Abstract
High-throughput sequencing of cDNA (RNA-seq) is a widely deployed transcriptome profiling and annotation technique, but questions about the performance of different protocols and platforms remain. We used a newly developed pool of 96 synthetic RNAs with various lengths, and GC content covering a 220 concentration range as spike-in controls to measure sensitivity, accuracy, and biases in RNA-seq experiments as well as to derive standard curves for quantifying the abundance of transcripts. We observed linearity between read density and RNA input over the entire detection range and excellent agreement between replicates, but we observed significantly larger imprecision than expected under pure Poisson sampling errors. We use the control RNAs to directly measure reproducible protocol-dependent biases due to GC content and transcript length as well as stereotypic heterogeneity in coverage across transcripts correlated with position relative to RNA termini and priming sequence bias. These effects lead to biased quantification for short transcripts and individual exons, which is a serious problem for measurements of isoform abundances, but that can partially be corrected using appropriate models of bias. By using the control RNAs, we derive limits for the discovery and detection of rare transcripts in RNA-seq experiments. By using data collected as part of the model organism and human Encyclopedia of DNA Elements projects (ENCODE and modENCODE), we demonstrate that external RNA controls are a useful resource for evaluating sensitivity and accuracy of RNA-seq experiments for transcriptome discovery and quantification. These quality metrics facilitate comparable analysis across different samples, protocols, and platforms.
High-throughput sequencing applications are revolutionizing genome-wide analysis (Mardis 2008; Mortazavi et al. 2008; Celniker et al. 2009; Morozova et al. 2009; Gerstein et al. 2010; Metzker 2010; Roy et al. 2010). RNA-seq offers single-nucleotide resolution, strand specificity, and short-range connectivity through paired-end sequencing. Because of these strengths, there has been great interest in using RNA-seq to distinguish isoforms, calculate expression levels for transcripts, and uncover low abundance RNAs (He et al. 2008; Mortazavi et al. 2008; Nagalakshmi et al. 2008; Sultan et al. 2008; Wang et al. 2008, 2010; Passalacqua et al. 2009; Gerstein et al. 2010; Roy et al. 2010; Trapnell et al. 2010; Berezikov et al. 2011; Graveley et al. 2011).
While there are clear advantages to RNA-seq, it is less clear how well the procedure performs, as several studies have reported conflicting RNA-seq accuracy results. RNA-seq–determined concentrations of six in vitro synthetic transcripts show good linearity (Mortazavi et al. 2008), and in a study using quantitative PCR as the benchmark, RNA-seq showed better performance for genes with high expression, while two-channel microarrays were more sensitive in identifying differential expression between genes with low expression (Bloom et al. 2009). Using measurements on a pool of synthetic miRNAs, microarrays showed better correlation with input than RNA-seq (Willenbrock et al. 2009), suggesting that RNA-seq is inferior in this application. However several other studies have shown good correlation between microarray and RNA-seq results (Agarwal et al. 2010; Zhang et al. 2010). As these somewhat contradictory reports suggest, determining the accuracy, detection limits, reproducibility, dynamic range, and other performance measures of RNA-seq assays and establishing best practices are critical. Standardized objective benchmarks provide quantitative measures of system performance and can be used routinely for quality control or for verification or optimization of system performance when changes are made in reagents or instrumentation.
RNA standards allow one to determine if an RNA-seq assay accurately represents the composition of known input and to derive standard calibration curves that relate read counts to RNA concentration in the studied sample. In addition, using fixed controls of known exogenous sequences allows for the direct measurement of sequencing error rates, coverage biases, and other variables that affect downstream analysis, such as quantification of alternative isoforms. The use of RNA standards to compute these values rather than using endogenous transcripts (e.g., actin and other “housekeeping” gene transcripts) is easier and more reliable since the standards are identical across samples (e.g., constant expression, single isoform, not subject to misannotation, sequence polymorphism between the sample and reference genome, or other biological variation). RNA standards, as opposed to the usual DNA controls, undergo more steps of library preparation and hence reflect performance of the endogenous sample more closely. The External RNA Control Consortium (ERCC) is developing a set of RNA standards for use in microarray, qPCR, and sequencing applications (Baker et al. 2005; ERCC 2005; Devonshire et al. 2010). Here we present Illumina GAII–generated RNA-seq data from several modENCODE and ENCODE experiments that contain the Phase IV test set of ERCC RNA standards. Our objective was twofold: First, determine how RNA-seq performs on known inputs and, second, evaluate spike-in controls as a tool for determining the sensitivity and biases in current and future experimental and computational methods for RNA-seq.
Results
The ERCC is working to develop and disseminate a standard set of exogenous RNA controls for use in gene expression assays. These controls, and methods that apply them, will support confidence in measurement results by enabling objective, quantitative assessment of assay performance. In this study, we used a Phase IV test set of ERCC RNAs in a combinatorial design, where some RNA concentrations were constant across pools and others vary in a Latin-square design (see Supplemental Methods).
The ERCC consortium synthesized control RNAs by in vitro transcription of synthetic DNA sequences or of DNA derived from the Bacillus subtilis or the deep-sea vent microbe Methanocaldococcus jannaschii genomes. They also contain a poly-A+ tail mimic in the DNA template. These diverse sequences show at least some of the properties of endogenous transcripts, such as diversity in the GC content and length (Supplemental Table S1; Supplemental Fig. S2). Importantly, ERCC RNAs show minimal sequence homology with endogenous transcripts from sequenced eukaryotes. In RNA-seq experiments, this minimizes confounding alignment of ERCC reads to the target genome. Indeed, when we constructed a library (for all libraries used in this study, see Supplemental Table S2) from 50 ng of ERCC RNA (100% ERCC library, library 6) and sequenced it on an Illumina GAII using 36-nt reads, we found that only 0.5% of reads aligned (for parameters, see Methods) to the Drosophila melanogaster genome, and the vast majority of these reads were due to polyA/T alignments to unassembled portions of the genome. Less than 0.01% of reads in the library mapped to the human genome (hg19). Any spurious alignments of the ERCC reads to genes result in density spikes that are easily distinguished from reads derived from endogenous transcripts. We therefore concluded that ERCC RNAs are distinct from Homo sapiens and D. melanogaster transcripts and are unlikely to interfere with transcript discovery and quantification when used as spike-in controls in these genomes.
