Abstract
The daf-12 gene acts at the convergence of pathways regulating larval diapause, developmental age, and adult longevity in Caenorhabditis elegans. It encodes a nuclear receptor most closely related to two C. elegans receptors, NHR-8 and NHR-48, Drosophila DHR96, and vertebrate vitamin D and pregnane-X receptors. daf-12 has three predicted protein isoforms, two of which contain DNA- and ligand-binding domains, and one of which contains the ligand-binding domain only. Mutations cluster in DNA- and ligand-binding domains, but correspond to distinct phenotypic classes. DAF-12 is expressed widely in target tissues from embryo to adult, but is upregulated during midlarval stages. In the adult, expression persists in nervous system and somatic gonad, two tissues that regulate adult longevity. We propose that DAF-12 integrates hormonal signals in cellular targets to coordinate major life history traits.
Keywords: Nuclear receptor, dauer formation, heterochronic gene, aging, diapause, endocrine regulation
At distinct points in a species life history, cellular programs across various tissues require global coordination. In particular, widespread changes in gene expression are associated with stage transitions and diapause, where development either advances or arrests throughout the organism. In many species, such transitions are often coupled closely to environmental conditions, such as food availability, to maximize reproductive success (Finch 1994). Endocrine mechanisms ensure such global coordination from the cellular to organismal level, coupling complex environmental cues to metabolic, homeostatic, and developmental processes.
In favorable environments, the nematode C. elegans develops rapidly from embryo, through four larval stages L1–L4, to reproductive adults. In unfavorable environments, they arrest prior to reproductive maturity, either as an L1 larva, or as a specialized L3 diapause form, the dauer larva. They resume development to reproductive adults upon return to favorable environments (Riddle and Albert 1997). Stage transitions and diapause both entail organism-wide commitments that coordinate cellular programs throughout nematode tissues. For example, during reproductive growth, epidermal seam cell division, intestinal cell endoreplication, gonadal leader cell migrations, and various programs of gene expression are synchronized with the molt cycle (Sulston and Horvitz 1977; Hedgecock and White 1985; Johnstone and Barry 1996; Antebi et al. 1997, 1998; Jeon et al. 1999). Stage commitments themselves are perhaps defined most clearly by the start of cuticle synthesis during the molt cycle (Antebi et al. 1998), and precede ecdysis by ∼6–7 hours (Johnstone and Barry 1996). Post-embryonic stage-appropriate cellular programs, denoted S1, S2, S3, S4, and SA, are likely initialized at these times (Antebi et al. 1998). Similarly, during L1 and L3 diapauses, cellular programs of growth, division, and migration coordinately arrest. Animals committed to dauer diapause further undergo a morphogenetic program termed S3d.
Genes in heterochronic and dauer pathways control distinct aspects of nematode life history. Heterochronic activities record elapsed stages within cells and control stage-specific cellular programs (Ambros and Horvitz 1984; Ambros 1989). At the organismal level, mutants delete or add life stages; at the cellular level, mutants advance or delay expression of stage-appropriate programs (Ambros 1997). Identified gene products include presumed trancriptional and translational regulators (Ruvkun and Giusto 1989; Lee et al. 1993; Wightman et al. 1993; Rougvie and Ambros 1995; Moss et al. 1997; Jeon et al. 1999; Reinhart et al. 2000), which affect temporal identity of stage programs, not progression per se.
The products of dauer formation genes integrate environmental signals (food, pheromone, and temperature), and mediate commitment to reproductive development or dauer arrest (Golden and Riddle 1984). Dauer-constitutive mutants (Daf-c) always form dauer larvae, whereas dauer-defective mutants (Daf-d) bypass dauer formation regardless of environmental cues (Riddle et al. 1981). Identified gene products include components of cGMP and TGF-β signaling that relay information from neurosensory cells to other parts of the soma (Georgi et al. 1990; Bargmann and Horvitz 1991; Estevez et al. 1993; Vowels and Thomas 1994; Ren et al. 1996; Schackwitz et al. 1996; Patterson et al. 1997; Coburn et al. 1998; Inoue and Thomas 2000) and an insulin/IGF-I-like receptor signaling pathway whose focus is more widely distributed (Morris et al. 1996; Kimura et al. 1997; Lin et al. 1997; Ogg et al. 1997; Apfeld and Kenyon 1998; Ogg and Ruvkun 1998). Apparently, these endocrine mechanisms regulate choice of alternative programs within rather than between stages. Interestingly, the insulin branch of the pathway also influences adult life span. For example, mutations in the daf-2 insulin/IGF-I-like receptor and the age-1 PI3 kinase extend life span twofold or more (Friedman and Johnson 1988; Kenyon et al. 1993; Dorman et al. 1995; Larsen et al. 1995; Morris et al. 1996; Kimura et al. 1997) and this extension depends on the daf-16(+) forkhead/AFX transcription factor (Kenyon et al. 1993; Lin et al. 1997; Ogg et al. 1997).
daf-12 lies at the convergence of dauer and heterochronic pathways, regulating third stage and later temporal fates. In the dauer pathway, Daf-d mutations in daf-12 are epistatic to most known Daf-c mutants with respect to dauer formation (Riddle et al. 1981; Vowels and Thomas 1992; Thomas et al. 1993; Gottlieb and Ruvkun 1994; Larsen et al. 1995; Gems et al. 1998). The isolation of both Daf-d and Daf-c alleles suggests that daf-12 instructs alternate third stage options of dauer formation (S3d) or reproductive growth (S3) (Antebi et al. 1998). In addition, mutants fail to express S3 and later larval stage programs on schedule, often repeating S2 programs instead. Alleles fall into six classes based on dauer phenotypes and heterochronic phenotypes in gonadal and extragonadal tissues (Antebi et al. 1998), and suggest at least two selectively mutable activities: daf-12a required for advance from S2 to S3 reproductive programs and daf-12b required for advance from S2 to S3d dauer programs. Genetic epistasis tests with other heterochronic regulators place daf-12 downstream of lin-14, but upstream of lin-28. Of the alleles that have been tested, daf-12 mutants have a modestly shortened life span. Interestingly, these weakly suppress adult life span extension of some class 1 daf-2 mutants, but enhance median adult life span of class 2 daf-2 mutants twofold (Larsen et al. 1995; Gems et al. 1998).
Here we report that daf-12 encodes a member of the nuclear receptor family, implicating a hormonal mechanism in regulatory circuits that control dauer diapause, developmental age, and adult longevity. We find that molecular lesions cluster in DNA- and ligand-binding domains of the receptor. Genetic evidence suggests daf-12 functional complexity arises from the interplay of modular domains of the receptor, various isoforms, and crosstalk with other loci. DAF-12 is expressed in phenotypically affected cellular targets, with stage- and tissue-specific dynamics consistent with its various roles. It is most closely related to vertebrate vitamin D and pregnane-X receptors (Baker et al. 1988; Bertilsson et al. 1998; Kliewer et al. 1998) and the Drosophila DHR96 (Fisk and Thummel 1995). Homologs in other phyla raise the possibility that similar regulatory circuits may be conserved in metazoans.
Results
Cloning daf-12
We cloned the daf-12 gene by Tc1 transposon-insertion mutagenesis in screens for dauer-defective mutants (see Materials and Methods). After backcrossing to wild type (Bristol, N2), Tc1 polymorphisms linked to daf-12 were visualized by Southern analysis (Fig. 1). The polymorphic Tc1-containing fragment was isolated from a size-fractionated library, and the sequence of the flanking genomic DNA revealed homology to the DNA-binding domain of nuclear receptors. Cosmid F11A1, which contains the complete daf-12 gene, was sequenced by the Genome Consortium (C. elegans Sequencing Consortium 1998). From sequenced cDNAs (below), the deduced daf-12 coding region consisted of 17 exons spanning 21 kb. A 14.9-kb intron separated exons 2 and 3, whereas exons 3–17 were densely packed within 4.7 kb (Fig. 2B).
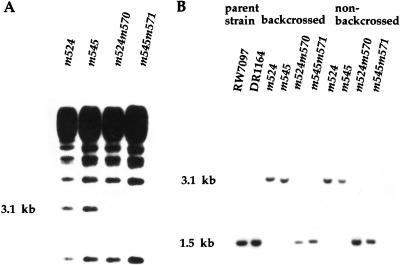
Figure 1.
