Centrobin recruitment to the centriole biogenesis site and its function during elongation and stabilization of centrioles depend on tubulin interaction.
Abstract
Centrobin is a daughter centriole protein that is essential for centrosome duplication. However, the molecular mechanism by which centrobin functions during centriole duplication remains undefined. In this study, we show that centrobin interacts with tubulin directly, and centrobin–tubulin interaction is pivotal for the function of centrobin during centriole duplication. We found that centrobin is recruited to the centriole biogenesis site via its interaction with tubulins during the early stage of centriole biogenesis, and its recruitment is dependent on hSAS-6 but not centrosomal P4.1–associated protein (CPAP) and CP110. The function of centrobin is also required for the elongation of centrioles, which is likely mediated by its interaction with tubulin. Furthermore, disruption of centrobin–tubulin interaction led to destabilization of existing centrioles and the preformed procentriole-like structures induced by CPAP expression, indicating that centrobin–tubulin interaction is critical for the stability of centrioles. Together, our study demonstrates that centrobin facilitates the elongation and stability of centrioles via its interaction with tubulins.
Introduction
Centrosomes constitute two symmetrical barrel-shaped centrioles that are embedded in the pericentriolar material. The centrioles are ∼200 nm in diameter and ∼500 nm in length (Doxsey, 2001; Doxsey et al., 2005; Bornens and Azimzadeh, 2007; Lüders and Stearns, 2007; Loncarek and Khodjakov, 2009; Nigg and Raff, 2009). The centriole barrel contains nine sets of microtubule triplets composed of heterodimers of α/β-tubulin in humans (Bornens, 2002; Bornens and Azimzadeh, 2007; Nigg, 2007).
Centriole duplication is tightly coupled to the cell cycle (Hinchcliffe et al., 1999; Lacey et al., 1999; Meraldi et al., 1999; Hinchcliffe and Sluder, 2001; Tsou and Stearns, 2006b; Strnad and Gönczy, 2008). Once the cell enters the S phase, centriole duplication begins with two procentrioles emerging from the proximal end of the existing centrioles. The centrosome in this phase has a mature mother centriole with appendages that was assembled two cell divisions prior, an immature mother without appendages that was daughter in the previous cycle, and two new emerging procentrioles. During mitosis, each centriole pair moves to either end of the cell to form the spindle poles (Lange and Gull, 1995; Gromley et al., 2003; Anderson and Stearns, 2009). After mitosis and before reentry into G1, the two centrioles disengage in response to activation of the enzyme separase (Tsou and Stearns, 2006a,b). Complete maturation of daughter centriole to mother centriole requires passage through the second mitotic cycle, during which it acquires appendages (Robbins and Gonatas, 1964; Robbins et al., 1968; Kuriyama and Borisy, 1981; Vorobjev and Chentsov, 1982; Lange and Gull, 1995; Anderson and Stearns, 2009). Uncoupling of the centrosome duplication process from the cell cycle can result in cells with more than two centrosomes, leading to aberrant centrosome amplification, genetic instability, and tumor progression (Pihan et al., 1998; Doxsey, 2001; Hinchcliffe and Sluder, 2001; Pihan et al., 2003).
The pathway for centriole biogenesis has been best delineated using Caenorhabditis elegans (Delattre et al., 2006; Pelletier et al., 2006; Dammermann et al., 2008). In C. elegans, a central tube is formed first, followed by assembly of nine singlet microtubules on the central tube (O’Connell et al., 2001; Kirkham et al., 2003; Leidel and Gönczy, 2003; Kemp et al., 2004; Pelletier et al., 2004; Rodrigues-Martins et al., 2007; Dammermann et al., 2008; Kitagawa et al., 2009). In mammalian cells, the composition of centrosomes is much more complex (Andersen et al., 2003). The homologues of a small number of mammalian centrosomal proteins have been identified in lower eukaryotes; i.e., hSAS-6 as the homologue of C. elegans SAS-6 (Leidel and Gönczy, 2003; Leidel et al., 2005), centrosomal P4.1–associated protein (CPAP)/hSAS-4 of SAS-4 (Hung et al., 2000), CEP192 of SPD-2 (Andersen et al., 2003), and PLK4 of ZYG-1 (Bettencourt-Dias et al., 2005; Habedanck et al., 2005). The homologue for SAS-5 has not yet been identified.