Library QC
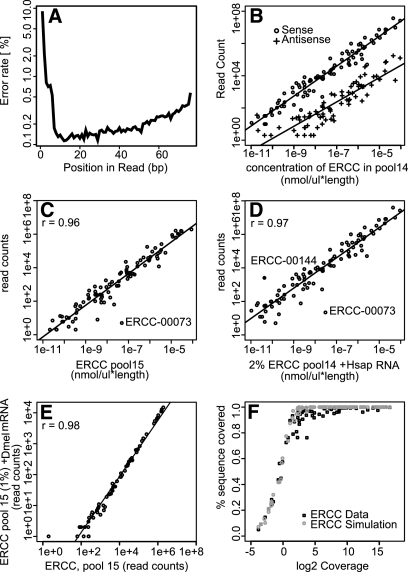
We then used ERCC RNAs to characterize parameters of RNA-seq data for downstream applications, including quantification and transcript annotation. In a set of libraries made from 2% mixtures of ERCC RNAs with H. sapiens mRNAs (libraries 7–50), we observed by far the highest sequence error rate at the first 6 nucleotides (nt) corresponding to the random hexamer priming site for the reverse transcriptase reaction during library preparation (Fig. 1A). We do not see such an increased sequence error rate at the paired-end read not corresponding to the random priming site, and previous studies have not reported it for Illumina DNA sequencing controls (Dohm et al. 2008), suggesting that these mismatches were due to imperfect hybridization between the primer and the RNA template. Error rates along the rest of the read increased with read length, as occurs for all Illumina sequencing runs due to the decline in the quality of sequencing chemistry over time.
Figure 1.
Library characteristics, ERCC quantification, and coverage. Quality control plots for a stranded ENCODE RNA-seq library of K562 cell Poly-A+ RNA with ERCC spike-ins (library 7). (A) Mismatch rate along reads mapped to all ERRC RNAs. The first 6 bp correspond to the random reverse transcription hexamer-priming site. (B) Scatter plot for sense and antisense read counts per ERCC. (C,D) Scatter plots of read counts versus mass (concentration times length) per ERCC: (C) 100% ERCC library (library 6) and (D) pool of 44 2% ERCC spike-in H. sapiens libraries (libraries 7–50). ERCC-00073 showed aberrant abundance patterns in multiple RNA-seq experiments, as did ERCC-00144 in ERCC pool 14. They may have been inaccurately quantified in our ERCC test set due to errors during the complex mixing scheme used to generate the pools, as they are also suspect in RT-PCR and array experiments on these ERCC pools (M Salit, unpubl.). (E) Scatter plot of read counts in the 100% ERCC library versus a 1% ERCC spike-in D. melanogaster library (library 5). (F) Average sequencing depth and percentage of primary sequence covered for all ERCC transcripts for real data (black) and simulated data (gray).
Antisense transcripts are of growing interest and are particularly challenging to annotate using RNA-seq in part due to strand errors introduced into libraries. The H. sapiens ENCODE libraries were prepared using a “dUTP” protocol to maintain strandedness (see Methods), where incomplete UNG digestion results in false antisense strand calls. One can estimate this global error rate in any library by quantifying the rate at which reads map to the complementary strand at annotated splice junctions, where the rules of splicing provide strand information. However, it is useful to uncouple this estimate from these limited sequence contexts, mapping uncertainties, and the poorly understood biology of antisense transcription. We measured the rate of this confounding effect directly by assessing the percentage of reads mapping in the antisense orientation to each ERCC RNA in these libraries (Fig. 1B). We found that 0.7% (±0.6%) of the inserts map to ERCC RNAs on the wrong strand (with one outlier at 3%). These measurements provide global false-positive rates and threshold levels for distinguishing endogenous antisense transcripts levels for each library, as bona fide endogenous antisense transcripts should occur in RNA-seq significantly more often than in ERCC controls.
Standard curves and detection limits
An understanding of the signal response in relation to input amount is critical for quantification, and spike-in controls are valuable for this, as they allowed us to determine the relationship between RNA-seq read counts and known inputs (Fig. 1 C,D). For detected ERCC RNAs, the relationship between RNA input abundance and read density output was constant over the six orders of magnitude in the 100% ERCC library and in libraries containing ERCC RNAs and either D. melanogaster or H. sapiens mRNAs (Pearson's r > 0.96 on log transformed counts). Since log-log scales obscure nonlinear effects, we also determined the slope of the regression (0.95 ± 0.03) and the correlation between input and read depth in the 100% ERCC library (library 6) by a test that transforms data to van der Waerden scores (Pearson's r = 0.93) (Lehmann and D'Abrera 1988) and in linear space (Pearson's r = 0.91). These results show linear quantification of the ERCC RNAs over six orders of magnitude. We found that ERCC read counts were highly correlated (Pearson's r > 0.98) between the 100% ERCC library and libraries mixed with mRNA from either H. sapiens (data not shown) or D. melanogaster (Fig. 1E), indicating that RNA-seq quantification of ERCC RNAs is uninfluenced by the complexity of endogenous RNAs in different species, a critical requirement for effective spike-ins. In practical terms, these data indicate that one needs to only sacrifice around 2% of reads to ERCC RNAs in a RNA-seq experiment in order to obtain a standard curve for quantification.
