Abstract
As whole-genome sequencing becomes commoditized and we begin to sequence and analyze personal genomes for clinical and diagnostic purposes, it is necessary to understand what constitutes a complete sequencing experiment for determining genotypes and detecting single-nucleotide variants. Here, we show that the current recommendation of ∼30× coverage is not adequate to produce genotype calls across a large fraction of the genome with acceptably low error rates. Our results are based on analyses of a clinical sample sequenced on two related Illumina platforms, GAIIx and HiSeq 2000, to a very high depth (126×). We used these data to establish genotype-calling filters that dramatically increase accuracy. We also empirically determined how the callable portion of the genome varies as a function of the amount of sequence data used. These results help provide a “sequencing guide” for future whole-genome sequencing decisions and metrics by which coverage statistics should be reported.
Whole-genome sequencing and analysis is becoming part of a translational research toolkit (Lupski et al. 2010; Sobreira et al. 2010) to investigate small-scale changes such as single-nucleotide variants (SNVs) and indels (Bentley et al. 2008; Wang et al. 2008; Kim et al. 2009; McKernan et al. 2009; Fujimoto et al. 2010; Lee et al. 2010; Pleasance et al. 2010) in addition to large-scale events such as chromosomal rearrangements (Campbell et al. 2008; Chen et al. 2008) and copy-number variation (Chiang et al. 2009; Park et al. 2010). For both basic genome biology and clinical diagnostics, the trade-offs of data quality and quantity will determine what constitutes a “comprehensive and accurate” whole-genome analysis, especially for detecting SNVs. As whole-genome sequencing becomes commoditized, it will be important to determine quantitative metrics to assess and describe the comprehensiveness of an individual's genome sequence. No such standards currently exist.
For several reasons (sample handling, platform biases, run-to-run variation, etc.), random generation of sequencing reads does not always represent every region in the genome uniformly. It is therefore necessary to understand what proportion of the whole genome can be accurately ascertained, given a certain amount and type of input data and a specified reference sequence. The 1000 Genomes Project (which aims to accurately assess genetic variation within the human population) refers to this concept as the “accessible” portion of the reference genome (1000 Genomes Project Consortium 2010). While population-scale sequencing focuses on low-coverage pooled data sets, here we focus on requirements for highly accurate SNV calls from an individual's genome, a question that is extremely important as whole-genome sequencing and analysis of individual genomes transitions from primarily research-based projects to being used for clinical and diagnostic applications. Additionally, we seek to understand the relationship between the amount of sequence data generated and the resulting proportion of the genome where confident genotypes can be derived—we refer to this as the “callable” portion, a term that is roughly equivalent to the 1000 Genomes Project's “accessible” portion. Using these sequencing metrics and genotype-calling filters will help obviate the need for costly and time-consuming validation efforts. Currently, no empirically derived data sets exist for determining how much sequence data is needed to enable accurate detection of SNVs.
To address this issue, we sequenced a blood sample from a male individual with an undiagnosed clinical condition on two related platforms—Illumina's GAIIx and HiSeq 2000—to a total of 359 Gb (equivalent to ∼126× average sequenced depth). Here we focus on the technical aspects of analyzing these data generated as part of the expanded whole-genome sequencing efforts of the National Institutes of Health (NIH) Undiagnosed Diseases Program (UDP). We leveraged the ultra-deep coverage of this genome to identify sources of incorrect genotype calls and developed approaches to mitigate these inaccuracies. We generated incremental data sets of the deep-sequenced genome to answer the following important questions: Given a specific amount of sequence data, what fraction of the genome is callable? and how many SNVs are detected? Ultimately, we seek to understand how much sequence data is needed for adequate representation of the whole genome for genotype calling and to develop standards by which all whole-genome data sets can be evaluated with respect to comprehensiveness.
Answers to these questions will help us make more informed decisions for designing whole-genome sequencing experiments to study genome biology and for clinical analyses, specifically in light of accurately detecting variants that directly modify phenotypes and cause disease.
Results
Summary of whole-genome sequencing data sets
Paired-end 100-bp reads were generated from a single human genomic sample on two Illumina GAIIx flowcells (14 lanes) and two HiSeq 2000 flowcells (16 lanes) (see Methods). Sequenced libraries had mean estimated insert sizes of 378 bp (±24 SD) and 436 bp (±26 SD) (Supplemental Fig. 1) and estimated diversities of 5.7 and 4.6 billion distinct molecules, respectively (see the Supplemental Material). A total of 359 Gb of data passing filters (see below) (Table 1) was collectively generated across the four flowcells. We defined the average sequencing depth as the mean coverage of a haploid genome with reads that (1) pass the Illumina Chastity filter and (2) contain at least 32 bases with phred-scaled qualities Q20 or higher, which removes an additional 6.6% of the reads (on average) beyond the Chastity filter (see Table 1). All reads that pass sequence-level filters were aligned to the hg18 reference (NCBI build 36) using BWA (Li and Durbin 2009). We then defined the average mapped depth based on all the aligned reads after duplicate read-pairs were removed (8.8% on average) (see Table 1). We combined the two GAIIx flowcells and report them as a single data set, and report the two HiSeq 2000 flowcells individually (Table 1). Each of these three data sets yielded an amount of data fairly typical compared with previously published whole-genome studies that use massively parallel short-read sequencing technology (Fujimoto et al. 2010; Lee et al. 2010).