The centriole duplication process can be classified into initiation, elongation, and maturation (Azimzadeh and Bornens, 2007). In humans, the initiation of procentriole biogenesis happens upon activation of PLK4, followed by recruitment of hSAS-6 to the proximal end of the existing centriole (Strnad et al., 2007). Although PLK4 and hSAS-6 can be recruited to the biogenesis site in the absence of CPAP, CEP135, and γ-tubulin, the biogenesis process does not progress beyond initiation. CP110 functions as a capping protein at the distal end of the procentriole, below which tubulin dimers are added to elongate the centriole wall (Kleylein-Sohn et al., 2007). Overexpression of CPAP and down-modulation of CP110 expression result in uncontrolled elongation of centrioles, highlighting the role of these proteins in controlling the length of centrioles (Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009). Maturation of the centrioles occurs after elongation when the centrioles acquire appendage proteins such as Ninein and Odf-2 (Ou et al., 2002; Ishikawa et al., 2005).
Our group previously identified the coiled-coil protein centrobin and demonstrated that it preferentially localizes to the daughter centriole and is required for centriole duplication (Zou et al., 2005). Centrobin is recruited to the procentrioles at the beginning of S phase. During S, G2, and M phases, there are two centrobin-positive centrioles, the newly assembled procentrioles. After cell division, most G1 phase cells have one centrobin-positive centriole, the daughter centriole assembled in the previous cell cycle. Upon reentering S phase, centrobin on the daughter centriole assembled in the previous cycle becomes undetectable in the majority of the cells. In the absence of centrobin, no discernible centriole structures were assembled as demonstrated by EM analysis (Zou et al., 2005). Centrobin has also been reported to be a substrate of the kinase Nek2 and plays a role in stabilizing the microtubule network (Jeong et al., 2007). In addition, centrobin was found to regulate the assembly of functional mitotic spindles (Jeffery et al., 2010).
In this study, we elucidate the molecular mechanism of centrobin function during centriole duplication. We found that centrobin is recruited to centrosomes early during centriole duplication, interacts with α/β-tubulin dimers, and promotes the elongation and stability of centrioles.
Results
Centrobin is recruited to the centriole biogenesis sites after hSAS-6
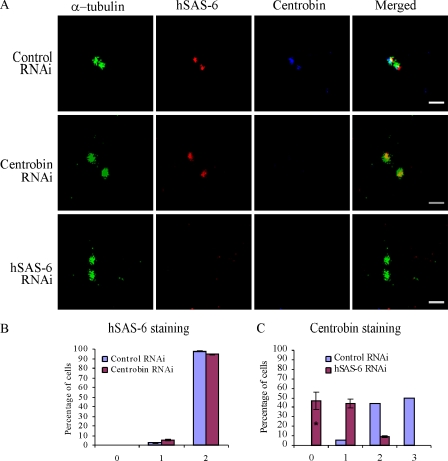
Recent studies have identified hSAS-6 as the critical protein to initiate centriole biogenesis. Upon PLK4 activation, hSAS-6 is recruited to the proximal end of the existing centrioles (Habedanck et al., 2005; Kleylein-Sohn et al., 2007; Strnad et al., 2007). Here, we first examined whether centrobin is recruited to the site of centriole biogenesis before or after hSAS-6. For this purpose, we depleted centrobin in HeLa cells using siRNA and treated the cells with hydroxyurea (HU) for 16 h. Similar to control cells, >90% of the centrobin-depleted cells still had two hSAS-6 dots, indicating that hSAS-6 can be recruited to the centriole biogenesis site in the absence of centrobin (Fig. 1, A [first and second panels] and B).
Figure 1.