Random sampling of reads and overall library complexity always limit RNA-seq detection. Of the six ERCC RNAs that we failed to detect in the 100% ERCC RNA-seq experiment, five were among the least abundant, suggesting that failure to detect RNA was a consequence of low input abundance, random sampling, and sequencing depth. In this case, we loaded ∼11 μL of a 10−8 nmol/μL solution during GAII clustering, corresponding to 107 molecules, which represents an upper boundary on the number of reads in this lane. The five least abundant molecules in the 100% ERCC library (library 6) were present between 0.6 and 2.5 molecules in 107. Even under ideal conditions, if library preparation and clustering followed an unbiased Poisson distribution, the detection probabilities for these least abundant RNAs were 0.3 < P < 0.9. The final undetected ERCC-00134 RNA in the 100% ERCC library was input at 8.3 molecules in 107 and hence should have been detected (P > 0.99). However, this is one of the shortest ERCC RNAs in the pool (274 nt) and showed a high probability of secondary structure (data not shown). Both of these features could have altered gel mobility during size selection (∼200 bp) and resulted in exclusion during library construction.
High transcript coverage is critical for building transcript models from RNA-seq data, since ideally the entire length of a transcript needs to be covered by reads. Based on simulations, we expected that 5× sequencing coverage is required to cover 99% of a transcript with at least one read. In the real data, where reads are not perfectly distributed, at least 8× coverage of an ERCC was required to cover 99% of its primary sequence (Fig. 1F). We suggest that gene models derived from regions with greater than 8× coverage should be considered as high-confidence annotations. Measuring for which ERCC spike-ins this coverage has been achieved provides a benchmark for the sensitivity of an RNA-seq transcript discovery experiment.
Quantification and rare transcripts
To estimate transcript abundances, we used the spike-in data to infer transcript copies per D. melanogaster S2 cell in three libraries, one (library 3) with 5% and-two libraries (libraries 1, 2) with 2.5% ERCCs added to S2 poly-A+ mRNA. We used Tophat (Trapnell et al. 2009) and Cufflinks (Trapnell et al. 2010) to align, assemble, and estimate the mRNA isoform and ERCC RNA abundance. We fit the S2 cell output to a regression of ERCC abundance input and output (for detected ERCCs only) to derive a standard curve with confidence intervals of quantification (Fig. 2A). We used this calibration to determine the concentration of S2 cell mRNAs in the RNA extract relative to the known concentrations of ERCC standards. Since we also determined the yield of RNA (nanograms per cell) extracted from S2 cells, we estimated the average recovered transcript number per cell. In these libraries, a yield of one copy/cell corresponded to 4.4 fragments per kilobase per million mappable fragments (FPKM) (95% confidence interval 3.3–5.7 FPKM).
Figure 2.
Estimation of cellular transcript abundance in a S2 cell. (A) This plot shows results from a library (library 3) made of 100 ng S2 polyA+ RNA (mRNA yield for this extraction is 0.175 pg/cell) and 5 ng of pool 15 ERCC RNAs. A linear regression of abundance estimated from RNA-seq and the known input amounts. Dashed lines represent 95% confidence intervals for the regression fit. (B) Distribution of S2 transcript abundance estimated from RNA-seq.
Of the 15,111 annotated transcripts (Tweedie et al. 2009) we detected, 6720 (44%) had an FPKM < 4.0 (Fig. 2B), strongly suggesting that a large portion of the transcriptome in a given cell type is rare in our preparations. However, these rare transcripts, estimated at less than one copy per cell, are nevertheless observed reproducibly between the two replicate RNA-seq libraries (Pearson r = 0.45, P < 2.2 × 10−16). How many of these transcripts are biologically relevant remains an open question.
The extended dynamic range of the transcriptome creates a familiar problem for discovering rare transcripts. The most abundant 1.5% of RNAs (more than 100 copies/cell) accounted for 43% of the mapped reads, while the least abundant 44% of RNAs accounted for just 1% of the reads. Only 52 out of the 551 mRNAs encoding transcription factors were present at over 10 copies per cell. To achieve 99% coverage of an mRNA, we estimated that at least an 8× sequencing depth is required. Achieving this standard for D. melanogaster S2 transcripts present at one copy per cell requires at least 68 million uniquely aligned single-end 36-bp reads (see Supplemental Methods). Additionally the underrepresentation of certain sequences and short transcripts in RNA-seq protocols means that significantly more overall reads and possibly different library preparation methods are needed to make up for biases and to cover most transcripts.
RNA-seq quantification accuracy
While there is clearly a linear relationship between RNA concentration and read density in the ERCC RNA collection over six orders of magnitude, there were significant deviations from a perfect fit. We explored these deviations to better understand the noise and systematic biases in RNA-seq, which are important for downstream analysis. To quantify noise, we compared the read densities for each ERCC RNA between two ENCODE libraries constructed with the same pool of ERCC RNAs (libraries 7, 8) (Fig. 3A). Any differences in the relative read counts of these ERCCs represent variation introduced during the independent library preparations or sequencing of the samples. Overall, we observed good correlation between the libraries (Pearson's r = 0.99). However given the huge dynamic range of RNA concentrations, even a very high r-value can obscure significant variation, uncovered when looking at fold deviation of individual transcripts between the replicates (Fig. 3B). For low abundance RNAs, we found that Poisson sampling noise due to finite read depth was the dominant source of error, such that the fold deviation between technical replicates decreased with increasing abundance as reported previously (Marioni et al. 2008; Bullard et al. 2010). However, we observed a significantly greater variation than expected from a pure Poisson sampling model among all the ERCC RNAs (P < 2.2 × 10−16, likelihood ratio test for over-dispersion). To further quantify this effect, we looked at the variation in relative read counts for individual ERCC RNAs across 44 (libraries 7–50) independent ENCODE RNA-seq libraries (Fig. 3C,D). This fits the expectation of a negative binomial error model (Lloyd-Smith 2007; Robinson and Smyth 2007) and shows significant over-dispersion of read counts, even in the absence of biological variation within the ERCC controls. This error is introduced during library preparation, as we did not observe similar over-dispersion between read counts from individual sequencing lanes of the same library (Supplemental Fig. S1), even when run on different flowcells. Comparing the ERCC counts between two libraries measures the technical variability (measurement imprecision) between them, which can be used as a parameter when testing for differential expression.