Table 1.
Sequencing and alignment summary
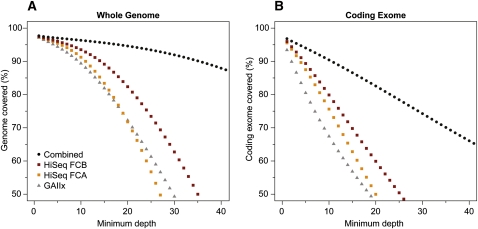
We first looked at the uniformity of coverage for each data set. To do this, we assessed what proportion of the genome and coding exome was represented at different minimum depths (Fig. 1A,B, respectively) of high-quality bases (≥Q20) from confident alignments (see below for definition and explanation). Theoretically, for a data set with perfectly uniform coverage, a large percentage of the genome will be covered at depths that approach its average mapped depth. However, Lander-Waterman statistics dictate that genomic coverage follows a Poisson distribution (Lander and Waterman 1988), and factors such as sample handling and library preparation may introduce biases in sequencing. It is also known that variation in data uniformity arises due to varying G+C content of the genome, specifically under-representation of regions with high G+C% (Bentley et al. 2008; Teer et al. 2010) and has been attributed to various amplification biases (Kozarewa et al. 2009; Aird et al. 2011). Regions with high G+C% are known to be correlated with high gene content (Mouchiroud et al. 1991; Zoubak et al. 1996; Vinogradov 2003) and explain the difference in slope we observe between coverage of the whole genome versus just the coding exome (Fig. 1, cf. A and B). Overall, we note that the three data sets have relatively similar coverage uniformity. HiSeq 2000 flowcell B (FC-B) performs slightly better and is likely due to the higher yield from this flowcell. As expected, combining all three data sets (Table 1, row “HiSeq 2000 FC-AB + GAIIx”) boosts the proportion of the genome with higher minimum depths.
Figure 1.
Breadth versus depth of whole-genome coverage. The x-axis represents the minimum number of high-quality bases (≥Q20) from high-quality alignments (≥MapQ30), and the y-axis represents the proportion of genome (A) or coding exome (B) covered at that depth. To calculate percentages, the total size of hg18 build and the total number of non-redundant coding bases from the UCSC Known Genes table (2,852,680,119 bp and 34,068,542 bp, respectively) were used. Gaps and pseudo-autosomal regions (PAR) were excluded. Values were plotted for GAIIx (triangle), HiSeq flowcell A (orange square), HiSeq 2000 flowcell B (dark red square), and all data sets combined (circle).
While coverage statistics reported in this manner convey how uniform a sequencing experiment is, it does not illustrate if the data generated are sufficient to make confident genotype calls and detect SNVs genome-wide. Many studies report the fraction of the genome and exome covered in terms of minimum 1×, 5×, or 10× depths, sometimes without base and alignment quality filters (Wang et al. 2008; Kim et al. 2009; McKernan et al. 2009; Fujimoto et al. 2010; Pelak et al. 2010; Sobreira et al. 2010). However, there is a finite probability associated with SNV detection such that all alleles might not be observed even at 10× depth, which can lead to an erroneous reference (or variant) genotype call. Keeping this in mind, a more informative metric about a whole-genome data set is what proportion of the genome is callable based on various genotype-calling filters (delineated below). Of the 2,852,680,119 positions (excludes gaps and pseudo-autosomal bases) examined in the hg18 build, 88.82% of the positions were callable in the GAIIx data set, 90.99% in HiSeq FC-A, and 93.10% in HiSeq FC-B. While these differences are largely due to the amount of sequence in each data set, we note that analyzing an equivalent amount of HiSeq 2000 data, compared with the GAIIx, still results in a slightly greater callable portion of the genome (by 2.92%) (see Supplemental Material). It is for this reason that we advocate here that, for the purpose of evaluating the completeness of “personal” individual genomes, the callable proportion should always be reported, as it is more reflective of the downstream usability of the data set.
In addition to the above analysis of uniformity, we also found that GAIIx and HiSeq 2000 runs were relatively equivalent with respect to SNP-chip concordance rates, alignment error rates, and G+C bias (see the Supplemental Material). We were therefore able to treat the reads as if originating from a single platform. This allowed us to pool all mapped reads from the GAIIx and HiSeq 2000 runs to create an extremely deep-sequenced data set (126×) with an average mapped depth totaling 102×, with which we could perform a number of informative downstream analyses.