Centrobin is recruited to the centriole biogenesis site after hSAS-6. (A [first and second panels] and B) hSAS-6 recruitment to the centriole biogenesis site is not dependent on the presence of centrobin. HeLa cells transfected with control or centrobin siRNA were treated with HU and stained with anti-centrobin, hSAS-6, and α-tubulin antibodies. The percentage of hSAS-6–positive centrioles is shown in B. (A [first and third panels] and C) Centrobin recruitment to the centriole biogenesis site is dependent on the presence of hSAS-6. HeLa cells treated with control or hSAS-6 siRNAs and HU were stained as previously described for A, first and second panels, and the percentage of cells with centrobin-positive centrioles is plotted in C. Histograms are plotted as mean ± SEM (n = 3). The asterisk denotes that the difference is significant, and P < 0.001. Bars, 1 µm.
Next, recruitment of centrobin to the centrioles was assessed in HU-treated HeLa cells transfected with control and hSAS-6 siRNAs by centrobin staining. It should be noted that HU treatment inhibited the decrease of centrobin from the daughter centriole (Fig. 1, A and C, control cell). Therefore, in these experiments, cells with two or three centrobin staining dots are the cells that have already recruited centrobin to the centriole biogenesis sites and are in the process of assembling the procentriole as opposed to untreated cells that have only one centrobin staining dot at G1 phase and two centrobin staining dots at S phase (see Fig. 8 L; Zou et al., 2005). The molecular mechanism of displacement/degradation of centrobin from the daughter centriole is currently under investigation in our laboratory. As shown in Fig. 1 C, the control cells had either three or two centrobin staining dots (50% and 45%, respectively), indicating that centrobin has been recruited and centriole biogenesis is in progress. However, 90% of hSAS-6–depleted cells had either one or zero centrobin dots (Fig. 1, A [first and third panels] and C). Therefore, it can be concluded that centrobin recruitment to the daughter centrioles depends on prior hSAS-6 recruitment, whereas the recruitment of hSAS-6 is not dependent on the presence of centrobin.
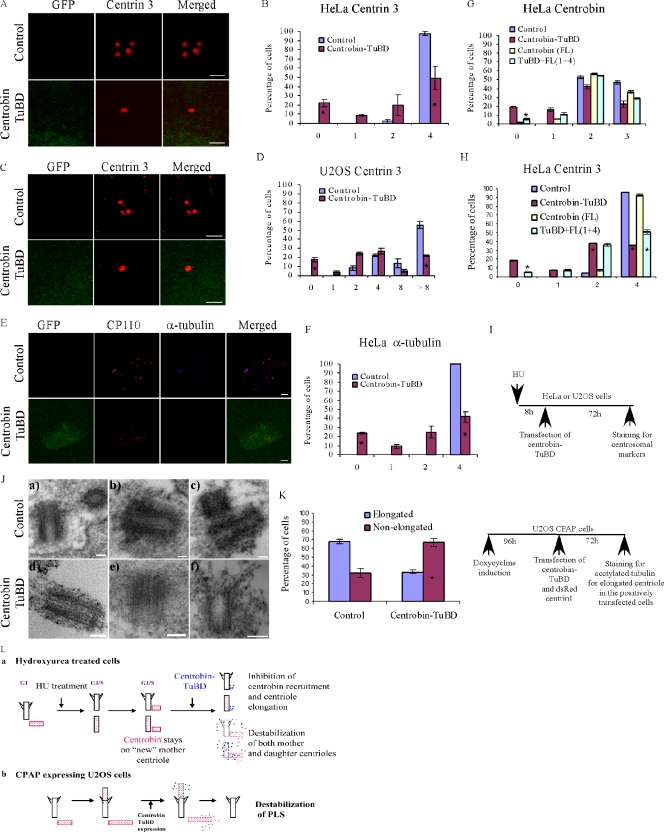
Figure 8.
