripts per million) (Meyers et al. 2004a). Thus, we used 35,675 unique tags in the analysis. Among these 11,248 genes, only 530 of them have all their signatures localized in the annotated 3′ UTR (based on Arabidopsis genome annotation version 8), suggesting a wide distribution of signatures in other parts of the transcripts. On average, each gene has about three distinct signatures (35,675 divided by 11,248). Most of the genes have a relatively small number of signatures (76% of genes have fewer than four signatures), but some genes have numerous signatures, such as one gene with 27 different signatures (AT5G40450) (Table 1).
Table 1.
Numbers of genes with alternative polyadenylation sites as indicated by number of signatures per gene
In rice, we used 17-bp signatures from 22 MPSS-DGE libraries (Nobuta et al. 2007) and searched for genes with more than one signature. The number of genes with multiple signatures is 12,075 (47% of 25,500 genes) (Table 1), slightly less than what was found in Arabidopsis. There are 1681 genes with all their signatures in the 3′ UTR, which might be due to the fact that 3′ UTRs in rice are generally longer than those of Arabidopsis (Shen et al. 2008a). The number of genes with APA is also slightly higher than our previous calculation based on ESTs (8596 genes, or 33% of 25,500 genes) (Shen et al. 2008a), suggesting that MPSS-DGE is a more powerful tool to detect rare APA events compared with ESTs.
In addition to the MPSS-DGE data, we obtained 5,522,207 distinct 20-bp tags (out of 114 million total reads) from 48 rice libraries sequenced by the Illumina's SBS-DGE method. These data are comparable to MPSS-DGE because they are 3′ tags anchored by a restriction site, but the data were independently obtained using a different technology. After we applied the same filter (uniquely mapped to genome and TPM > 3), 168,223 tags (from 30,288 genes) were used for further analysis. Among these genes, 24,788 (82%) (Table 1) of them have multiple tags, suggesting that SBS-DGE is a very sensitive method to detect APA and that APA is a universal phenomenon in rice.
To confirm that signatures from MPSS-DGE and DGE are consistent with known and validated poly(A) sites of mRNAs, we examined 55,742 previously authenticated rice poly(A) sites (Shen et al. 2008a) to see whether or not these poly(A) sites are represented by SBS-DGE signatures and whether or not partial digestion during SBS-DGE signature preparation was an issue. Among all rice poly(A) sites examined, 54,621 (98% of 55,742) poly(A) sites that were sequenced by ESTs were verified by the rice SBS-DGE signatures. In contrast, only 1121 (2%) EST-verified poly(A) sites located downstream from a DpnII site were not identified by SBS-DGE signatures. This result suggests that DGE-based methods are reliable in defining poly(A) sites, and the signatures resulting from incomplete digestion are very rare (<2%). Furthermore, we cross-checked the validity of MPSS-DGE and SBS-DGE tags studied in this paper, since both methods use similar sample preparing procedures. About 85.8% (28,994 out of 33,794) of rice MPSS-DGE signatures can also be detected in the rice SBS-DGE data set, indicating that most MPSS-DGE tags were derived from real poly(A) sites.
Locations of potential poly(A) sites on transcripts
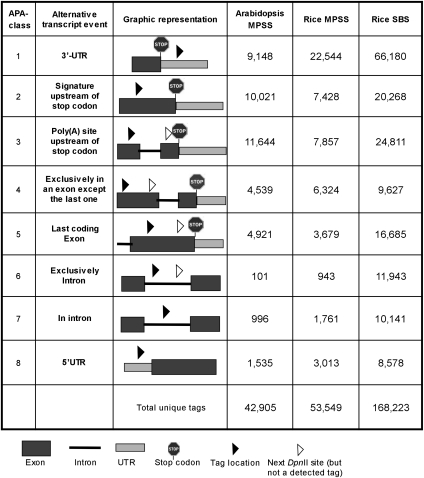
While the location of an MPSS-DGE or SBS-DGE signature does not directly coincide with a poly(A) site at the nucleotide level, it does indicate the presence of a poly(A) site within a tightly defined region. Both MPSS-DGE and SBS-DGE signatures from Arabidopsis and rice were derived from restriction enzyme DpnII sites immediately upstream of the poly(A) sites (Fig. 1; Meyers et al. 2004a). Therefore, the poly(A) site must be located between the sequenced signature mapping site and the immediately downstream DpnII site. Based on this notation, we developed a method to further categorize MPSS-DGE signatures according to both their locations and the poly(A) sites from which they were derived. To differentiate these signatures (derived from genomic sequences) from a method used in a previous analysis of MPSS-DGE signatures (Meyers et al. 2004c), we call this form of categorization “APA-class signatures” because our classification focused on signatures associated with poly(A) sites. To reduce the complexity of this analysis, we only studied signatures found in genes with multiple signatures, and they were grouped into eight different classes for discussion purposes (Fig. 2).