Figure 3.
Quantification errors and biases. (A) Scatter plot of read counts for each ERCC transcript in two different libraries of human RNA-seq with 2% ERCC spike-ins (K562 A+ Repl.1 and K562 A+ Repl.2, libraries 7, 8). (B) Scatter plot of fold deviation between replicates versus read counts for a given ERCC RNA. (C,D) Read counts for two example ERCCs relative to the total number of ERCC reads across 44 different libraries (libraries 7–50) with ERCC spike-in H. sapiens RNA samples (black line), the negative binomial distribution (solid gray), and random samples (n = 44) from the negative binomial distribution (dotted gray). The observed distribution fits a negative binomial model over a Poisson model (P < 2.2 × 10−16, likelihood ratio). (E–G) Scatter plots of the fold deviation between observed and expected read count for each ERCC in the 100% ERCC library (library 6) compared with read count (E), GC content (F), and ERCC RNA length (G).
Given that we use many enzymes in RNA-seq experiments (e.g., reverse transcriptase, Taq polymerase, and Klenow) as well as chemical hydrolysis to fragment RNAs (in the 100% ERCC and D. melanogaster libraries; libraries 1–6) or cDNA shearing (in the H. sapiens libraries; libraries 7–50), measurement accuracy can be influenced by sequence-specific properties in different transcripts. Indeed, we saw better agreement between ERCC read counts from replicates than between the observed read counts and expected concentration of the ERCC RNAs within a given library (Figs. 1D; 3A), suggesting the presence of systematic biases. To explore transcript-specific sources of error, we tested if the ratio between the expected and observed read counts of each ERCC (library 6) correlated with characteristics of the ERCC RNAs. Accuracy in the observed read count values improved with read depth, higher GC content, and RNA length (Fig. 3 E–G). Including GC content and transcript length in addition to read depth in a component regression model produced a highly significant score (ΔBIC [Bayesian information criteria] = 12; see Methods). These results show that transcript-specific biases affect comparisons of RNA-seq read counts between different RNAs in one library, which are less accurate than comparisons of read counts for the same transcript in different samples.
Read coverage biases
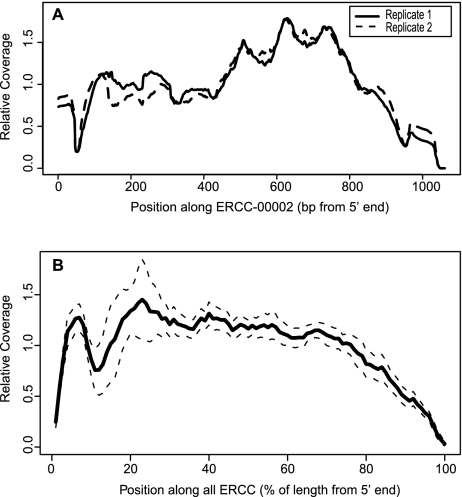
In addition to the global deviations outlined above, we observed significant reproducible unevenness in read coverage along transcripts similar to previous reports (Mortazavi et al. 2008; Li et al. 2010) both on the ERCC RNAs (Fig. 4A) and similarly on endogenous transcripts (data not shown). This pernicious effect is especially problematic for the task of isoform quantification, where one would like to use changes in read depth in a particular exon to estimate the abundance of an alternative isoform (Jiang and Wong 2009). The ERCC RNAs are all single isoform with well-defined ends and are therefore ideal for measuring read heterogeneity without complications from alternative or unknown transcript structures. Reproducible biases in coverage could have been due to effects common to all ERCC RNA, such as the position relative to transcript termini or to transcript-specific effects such as RNA sequence. We found clear evidence of common end-effects by averaging coverage along all 96 ERCC RNAs (Fig. 4B). We suggest that the drop in coverage at the 3′ end of ERCC RNAs was due to the inherently reduced number of priming positions at the end of the transcript. The central portion of the averaged ERCC transcript coverage was smoother than we observed in any individual ERCC RNA, where distinct, transcript-specific peaks and valleys were reproducibly observed between different libraries (Fig. 4A). These data confirm that both position and local sequence contribute to stereotypic heterogeneity.
Figure 4.
Stereotypic read density heterogeneity in ERCC RNA-seq. (A) Traces of relative coverage along ERCC-0002 in two different ENCODE libraries (libraries 7, 8). The pattern is highly reproducible (Pearson's r = 0.96). (B) Average relative coverage along all control RNAs for ERCC spiked in the H. sapiens libraries (libraries 7–50). Dashed lines represent 1 SD around the average across different libraries.