(A) Tc1 transposon-induced mutations in daf-12 reveal a polymorphic 3.1-kb BglII fragment. Genomic DNA from backcrossed Tc1-induced alleles, m524 and m545, and their respective non-Daf revertants, m524m570 and m545m571, was digested with BglII, separated in 1% agarose, blotted onto nitrocellulose, and probed with radiolabeled Tc1. (B) Detection of a Tc1-associated mobility shift at the daf-12 locus. A Southern blot of BglII digested DNA was prepared as in A and hybridized with a radiolabeled 1.2-kb daf-12 probe. A 1.6-kb increase in size, due to Tc1 insertion, is seen in backcrossed and nonbackcrossed Tc1-tagged alleles (m524 and m545) but absent in parental mutator strains (RW7097, DR1164) and Tc1-excised nonDaf revertants (m524m570 and m524m571).
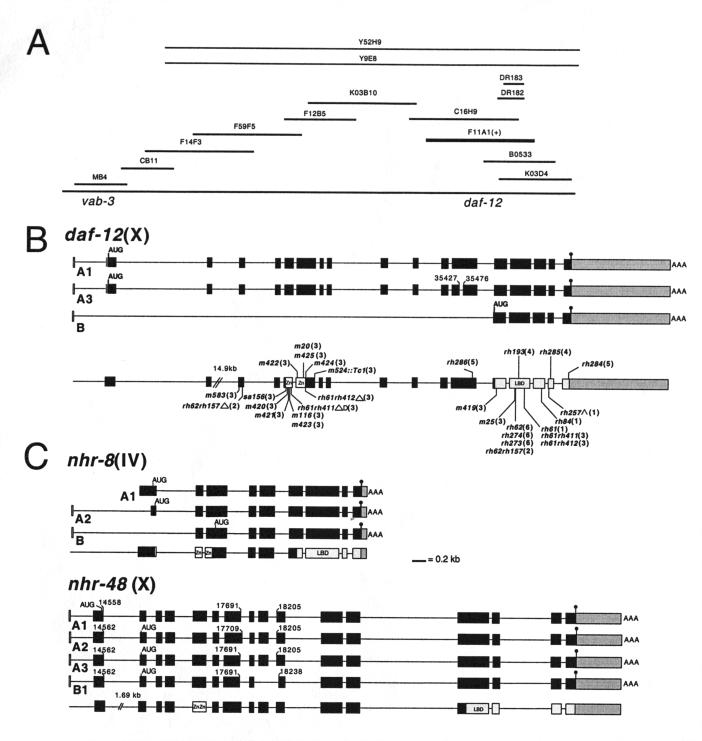
Figure 2.
(A) Physical map of the daf-12 region. Rescuing cosmid, F11A1 (+). (B) cDNA structure (filled boxes) of 12A1, 12A3, and 12B isoforms, genomic organization, and corresponding positions of identified mutations at the daf-12 locus. Each isoform begins with the SL1 spliced leader (in white). Paired zinc fingers (Zn) of the DNA-binding domain (in white) and the ligand-binding domain (LBD) are indicated (in light gray). The 3′ untranslated region is shown in dark gray. Positions of the alternate splice donor and acceptor sites are shown for isoform 12A3 (numbering according to cosmid F11A1). Deletion (Δ), deletion/duplication (ΔD), splice acceptor mutation (^). Proposed initiator codons (AUG) and stop codons (●), and sites of polyA addition (AAA) are shown for each isoform. Allele names are followed by class in parentheses. The position of both lesions in alleles rh61rh411, rh61rh412, and rh62rh157 are shown. (C) cDNA structure (filled boxes) and genomic organization of daf-12 homologs nhr-8 and nhr-48. For nhr-8, full-length isoform 8A1 (clone cm14e12) lacks the SL1 spliced leader. Isoform 8A2 contains the SL1 spliced to an internal site in exon 1. Isoform 8B contains SL1 spliced to exon 2. For nhr-48, alternate exon 1 (14558/14562) and exon 7 (17691/17709) splice donors, and alternate exon 10 (18205/18238) splice acceptors for the various isoforms are indicated. A fifth 48B2 isoform (not shown), inferred from a partial cDNA clone, arises from the use of the exon 7 donor (17709) joined to the middle of exon 10 (18238). (Numbering is according to the ZK662 cosmid).
Cosmid F11A1 (Fig. 2A) rescued the dauer-defective phenotypes of rh61rh411 (a candidate null allele). Rescue was scored as the ability to form dauer larvae among populations grown to exhaustion on nematode growth media. Although many transgenic animals formed normal dauer larvae, rescue was incomplete in some animals. Interestingly, 29% of the dauer larvae (n = 21) were phenotypic mosaics composed of cells expressing dauer and nondauer programs. Phenotypic mosaicism may correspond directly to genotypic mosaicism or result from incomplete rescue in individual cells. Notably, however, this property differs from wild type, or daf-2 and daf-4 genotypic mosaics in which all cells express either dauer or non-dauer programs (Apfeld and Kenyon 1998; Inoue and Thomas 2000).
daf-12 encodes multiple isoforms
We determined the sequence of the daf-12 coding region from cDNA clones isolated from mixed-stage libraries and by RT–PCR (Figs. 2B, 3). Analysis of the products revealed three SL1 spliced transcripts of 3.7 kb (12A1), 3.6 kb (12A3), and 2.2 kb (12B), which encoded predicted proteins of 753, 737, and 267 amino acids, respectively. All identified cDNA products contained a long 1.4 kb 3′ untranslated region. The 12A and 12B isoforms differed at their 5′ ends as a result of the SL1 leader splicing to exons 1 and 13, respectively. These isoforms may arise either from different promoters, differential splicing, or both. 12A1 and 12A3 are identical, except that 12A3 has an internal portion of exon 12 spliced out, removing 16 amino acids (Figs. 2B, 3). Notably, these isoforms encode different modules of receptor function. Isoforms 12A1 and 12A3 contain DNA- and ligand-binding domains, whereas isoform 12B contains the ligand-binding domain only.
Figure 3.
cDNA sequence, predicted protein products, and mutations in daf-12. Arrows indicate the starts of 12A1, 12A3, and 12B isoforms. The SL1 spliced leader is not shown. (▿) The position of introns; (v) bracket the amino acids missing from isoform 12A3. Base substitutions are indicated above the sequence, corresponding amino acid changes below, and premature stop codons indicated by a filled circle (●). The position of Tc1 insertion, m524::Tc1, is between T746 and A747 highlighted in bold. Underlined “M”, the predicted initiator methionines for the three isoforms. Deletion (Δ68) rh62rh157 deletes nucleotides 32347-32414. Deletion/duplication (ΔD) rh61rh411 deletes nucleotides 33058–33066, duplicates 33069–33090, and continues with 33067 (numbering according to cosmid F11A1). rh61rh412 deletes the underlined bases. rh257 splice acceptor mutation (^).
DAF-12 and homologs define an ancient branch of the nuclear receptor family
The predicted full-length DAF-12 gene product (753 amino acids) retains the modular organization typical of ligand-activated nuclear receptors (Mangelsdorf et al. 1995): a hypervariable amino terminus, a highly conserved DNA-binding domain (DBD), a variable hinge region, and a conserved carboxy-terminal ligand-binding domain (LBD). We have aligned DAF-12 and related receptors (Fig. 4) with three receptors (thyroid receptor, retinoic acid receptor α, and retinoid-X receptor γ) whose LBD crystal structures are known. The LBD structure reveals 12 α helices organized into a three layered sandwich with various ligand contact sites (Bourguet et al. 1995; Renaud et al. 1995; Wagner et al. 1995; Wurtz et al. 1996). DAF-12 shows greatest conservation in helices 3–5, helix 8, and helix 12 comprising the AF-2 activation domain core, a transactivation helix that closes the ligand-binding pocket on one face and interacts with coactivators on the other.
Figure 4.
(A) Sequence alignments of the DBD and LBD of the ESCKA receptors with known nuclear receptors. Conserved cysteines that comprise Zn coordination sites of the DBD are starred. DAF-12 mutations are shown above the alignment (−1 = rh61rh412 frameshift). In the LBD, alignment with RAR-γ and RXR-α shows the positions of the 12 α helices (H1–H12) and β strands for comparison (Bourguet et al. 1995; Renaud et al. 1995; Wurtz et al. 1996). Helices 9–10 comprise a homodimerization interface in RXR (Bourguet et al. 1995). The predicted AF-2 domain with consensus hhXEhh (Danielian et al. 1992) lies in helix 12 (h = hydrophobic residue). In vitamin D and PXR receptors, forty amino acids that form an extended loop between helices 2 and 3 are omitted (indicated by ++). Boxed residues are ligand contact sites in TR and RAR receptors, as determined by X-ray crystallography (Renaud et al. 1995; Wagner et al. 1995; Wurtz et al. 1996). (Black shading) Residues conserved in nearly all nuclear receptors; (dark gray shading) residues conserved in a majority of the group shown; (light gray shading) residues matching DAF-12. The alignment includes Strongyloides stercoralis DAF-12 (Siddiqui et al., unpubl.; accession no. AAD37372), DHR96 (Fisk and Thummel 1995), human vitamin D (Baker et al. 1988), human PXR (Bertilsson et al. 1998), Drosophila ecdysone (Koelle et al. 1991), rat thyroid α1 (Izumo and Mahdavi 1988), human RAR-γ (Krust et al. 1989), and human RXR-α (Mangelsdorf et al. 1990). (B) Phylogenetic tree showing relationships between various nuclear receptors in the DBD. The tree was generated using the Megalign program of DNAStar, which derives a tree based on distance and then evaluates the tree based on parsimony criteria.