Figure 2.
APA classes and illustrations of the locations of MPSS-DGE and SBS-DGE tags. The symbols are annotated on the bottom. Tag locations mark where the tag is mapped on the gene. The nearest DpnII site (open triangle) indicates a potential tag location that is not found in the pool of experimentally sequenced tags, meaning that a poly(A) site should be located between the tag and the next DpnII site (Meyers et al. 2004b). The numbers of the three columns on the right side are the distributions of the MPSS-DGE or SBS-DGE tags in each APA-class.
In this APA-class system, signatures of APA-class 1 are located in the 3′ UTR of the gene (Fig. 2). Signatures of APA-class 2 are located upstream of the stop codon with no other possible DpnII site found between the signature and the stop codon. Signatures in this class could be derived from two types of poly(A) sites: poly(A) sites located upstream of the stop codon (an APA site) or conventional poly(A) sites located in the 3′ UTR. The reason why APA-class 2 signatures could include poly(A) sites in the 3′ UTR is that there are no DpnII sites between the stop codon and terminal poly(A) site. It is currently not possible to differentiate APA sites from conventional sites in APA-class 2 in this data set, but the higher expression levels and the similarity to APA-class 1 signatures based on the fact that they are more frequently used than signatures from other APA classes (see details below) made us believe that most signatures in this class are derived from conventional poly(A) sites. In Arabidopsis and rice, ∼45% and 56% of MPSS-DGE signatures and 51% of SBS-DGE rice signatures belong to APA-classes 1 and 2 of conventional polyadenylation events. We further tested how often these poly(A) sites are used based on their relative TPM values. In both species, ∼90% of all sequenced signatures are from the first two APA classes, suggesting that mRNAs with their poly(A) sites in the 3′ UTR are still dominant over other alternative transcripts.
Signatures from all APA-classes 3 to 8 positively locate poly(A) sites upstream of stop codons because there is at least one DpnII site upstream of stop codons in each case. Therefore, each signature in these six classes represents a possible APA event leading to the production of a truncated transcript. All signatures in APA-classes 3, 4, and 5 are located in exons. The difference among the three classes is that the poly(A) sites derived from class 3 extend more than one exon, while the poly(A) sites of classes 4 and 5 can be limited to one exon. Class 5 is more specifically indicative of poly(A) sites located in the last exon because many of the truncated transcripts could still produce functional proteins. Signatures located in the exons are one of the most abundant APA groups in our analysis, mainly because MPSS-DGE signatures are collected from mature mRNA where introns might have been spliced out. In Arabidopsis and rice, ∼49% and 33% of MPSS-DGE signatures and 30% of rice SBS-DGE tags belong to these three exon-located APA classes (Fig. 2).
Signatures from APA-classes 6 and 7, however, are located in introns, where poly(A) sites of APA-class 6 signatures can be positioned in just one intron, and poly(A) sites of class 7 might be located outside the particular intron where the signature is located. Signatures from these two classes indicate the association of two tightly coupled mRNA processing events, alternative splicing and APA. Since MPSS-DGE signatures were collected from mostly mature mRNA where intron sequences were less expected, signatures from classes 6 and 7 suggest that introns were retained in some mature mRNA transcripts. This type of intron retention has been found to be common in plants (Ner-Gaon et al. 2004). The question remains as to whether the retention of unspliced introns induced the APA events or the cleavage of transcripts in the introns [for generating poly(A) sites] that blocked the splicing process. About 2% and 5% of Arabidopsis and rice signatures, respectively, belong to these two classes. Signatures sequenced by SBS-DGE technology, however, suggest a bigger role of introns in APA. Up to 13% of APA events could be associated with introns, correlating with our previous finding using ESTs (Shen et al. 2008a) and suggesting that introns are a major player in APA events. The difference between MPSS-DGE and SBS-DGE could be due to the greater depth of sequencing of the tags with SBS-based sequencing methods.
Signatures of APA-class 8 are located in the 5′ UTR of the genes, which can be found in ∼4% and 6% of Arabidopsis and rice signatures, respectively. This further confirms our previous finding that some transcripts contain poly(A) sites in the 5′ UTR (Shen et al. 2008a,b), although the significance of these events is not yet fully known.