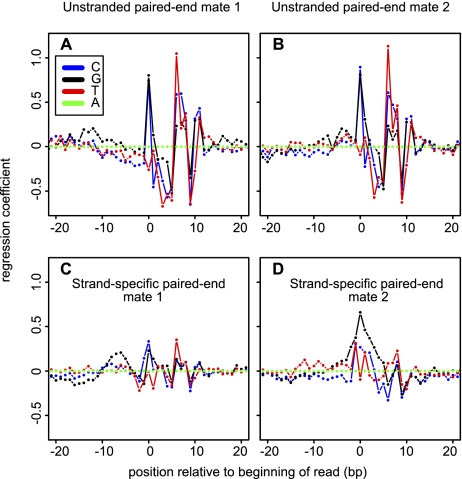
Sequence-specific coverage heterogeneity could be due to RNA structure (e.g., single- versus double-stranded template regions) and/or the preparation of the RNA (e.g., nonrandom hydrolysis) or the cDNA synthesis (e.g., reverse transcriptase priming site sequence preference or slight nonrandomness in “random” hexamer primers) and/or library construction (e.g., PCR biases). The ENCODE libraries (libraries 7–50) were prepared in a stranded and paired-end manner, giving the reads a fixed orientation relative to the original mRNA. This allows us to separate out the effects introduced at different parts of the library construction and the sequencing procedure according to the effect they cause on specific parts of the reads (Fig. 5 C,D). We confirmed that the strongest predictor of coverage was the sequence around the reverse-transcriptase priming site (Hansen et al. 2010), in our case exclusively at the read positions corresponding to the 3′ end of the RNA fragment, where we observed a strong G preference (C preference in the original mRNA). The 5′ end is generated by second-strand synthesis and cDNA fragmentation. There we found a completely different pattern, i.e., a C/G preference at the terminus and a T preference at +6 nt. The unstranded modENCODE D. melanogaster libraries (Fig. 5A,B) show a different sequence pattern, which is symmetric at both ends of read pairs. These patterns are thus strongly protocol dependent, highlighting the importance of assessing each RNA-seq protocol independently. We conclude that RNA-seq library construction and sequencing protocols introduce specific signatures that are quantifiable with the ERCC RNAs.
Figure 5.
Sequence patterns predictive of overrepresentation in RNA-seq. Patterns in the single-end 100% ERCC library (library 6) and ENCODE strand-specific pair end libraries (libraries 7–50) based on coefficients from the glm model (Li et al. 2010) (see Methods). (A,B) Regression coefficient for each base at positions around the beginning of reads mapped to the forward (A) and reverse (B) strands of ERCC-transcripts in the unstranded 100% ERCC library (library 6). (C,D) Regression coefficient for each type of nucleotide at different relative position to the upstream (C) or downstream (D) read of read pairs mapped to ERCC in the stranded ENCODE libraries. Adenosine is treated as base level in the regression model; i.e., the coefficient for “A” is always 0, while the other coefficients represent the predicted overrepresentation due to the presence of this nucleotide at this position, relative to an adenosine.
Previous work has used statistical models to smooth sequence-dependent stereotypic heterogeneity in coverage (Li et al. 2010). We wanted to use the strict single isoform nature of the ERCC spike-ins and their known input concentration to benchmark the performance of such a model and to test if its use can help better ascertain alternative isoform quantification. Li et al. (2010) used a Poisson general linear regression model (GLM), in which the number of reads mapping to a given position in a transcript was modeled as a log-linear function of the transcript abundance (the quantification signal) and the local sequence around the position (the sequence bias). For our 100% ERCC library (library 6), this model explained 50% of the variation in coverage (Fig. 6A–C). A more complex multiple additive regression trees model (MART) (Li et al. 2010) explained 67% of the variation. Smoothing with these models greatly improved the evenness of sequence coverage (Fig. 6A–C).
Figure 6.
Smoothing read densities. (A,B) Local read heterogeneity of a single ERCC RNA in the 100% ERCC library (library 6). Smoothing read density using the GLM linear model (B), and the more complex MART model (C; see text). (D) Variance in read depth of randomly drawn 50-bp windows from all ERCC RNAs based on an unbiased simulation, raw data, and the smoothed coverage from the sequence specific models. (E) The effect of coverage on read depth variance in simulated data. For the most abundant quartile of transcripts (Q1, mean coverage >19.9), the ratio of the read depth of 50-bp windows to the average depth is between 0.2853 and 1.2360. For Q2, (mean coverage >1.5), inner quartile range for the ratio is between 0.1917 and 1.2540. For Q3, (mean coverage >0.09), it is 0 with moderate large outliers (>21). For Q4 (mean coverage <0.08), it is 0 with very large outliers (>400).
Exons in higher eukaryotes are often short and, therefore, susceptible to strong bias from local read depth heterogeneity. Therefore, we were especially interested to see if this correction improved the accuracy of quantifying short regions of a transcript. To model the effect of sequence biases on mRNA isoform quantification, we binned ERCC data (library 6) into small exon-sized fragments (50 nt) and asked how well the read density of those fragments agreed with the overall read density of the ERCC, compared with similarly binned data from unbiased simulated reads (Fig. 6D). The real data showed significantly increased variation relative to the simulation (P < 10−9, unpaired Wilcoxon rank sum test). We then used the GLM and MART bias models to smooth read coverage and compared the read depth heterogeneity to the simulations and unadjusted numbers. Both models improve homogeneity (GLM P = 0.06; MART P = 7 × 10−6 unpaired Wilcox rank sum test). The agreement between read counts in windows and mean read count across the RNA drops precipitously in the third quartile of coverage in the simulation (less than 1.5× coverage), and scatter is greater (Fig. 6E). We conclude that both correction for heterogeneity and sufficient read depth are important for quantifying transcript isoforms generated by alternative splicing, promoters, and termination sites.
Discussion
Here we characterized a complex pool of synthetic control RNAs for use in RNA-seq experiments. We assessed the precision (repeatability and reproducibility), dynamic range, and linearity of RNA-seq experiments using the RNA standards in a library constructed solely of ERCC RNAs, and we demonstrated their utility as spike-ins in complex D. melanogaster and H. sapiens samples to derive standard curves. Our experiments on 100% ERCC and spike-in libraries provide definitive evidence that RNA-seq provides useful input/output response over the entire measurement range.