DAF-12 represents an ancient branch of the nuclear receptor superfamily (Fig. 4). In the DBD, the closest nematode relatives include a DAF-12 ortholog from the closely related species, Strongyloides stercoralis (96% identical) and two C. elegans receptors, NHR-8 (59%) and NHR-48 (73%) (Sluder et al. 1999). We isolated cDNAs encoding NHR-8 and NHR-48 to verify experimentally their gene structure and predicted amino acid sequence (Fig. 2C, see Materials and Methods). Outside of the nematode, the closest relatives are the Drosophila DHR96 (63%) (Fisk and Thummel 1995), and vertebrate PXR (57%) (Bertilsson et al. 1998; Kliewer et al. 1998) and vitamin D receptors (52%) (Baker et al. 1988). DAF-12, NHR-8, NHR-48, and DHR96 contain 13 contiguous conserved residues that comprise the predicted DNA-recognition α helix (Luisi et al. 1991; Schwabe et al. 1993), suggesting they recognize similar or identical hormone response elements. Notably, the base-contact residues (ESCKAFFR) are unique among the nuclear receptor family. When we examined the sequence relationships in the DBD among the ESCKA family members by the methods of distance and parsimony, DHR96 is not more significantly related to DAF-12 than NHR-8 or NHR-48 (Fig. 4B).
The nearest relatives to DAF-12 within the LBD (helix 3–5, residues 546–604) are the S. stercoralis DAF-12 (64% identical), Drosophila DHR96 (40%), NHR-8 (34%), Drosophila ecdysone (34%), and vertebrate thyroid hormone (31%) receptors. In contrast to the DBD, the LBD is rather diverged from PXR (22% identical) and vitamin D receptors (20%). Also, the predicted LBD of NHR-48 is highly diverged from most known nuclear receptors.
Molecular lesions
daf-12 alleles have been grouped previously into six classes based on separable heterochronic and dauer phenotypes (Antebi et al. 1998; Table 1). Moreover, heterochronic phenotypes show independent regulation in gonadal and extragonadal tissues. To understand the molecular basis of these allele classes, we identified and sequenced 26 molecular lesions. The functional consequences of mutations can be inferred, since nuclear receptors display a conserved architecture in DBDs and LBDs, including conserved positions for some DNA and ligand contact sites (Luisi et al. 1991; Schwabe et al. 1993; Bourguet et al. 1995; Renaud et al. 1995; Wagner et al. 1995; Wurtz et al. 1996). Generally, daf-12 alleles within a class cluster in specific domains of the protein, particularly in the DBD (class 2, 3) and LBD (class 1, 4, 5, 6). Only one mutation lies within the hinge (rh286). All sequenced alleles generated by ethyl-methanesulfonate mutagenesis exhibited GC-to-AT transitions, with the exception of m422, which contained an AT-to-TA transversion.
Table 1.
Sites of mutations, and summary of dauer and heterochronic phenotypes of daf-12 alleles
| Class
|
Allele
|
Mutation
|
Location
|
Dauer phenotype
|
Heterochronic phenotypes
|
|
|---|---|---|---|---|---|---|
| Extra-gonadal (L3 seam)a
|
Gonadal (L4 distal tip)b
|
|||||
| WT | + | Normal | ++++ | ++++ | ||
| Class 1 | rh61 | Q618stop | LBD beta turn | Defective | + | + |
| rh84 | R688stop | LBD H9 | ++ | |||
| rh257 | Y699splice acceptor | LBD H9 | + | |||
| Class 2 | rh62rh157 | S70stop, R564C | before DBD, LBD, H3 | Defective | + | ++++ |
| Class 3 | m583 | Q59stop | before DBD | Defective | +++ | ++++ |
| sa156 | C121Y | DBD Zn1 | ||||
| m421 | S137F | DBD Zn1 | ||||
| m116/m423 | R143K | DBD Zn1 | ||||
| rh61rh411 | L147stop, Q618stop | DBD after Zn1, LBD beta turn | ||||
| m422 | C154stop | DBD Zn2 | ||||
| m20/m425 | W187stop | DBD after Zn2 | ||||
| rh61rh412 | R195frameshift,Q618stop | DBD after Zn2, LBD beta turn | ||||
| m524::Tc1 | Y239insertion | DBD after Zn2 | ||||
| m419 | Q501stop | LBD before H1 | ||||
| rh258 | n.d. | |||||
| cm420 | A125V | DBD Zn1 | ++++ | ++++ | ||
| m424 | R197K | DBD Zn1 | ||||
| m25 | M562I | LBD H3 | ||||
| Class 4 | rh193 | G582K | LBD H4 | Normald | ++ | ++ |
| rh285 | Q707stop | LBD after H9 | + | |||
| Class 5 | rh284 | P746S | LBD H12 | Normal | ++++ | + |
| rh286 | C461Y | Hinge | +++ | +++ | ||
| Class 6 | rh62/rh274 | R564C | LBD H3 | Constitutive | ++++ | + |
| rh273 | R564H | LBD H3 | +++ | |||
(+) 0%–33% wild type; (++) 34%–66% wild type; (+++) 67%–99% wild type; (++++) 100% wild type.
Percentage of seam cells at larval stage L3 that execute stem cell division (S3) versus equational division followed by stem cell division (S2) as described in Fig. 5 legend. ca. 10 animals (120 cells) were scored by DIC microscopy as described (Antebi et al. 1998).
Percentage of distal tip cells that migrate dorsally by larval stage L4 as described (Antebi et al. 1998). ca. 100 animals (200 cells) were scored by DIC microscopy or dissecting scope.
Missense alleles m420 and m424 have no observable heterochronic phenotype in seam cells, but have impenetrant heterochronic phenotypes in the intestine (not shown). m25 had no observable heterochronic phenotypes.
A small fraction of partial dauer animals formed at 25° and 15°C for rh193 and rh285, respectively. Phenotypes for alleles rh61, rh84, rh257, rh62rh157, rh258, rh61rh411, m20, m583, rh193, rh285, rh284, rh286, rh62, rh274, and rh273 are adapted from Antebi et al. 1998.
Class 3
Most class 3 alleles are 100% dauer-defective and have impenetrant heterochronic phenotypes (Table 1, Fig. 5B) (Antebi et al. 1998). Here we have characterized an additional 11 alleles (m25, m116, m419, m420, m421, m422, m423, m424, m425, m524, rh61rh412) and found that all are class 3 mutants (Table 1). m420 and m424 differ from canonical class 3 alleles, exhibiting no observable heterochronic phenotypes in epidermal seam cells, but impenetrant heterochronic phenotypes in the intestine. In addition, m25 has no observable heterochronic phenotypes, but forms mosaic dauer larvae (Fig. 5D, E).
Figure 5.
(A–C) Comparative heterochronic phenotypes at larval stage L3 in seam cells of wild type, class 3 allele rh61rh411, and class 1 allele rh61. Left lateral aspect. Scale bar, 10 μm. Seam cells and their immediate daughters express the GFP fusion to the Seam Cell Marker in their nuclei (Terns et al. 1997; J. Rothman, pers. comm.). (A) In wild type, epidermal seam cells along the lateral midline undergo a single stem cell division giving rise to two daughters (S3 program) at larval stage L3. Later the anterior daughter fuses with the hypodermal syncitium and the posterior daughter divides in subsequent stages. At larval stage L2 (not shown), seam cells undergo equational division followed by stem cell division, giving rise to 4 daughters (S2 program). (B) In rh61rh411 at larval stage L3, seam cells exhibit impenetrant repetition of S2 programs. (C) In rh61 seam cells exhibit penetrant repetition of S2 programs. (D–F) Mosaic dauer larvae form in some daf-12 mutants. (D) Wild-type dauer larva with full-length dauer alae, which are lateral cuticular ridges (arrows), and complete radial constriction. Cellular mosaicism is not seen. (E) daf-12(m25) and (F) daf-12(rh193) mosaic dauer larvae (grown at 15°), which occasionally show dauer alae and radial constriction in some cells (arrows) and partial dauer or non-dauer programs in others (arrowheads).