Accurate genotype calling and SNV detection
We next sought to establish appropriate filters to produce accurate SNV calls from whole-genome sequencing. Specifically, we wanted to reduce incorrect genotype calls while maintaining a high overall sensitivity. To do this, we created two identical genomes by splitting the deep-sequenced data set described above into two equal-sized data sets (each with an average mapped depth of 50×) and attempted to minimize the apparent discordance between them; any discordant genotypes between the two data sets would likely represent incorrect calls.
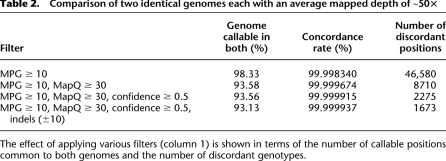
Genotype calls were made on unfiltered BWA alignments using the Bayesian genotype caller—Most Probable Genotype (MPG) (Teer et al. 2010)—and were then compared at overlapping positions. We first required bases to have qualities Q20 or greater and a genotype call to have a MPG score of 10 or greater, signifying that the theoretical probability of the call being incorrect is 1/e10 or 1/22,026. This is the lowest score that exceeds the minimum operational accuracy of the human reference genome assembly (1 error in 104 bases) (International Human Genome Sequencing Consortium 2004; Schmutz et al. 2004). Out of 2,805,179,303 positions that met or exceeded this score in both 50× genomes (98.33% of hg18 non-N and non-PAR bases), there were 46,580 discordant genotypes between the two data sets (Table 2). This equates to 1 in 60,223 positions that do not agree.
Table 2.
Comparison of two identical genomes each with an average mapped depth of ∼50×
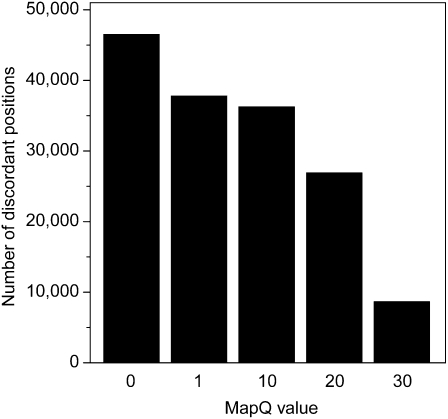
We and others have observed that a substantial non-trivial source of incorrect genotype calls arises from the improper alignment of sequenced reads (both read placement and basewise alignment). To minimize incorrect calls arising from such artifacts, we restricted reads to only those that were mapped with quality scores of 30 or higher. While this eliminated 11.7% of the mapped reads (see Supplemental Fig. 4), it also reduced the number of discordant genotypes by 81.31% to 8710 (Table 2). A mapping quality of 30 also showed the greatest reduction in the number of discordant genotypes compared with lower alignment quality values (Fig. 2).
Figure 2.
Effect of alignment filter on the discordance rate of identical genomes. The number of discordant positions (y-axis) was observed by varying MapQ values (x-axis). A MapQ value of 0 indicates that no mapping quality filter was applied.
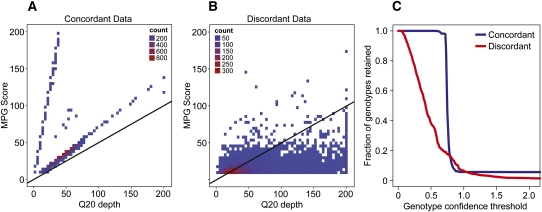
We further examined the remaining discordant positions and noted that their genotype confidence scores were, on average, lower than the concordant ones, particularly in high-coverage regions (Fig. 3, cf. A and B). Rather than using the same score threshold for all positions, we scaled this score based on the depth of coverage at any given position, which dramatically reduced the number of discordant genotypes. Specifically, we determined a confidence measure such that at higher depths of coverage, a higher genotype score is required. This filter reduced the number of discordant genotypes by 61.47% while retaining all of the sampled concordant positions (Fig. 3C) from one of the 50× genomes. By applying this filter to both 50× genomes, the number of discordant genotypes was reduced to 2275, and the proportion of callable positions in hg18 was only reduced by 0.02% to 93.56% (Table 2). We also demonstrate that the confidence filter is more broadly applicable and not specific to MPG. Specifically, we show that this filter also works with samtools/bcftools, resulting in a substantial reduction of discordant calls in a similar comparison of two identical genomes (see the Supplemental Material). A recent report also uses a similar variant quality-to-depth measure to recalibrate and improve variant calls (DePristo et al. 2011). Additional filtering for nearby indels further reduced the number of discordant calls to 1673 (out of 93.13% of the positions callable in both genomes), or a discordance rate of 1 in 1,588,046 (Table 2).
Figure 3.
Determination of genotype confidence threshold for genotype calls. (A,B) The x-axes represent Q20 depth for genotype calls from one of the 50× genomes, and the y-axes represent corresponding MPG scores. (A) A random set of ∼8700 concordant genotypes; (B) 8710 discordant genotypes. Black lines represent a line with slope of 0.5, which is the confidence threshold used to filter genotypes. (C) The fraction of genotypes retained by varying the confidence threshold; (blue curve) the fraction of concordant genotypes retained; (red curve) the fraction of discordant genotypes retained.