More generally, we suggest external RNA standards are a powerful tool for routine assessment of RNA-seq experiments and during experimental and computational protocol development. Many values that are commonly computed on endogenous transcripts for this purpose, such as sequence base call error rates, insert size distributions, library complexity, average transcript coverage, or inconsistent mapping of read-pairs can be confounded by incomplete annotation, alternative isoforms, or sequence divergence between the reference genome and other inherently variable aspects of biology. ERCC RNAs provide more reliable and consistent measurements and greatly facilitate comparisons of data quality across different biological samples. We have also used them as benchmarks to estimate the precision of RNA-seq quantification, tested the common assumptions about noise distributions, and estimated confidence in quantification by RNA-seq. Several different protocols for RNA-seq are available on different sequencing platforms, which differ in the errors and biases they introduce into the data. We have used a couple of different library construction protocols, alignment methods, and species, yet the comparable results obtained on the ERCC controls allowed us to have confidence in these methods and in data compatibility for future meta-analysis.
Several studies have identified biases and errors resulting from library construction and sequencing chemistry on the Illumina instruments (Dohm et al. 2008; Bullard et al. 2010; Hansen et al. 2010; Li et al. 2010). ERCC RNAs allowed us to quantify the systematic biases in quantification, such as underrepresentation of short transcripts, and the read coverage heterogeneity. We extend the previous work (Hansen et al. 2010) showing directly that random hexamer reverse transcription priming sites contribute strongly to both qualitative (mismatches) and quantitative (density) errors. More generally, these results highlight that while the reproducibility of transcript quantification by RNA-seq is very high, significant transcript-specific biases affect the ability to compare read-densities (FPKMs) between different RNAs.
Over the past decade, we have become accustomed to questioning low-end expression in microarray experiments due to the challenge of interpreting signals that approach the cross-hybridization signal background (van Bakel et al. 2010). In RNA-seq experiments, low-abundance expression is subject to sampling noise during library construction (complexity) and the surface area available on the sequencing flow cell. In general, we found that detection limits of low abundance transcripts in RNA-seq experiments behave as expected from random sampling, while the highly abundant transcripts show no sign of saturation. Specific features of individual transcripts, however, especially length and GC content, can lead to significant underrepresentation or failure of detection.
There are important consequences of sampling that have not been widely addressed. Some transcripts show abundances below one copy per cell, as a stable protein that is present at about one per cell in yeast, can be produced by a transcript expressed every few cell divisions (Ghaemmaghami et al. 2003). While deeper sequencing from suitable libraries might generate enough reads to discover even the rarest transcripts in a cell culture line, this is unlikely to be the case for tissues, organs, and organisms. For example, if a transcript present at one copy per cell is expressed in 1% of cells in a D. melanogaster tissue, then we estimate that more than 6.8 billion 36-bp reads would be required for 8× coverage of that transcript (see Supplemental Methods). Even such extremely deep sampling will not be helpful if the levels are below background, as determined by modeling read errors. Additionally, there are limits to library complexity, which will tend to make rare transcripts in complex tissues mixtures appear stochastically (and thus fail in replicates). Rare transcripts are clearly a challenge. For example, in the modENCODE D. melanogaster developmental RNA-seq profile (Graveley et al. 2011), genes such as dsx, which have transcripts expressed in a few cells only in the male embryos (Hempel and Oliver 2007), are not detected despite read depths of over 100 million uniquely mapped reads. Such transcripts are beyond reliable detection using the types of libraries and methods we report here. Additional methods such as subcellular fractionation, cell type isolation, and library normalization or targeted enrichment will be required to reach the bottom of the transcriptome (Kapranov et al. 2007; Bogdanova et al. 2008).
To control for most steps of RNA-seq library preparation, it is preferable to add spike-in controls as early in the protocol as possible. In the current test version of the ERCC pool, the short Poly-A tail mimics preclude their addition prior to oligo-dT selection. Future versions could also be extended to cover longer transcripts, possibly with multiple isoforms as well as a set of short RNAs with different 5′ and 3′ ends to make them useful in different protocols.
As the strengths and weaknesses of RNA-seq become better explored in experiments with known input RNAs, we will be able to identify problems and devise more powerful strategies. The demonstrated robust linear relationship between input and output in RNA-seq is clearly a major strength. The identified biases can be tracked and corrected through further experimental and computational protocol development. In addition to helping benchmark RNA-seq experiments, especially during protocol development, widespread adoption of external RNA standards by researchers and those in the biomedical community provides robust quality metrics for all steps following their addition and will facilitate meta-analysis of deposited data sets with radically differing protocols and data handling pipelines.
Methods
ERCC control RNA pools
The ERCC consortium synthesized RNAs by in vitro transcription of de novo DNA sequences or of DNA derived from the B. subtilis or the deep-sea vent microbe M. jannaschii genomes. The pools used in this study were prepared for the ERCC Phase IV testing process from individually purified RNAs using a series of subpools and dilutions (see Supplemental Methods). These ERCC pools are available from several commercial vendors under the names ERCC spike-in control mixes or NIST RNA controls.
ERCC pools were stored in Ambion's citrate buffer RNA Storage Solution, THE RNA Storage Solution. To test for stability after preparation by in vitro transcription, the individual RNA species were incubated at 37°C overnight, before and after spectrophotometric scans and Bioanalyzer electropherograms; the NIST specification was no observable change in the electropherogram or spectrum. All ERCC pools used in this study were prepared in a large batch, and no systematic changes in RNA structure or relative abundances over time were observed.
RNA-seq libraries
All libraries used in this study, their RNA sources, identifications, accession numbers, and summary statistics are presented in Supplemental Table S2.