Although the last six APA classes consist of ∼50% of all unique signatures, analysis of the frequency of these signatures (representing the expression levels) suggests that they consist of only 10% of all sequenced signatures (Supplemental Fig. S1). In addition to lower abundances, signatures from these APA classes are more library-specific (described in more detail below), suggesting that their regulatory roles might be limited to certain tissues or development stages.
Library/tissue-specific APA events
To determine whether or not APA events are associated with certain libraries (made from specific tissues, developmental stages, mutants, or certain stress treatments) for each APA class, we assessed the number of signatures expressed only in one tissue compared to the number of all signatures. In both species, the percentages of library-specific signatures in APA-classes 1 and 2 are much lower compared with the other six classes (10% compared to 36% in Arabidopsis, 26% compared to 40% in rice) (see Supplemental Data S1). This suggests that conventional poly(A) sites represented by the first two classes are more likely to be ubiquitously used and less tissue-specific.
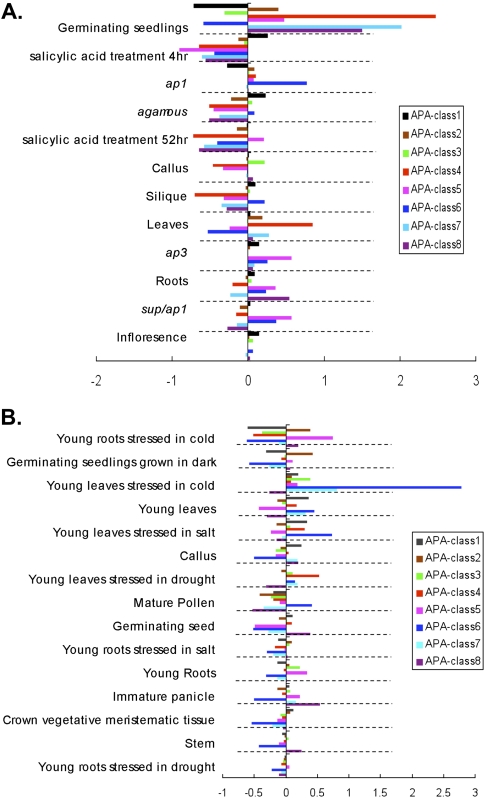
To further study the usage of alternative poly(A) sites in each library, we used a method similar to GAUGE (Zhang et al. 2005; Lutz 2008) with slight modifications. In each library, the expression value of each signature (TPM value) was grouped based on its APA class. To normalize the differences between libraries, TPM values were divided by the total number of signatures sequenced in that library. The percentage of usage of an APA class in a library can then be measured by the sum of all signatures in that APA class. For each poly(A) site type in a library, its usage percentage was compared to the average usage percentage of all libraries. The difference was then normalized to the mean and called the “relative distance.” Figure 3 shows libraries and their relative preferences to usage of APA, where a positive value means that the library uses more APA, and vice versa. A complete list of values for all libraries is provided in Supplemental Data S2. The significant differences from the mean usage of APA were further calculated by a χ2 test, and the libraries were ranked based on P-value with the more significant group on the top (Fig. 3).
Figure 3.
Tissue-specific usage of different APA classes. The x-axis shows the relative distance (or difference) to the average usage of all libraries analyzed. The library designations are on the left. (A) Libraries from Arabidopsis. (B) Libraries from rice. Libraries most significantly different from average usage of APA are on top (χ2 test).
Among all libraries of Arabidopsis, germinating seedlings (GSE in Fig. 3A) show a significant bias toward usage of APA sites (χ2 test, p < 0.05), particularly for APA-classes 4, 7, and 8. This suggests that APA might play a more important role in fast-growing tissues. Another possible interpretation is that the polyadenylation machinery is less tightly controlled when it comes to selecting a poly(A) site due to large amounts of mRNAs produced in these actively growing tissues. Interestingly, some libraries show a preference for a certain APA group. There are two library samples with a single prominent APA class, such as 21-d leaves that have a strong preference for APA-class 4, and an ap1-10 mutant with a preference for APA-class 6. This suggests that some tissues or mutants respond differently in each library, potentially under library-specific regulations. Besides the dominant utility of different classes of APA sites, there may be general avoidance of APA (more negative values). As an example, plants treated with salicylic acid tend to use less APA (Fig. 3) in APA-classes 3 to 8. While not as drastic as in the case of germinating seedlings in terms of preference on certain types of APA classes, such a broad range reduction of APA would warrant further investigation.