Since systematic errors in genotype calling cannot be identified by comparisons between two identical genomes, we compared our calls with those generated by genotyping the same sample on the Illumina Infinium HD Assay using the Human1M-Duo BeadChip. We compared the calls between one 50× data set and those made on the “clean” BeadChip positions (see Methods). We were able to call 1,068,010 out of 1,096,530 positions (97.40%), of which 1,067,563 agreed, yielding an overall concordance rate of 99.958% (or 1 in 2389 positions disagreeing with a call on the BeadChip) and 99.928% for positions called as heterozygous from sequencing. Similar results were also observed for genotype calls on the other 50× data set, the individual HiSeq 2000 flowcells, and the GAIIx data set (Supplemental Tables 3, 4). These results show that genotypes called over the whole genome using these methods are remarkably concordant with an independent assay.
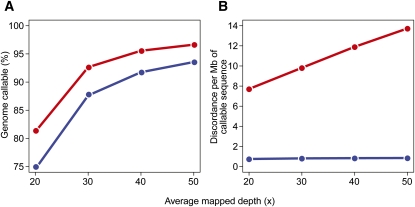
As a final illustration of the effect of these filters on sensitivity and discordance, we extended the analysis by comparing the identical genomes at various lower mapped depths (20×–40×) with and without genotype-calling filters (Fig. 4). When applying these filters, more sequence data were required to obtain the same sensitivity as a 30× genome analyzed using all unique (MapQ > 0) alignments (Fig. 4A). However, discordance between the identical genomes was an order of magnitude lower with these filters (Fig. 4B).
Figure 4.
Comparison of identical genomes at various mapped depths. (A,B) The x-axes represent the average mapped depths at which two identical genomes were compared. The y-axes represent the proportion of hg18 callable in both genomes (A) and the discordance per megabase of callable sequence (B). Analyses were done on all unique alignments (MapQ > 0) without applying any filters (red curve) and after applying mapping quality and genotype confidence filters as explained in the text (blue curve).
Whole-genome coverage and SNV detection as a function of sequencing depth
With a firm understanding of appropriate alignment and genotype score filters from the previous sections, we next performed a depth-of-coverage analysis to determine the relationship between the amount of sequence data generated and the proportion of the genome callable for confident genotype calls. We used these results to provide estimates of how much sequence data is needed to reach specific levels of completeness for whole-genome analyses. To the best of our knowledge, this is the first comprehensive study of the relationship between effect of sequence depth and the proportion of genome callable to determine how much sequence data is sufficient for a given study. In addition, we specifically sought to clarify ambiguous metrics of coverage currently being reported in the literature that do not necessarily correlate with what is callable, and propose the establishment of objective parameters by which one can describe the completeness of a whole-genome analysis.
To address these questions, we used the deep-coverage data set to create 20 different data sets that each represented an incremental coverage of the genome from ∼5× to 100× average mapped depth (before alignment quality filters). For each data set, we first calculated the percentages of the genome and CDS regions that were callable, which, based on the above results, we define as positions that satisfy the following criteria on MapQ ≥ 30 filtered alignments: (1) MPG score is ≥10 for the genotype call; (2) MPG score exceeds the confidence threshold based on depth of coverage at the called position (i.e., a higher minimum score at higher depths of coverage based on a simple formula of MPG/Q20 coverage ≥ 0.5) (see Methods); and (3) positions are not within 10 bp of a called indel.
At lower average mapped depths, the fraction of genome callable increased exponentially, followed by a linear increase until 30×. At this depth, 89.7% of the non-N and non-PAR bases genome-wide were callable (Fig. 5A; Supplemental Table 5) with high-quality bases. Beyond 50×, less than an additional 0.5% of the genome was callable with every successive data set, which suggested that these are likely regions where sequencing and/or alignments pose a challenge. Indeed, when we examine the 2.3 million positions (<0.1% of the genome) that were callable only at 100× (but not 95×), we noted that the median number of high-quality bases (Q20 or more) was ≤15 (Supplemental Fig. 7). When considering only CDS bases in the genome, only 70% were callable with the 30× data set, increasing to 81.36% with the 50× data set (Fig. 5A; Supplemental Table 5). While we believe that the callable portion of the genome should be used to report whole-genome sequencing studies, we also calculated the proportion of the genome and coding exome that are covered by 5, 10, and 20 or more high quality bases (Supplemental Fig. 6). We noticed a similar paucity of coding exome covered by 20 or more Q20 bases even at average mapped depths >50×. This may be due to GC-biases known to be platform-specific or library-specific high cluster densities that were observed for these flowcells (see GERALD summaries in the Supplemental Material). We show that the disparity between the proportion of genome and coding exome callable or covered may be reduced when these biases are mitigated (see below).