ENCODE
H. sapiens cells were grown according to ENCODE growth protocols and standards (for a list of the cell types used, see Supplemental Table S2). Briefly, we lysed cells in QIAzol (Qiagen) and extracted RNA with miRNeasy (Qiagen), which we then treated with RNase-free DNase (Roche) in the presence of RNasin (Ambion). Total RNA was run on a BioAnalyzer to check for integrity and to determine the concentration. Only RNA with a RNA integrity number (RIN) >9.5 was used for library construction. Poly-A+ RNA was isolated with Oligotex (Qiagen) and depleted of rRNA using Ribominus (Invitrogen). Stranded libraries were prepared using the dUTP protocol (Parkhomchuk et al. 2009). Briefly, 100 ng of human Poly-A+ RNA >200 nt and 2 ng of ERCC pool 14 RNA were used in a random hexamer (Invitrogen) and oligo-dT (Invitrogen) primed reverse transcription with Superscript III (Invitrogen) reaction carried out in the presence of actinomycin D (Invitrogen). Second-strand synthesis was carried out by Escherichia coli DNA polymerase 1 (Invitrogen) from RNAse H (Invitrogen)–generated priming sites. dTTP was replaced with dUTP (Roche) during second-strand synthesis. cDNAs were fragmented by sonication (Covaris). Illumina Y-adapters were added to end-repaired cDNA fragments (Illumina genomic DNA protocol). The library was then rendered directional by eliminating the second strand using UNG digestion. Fragments with insert sizes at 200 bp (±50 bp) were size-selected on an agarose gel and used as templates in a PCR reaction to append Illumina p5 and p7 sequences to facilitate cluster formation and pair-end sequencing. In this study, data from 44 different human ENCODE samples (libraries 7–50) were used. Each library was sequenced to an average depth of 100 million read-pairs with 2 × 76 bp read length on the Illumina GAIIx.
modENCODE
Fly libraries were prepared solely from 50 ng of pool 15 ERCC RNAs or from mixture of 1 ng, 2.5 ng, 5 ng, or 10 ng ERCC RNAs with 100 ng D. melanogaster S2 cell line mRNA as described (Zhang et al. 2010). The D. melanogaster S2 cells were from RNAi titration experiments (sham, msl2, or mof RNAi) that supported a previous set of experiments but were not previously published. The yield of poly-A+ RNA, as determined by NanoDrop and cell number by hemocytometry, resulted in calculation of 0.175 pg/cell (sham, 5% ERCC, library 3), 0.155 pg/cell (msl2, 2.5% ERCC, library 4), and 0.142 pg/cell (mof, 1% ERCC, library 5). Another two libraries (libraries 1, 2) were made from a mixture of mRNA extracted from untreated S2 cell (different biological repeat as described above with 0.165 pg/cell mRNA). We made a master mix with 7.5 ng of ERCC and 300 ng of mRNA extracted from untreated S2 cells. Libraries were made with 100 ng of input mRNA with the gel isolation preceding or following the PCR step in library construction. Briefly, poly-A+ RNA was fragmented with zinc buffer (Ambion) and used for first-strand cDNA synthesis with random hexamer primers (Invitrogen) and reverse transcriptase (Invitrogen). This was followed by second-strand DNA synthesis, end repair (Illumina genome DNA sample preparation kit), poly-A addition (Illumina genome DNA sample prep. kit), and adaptor ligation (Illumina genome DNA sample prep. kit). cDNAs at 200 bp (±50 bp) were isolated using agarose gel electrophoresis and amplified by 15 cycles of PCR (Illumina genome DNA sample prep. kit). We obtained 36-bp reads on the Illumina GA II platform.
Data handling
For human libraries mapping was done using STAR software (available at http://gingeraslab.cshl.edu/STAR) (A Dobin, C Davis, F Schlesinger, J Drenkow, C Zaleski, S Jha, P Batut, and T Gingeras, in prep.) which allows for split mapping of reads against known and novel splice junctions. We only used read-pairs that consistently map to a single locus against the human genome (hg19) and the ERCC reference sequences simultaneously. For fly and 100% ERCC libraries, reads were compiled from Illumina pipeline (1.4.0). Reads passing the Illumina quality filtering were retained for the downstream data analysis and mapped with Bowtie version 0.11.3 (Langmead et al. 2009) to the ERCC reference and/ or D. melanogaster genome FlyBase Release 5 (dm3) with parameters –v 2 –m 1. For transcript level abundance estimates, we mapped with TopHat (1.0.13) (Trapnell et al. 2009) and used the ERCC and FlyBase annotation 5.12 in Cufflinks (0.8.2) (Trapnell et al. 2010) to calculate FPKM.
Simulated reads
To simulate ideal uniform coverage, perl scripts were used to simulate reads of 36 bp following a Poisson distribution. To generate simulated reads, probability distributions were first obtained from real data to model mismatches and quality scores, as well as a frequency table of mismatch types. Simulated ERCC reads proportional to the pool input concentrations were mapped against the ERCC reference with Bowtie, and results for all mapped reads were parsed. To quantify heterogeneity of reads distribution and its effect, we matched coverage of ERCC in the simulation with the real data. Simulated reads were then mapped with Bowtie (0.11.3) with parameters –m 1 –v 2, which forces uniqueness, allowing up to two mismatches.
Sequence bias models
We made heavy use of the R environment of BioConductor (Gentleman et al. 2004). Adjustment based on local sequence preference was carried out with R package mseq (version 1.1) according to the method described by Li et al. (2010). Briefly, we used the 56 highest abundance ERCC RNAs from the ERCC library (library 6; mean coverage was more than 10 for all these 56 ERCC RNAs) as a training data set. We analyzed reads initiated from each position with the expandData function in R package mseq to extract the local sequences (extending bidirectionally 40 bp) of each position along ERCC RNAs. Then the training data set was applied as input for both the GLM and MART models to obtain a sequence preference model. Fivefold cross-validation was applied during training. A cross-validation score was obtained to evaluate the eligibility of the strategy to our data (GLM > 0.5; MART > 0.6). Then the sequencing preference model was applied to the whole data set to obtain adjusted reads count initiated at each position. For the pooled ENCODE, stranded paired-end libraries (libraries 7–50) reads were analyzed separately. Read 1 (upstream) was treated as above for the single-end data, while for read 2 (downstream), the end (3′) position was considered. To avoid the observed RNA edge-effects obscuring the sequence-specific patterns, we removed 50 bp from the 5′ end and 100 bp from the 3′ end of each ERCC from the analysis.