Nearly all class 3 mutations affect the DBD. These include nonsense mutations (m583, m422, m20, m425), a Tc1 insertion (m524), missense alleles within conserved residues (sa156, m420, m421, m116, m423, m424), as well as double utants (rh61rh411, rh61rh412). Many missense mutations identify critical residues in the DBD. For example, S137F (m421) and R143K (m116 and m423) affect predicted contact residues in the DNA-binding helix itself, and C121Y (sa156) affects one of four conserved Zn coordinating cysteines in the first Zn finger. Several independently isolated alleles carry the same mutations (e.g., m20/m425, m116/m423).
Two exceptional class 3 mutations are not in the DBD. Dauer-defective nonsense allele m419 (Q501stop) affects the amino-terminal boundary of the LBD. Missense allele m25 (M562I) is within the LBD itself, and gives rise to partial (Vowels and Thomas 1992) or mosaic dauer larvae (Fig. 5E), similar to a handful of daf-12 alleles (e.g., rh193, rh274) and daf-12 mutants harboring an unstable daf-12(+) transgene (above). Some class 3 alleles disrupt or alter only isoforms 12A1 and 12A3 (m20/m425, m116/423, m420, m421, m422, m424, m524, m583, sa156); others disrupt 12A1, 12A3, and B isoforms (m419, rh61rh411, and rh61rh412). The latter are candidate null mutants (see below).
Class 1
All three class 1 alleles are dauer-defective and have penetrant gonadal and extra-gonadal heterochronic phenotypes (Table 1, Fig. 5). They truncate the LBD at various positions between helices 5–9. Two (rh61 and rh84) generate stop codons, and a third (rh257) mutates the splice acceptor in exon 16. Class 1 alleles affect all isoforms.
Class 6
All three class 6 alleles, which are dauer-constitutive and exhibit gonadal heterochrony, affect the same amino acid within helix 3 of the LBD. Independent isolates rh62 and rh274 both generate an R564C substitution and rh273 generates an R564H substitution. These mutations cause lesions in all isoforms.
Class 2
A single class 2 allele, rh62rh157, which is dauer-defective with moderate heterochronic phenotypes in extragonadal tissues, arose as a spontaneous intragenic revertant of the Daf-c class 6 allele rh62. The double mutant contains the original rh62 missense allele R564C, and a second lesion rh157, a 68-bp deletion in exon 3 that creates a stop codon at S70, upstream of the DBD. This double mutant results in lesions in all isoforms.
Class 4
The two class 4 alleles, rh193 and rh285, have nearly normal dauer regulation, but exhibit penetrant, temperature-sensitive heterochronic phenotypes that affect the LBD. rh193 (G582K) alters a conserved region within helix 4. Nonsense allele rh285 truncates the protein near the carboxyl terminus, upstream of helix 10. Both alleles produce lesions in all isoforms.
Class 5
Class 5 alleles have normal dauer regulation, but exhibit gonadal heterochrony. rh284 (P746S) affects helix 12 of the LBD. Missense allele rh286 (C461Y), which has a very mild phenotype, affects the hinge. rh284 causes lesions in all isoforms; rh286 affects 12A1 and 12A3 only.
daf-12 loss of function
Class 3 alleles rh61rh411 and rh61rh412 are severe loss-of-function mutants that are candidate null alleles. Both arose as spontaneous non-Mig revertants of the gonadal heterochrony of class 1 allele rh61. In addition to the original rh61 nonsense codon in the LBD, rh61rh411 also contains a small deletion/duplication after the first Zn finger that creates an inframe stop. Similarly, rh61rh412 contains a small 5-bp deletion after the second Zn finger, creating a frameshift that truncates the receptor 12 amino acids downstream. With stop codons in DNA- and ligand-binding domains, both alleles presumably disrupt DNA- and ligand-binding activities of 12A1, 12A3, and 12B isoforms. rh61 exhibits penetrant heterochronic phenotypes, whereas the revertants are impenetrant (Fig. 5). Therefore, class 3 impenetrant heterochronic phenotypes, surprisingly, represent the more severe loss of function. All three alleles are completely dauer-defective.
daf-12 expression pattern
To gain insight into daf-12 regulation and cellular focus, we determined the daf-12 expression pattern. We made a fusion construct with GFP (Chalfie et al. 1994) inserted in-frame within exon 1 of the daf-12 gene. Because the gene is large, the gene fusion was reconstructed in cosmid F11A1 to include the entire regulatory region. The transgene rescued the Daf-d phenotype of rh61rh411 animals, suggesting that daf-12::GFP is functional. Several integrated transgenic lines were made and all gave a similar pattern of expression. DAF-12::GFP localized primarily to the nucleus (Fig. 6), except during mitosis, when expression became diffuse. DAF-12::GFP was expressed widely in most cells including tissues modified for dauer formation or by stage. In particular, it was expressed in phenotypically affected target tissues (e.g., epidermis, vulva, somatic gonad, intestine, pharynx, sex myoblasts) (Riddle et al. 1981; Antebi et al. 1998), as well as other tissues with no known phenotype (e.g., nervous system, body wall muscle). Expression was seen from embryo to adult, but was most elevated and widespread during L2. Below we highlight the dynamics within specific tissues.
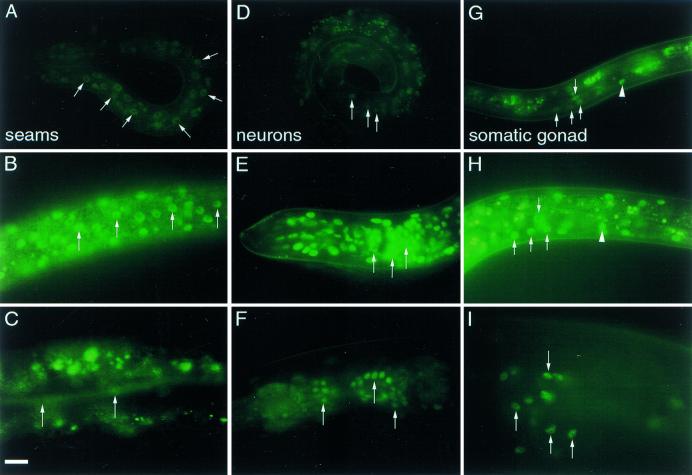
Figure 6.
DAF-12::GFP expression pattern. Expression in epidermal seam cell nuclei (arrows) of early L1 (A), late L2 (B), and adult animals (C). Expression in various head neurons (arrows) of early L1 (D), late L2 (E), and adult (F) animals. Expression in somatic gonadal tissues of L3 dauer (G), late L2 non-dauer (H), and adult somatic gonadal tissues (I). In G and H, the arrows indicate the nuclei of somatic gonadoblasts, arrowheads the distal-tip cell. In I, arrows indicate various spermathecal nuclei. Lateral aspect, anterior is left. Scale bar, 10 μm. All photographs were exposed for equal times.
Epidermis
In seam cells and hypodermis, DAF12::GFP expression was first seen at the threefold stage of embryogenesis, increased by late L1, peaked during L2, diminished by late L3, and was low or off in L4 and young adults (Fig. 6A–C). Expression was also seen in the ventral epidermal L1 P ectoblasts, L2 vulval precursors, and their L3 descendants. Expression continued during L4 vulval morphogenesis and persisted occasionally in the mature adult vulva at reduced levels.
Somatic Gonad
Faint expression was seen as early as L1 in Z1 and Z4 somatic gonadal precursors. By L2, their descendants, the somatic gonadoblasts, including the migratory distal-tip cell, strongly expressed DAF-12::GFP (Fig. 6H). Expression continued in somatic gonadoblast descendants and distal-tip cells in L3 and early L4. In the adult, expression was robust in the mature spermatheca and uterus (Fig. 6I).
Intestine
Expression in intestinal nuclei was diffuse during the larval stages, but became somewhat stronger in the adult.
Nervous System
Only a handful of head and tail neurons expressed GFP early in L1 (Fig. 6D). By mid-L2, DAF-12::GFP was expressed strongly throughout the nervous system (Fig. 6E), including the ventral cord and peripheral neuroblasts. Expression continued in many neurons in the adult, albeit at reduced levels (Fig. 6F).