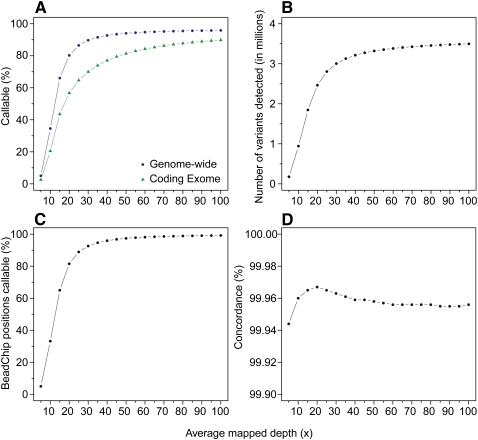
Figure 5.
Genotype calling as a function of average mapped depth. The x-axes represent the average mapped depth of each data set, and the y-axes represent the proportion of the whole genome (dark blue circles) and coding exome (green triangles) that is callable (A), the number of SNVs detected (B), the proportion of Illumina BeadChip positions callable (C), and the concordance rates with the BeadChip calls (D).
Next, we ran each data set through the above-established variant detection pipeline to assess the number of SNVs at every depth. We observed an exponential increase in the number of SNVs detected as we increased the average mapped depth to 25× before saturation at ∼40–45× (Fig. 5B; Supplemental Table 6). With the 50× data set, we detected 3,319,872 autosomal variants, 94.89% of what was observed at 100×. We also noted that 46,529 new variants were added at 55× compared with 50×, while 16,954 were found at 100× compared with 95× (Supplemental Table 6). Overall concordance of genotype calls with the BeadChip was at least 99.94% in all data sets (Fig. 5D) and >99.92% at heterozygote positions (data not shown), highlighting that our established alignment and variant detection filters are equally accurate at low depths of coverage and are only limited by sensitivity (Fig. 5C) with respect to the amount of input data. This is an improvement over other similar analyses where accuracy is affected at lower depths of coverage (Wang et al. 2008).
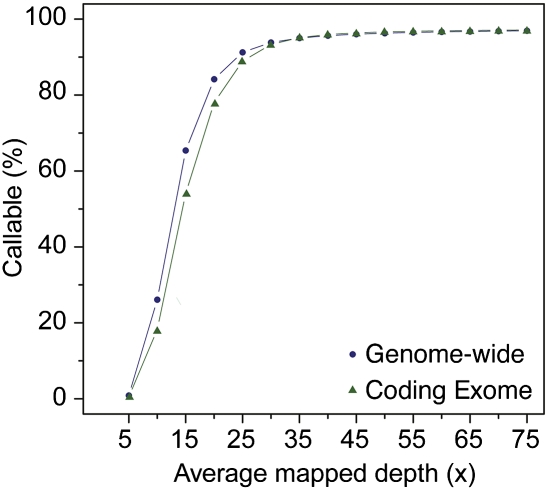
Using the methods developed here, an average mapped depth of 50× (or ∼45× using reads with MapQ ≥ 30) was required for this data set to produce confident genotype calls for >94% of the genome and >80% of the coding exome (which is what we minimally define as “comprehensive”). Given that an average of 85% passing-filter reads were mapped to the genome (Table 1), this equated to 170 Gb (or 60×) of passing-filter reads, taking into consideration the uniformity of data generated and the diversities of the libraries sequenced here. We note that the amount of sequence data required for confident calls is dependent on the uniformity of coverage, specifically the representation of all G+C fractions of the genome fairly equally. We recently sequenced an unrelated genome on the HiSeq 2000 platform using newer versions of chemistry and base-calling software. The average sequenced depth and mapped depth for this genome (aligned to hg19) was ∼90× and 75×, respectively. We repeated the sampling followed by genotype-calling experiment on this genome and noticed a much better representation of the coding exome even at 35× mapped depth (>40× sequencing depth), indicating a noticeable reduction in GC-bias (Fig. 6).
Figure 6.
Improved representation of genome with TruSeq v3 sequencing chemistry and software. The x-axis represents the average mapped depth of the data set, and the y-axis represents the proportion of the whole genome (dark blue circles) and coding exome (green triangles) that is callable.
Roughly 30× has been purported to be required for adequate whole-genome analyses (Koboldt et al. 2010), but we were unable to find any published quantitative data to support this level of coverage. To the best of our knowledge, the only relevant quantitative data that have been reported supporting this level of coverage is in Figure 5 from a recent publication (Bentley et al. 2008). However, their analysis highlighted the sensitivity and accuracy of identifying variants, not calling genotypes. Furthermore, it was limited to chromosome 2 with reduced stringency of detecting variants. While the Bentley et al. (2008) analyses highlighted the accuracy of the newly reported sequencing chemistry, it did not address genotype-calling accuracy for genome-wide studies, as we do here.
Discussion
In this study, we sequenced a clinical human sample on two separate Illumina platforms (GAIIx and HiSeq 2000) to a combined average mapped depth of 102×. Our analysis of the data generated from these platforms showed that sequence from both instruments were of comparable quality with the exception that HiSeq 2000 reads covered the genome slightly more uniformly. We then compared two identical 50× genomes to establish genotype-filtering methods, thereby generating genotype calls that were nearly identical (99.999937%) in both data sets and highly concordant (>99.955%) with those obtained by genotyping the same sample on the Illumina Human1M-Duo BeadChip. These filters were used to determine how much sequence data is required for a comprehensive study of an individual whole genome, particularly to call genotypes and detect SNVs with extremely high accuracy.