In rice, 15 libraries were examined by the same method (Fig. 3B). The most clearly demonstrated case is in the library of young leaves stressed in cold, which shows a very significant increase in the usage of poly(A) sites in introns (APA-class 6, χ2 test, p < 0.05), suggesting that APA may have a role in cold response in rice leaves through the use of poly(A) sites in an intron. In contrast to germinating Arabidopsis seedlings, germinating rice seedlings grown in dark generally avoid the use of APA sites. This could be due to the fact that many gene regulation pathways are not active in dark conditions. The rest of the 12 groups do not show significant differences in the average usage of APA sites. Overall, the difference in APA usage among different libraries, as well as preference over certain APA classes within a particular library, suggests that there is a complex mechanism regulating the degree of APA. To further explore the differentials between usages of APA sites in different tissue types, we examined the expression level of 43 known polyadenylation factors and splicing factors in each MPSS-DGE library. A detailed discussion of trans-acting genes' effect on APA is provided in the Supplemental Data S3.
To further examine if cells use specific APAs to fine-tune gene expression as a response to environmental changes, we identified 34 Arabidopsis genes that use APA only when treated with salicylic acid (Supplemental Data S4, S5). Interestingly, the functions of these genes are part of responses to abiotic or biotic stimuli (35% compared to 10% in the rest of Arabidopsis genes), suggesting that cells may use APA as a response mechanism to adjust the expression dynamic of certain genes.
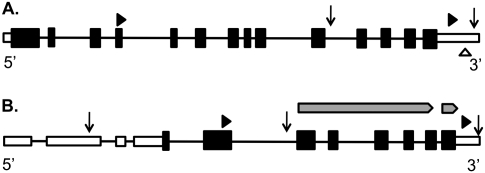
In rice, an examination of pollen-specific APA sites found that 102 genes only have APA sites in pollen (Supplemental Data S6). To further illustrate cases of library-specific APA in pollen, we used two genes as examples. One of them is LOC_Os02g02980, the full-length isoform of which encodes a 533-amino-acid protein (Fig. 4A) that has significant similarity to the Arabidopsis ENHANCED DISEASE SUSCEPTIBILITY 5 gene (EDS5 AT4G39030, E-value = 1.1×10−149). One tag located in the fourth protein-coding exon (APA-class 3) was sequenced 31 times, exclusively in pollen, while another tag in the 3′ UTR (APA-class 1) was found to be expressed ubiquitously in most of the tissues we sequenced. We further looked in the rice small-RNA database (Nobuta et al. 2007) and the 3′ UTR of this gene was enriched in small-RNA target sites. Ten distinct 17-bp small RNAs (Nobuta et al. 2007) are found within 100 bp, and two of them are derived from microRNAs (osa-miR1436). To test whether small RNA-induced cleavage happens in this location, we further queried the rice parallel analysis of RNA ends (PARE) data (German et al. 2008) for microRNA-induced cleavage and found more than 1000 tags mapped to this region, suggesting that mRNA fragments are generated through miRNA-induced cleavage. The shorter isoform, found only in pollen, might be able to escape the regulation of small RNAs. This example demonstrates that a gene might undergo APA in a tissue-specific manner and its possible association with other regulatory pathways.
Figure 4.
Examples of pollen-specific APA genes in rice. (Triangles) The locations of MPSS-DGE or SBS-DGE tags. (Arrowheads) The poly(A) sites from the full-length cDNA library. (A) LOC_OS02G02980, encoding a homolog of Arabidopsis EDS5, is regulated by small RNA and APA. The empty triangle in 3′-UTR indicates a small RNA target. (B) Structure of LOC_OS01G51754, an alpha-amylase precursor. (Empty boxes) Introns; (solid boxes) exons. Two pointed gray bars above the gene indicate two functional domains identified by Pfam search as the catalytic domain of alpha-amylase (longer bar) and the alpha-amylase C-terminal beta-sheet domain (shorter bar), respectively.
Another gene, LOC_Os01g51754 (alpha-amylase precursor, homologs of AT1G69830), that has a tag located in the second protein-coding exon (APA_class 4) was sequenced 17 times exclusively in pollen, while another tag in the 3′ UTR (APA-class 1) was found ubiquitously in most of the tissues sequenced. This APA site is further supported by a rice full-length cDNA (GenBank Accession AK120130) from which two transcripts (short and long) (Fig. 4B) were found. The full-length cDNA sequence also suggests that this gene has an alternative transcription start site, suggesting an interaction of transcription initiation and polyadenylation in generating transcriptome diversity (Fig. 4). Functional domain searches of this gene have shown that the shorter isoform does not carry two α-amylase domains in the 3′ end of its longer isoform (Finn et al. 2010). However, no functional domain is found in short transcripts produced by APA, suggesting that APA could add another layer of regulation to turn off gene production in a tissue-specific manner.