Musculature
Expression in body wall muscles became visible by late L1 and L2. Expression continued at later larval stages and in the adult at reduced levels. Expression in pharyngeal muscle was strong by L2 and downregulated by adult. DAF-12::GFP was also expressed in the L1 M-mesoblast, and its derivatives, including post-embryonic body wall muscles, sex myoblasts and their descendants.
Dauer formation
DAF-12::GFP was downregulated in dauer larvae in all tissues, but perdured in the somatic gonad and occasional neurons (Fig. 6G). Upon recovery from dauer diapause, DAF-12::GFP was expressed weakly in most tissues.
Discussion
Nuclear receptors in metazoan life history
In metazoans, development, metabolism, and homeostasis are coordinated by lipophilic hormones that act through members of the nuclear receptor family (Mangelsdorf et al. 1995). In the nematode C. elegans, daf-12 encodes a novel orphan nuclear receptor that controls various life history options during development. In particular, it regulates the advance from second to third and later larval stage fates, and mediates the choice between alternate third-stage programs of dauer diapause or reproductive development (Riddle et al. 1981; Antebi et al. 1998). In the adult, daf-12 influences the extent of adult life span in daf-2 mutant backgrounds (Larsen et al. 1995; Gems et al. 1998) and in germ cell ablated animals (Hsin and Kenyon 1999). Although a hormone has not yet been identified for daf-12 or any other nematode nuclear receptor, the global decisions regulated by daf-12 make a hormonal mechanism an attractive hypothesis.
The ESCKA family of receptors
Among 250 or more nuclear receptor genes found in the C. elegans genome (Sluder et al. 1999), DAF-12 and two DAF-12 paralogs, NHR-8 and NHR-48, define an ancient branch of the family with phyletically conserved relatives. The most conspicuous shared feature of the three nematode receptors is an identical thirteen amino acid sequence that comprises an unusual DNA recognition helix that we call the ESCKA helix for the unique predicted DNA contact residues. By inference, these receptors may recognize identical DNA half sites. Outside of the nematode, the Drosophila DHR96 (Fisk and Thummel 1995) is the closest DAF-12 relative. It is identical throughout the ESCKA helix and also exhibits significant similarity in the LBD, particularly in helices 3–5, helices 8–10, and helix 12. Though no DHR96 mutants have been described, the gene is expressed during third instar and prepupal development, and binds in vitro to the hsp27 ecdysone response element (Fisk and Thummel 1995). It will be interesting to see if DHR96 regulates stage-specific programs, diapause, or life span in insects. The closest vertebrate homologs, the vitamin D (Baker et al. 1988) and PXR receptors (Bertilsson et al. 1998; Kliewer et al. 1998), are highly related in the DBD, but differ in the DNA recognition helix (EGCKG) and LBD. Conceivably, the ESCKA family arose before the metazoan radiation, and a closer DAF-12 relative might be found in vertebrates.
Correlation between phenotypic classes and sites of mutations
daf-12 mutants have been categorized into six classes, grouped first on the basis of their dauer phenotype (Daf-d, non-Daf, Daf-c), and second on their heterochronic phenotypes in gonadal and extragonadal tissues (Antebi et al. 1998). One interpretation of these phenotypic classes is that daf-12(+) functions solely during dauer formation to promote S3d dauer programs and repress S3 programs of reproductive growth. In this view, daf-12 heterochrony would then arise from the inappropriate inhibition of S3 programs in the absence of the dauer-inducing signal. This model, however, does not explain how nearly all alleles, including candidate nulls, exhibit variable heterochronic phenotypes, suggesting that daf-12(+) is required to some degree for both reproductive growth and dauer formation. Hence, we currently favor the view that these allele classes represent at least two activities, daf-12a required under reproductive growth conditions to advance S3 programs (and inhibit S3d programs), and daf-12b required under dauer-inducing conditions to promote S3d dauer programs (and to inhibit S3 programs) (Fig. 7). Tissue-specific phenotypes in gonadal and extra-gonadal tissues represent a further subdivision of daf-12 activity, perhaps reflecting interaction with other regulators.
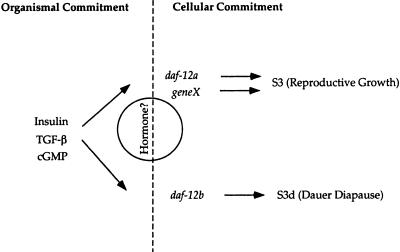
Figure 7.
A model for daf-12 action. Insulin-like, TGF-β, and cGMP signaling act upstream of organismal commitments to diapause or reproductive growth. In favorable environments, the activity of these pathways stimulate reproductive growth, while unfavorable environments lead to diapause. Proposed hormonal signaling acts through daf-12 to select S3 or S3d alternatives. daf-12 together with other heterochronic genes acts in cells downstream of organismal commitments. daf-12a and a proposed redundant function, geneX, are required for S3 reproductive programs, whereas daf-12b activity is required for S3d dauer programs.
There is a strong correlation between mutant class and the location of the sequence change within the predicted protein. Reference allele m20 and most other class 3 alleles (13) cluster upstream or within the DBD. They include nonsense as well as missense alleles within conserved residues that presumably disrupt DNA binding and transactivation. Only two class 3 alleles (m25 and m419) map in the LBD. By contrast, other classes, including those with penetrant heterochronic (classes 1, 4, 5) and dauer-constitutive phenotypes (class 6), cluster in the LBD, and likely uncouple DNA binding from LBD regulation. In particular, class 1 alleles truncate the LBD within or upstream of helix 9 (Fig. 4), presumably perturbing ligand binding, dimerization (helices 9–10), and transactivation (helix 12) (Bourguet et al. 1995; Renaud et al. 1995; Wagner et al. 1995).
Among the receptors with known structure, ligand contact residues are distributed throughout various helices and their positions are often conserved (Wurtz et al. 1996). Remarkably, several daf-12 missense alleles specifically perturb these predicted sites, consistent with the idea that daf-12 has a ligand. All three class 6 Daf-c alleles map to a single residue (R564) in helix 3; the analogous residue in the thyroid receptor (R228) makes important hydrogen bonds with ligand (Wagner et al. 1995). Conceivably, R564 mutations render the receptor either ligand insensitive or independent. All three alleles are recessive, suggesting that daf-12(+) may repress S3d dauer-specific genes under S3 reproductive growth conditions. By contrast, class 3 allele m25 (M562I), a neighboring missense mutation, is partially Daf-d and confers a mosaic dauer phenotype. Potentially, this mutation diminishes, but does not abolish, affinity for a ligand produced in dauer-inducing conditions. Finally, class 5 allele, rh284 (P746S), which exhibits gonadal heterochrony but near normal dauer regulation, aligns with a ligand contact site in helix 12, and presumably disrupts ligand interactions or transactivation.
Receptor transactivation via AF-2 depends in part on interactions of helices 4 and 12 (Renaud et al. 1995). Class 4 alleles (rh193, rh285), which have strong heterochronic phenotypes and near normal dauer regulation, could primarily affect AF-2 function or, more generally, LBD structure. rh285 (Q707stop) truncates the receptor upstream of helix 10, whereas rh193 (G582K) creates a missense mutation in helix 4 (Fig. 4). rh193 also aligns with conserved residues that stabilize the ligand binding core (Wurtz et al. 1996). Surprisingly, rh285 mutants can execute S3d but not S3 programs, suggesting that the very carboxyl terminus, including the putative AF-2 core, is dispensable for daf-12b (S3d-promoting) but not daf-12a (S3-promoting) activities. By contrast, class 1 allele rh84 (R688stop), which truncates the receptor farther upstream, disrupts both daf-12a and daf-12b activities. This suggests that the 19 intervening amino acids encompassing putative dimerization helix 9 are required for daf-12b activity. Biochemical characterization of receptor complexes should ultimately clarify how these mutations respectively perturb ligand binding, dimerization, repression, and activation.
daf-12 isoforms
RT–PCR revealed that daf-12 encodes 3 predicted isoforms that contain different modules of receptor function. Isoforms 12A1 and 12A3 contain the DBD and LBD, whereas isoform 12B contains the LBD only. Some class 3 alleles truncate 12A isoforms only (e.g., m583, m20, m422), whereas others truncate 12A and 12B isoforms (e.g., rh61rh411, rh61rh412). Nevertheless, these various class 3 alleles are nearly indistinguishable for dauer and heterochronic phenotypes (Antebi et al. 1998; this work). Hence, 12A1 and 12A3 must be limiting for daf-12 function. However, class 2 allele rh62rh157 (12A1–12A3–12B missense) gives rise to stronger heterochronic phenotypes than any class 3 alleles, suggesting a possible role for 12B independent of 12A. No identified mutation selectively disrupts 12B, since it completely overlaps 12A1 and 12A3. Apparently, proposed daf-12a and daf-12b activities do not correlate directly with isoforms 12A1, 12A3, and 12B, but tests of minigenes may expose isoform-specific functions. A fourth isoform reported in public databases (Accession no. AF136239; P. Larsen and M. Snow, pers. comm.), called DAF-12A2, contains exons 3–17. For unknown reasons, we have not detected this isoform.