We also believe that an objective measure of completeness of genome sequencing is the proportion of reference bases where genotypes can be accurately determined using high-quality sequence data from confidently aligned reads. For example, with the data reported here (generated at the beginning of 2010), if we require that this proportion be at least 95% of the genome, we showed that a genome data set with an average mapped depth of 50× (before alignment filters) or average sequenced depth of 60× was necessary. We could have obtained similar sensitivity at lower mapped depths without any of the genotype filters, but at the great sacrifice of accuracy. Conversely, lower mapped depths (with these additional filters) may be sufficient to reach the level of completeness recommended above with improvements in sequencing technologies that produce more uniform coverage of the genome. Indeed, using newer chemistry and software, we sequenced an unrelated genome and determined that 95% of the genome and 95% of the coding exome were callable at 35× average mapped depth (>40× average sequencing depth assuming 85% read-pairs align).
It is important to note that the high sequencing depth needed to reach the level of completeness is more applicable for personal genomes (especially in a clinical setting) rather than population-scale sequencing studies. We also anticipate that it will be increasingly easier in the near future to generate greater amounts of data in an economical and timely manner, such that sequencing cost will be a small factor when designing whole-genome studies. This then lends to making highly confident genotype calls thereby minimizing the substantial time and cost needed to perform secondary validations and analysis of a small number of positions. Today, an average sequencing run spans 8–10 d and can produce two to four genomes. Secondary alignment, genotyping, and variant detection analyses can be completed in ∼3–4 d when using several hundred CPUs concurrently. However, the interpretation of these results is open-ended—the cost of these various tertiary analyses of whole-genome data will greatly overshadow that of sequencing (Mardis 2010), underscoring the importance of spending a bit more for increased accuracy in the initial data-generation phase.
As whole-genome sequencing becomes routine, it is important to understand what level of coverage is required to perform various base-pair-level analyses. With different metrics being reported currently in whole-genome studies, both in terms of quantity and quality of bases that cover the genome, there is no established measure of the completeness of a genome sequencing experiment. We, therefore, outline objective parameters that may be used to characterize such studies. We first define the “average sequenced depth” as the mean coverage of a haploid genome with usable sequence data that are obtained after applying various read-level filters. In our analysis of the sequenced genome, we use two filters—Illumina Chastity filter (PF) and a requirement that each read in a pair must contain at least 32 high-quality (≥Q20) bases (by which we eliminate ∼5% of the PF reads). We then define the “average mapped depth” as the mean coverage of a haploid genome after all usable read-pairs are mapped to the reference and duplicate read-pairs are removed. The average sequenced depth and average mapped depth, when taken together, then give a fair indication of the amount of “data loss” associated with (1) unmapped reads and (2) redundant sequence attributed to molecular and/or optical duplicates. To make accurate genotype calls, it is further necessary to filter out reads that are placed ambiguously on the genome, resulting in additional data loss (see Supplemental Fig. 4; Supplemental Table 5).
The proportion of genome covered by a specified minimum number of high-quality bases has been used as a metric to judge the comprehensiveness of a whole-genome sequencing experiment and is currently being reported in many such studies. While reporting the fraction covered at a 10× minimum depth might be informative if the uniformity of the sequence data were reproducible and predictable, this does not translate to a consistent proportion of the genome where high-confidence genotype calls can be made. From a probabilistic standpoint, if we assume a binomial model, there is a 22.6% chance of not identifying a known heterozygous position where the depth is 10× (using MPG) (see Supplemental Material) owing to events where (1) only one allele is observed leading to an incorrect homozygous call or (2) the heterozygous genotype call does not meet score requirement (MPG ≥ 10, confidence ≥ 0.5). Incidentally, at 10× depth, the MPG score of a homozygous genotype call is always <10 regardless of the allele observed. This probability and the number of less-confident genotypes is much higher at 1×–5× depths, such that reporting the fraction of genome covered at these minimum depths ceases to be useful for a personal genome. Therefore, in addition to the average sequenced and mapped depths, we report the proportion of bases in the genome and CDS where genotypes are callable with an accuracy that exceeds the minimum human genome reference base accuracy (1 error in 104 bases). In our analysis of different incremental data sets of the deep-sequenced genome, we show how the callable proportions of the genome and CDS vary as a function of average mapped depth (Fig. 4A; Supplemental Table 5). Importantly, these metrics are derived from an empirical analysis and can be used as a “sequencing guide” with respect to sequencing technology (circa spring 2010) noting that sample preparation, quality of input DNA, and other factors can dramatically affect the numbers reported.
As technologies improve, we anticipate a reduction in sequencing biases thus producing more uniform coverage of the human genome (as we show above) and thereby reducing the input data required to achieve the same level of genome representation. One approach to alleviate biases specific to a particular technology is to evaluate and adopt a multi-platform sequencing methodology to possibly increase the accessible portion of the genome (Sampson et al. 2011). Longer read lengths, improved experimental protocols, newer single-molecule sequencing technologies, improvements in alignment algorithms, and localized assembly techniques will also contribute to increasing the callable proportion of the genome.