Conservation of APA between Arabidopsis and rice
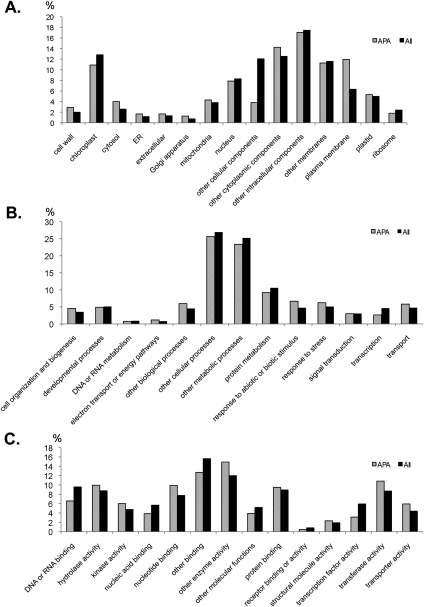
If the APA events identified herein play a role in gene expression regulation or proteome diversity, they would be expected to be conserved in homologous genes in closely related plant species. To test this hypothesis, we compared orthologous genes with APA sites in exons or introns (APA-classes 3 through 7) since they are likely to produce shorter-than-normal transcripts. Because protein sequences are more conserved, we used the percentage of proteins produced by these transcripts as a measurement. We consider that a ±10% difference of the final polypeptide length may be tolerated in the homolog group of APA. Using known orthologous groups downloaded from OrthoMCL (Chen et al. 2006), a total of 3034 genes (listed in Supplemental Data S7; 20% of the orthologous gene groups) were found to have conserved APA sites shared by orthologous genes. This result is indicative of a large amount of conservation among related genes between these two species, representing two groups of evolutionarily divergent plants. To further study whether certain functional gene groups are biased toward APA, we examined the relationships between gene functions based on Gene Ontology and their APA configurations. We included genes that have conserved poly(A) sites between two species and compared each functional group's proportion to all Arabidopsis genes with APA. The most noticeable changes were observed in the category of “cellular location” (Fig. 5A). Genes localized in the plastid and plasma membrane are rich in conserved APA, suggesting that membrane proteins are more likely to undergo APA. In the category of biological processes, many conserved genes are stress-related, suggesting a potential role for APA in stress responses (Fig. 5B). In terms of molecular functions, genes that have conserved APA events show a slight enrichment in transferase and kinase activity (Fig. 5C). These results suggest that there are certain conserved groups of genes prone to APA regulation.
Figure 5.
Functional distributions of 3034 genes identified to have conserved APA between rice and Arabidopsis. Comparisons were made using the functional distributions of these genes against the general distribution of 27,235 protein-coding genes (All) in Arabidopsis TAIR8 with known GO categories. (A) Cellular or subcellular localizations. (B) Biological processes. (C) Molecular functions.
Discussion
Advancements in DNA sequencing technologies provide us with a vast amount of cDNA sequence data for targeted interrogation of genome-level transcriptome characteristics. We took advantage of the unique properties of MPSS-DGE and SBS-DGE-based transcript data; these restriction-enzyme-anchored tags are immediately 5′ of poly(A) sites and thus provide data specifically delimiting the 3′ ends of the transcripts. Those data could not be easily derived from fragmented RNAs, such as those measured by Illumina's RNA-seq method. The results of the analysis are the first in-depth examination of the full polyadenylation landscape dynamics of any plant species, with an emphasis on different developmental stages and responses to environmental stresses.
In this study, we used the unique characteristic of the DGE data to study the extent of APA in two plant species, which resulted in the identification of many novel APA sites. Based on signatures from MPSS-DGE, we found that 11,248 and 12,075 genes show evidence of APA, corresponding to 59% and 47% of rice and Arabidopsis genes, respectively. We then further refined the rice APA profile using tags from SBS-DGE and found that 24,788 (82%) rice genes undergo APA. This number is much higher than the previous estimated extent of APA using EST data, which was ∼50% (Shen et al. 2008a), demonstrating the power of current generation sequencing methods in finding new APA events. In addition, analyses based on MPSS-DGE and SBS-DGE provided us with more information on these APA events found in exons and introns. About 90% of APA genes found by ESTs have all their poly(A) sites located in the 3′ UTR, and most of them are only within tens of nucleotides apart. MPSS-DGE and SBS-DGE signatures, however, tend to overlook these closely located poly(A) sites and cluster them together based on their immediate 5′ DpnII site. In rice, the EST-based method found about 1000 APA events located in CDS, introns, or 5′ UTRs. In contrast, the MPSS-DGE and SBS-DGE methods found more than 20,000 and 97,000 events (APA-classes 3–8) respectively. This suggests that MPSS-DGE and SBS-DGE-based analyses are more sensitive in finding unconventional APA events.