Loss-of-function and evidence for functional redundancy
With stop codons in the DBD and LBD, class 3 alleles rh61rh411 and rh61rh412 severely truncate 12A and 12B isoforms. Although some coding regions remain, major functional moieties are disrupted, hence making these alleles candidates for complete loss-of-function or null mutations. Consistent with this, class 3 alleles constitute the major class and some class 1 alleles spontaneously mutate to class 3. Class 3 mutants are completely dauer-defective but have impenetrant heterochronic phenotypes. Hence, these alleles must be null for the proposed daf-12b, but only partially defective for daf-12a owing to an activity from at least one other locus (Fig. 7). By contrast, class 1 alleles, strongly defective for both daf-12a and daf-12b activities, may represent a recessive gain-of-function. A simple model is that class 1 mutant receptors interfere with a redundant activity. Apparently, wild-type protein overcomes this interference, since all mutations are recessive.
By inference, penetrant class 1 heterochronic phenotypes may require more than a functional DBD to cause transcriptional interference. A variety of receptors repress transcription in the absence of ligand but activate in their presence, through recruitment of various corepressor and coactivator complexes to partly overlapping surfaces of the receptor (Freedman 1999; Hu and Lazar 1999). Notably, class 3 allele m419 contains the DBD but not the LBD, suggesting that penetrant heterochronic phenotypes require a region between helix 1 (m419) and the β turn (rh61) (Fig. 4), a region which in other receptors recruits corepressors (Horlein et al. 1995; Burke et al. 1998; Hu and Lazar 1999). Although several interpretations are possible, we suggest that class 1 mutant proteins repress but cannot activate S3 and S3d programs. The failure to express S3 programs may also be indirect, and reflect, for example, a failure to downregulate S2 programs. Conversely, class 6 mutant proteins inappropriately activate but cannot repress S3d dauer programs during reproductive growth. Finally, class 3 severe loss-of-function alleles can neither repress nor activate S3 and S3d programs, but a redundant activity can partially activate S3 programs.
In humans, resistance to thyroid hormone, a dominantly inherited disorder, is caused by mutations in the LBD of the thyroid receptor (TR) β. Apparently, dominant negative receptors interfere with both wild-type TR-α and TR-β receptors, through constitutive recruitment of corepressor complexes (Yoh et al. 1997). Mutations in the daf-12 LBD, though recessive, could behave in an analogous manner.
DAF-12 is expressed in cellular targets and is temporally regulated
The DAF-12::GFP expression pattern suggests that DAF-12 acts directly in target tissues at appropriate stages, in concert with its proposed roles in dauer formation, developmental age, and aging. First, DAF-12::GFP is expressed widely in tissues that undergo modifications during developmental stages and diapause. In particular, it resides in the nuclei of all cells with previously observed phenotypes (e.g., epidermal seam cells, gonadal distal tip cells, intestinal cells, sex myoblasts, etc.) (Antebi et al. 1998), suggesting that DAF-12 acts within the target cells themselves. Moreover, DAF-12::GFP is temporally regulated, with peak expression in most cells during L2/L3, when heterochronic and dauer phenotypes first appear (Antebi et al. 1998), and when final commitments to reproductive growth or dauer formation are made (Golden and Riddle 1984). Temporal dynamics are most evident in the epidermis, the cellular focus for most heterochronic genes. Here, DAF-12::GFP first appears faintly by the threefold stage of embryogenesis, is upregulated by late L1/L2, and down-regulated by L4. This temporal modulation may reflect regulation by other heterochronic genes. In addition, DAF-12 is expressed in the adult, most conspicuously in intestine, nervous system, and somatic gonad tissues. Interestingly, the latter two tissues regulate longevity in nematodes (Apfeld and Kenyon 1999; Hsin and Kenyon 1999). In dauer larvae, DAF-12::GFP is downregulated in most tissues except the somatic gonad and occasional neurons, suggesting tissue- and dauer-specific regulation.
Notably, stage- and tissue-specific control of daf-12 expression does not exclude other mechanisms of regulation. In some cells, expression does not correlate with any visible phenotype, for example, various L1 blast cells. Conceivably, phenotypic effects may depend on temporal changes in other rate-limiting components (e.g., hormone, coactivators). Nor does expression in various target tissues exclude downstream nonautonomous action.
Endocrine mechanisms in nematode life history
The identification of daf-12 as a nuclear receptor suggests that developmental age and the dauer diapause may be controlled by a hormonal mechanism. This work in particular provides the first evidence of potential endocrine signaling in the heterochronic circuit of nematodes. Nuclear receptors in other metazoa are known to influence developmental age. Alterations in thyroid hormone signaling in amphibia give rise to classic cases of delayed heterochronic transformations in the soma (Frieden 1981). Similarly, mutations in distinct ecdysone receptor isoforms in Drosophila cause tissue- and stage-specific heterochrony (Bender et al. 1997). These receptors, though clearly not structural orthologs of daf-12, may indicate a fundamentally conserved role of regulating developmental age reserved for some nuclear receptors.
In the C. elegans dauer pathway, genetic epistasis experiments reveal that TGF-β, cGMP, and insulin/IGF-I-like signaling may ultimately converge on daf-12 (Riddle et al. 1981; Vowels and Thomas 1992, 1994; Thomas et al. 1993; Gottlieb and Ruvkun 1994; Larsen et al. 1995; Grenache et al. 1996). These inputs bias selection of S3 or S3d programs, possibly through regulation of daf-12 expression, activity or that of potential heterodimeric partners. In vertebrates, cross-talk between TGF-β and vitamin D receptor signaling is mediated by SMAD proteins, which act as ligand-dependent coactivators of the vitamin D receptor (Yanagisawa et al. 1999).
Alternately, TGF-β, insulin, or cGMP signaling pathways, singly or in combination, might control biosynthesis or availability of lipophilic hormone(s) that activate nuclear receptor transcriptional complexes. Consistent with this, both the DAF-2 insulin/IGF-I receptor and DAF-4 TGF-β Type II receptor act nonautonomously, most likely in neurons, to control dauer diapause, suggesting a downstream intercellular signaling mechanism (Apfeld and Kenyon 1998; Inoue and Thomas 2000). In both cases, genotypic mosaics are not phenotypic mosaics. Instead, all cells collectively express either dauer (S3d) or reproductive (S3) programs, suggesting that daf-2 and daf-4 lie upstream of organismal commitments to diapause. By contrast, daf-12 and other heterochronic loci likely act downstream of organism-wide decisions (Fig. 7). These mutants are often mosaics of cells expressing dauer/nondauer programs, or mosaics of cells of disparate developmental age (Fig. 5) (Ambros and Horvitz 1984; Liu and Ambros 1989; Antebi et al. 1998), suggesting an uncoupling between organismal and cellular commitments. With respect to aging, analysis of daf-2 mosaics also shows that life span is regulated nonautonomously, probably through the nervous system (Apfeld and Kenyon 1998). An additional pathway of life span regulation comes from opposing nonautonomous signals from the somatic gonad and germ line (Hsin and Kenyon 1999). Notably, ablation of germ-line precursors extends life span, and this extension is daf-12(+) dependent.
A unifying hypothesis is that lipophilic hormones acting through daf-12 and related receptors could coordinate various aspects of nematode life history. In particular, hormonal input at one or more stages could synchronize stage programs throughout the body. Absence of hormone or distinct hormones, acting through daf-12 in target cells, may regulate S3 and S3d alternatives. Heterochronic activities distributed in the various tissues could then select stage- and tissue-appropriate programs, and advance the heterochronic circuit. A hormonal mechanism could couple environmental cues to coordinate changes in development and metabolism throughout the organism. In the adult, this may control longevity. Ultimately, further characterization of signaling components upstream and downstream of daf-12 may reveal the mechanisms that govern nematode and perhaps higher metazoan life histories.