Transcript segment coverage
An in-house script was used to randomly sample contiguous 50-bp windows along the center (trimming 136 bp from both ends) of ERCC RNAs longer than 422 bp and detected in RNA-seq on the pure ERCC library (library 6). We sampled 100 windows for each ERCC RNA. The mean read counts (number of 5′ reads falling into a region) were calculated, and then the ratio of mean counts over mean read counts of the central portion of the ERCC was calculated. We compared the distribution of this ratio between real data, simulation data, and data after smoothing with mseq models.
Quantification error model
The Poisson and negative binomial models were fit as generalized linear regression models using the glm and glm.nb functions in R (package MASS version 7.3-5) with a logarithmic link function (i.e., general log-linear regression). glm.nb iterates estimation of the regression parameters and the over-dispersion parameter of the negative binomial error term until convergence. For replicate read counts, ERCCs with a count of zero in one replicate were excluded from model fitting. For the quantification standard curve, the raw number of reads of each ERCC is modeled as a function of the concentration of that ERCC (in molecules, i.e., its copy number) multiplied by its length in base pairs. To test for over-dispersion in these models, we compare the regression models with Poisson and Negative Binomial error models using a likelihood ratio test implemented in the odTest function in the R package pscl.
Quantification bias
We used the nlme (version 3.1-97) R package to fit the component regression models and to compute the BIC score for the models:
where Y denotes the read counts in the pure ERCC library; M indicates the number of molecules; L and G denote the length and GC content of the ERCC molecules; b0, b1, b2, and b3 are coefficients; and e is residual error. All of these variables are in log2 scale. We performed ANOVA tests of model 3 in R.
Data access
All sequencing data sets have been submitted to GEO under accession nos. GSM516588, GSM516589, GSM517059, GSM517060, GSM517061, GSM517062, and GSE26284.
Acknowledgments
We thank the members of our laboratories as well as the modENCODE, ENCODE, and ERCC consortia for valuable discussions. We thank Carlo Artieri and David Sturgill for pilot work on data analysis and comments on the manuscript. This work was supported in part by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Disease, and by the National Human Genome Research Institute Grant 5U54HG004557-05. Disclaimer: Certain commercial equipment, instruments, or materials are identified in this document. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.121095.111.
References
- Agarwal A, Koppstein D, Rozowsky J, Sboner A, Habegger L, Hillier LW, Sasidharan R, Reinke V, Waterston RH, Gerstein M 2010. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics 11: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SC, Bauer SR, Beyer RP, Brenton JD, Bromley B, Burrill J, Causton H, Conley MP, Elespuru R, Fero M, et al. 2005. The External RNA Controls Consortium: a progress report. Nat Methods 2: 731–734 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. 2011. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res 21: 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA 2009. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics 10: 221 doi: 10.1186/1471-2164-10-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanova EA, Shagin DA, Lukyanov SA 2008. Normalization of full-length enriched cDNA. Mol Biosyst 4: 205–212 [DOI] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 11: 94 doi: 10.1186/1471-2105-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devonshire AS, Elaswarapu R, Foy CA 2010. Evaluation of external RNA controls for the standardisation of gene expression biomarker measurements. BMC Genomics 11: 662 doi: 10.1186/1471-2164-11-662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H 2008. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res 36: e105 doi: 10.1093/nar/gkn425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ERCC 2005. Proposed methods for testing and selecting the ERCC external RNA controls. BMC Genomics 6: 150 doi: 10.1186/1471-2164-6-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science 330: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Brenner SE, Dudoit S 2010. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res 38: e131 doi: 10.1093/nar/gkq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW 2008. The antisense transcriptomes of human cells. Science 322: 1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel LU, Oliver B 2007. Sex-specific DoublesexM expression in subsets of Drosophila somatic gonad cells. BMC Dev Biol 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH 2009. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25: 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann EL, D'Abrera HJM 1988. Nonparametrics: Statistical methods based on ranks. McGraw-Hill, New York [Google Scholar]
- Li J, Jiang H, Wong WH 2010. Modeling non-uniformity in short-read rates in RNA-Seq data. Genome Biol 11: R50 doi: 10.1186/gb-2010-11-5-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Smith JO 2007. Maximum likelihood estimation of the negative binomial dispersion parameter for highly overdispersed data, with applications to infectious diseases. PLoS ONE 2: e180 doi: 10.1371/journal.pone.0000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER 2008. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9: 387–402 [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML 2010. Sequencing technologies—the next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Morozova O, Hirst M, Marra MA 2009. Applications of new sequencing technologies for transcriptome analysis. Annu Rev Genomics Hum Genet 10: 135–151 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123 doi: 10.1093/nar/gkp596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passalacqua KD, Varadarajan A, Ondov BD, Okou DT, Zwick ME, Bergman NH 2009. Structure and complexity of a bacterial transcriptome. J Bacteriol 191: 3203–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK 2007. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23: 2881–2887 [DOI] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371 doi: 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wu Z, Zhang X 2010. Isoform abundance inference provides a more accurate estimation of gene expression levels in Rna-Seq. J Bioinform Comput Biol 8: 177–192 [DOI] [PubMed] [Google Scholar]
- Willenbrock H, Salomon J, Sokilde R, Barken KB, Hansen TN, Nielsen FC, Moller S, Litman T 2009. Quantitative miRNA expression analysis: comparing microarrays with next-generation sequencing. RNA 15: 2028–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, Oliver B 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol 8: e1000320 doi: 10.1371/journal.pbio.1000320 [DOI] [PMC free article] [PubMed] [Google Scholar]