Methods
Library preparation and sequencing
Two libraries with slightly different insert mean sizes, 378 bp and 436 bp (see Supplemental Fig. 1), were prepared from 5 μg of genomic DNA purified from a blood sample following the standard protocol provided by Illumina with the following modifications. After shearing with a Covaris instrument, but before adapter ligation, we performed an initial gross size selection by gel electrophoresis, cutting out an ∼200-bp band around the final desired size range. Also, to minimize any biases introduced in the final PCR amplification, we took an aliquot of the final library template and performed a series of PCRs with different numbers of cycles to determine the minimum amount of amplification needed to visually observe the template by gel electrophoresis. After a suitable number of cycles was determined, in this case six cycles, two tubes were amplified, pooled together, and purified using Agencourt Ampure SPRI beads.
One GAIIx run (378-bp library) was performed with version 4 chemistry, and the images were saved and subsequently analyzed offline with OLB v1.8 (equivalent to the image analysis algorithm of RTA v1.8). The other GAIIx run (436-bp library) was with version 5 chemistry and RTA v1.8. Sequence data from a single HiSeq 2000 run (378-bp library) with two flowcells were obtained from the on-rig RTA software. An unrelated human genome was sequenced on the HiSeq 2000 with TruSeq v3 chemistry and updated RTA (v1.10.36) to illustrate the effect of the improved chemistry/software. The remainder of our analyses were initiated from the FASTQ files provided by Illumina's downstream analysis CASAVA software suite.
Alignment of short reads
Paired-end fragments from each flowcell were pre-processed to eliminate read-pairs that did not pass Illumina Genome Analyzer Pipeline's chastity filter. In addition, both reads in a pair were required to have at least 32 Q20 bases. We then aligned read-pairs meeting these criteria using BWA against the reference human genome (hg18). Alignments were further processed to remove duplicate read-pairs using the rmdup utility from the samtools suite of programs. This resulted in an average mapped depth of 30×–40× of the haploid genome for the individual data sets (GAIIx, HiSeq A, and HiSeq B). We derived larger data sets by merging smaller ones and further removing duplicate read-pairs that may have been added.
Single-nucleotide variant detection
Genotype calls were made using Most Probable Genotype (MPG), a Bayesian algorithm that produces single-nucleotide and small-scale insertions and deletions (of the size that can be aligned across with BWA and 100-bp reads). The algorithm was originally published recently (Teer et al. 2010), and the most recent version and description can be found at http://research.nhgri.nih.gov/software/bam2mpg/. In the case of autosomes, genotypes were called in diploid mode, while non-PAR regions on ChrX and ChrY were called in haploid mode. We further imposed several filters as outlined below:
Base qualities
Bases were considered for genotype calling only if their phred-scaled qualities were 20 or greater.
Mapping qualities
Reads were considered only if their mapping qualities (see BWA MapQ description) were 30 or greater. This serves to eliminate reads that are (1) placed in repetitive regions on the genome, or (2) mapped with multiple mismatches or clipped bases, which occurs when sequences in the sample are aligned to paralogous regions in the reference.
Genotype score and confidence filters
For each genotype call made, a score is assigned to represent the difference between natural log-scaled probabilities of the most probable genotype and the next most probable genotype. We used a MPG score cutoff of 10 that was determined empirically but theoretically represents the first score at which the probability of an incorrect genotype call (e10 ≈ 1/22,026) exceeds the probability of an incorrect base in the reference sequence (one error in 104 bases). The MPG score was also required to be no less than half the number of Q20 bases that were observed at that position, that is, MPG:Q20-coverage ≥ 0.5. This filter allows us to scale up the MPG score threshold in regions where coverage is higher.
Indel filter
Indel calls identified by MPG were subjected to the same thresholds and criteria as variants (MPG ≥ 10 and confidence measure ≥ 0.5). SNVs that were within 10 bp of indel positions were then discarded.
BeadChip validation
We genotyped the clinical sample on the Human1M-Duo BeadChip, part of Illumina's Infinium HD assay. To prevent “sample effects,” we did not consider array positions that were (1) within 25 bp of an indel as determined by MPG and/or (2) within 25 bp of a variant as determined by sequencing. These indels and so-called hidden SNPs in the sample can likely result in nearby array positions being incorrectly genotyped. All filters were imposed on the largest data set (“HiSeq AB + GAIIx” or 100× data set in the depth-of-coverage analysis) to allow for between-data set comparisons using a common denominator.