These sequencing methods, however, do have some disadvantages compared with classical EST collections. First, MPSS-DGE and SBS-DGE signatures do not provide information on the exact locations of the poly(A) sites. This makes it hard to determine polyadenylation signals based on the location of poly(A) sites. Second, neighboring poly(A) sites are likely to be detected by the same signatures, thus slightly masking the full complexity of APA. The poly(A) sites could span a few nucleotides to a few hundred nucleotides over the genome and still be detected by the same signatures. Ultimately, even more detailed sequencing data are required to determine the precise nucleotides where poly(A) sites could be located. Third, since tags from our MPSS-DGE and SBS-DGE data sets are usually short sequences, they are more easily confounded due to sequence error or genomic repeat sequences compared to longer EST sequences. To overcome these limitations, we used strict filters (signatures must be uniquely mapped to the genome and have been sequenced at least three times) to eliminate unreliable signatures. MPSS-DGE and SBS-DGE data sets agree well with each other and EST-based data, indicating that tags we analyzed were derived from authentic poly(A) sites. However, just as in any large-scale data set analysis, it is unavoidable that there may be some unfiltered background noise in the polyadenylation profile of particular genes. Thus, confirmation of individual poly(A) sites should be done before a definitive APA is called for.
Previous MPSS-DGE-based analysis found that ∼25% of Arabidopsis genes were alternatively polyadenylated (Meyers et al. 2004c). Our present analysis found that more genes have more than one signature than previously calculated. This is mainly due to the fact that more libraries (17 compared to the original set of five) and more reads were taken into consideration, so that more library-specific APA events are included in this analysis. In addition, in this study, we focused on the location of poly(A) sites instead of the location of signatures. All signatures located in exons were grouped in one class, while we refined signatures in the class into several different APA classes. Furthermore, we categorized signatures based on their locations and the regions of poly(A) sites located to find possible APA events that would produce truncated transcripts.
The determination of a poly(A) site is influenced by the interactions between cis-elements in the pre-mRNA sequence and trans-acting factors—the polyadenylation machinery—in the cell (Danckwardt et al. 2008). Many APA events are usually limited to certain libraries and thus behave in a tissue/library-specific manner (Fig. 3). The components in the polyadenylation apparatus may therefore play an important role because sequences in pre-mRNAs in different libraries were transcripts from the same genome. The current data provide us an opportunity to study how polyadenylation trans-acting factors affect the choices of poly(A) sites. First, expression levels of each polyadenylation factor were obtained simultaneously as an APA gene, thus reflecting the real-time conditions in the cells. Second, all libraries have been normalized so that the comparison between libraries would be robust. This is a novel observation of how plant polyadenylation factors affect APA at a large scale, the results of which suggest that perhaps the expression levels of several factors affect APA. To our surprise, some splicing factors were also found to be important contributors to the “decision making” of APA sites. The presence of these factors increases the likelihood of cells undergoing APA. Of course, our data have only suggested consideration of this relationship of the expression levels of these trans-acting factors and APA. Further studies are needed to support such a hypothesis.
To better understand APA in an evolutionary sense, we compared APA genes between Arabidopsis and rice and found that more than 3000 gene-pair orthologs have APA sites in similar regions of the genes. This means that these regulation pathways are conserved across different species, suggesting that some APA events play important regulatory roles, as seen in flower development (Simpson et al. 2003). Analysis based on GO indicates that in some subcellular compartments, genes with conserved APA are over-represented. It is possible that a local concentration of trans-acting factors or activation of these factors to certain responses determines the choice of mRNA 3′ ends. The utility of these different poly(A) sites may alter the inclusion or exclusion of some regulatory elements that may have an impact on transcript stability, among other possibilities. Examples have been recently demonstrated in the APA of cancer cells (Mayr and Bartel 2009).
Finally, data on the functional relevance of non-canonical polyadenylation and its importance in the regulation of gene expression have recently emerged (Hamill et al. 2010). However, very little information is known about the role of non-canonical polyadenylation in plants. The deep sequencing methods used in this study could be employed to detect the evidence of non-canonical polyadenylation, but the functional importance of this phenomenon in plants is yet to be determined. The position of polyadenylation sites as measured on a genome-wide scale might provide advances in the study of non-canonical polyadenylation.