Materials and methods
Nematode strains
Except where noted otherwise, nematodes were cultured at 20° on NG agar inoculated with Escherichia coli strain OP50 (Sulston and Hodgkin 1988). Alleles used in this study were wild-type N2, daf-12 (m20, m25, m116, m419, m420, m421, m422, m423, m424, m425, m524, m545, m524m570, m545m571, rh61, rh61rh411, rh61rh412, rh62, rh62rh157, rh84, rh193, rh257, rh258, rh273, rh274, rh284, rh285, rh286, a156)X; lin-15(n765)X; daf-1(m40)IV; mut-6(st702)IV; unc-22(st192st527)IV; daf-2(m41 or e1370)III; daf-4(e1364)III; mut-2(r459)I.
Mutant isolation
Revertants of daf-1/daf-2 double mutants were selected because daf-12 mutations suppress both daf-1 and class 1 daf-2 mutants (Gems et al. 1998). EMS-mutagenized daf-1(m40)IV;daf-2(m41)III hermaphrodites were placed individually onto 40, 10 cm diameter plates and grown one generation at 15°. Cultures were shifted to 25° and screened for F2 revertants that escaped dauer arrest. Alleles m419, m420, m421, m424, and m425 were isolated after screening 12,000 F2 animals. m422 and m423 were obtained in a similar screen. Likewise, 1300 daf-1(m40);daf-2(e1370) EMS-mutagenized hermaphrodites were distributed on 358, 10-cm plates, grown at 15° for one generation, and shifted to 23.5° to screen for revertants. Allele m583 was isolated after screening 110,000 F2 animals.
Tc1 mutagenesis
For isolation of m524::Tc1, eggs were collected by alkaline hypochlorite treatment of mutator strain RW7097, mut-6(st702); unc-22(st192st527)IV, and incubated overnight in M9 buffer to obtain synchronized L1 larvae. These larvae were incubated with shaking for 3–4 days at a final concentration of 3500–4000 larvae per ml (3 million L1) in S-medium (Sulston and Hodgkin 1988) containing 0.2% OP50 and dauer-inducing pheromone. The pheromone preparation was diluted from concentrated extract to an activity approximately tenfold greater than that found in depleted culture media (Golden and Riddle 1984). Only dauer-defective mutants were able to escape developmental arrest (Starich et al. 1995).
Four DR1164, mut-2(r459);daf-4(e1364) adults were placed on each of 132 plates and grown 2–3 generations at 15°. Plates were then shifted to 23.5° for 14–19 days and screened for the presence of revertant L4 larvae or adults. One million worms were screened to yield m545.
rh61rh412 was isolated as a spontaneous revertant of gonadal heterochrony from rh61. Other daf-12 alleles are described elsewhere (Riddle et al. 1981; Thomas et al. 1993; Larsen et al. 1995; Antebi et al. 1998).
daf-12 cloning and cosmid rescue
m524::Tc1 was backcrossed 10 times to N2, and the polymorphic 3.1-kb BglII fragment was isolated from size-fractionated libraries. The genomic flanking DNA was used as a probe to isolate genomic clones DR182 and DR183 from a lambda EMBL4 genomic library (courtesy of C. Link, U. Colorado, Boulder). DR182 and DR183 were fingerprinted to YACS Y52H9 and Y9E8 and cosmids C16H9, B0533, and K03D4 (courtesy of A.R. Coulson, the Sanger Center, U.K.). The sequence of the daf-12 genomic region on cosmid F11A1 was completed by the Genome Consortium. For cosmid rescue, cosmid F11A1 (10 ng/μl) and pRF4 rol-6(su1006) marker (75 ng/μl) (Mello et al. 1991) were microinjected into the oogonia of daf-12(rh61rh411) dauer-defective animals. One of three roller lines gave rescue, as indicated by the formation of dauer larvae among populations grown to exhaustion on NG agar plates.
cDNA cloning
daf-12
To isolate cDNAs, 8 × 105 plaques from a λ gt10 mixed-stage library (courtesy of J. Ahringer and J. Kimble, U. Wisconsin, Madison) were screened using cosmid KO3D4, and positives rescreened with a 232-bp SalI daf-12 probe to obtain clones DR187 and DR188. Two partial cDNA clones were isolated that extended from the 3′ half of exon 1 to the 5′ half of exon 11. To obtain daf-12 mRNA 5′ ends, we performed nested RT–PCR (Frohman et al. 1988). Total RNA from mixed-stage cultures was used for cDNA synthesis. For first-round PCR, a forward primer containing the SL1 splice leader coupled to an external adapter (5′-aactgcagccgcggctcgagGGTTTAATTACCCAAGTTTGAG-3′) and a daf-12 reverse primer downstream of the predicted stop codon
(5′-GGCTAGATTGAGTTTGTTGGATGGAGGC-3′) were used. For second-round PCR, external adapter alone (5′-AACTGCAGCCGCGGCTCGAG-3′) and a nested daf-12-specific reverse primer (5′-CGGGATCCGTTGTGAGGTATTGAAGGCAT-3′) were used. Several clones for the isoforms 12A1(one), 12A3 (five), and 12B (four) were fully sequenced. For amplifying 3′ ends, forward primer (5′-cgggatccGAGGCGTTTCGTCAAAGTTGC-3′) and the oligo-dT-external adapter primer 5′-AACTGCAGCCGCGGCTCGAGT18-3′ were used in first-round PCR. Forward primer 5′-CTTCGGTTTCTTCGACGAGG-3′ and the external adapter were used in second-round PCR. The sequence of three 3′ end clones revealed that the polyA addition site was polymorphic, following 1311, 1316, and 1397 nucleotides after the stop codon. Three cDNA clones recently deposited in public databases, DAF-12A1 (no. AF136238), DAF-12A2 (no. AF136239), and DAF-12B (no. AF136240), match our 12A1 and 12B, but we found clone 12A3 instead of DAF-12A2.
nhr-8 and nhr-48
SL1 primer 5′-GGAATTCGGTTTAATTACCCAAGTTTGAG-3′ and a reverse primer located downstream of the predicted stop codon were used to amplify clones by PCR. The reverse primers for nhr-8 and nhr-48 were, respectively, 5′-GGAATTCGCAATATGTGCAATAATTCATGG-3′ and 5′-GGAATTCATAACAGTGAACGCTAGGTCTGG-3′. In addition, a full-length nhr-8 cDNA clone from the Chris Martin library (Waterston et al. 1992) encoding isoform 8A1 (cm14e12) was sequenced. The 3′ ends of nhr-48 were amplified as above using the primer pairs 5′-GCATGTGCGGACGCTATCCACC-3′/external adaptor-dT18 and 5′-GCTGAAAATGAGTCTCGGGAAT-3′/external adaptor for first- and second-round PCR, respectively.
Mapping mutations
Mutations were scanned using chemical mismatch detection (Cotton et al. 1988). For all reactions, wild-type DNA was end-labeled and hybridized with unlabeled mutant DNA. First, genomic PCR fragments spanning the DBD (F11A1, 32212–33613) were scanned for alleles m20, m25, m116, m419, m420, m421, m422, m423, m424, m425, m583, sa156, rh61, rh62, rh62rh157, rh84, rh193, rh257, rh258, rh273, rh274, rh284, rh285, and rh286. Chemical cleavage was detected in m20, m116, m420, m421, m422, m423, m424, m425, m583, sa156, and rh62rh157. These mutations, as well as alleles rh61rh411 and rh61rh412 were identified by direct DNA sequencing of PCR fragments. Second, a fragment spanning the amino-terminal half of the LBD (F11A1, 34818-36548) was scanned with the remaining unmapped mutations. Chemical cleavage was detected in m25, m419, rh61, rh61rh411, rh62, rh62rh157, rh193, rh273, rh274, and rh286, and the mutations were sequenced. Third, PCR fragment 35872–37027, corresponding to the carboxy-terminal half of the LBD, was sequenced directly for the remaining mutants. Mutations were identified in rh84, rh257, rh284, and rh285. rh258 was not found.
daf-12::GFP
A daf-12::GFP gene fusion was generated by GFP-tagging exon 1 of the daf-12 gene in cosmid F11A1. First, the SgrA1 fragment from F11A1 (pos. 8613–18314), containing the first two exons was subcloned into the AgeI site of pLITMUS 28 (New England BioLabs). A unique ApaI restriction site was created in exon 1 (pos. 16028) by site-directed mutagenesis (GAGCCC → GGGCCC) (ExSite PCR-Based Site Directed Mutagenesis Kit, Stratagene), and a GFP fragment cloned in-frame into the new ApaI site. Then the SrgA1 F11A1 fragment containing GFP was cloned back into the original cosmid using phage packaging procedure (Gigapack III Gold, Stratagene). Cosmid F11A1 containing daf-12::GFP (10–20 ng/μl) and lin-15(+) pL15.EK coinjectible marker (75 ng/μl) (Clark et al. 1994) were microinjected into the oogonia of daf-12(rh61rh411);lin-15(n765) dauer-defective animals. Transgenic lines containing lin-15(+) extrachromosomal arrays were scored for rescue of the Daf-d phenotype. Chromosomal integrants were generated by UV mutagenesis (30,000 μjoules/cm2) with a UV Stratalinker (Stratagene). F2 progeny were screened for the stable inheritance of lin-15(+) and daf-12::GFP. Integrants were outcrossed twice. Fluorescence photographs were taken with a Sony DXC9100P video camera at 0.5 sec exposure.