Depth of coverage analysis
From the merged data set (102×) that was created, we sampled mapped read-pairs at random to create 20 different genomic data sets each with 5× more data than the previous. Only properly mapped read-pairs were considered using the “samtools view” command with the “−f 2” option. To parallelize this process on a computing cluster, we sampled read-pairs from 10-Mb segments of the genome. By doing so, read-pairs that span the boundaries of adjacent 10-Mb segments do not get sampled. We believe that this loss is not significant compared with the speedup that was achieved by parallelization. Each data set, N×, thus created contained all read-pairs in the data set (N-5)× and an additional 5× worth of data. For example, all the data in the 25× data set was also present in the 30× data set.
Acknowledgments
We thank J.C. Mullikin, N.F. Hansen, J.K. Teer, and P. Cruz for valuable advice; A.C. Young, R.W. Blakesley, and the NIH Intramural Sequencing Center for sequencing support; T.G. Belgard for critical review of the manuscript; the NHGRI genomics core for help with SNP-chip validation; J. Becker, M. Lesko, and B. Stanfield for IT support; and D.R. Bentley and K. Hall for early access to data generated on the HiSeq2000 platform. The data analyzed here were primarily generated for a continuing fruitful interdisciplinary collaboration with Drs. D. Adams, G. Golas, A. Gropman, C. Groden, R. Fisher, and W.A. Gahl, and other members of the NIH Undiagnosed Diseases Program. This work was funded by the Intramural Research Program of the National Human Genome Research Institute at the National Institutes of Health.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.123638.111.
Data access
Raw sequence data are publicly available from the European Nucleotide Archive's Sequence Read Archive (http://www.ebi.ac.uk/ena/) through accession number ERP000765.
References
- 1000 Genome Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird D, Ross MG, Chen W-S, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, Gnirke A 2011. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol 12: R18 doi: 10.1186/gb-2011-12-2-r18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kalscheuer V, Tzschach A, Menzel C, Ullmann R, Schulz MH, Erdogan F, Li N, Kijas Z, Arkesteijn G, et al. 2008. Mapping translocation breakpoints by next-generation sequencing. Genome Res 18: 1143–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang DY, Getz G, Jaffe DB, O'Kelly MJT, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES 2009. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods 6: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A, Nakagawa H, Hosono N, Nakano K, Abe T, Boroevich KA, Nagasaki M, Yamaguchi R, Shibuya T, Kubo M, et al. 2010. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nat Genet 42: 931–936 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Kim JI, Ju YS, Park H, Kim S, Lee S, Yi J-H, Mudge J, Miller NA, Hong D, Bell CJ, et al. 2009. A highly annotated whole-genome sequence of a Korean individual. Nature 460: 1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Ding L, Mardis ER, Wilson RK 2010. Challenges of sequencing human genomes. Brief Bioinform 11: 484–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ 2009. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods 6: 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Waterman MS 1988. Genomic mapping by fingerprinting random clones: A mathematical analysis. Genomics 2: 231–239 [DOI] [PubMed] [Google Scholar]
- Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, et al. 2010. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465: 473–477 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DCY, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, et al. 2010. Whole-genome sequencing in a patient with Charcot-Marie-Tooth Neuropathy. N Engl J Med 362: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER 2010. The $1,000 genome, the $100,000 analysis? Genome Med 2: 84 doi: 10.1186/gm205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, et al. 2009. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res 19: 1527–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud D, D'Onofrio G, Aïssani B, Macaya G, Gautier C, Bernardi G 1991. The distribution of genes in the human genome. Gene 100: 181–187 [DOI] [PubMed] [Google Scholar]
- Park H, Kim JI, Ju YS, Gokcumen O, Mills RE, Kim S, Lee S, Suh D, Hong D, Kang HP, et al. 2010. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet 42: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelak K, Shianna KV, Ge D, Maia JM, Zhu M, Smith JP, Cirulli ET, Fellay J, Dickson SP, Gumbs CE, et al. 2010. The characterization of twenty sequenced human genomes. PLoS Genet 6: e1001111 doi: 10.1371/journal.pgen.1001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin M-L, Ordonez GR, Bignell GR, et al. 2010. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J, Jacobs K, Yeager M, Chanock S, Chatterjee N 2011. Efficient study design for next generation sequencing. Genet Epidemiol 35: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Wheeler J, Grimwood J, Dickson M, Yang J, Caoile C, Bajorek E, Black S, Chan YM, Denys M, et al. 2004. Quality assessment of the human genome sequence. Nature 429: 365–368 [DOI] [PubMed] [Google Scholar]
- Sobreira NLM, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, Ge D, Shianna KV, Smith JP, Maia JM, et al. 2010. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet 6: e1000991 doi: 10.1371/journal.pgen.1000991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Bonnycastle LL, Chines PS, Hansen NF, Aoyama N, Swift AJ, Abaan HO, Albert TJ, NISC Comparative Sequencing Program, Margulies EH, et al. 2010. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res 20: 1420–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE 2003. DNA helix: the importance of being GC-rich. Nucleic Acids Res 31: 1838–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. 2008. The diploid genome sequence of an Asian individual. Nature 456: 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubak S, Clay O, Bernardi G 1996. The gene distribution of the human genome. Gene 174: 95–102 [DOI] [PubMed] [Google Scholar]