Methods
Sequencing data retrieval and processing
For MPSS-DGE analysis in Arabidopsis, we used the data of 17-base signatures (17bp_summary.txt) available from our “MPSS Plus” database at http://mpss.udel.edu/at/, which contains 297,313 rows of genome mapping and expression information (Meyers et al. 2004a). To remove unreliable sequencing data, we retained only signatures uniquely mapped to the genome and having an expression abundance larger than 3 TPM (normalized transcripts per million). Then the number of signatures in each gene was calculated, and only signatures from genes with more than one signature were used in further analysis. A total of 36,403 signatures from 11,263 genes were used in this analysis. For rice MPSS-DGE data, we used the same procedure to filter signatures, and 34,195 distinct signatures from 12,091 rice genes were analyzed.
For rice SBS-DGE data (GEO ID: GSE25596), we prepared samples for digital gene expression-tag profiling with DpnII using protocol recommended by Illumina (detailed protocol can be viewed from http://illumina.ucr.edu/ht/documentation/molbiol-docs/DGE-DpnII-Sample-Prep.pdf/view). Briefly, 2 g of rice tissue was ground into a fine powder, and RNA was extracted by TRIzol. One to 6 μg of total RNAs were mixed with 50 μL of oligo(dT) beads, where double-stranded cDNAs were synthesized using SuperScript II Reverse Transcriptase and DNA polymerase I. The double-stranded cDNAs were then digested with DpnII for 1 h at 37°C, and the beads were washed twice so only the sequences adjacent to poly(A) tail remained for ligation. The GEX MmeI adapter was ligated to the 5′ end of the DpnII restriction site followed by MmeI restriction digest for 2 h at 37°C. The supernatant was collected and DNA was purified for Illumina GAII sequencing. In total, we sequenced RNAs from 48 rice libraries, and 5,522,207 unique signatures were obtained (http://mpss.udel.edu/rice_sbs/) (Venu et al. 2011). To improve the quality of our analyses, we used the same set of filters described above, which resulted in 178,555 tags for further analysis. All data were saved in an MySQL database, and a series of Perl scripts was used for the following analyses. To assess the extent of falsely identified poly(A) sites due to internal priming, we searched for potential A-rich motifs (more than eight As in a 10-nt region) in sequences between DGE tags and a downstream DpnII site in the genome. The result shows that 859, 377, and 4948 tags were located near the A-rich regions in Arabidopsis MPSS-DGE, rice MPSS-DGE, and rice SBS-DGE, respectively. Thus, the rates of potential false negatives caused by possible internal priming are 2%, 1%, and 3%, respectively. All of these tags were removed from further analyses. To verify the reliability of DGE methods in defining poly(A) sites in the genome, we tested if poly(A) sites authenticated from the previous EST sequencing method are also found in our new data sets. For this, 55,742 rice poly(A) sites (Shen et al. 2008a) were used, and positions of their immediate upstream DGE signatures (sequenced at least three times) were recorded. Sequences between sequenced DGE signatures and poly(A) sites were further examined to see if any other DpnII sites exist. If there is no DpnII site between a poly(A) site and its upstream sequenced DGE signature, this poly(A) site was considered to be confirmed by DGE.
For all of the signatures we analyzed, most were previously assigned as classes within exons, on the same DNA strand as the coding sequence (Meyers et al. 2004a). To better categorize MPSS-DGE signatures according to both their locations and the poly(A) sites from which they were derived, we developed a method to differentiate these MPSS-DGE signatures and grouped them based on APA classes as detailed in Figure 2 and described in the text and figure legend.
Library-specific data analysis
To further study the usage of alternative poly(A) sites in each library, we used a method similar to GAUGE (Zhang et al. 2005), with small modifications. In each library, the expression values of each signature (TPM value) were grouped based on their APA classes. To normalize the differences between libraries, TPM values were first divided by the total number of signatures in that library. The percentages of usage of an APA class were calculated by comparing to the sum of all signatures in that library. For each APA class in a library, its percentage of usage was compared with the average percentage of usage of this particular APA class in all libraries. The difference was normalized to the average and called the “relative distance.” A χ2 test was performed in each library against the null hypothesis that the usage of a given APA class in this library is not different from the mean usage (complete lists of values for all libraries are provided in Supplemental Data S2).