Acknowledgments
We thank J. Rothman for providing JR667 Seam Cell Marker GFP strain, Y. Kohara for yk333f9 and yk226g12 cDNA clones, A. Fire for GFP vectors, P. Larsen for some daf-12 primers and for communication of unpublished results, C. Weitzel and C. Kober-Eisermann for technical assistance, and K. Nishiwaki for critical reading of the manuscript. We also thank the Genome Consortium for completing the daf-12 genomic sequence and the CGC for strains. This work was supported by NIH grant NS26295 and NSF IBN 9604145 to E.M.H, the MPG for A.A., and NIH grants GM60151 and AG12689 to D.L.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL antebi@molgen.mpg.de; FAX 49 30 8413 1130.
References
- Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Ambros V. Heterochronic genes. In: Riddle DL, Blumenthal T, Meyer B, Priess J, editors. C. elegans II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 501–518. [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Antebi A, Culotti JG, Hedgecock EM. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development. 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- Antebi A, Norris C, Hedgecock E, Garriga G. Cell and growth cone migrations. In: Riddle DL, Blumenthal T, Meyer B, Priess J, editors. C. elegans II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 583–609. [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Regulation of life span by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- Baker AR, McDonnell DP, Hughes M, Crisp TM, Mangelsdorf DJ, Haussler MR, Pike JW, Shine J, O'Malley BW. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc Natl Acad Sci. 1988;85:3294–3298. doi: 10.1073/pnas.85.10.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- Bender M, Imam FB, Talbot WS, Ganetzky B, Hogness DS. Drosophila ecdysone receptor mutations reveal functional differences among receptor isoforms. Cell. 1997;91:777–788. doi: 10.1016/s0092-8674(00)80466-3. [DOI] [PubMed] [Google Scholar]
- Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Bäckman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- Burke LJ, Downes M, Laudet V, Muscat GE. Identification and characterization of a novel corepressor interaction region in RVR and REV-erbA alpha. Mol Endocrin. 1998;12:248–262. doi: 10.1210/mend.12.2.0061. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Corsortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Clark SG, Lu X, Horvitz HR. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics. 1994;137:987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn CM, Mori I, Ohshima Y, Bargmann CI. A cyclic nucleotide-gated channel inhibits sensory axon outgrowth in larval and adult Caenorhabditis elegans: A distinct pathway for maintenance of sensory axon structure. Development. 1998;125:249–258. doi: 10.1242/dev.125.2.249. [DOI] [PubMed] [Google Scholar]
- Cotton RG, Rodrigues NR, Campbell RD. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci. 1988;85:4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, White R, Lees JA, Parker MG. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the life span of Caenorhabditis elegans. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez M, Attisano L, Wrana JL, Albert PS, Massague J, Riddle DL. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature. 1993;365:644–649. doi: 10.1038/365644a0. [DOI] [PubMed] [Google Scholar]
- Finch CE. Longevity, senescence and the genome. Chicago, IL: University of Chicago Press; 1994. [Google Scholar]
- Fisk GJ, Thummel CS. Isolation, regulation, and DNA-binding properties of three Drosophila nuclear hormone receptor superfamily members. Proc Natl Acad Sci. 1995;92:10604–10608. doi: 10.1073/pnas.92.23.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman LP. Increasing the complexity of coactivation in nuclear receptor signaling. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- Frieden E. The dual role of thyroid hormones in vertebrate development and calorigenesis. In: Gilbert L, Frieden E, editors. Metamorphosis: A problem in developmental biology. New York: Plenum; 1981. pp. 545–564. [Google Scholar]
- Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi LL, Albert PS, Riddle DL. daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell. 1990;61:635–645. doi: 10.1016/0092-8674(90)90475-t. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: Developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102:368–378. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G. daf-2, daf-16, and daf-23: Genetically interacting genes controlling dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenache DG, Caldicott I, Albert PS, Riddle DL, Politz SM. Environmental induction and genetic control of surface antigen switching in the nematode Caenorhabditis elegans. Proc Natl Acad Sci. 1996;93:12388–12393. doi: 10.1073/pnas.93.22.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol. 1985;107:128–133. doi: 10.1016/0012-1606(85)90381-1. [DOI] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the life span of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Inoue T, Thomas JH. Targets of TGF-β signaling in Caenorhabditis elegans dauer formation. Dev Biol. 2000;217:192–204. doi: 10.1006/dbio.1999.9545. [DOI] [PubMed] [Google Scholar]
- Izumo S, Mahdavi V. Thyroid hormone receptor alpha isoforms generated by alternative splicing differentially activate myosin HC gene transcription. Nature. 1988;334:539–542. doi: 10.1038/334539a0. [DOI] [PubMed] [Google Scholar]
- Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- Johnstone IL, Barry JD. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 1996;15:3633–3639. [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, Hogness DS. The Drosphila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- Krust A, Kastner P, Petkovich M, Zelent A, Chambon P. A third human retinoic acid receptor, hRAR-γ. Proc Natl Acad Sci. 1989;86:5310–5314. doi: 10.1073/pnas.86.14.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Liu ZC, Ambros V. Heterochronic genes control the stage-specific initiation and expression of the dauer larva developmental program in Caenorhabditis elegans. Genes & Dev. 1989;3:2039–2049. doi: 10.1101/gad.3.12b.2039. [DOI] [PubMed] [Google Scholar]
- Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Ong ES, Dyck JA, Evans RM. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- Patterson GI, Koweek A, Wong A, Liu Y, Ruvkun G. The DAF-3 Smad protein antagonizes TGF-β-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes & Dev. 1997;11:2679–2690. doi: 10.1101/gad.11.20.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz H, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-β homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Albert PS. Genetic and environmental regulation of dauer larva development. In: Riddle DL, Blumenthal T, Meyer B, Priess J, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 739–768. [PubMed] [Google Scholar]
- Riddle DL, Swanson MM, Albert PS. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Ambros V. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development. 1995;121:2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- Ruvkun G, Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- Schackwitz WS, Inoue T, Thomas JH. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron. 1996;17:719–728. doi: 10.1016/s0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: How receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- Sluder AE, Mathews SW, Hough D, Yin VP, Maina CV. The nuclear receptor superfamily has undergone extensive proliferation and diversification in nematodes. Genome Res. 1999;9:103–120. [PubMed] [Google Scholar]
- Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, Schuyler MW, Collet J, Thomas JH, Riddle DL. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139:171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The nematode, Caenorhabditis elegans. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Terns RM, Kroll-Conner P, Zhu J, Chung S, Rothman JH. A deficiency screen for zygotic loci required for establishment and patterning of the epidermis in Caenorhabditis elegans. Genetics. 1997;146:185–206. doi: 10.1093/genetics/146.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Birnby DA, Vowels JJ. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics. 1993;134:1105–1117. doi: 10.1093/genetics/134.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels JJ, Thomas JH. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics. 1994;138:303–316. doi: 10.1093/genetics/138.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RL, Apriletti JW, McGrath ME, West BL, Baxter JD, Fletterick RJ. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- Waterston R, Martin C, Craxton M, Huynh C, Coulson A, Hillier L, Durbin R, Green P, Shownkeen R, Halloran N, et al. A survey of expressed genes in Caenorhabditis elegans. Nat Genet. 1992;1:114–123. doi: 10.1038/ng0592-114. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- Yanagisawa J, Yanagi Y, Masuhiro Y, Suzawa M, Watanabe M, Kashiwagi K, Toriyabe T, Kawabata M, Miyazono K, Kato S. Convergence of transforming growth factor-β and vitamin D signaling pathways on SMAD transcriptional coactivators. Science. 1999;283:1317–1321. doi: 10.1126/science.283.5406.1317. [DOI] [PubMed] [Google Scholar]
- Yoh SM, Chatterjee VKK, Privalsky ML. Thyroid hormone resistance syndrome manifests as an aberrant interaction between mutant T3 receptors and transcriptional corepressors. Mol Endocrin. 1997;11:470–480. doi: 10.1210/mend.11.4.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]