Homologous analysis between rice and Arabidopsis
The known orthologous groups were downloaded from orthoMCL (Chen et al. 2006) and examined for APA events leading to the production of truncated proteins losing approximately similar lengths of amino acid sequence in rice and Arabidopsis. A 10% difference in percentage of final protein length was tolerated, since MPSS-DGE signatures do not discern the exact location of the poly(A) sites. The relationship between gene functions (Gene Ontology data) and their APA configuration was identified by comparing the function of these 5063 genes with all Arabidopsis genes using TAIR's GO web portal (http://www.arabidopsis.org/tools/bulk/go/index.jsp) (Swarbreck et al. 2008).
Data access
The rice SBS-DGE data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under access no. GSE25596. Other data sets have been published before.
Acknowledgments
We thank Jianti Zheng and Michael Hughes for their help with statistical analyses and Tommy Li for editing the manuscript. This project was funded in part by U.S. National Science Foundation grants (IOS-0817829 to Q.Q.L., DBI-0321437 and DBI-0701745 to B.C.M. and G.L.W.); by a U.S. National Institutes of Health grant (GM07719201A1 to Q.Q.L.); by a grant from the Committee for Faculty Research, and Academic Challenge grants from the Botany Department, Miami University. Y.S. was an awardee of the Cell, Molecular and Structural Biology graduate program Research Assistantship, and the Botany Department's dissertation fellowship, Miami University. X.W. was supported by a scholarship from Xiamen University, grants from the National Natural Science Foundation of China (no. 60774033), and Special Research Funds for the Doctoral Program of Higher Education (no. 20070384003 and no. 20090121110022).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.114744.110.
References
- Chen F, Mackey AJ, Stoeckert CJ Jr, Roos DS 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34: D363–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE 2008. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 27: 482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. 2010. The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis L, Nobuta K, German R, et al. 2008. Novel microRNA-target RNA pairs revealed by Parallel Analysis of RNA Ends (PARE). Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Hamill S, Wolin SL, Reinisch KM 2010. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci 107: 15045–15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AG, Xu R, Addepalli B, Rao S, Forbes KP, Meeks LR, Xing D, Mo M, Zhao H, Bandyopadhyay A, et al. 2008. Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein–protein interactions and gene expression profiling. BMC Genomics 9: 220 doi: 10.1186/1471-2164-9-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP 2008. Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res 18: 780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B 2009. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE 4: e8419 doi: 10.1371/journal.pone.0008419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K 2009. SNP detection for massively parallel whole-genome resequencing. Genome Res 19: 1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS 2008. Alternative polyadenylation: A twist on mRNA 3′ end formation. ACS Chem Biol 3: 609–617 [DOI] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak S, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD 2004a. Arabidopsis MPSS. An online resource for quantitative expression analysis. Plant Physiol 135: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S 2004b. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res 14: 1641–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD 2004c. Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 22: 1006–1011 [DOI] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R 2004. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39: 877–885 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Belo A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, et al. 2007. An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol 25: 473–477 [DOI] [PubMed] [Google Scholar]
- Shen Y, Ji G, Haas BJ, Wu X, Zheng J, Reese GJ, Li QQ 2008a. Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation. Nucleic Acids Res 36: 3150–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Liu Y, Liu L, Liang C, Li QQ 2008b. Unique features of nuclear mRNA poly(A) signals and alternative polyadenylation in Chlamydomonas reinhardtii. Genetics 179: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, Frank J, Manley JL 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SA, Zhai J, Nandety RS, McCormick KP, Zeng J, Mejia D, Meyers BC 2009. Short-read sequencing technologies for transcriptional analyses. Annu Rev Plant Biol 60: 305–333 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C 2003. FY is an RNA 3' end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787 [DOI] [PubMed] [Google Scholar]
- Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH 2009. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res 69: 9422–9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. 2008. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venu RC, Zhang Y, Weaver B, Carswell P, Mitchell TK, Meyers BC, Boehm MJ, Wang G-L 2011. Large scale identification of genes involved in host–fungal interactions using Illumina's sequencing-by-synthesis technology. Methods Mol Biol 722: 167–178 [DOI] [PubMed] [Google Scholar]
- Xing D, Li QQ 2010. Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip Rev RNA 2: 445–458 doi: 10.1002/wrna.59 [DOI] [PubMed] [Google Scholar]
- Zhang HB, Lee JY, Tian B 2005. Biased alternative polyadenylation in human tissues. Genome Biol 6: R100 doi: 10.1186/gb-2005-6-12-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Xing D, Wu X, Shen Y, Kroll D, Ji G, Li QQ 2011. Ratio-based analysis of differential mRNA processing and expression of a polyadenylation factor mutant pcfs4 using Arabidopsis tiling microarray. PLoS ONE 6: e14719 doi: 10.1371/journal.pone.0014719 [DOI] [PMC free article] [PubMed] [Google Scholar]