Disruption of centrobin–tubulin interaction destabilizes the centrioles. (A–I) Centrobin–tubulin interaction confers stability to the centrioles. HeLa (A, B, E, and F) and U2OS (C and D) cells pretreated with HU and transfected with either control or GFP–centrobin-TuBD were stained with anti–centrin 3 (A–D) or anti–α-tubulin and CP110 antibodies (E and F), and the percentage of cells with the indicated number of respectively stained centrioles is presented. (G and H) Increasing concentration of full-length centrobin partially rescued the inhibitory effect of centrobin-TuBD on centrobin recruitment and centriole elongation. HU-pretreated HeLa cells were transfected with centrobin-TuBD, full-length centrobin (pMini-myc-centrobin), or a combination as indicated. Cells were stained with anti-centrobin (G) and centrin 3 (H) antibodies, and the percentage of centrobin (G)- and centrin 3 (H)–positive centrioles is presented. (I) The schematic depicts the experimental method in A–H. (J) EM analysis of centrobin-TuBD cells. HeLa cells pretreated with HU and transfected with control or GFP–centrobin-TuBD were processed for EM, and 250-nm-thick sections were analyzed to detect centrosomes. (K) Overexpression of centrobin-TuBD destabilizes the preformed PLSs. U2OS-CPAP cells preinduced with doxycycline for 96 h were transfected with control or GFP–centrobin-TuBD along with dsRed–centrin 1 vector. dsRed-centrin–positive cells were evaluated for the presence of elongated or nonelongated PLSs by staining with anti-acetylated tubulin antibody. (L) The schematic illustrates the destabilization effect of centrobin-TuBD in HU-treated cells (panel a) or U2OS-CPAP–overexpressing cells (panel b). Histograms are plotted as mean ± SEM (n = 3). Asterisks denote that the difference is significant, and P < 0.001. Bars: (A and C) 1 µm; (E) 3 µm; (J) 100 nm.
Centrobin recruitment to the centriole biogenesis site is not dependent on the recruitment of CPAP and CP110
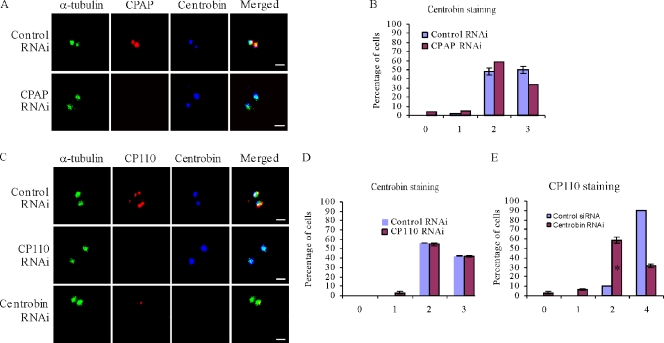
In addition to PLK4 and hSAS-6, CPAP and CP110 have also been demonstrated to be recruited to the centriole biogenesis site at a very early stage, and CPAP is also required for procentriole biogenesis (Kleylein-Sohn et al., 2007). Recruitment of hSAS-6 to the procentriole initiation site was not affected in the absence of CPAP and CP110 proteins (Kleylein-Sohn et al., 2007; Kohlmaier et al., 2009). To further delineate the order of centrobin recruitment during the centriole biogenesis process, we examined centrobin recruitment in the CPAP and CP110 RNAi cells. We found that centrobin was still recruited to the centriole biogenesis site in the CPAP- and CP110-depleted cells (Fig. 2, A and B and C and D, respectively), suggesting that centrobin recruitment is not dependent on the prior recruitment of CPAP and CP110. Knockdown efficacy of CPAP RNAi was assessed in parallel by its effect on centriole duplication using acetylated tubulin as a centriole marker. Although >90% of the control cells exhibited four centrioles, <30% of CPAP-depleted cells had four centrioles (Fig. S1, A and B), indicating that CPAP was efficiently depleted, thereby resulting in a block in centriole duplication. Our data indicate that centrobin recruitment is not dependent on CPAP, but its recruitment is not sufficient for centrobin duplication to progress in the absence of CPAP as expected. CP110 RNAi efficiency was evaluated by quantitating the percentage of cells having elongated centrioles. 40% of the CP110 RNAi cells had elongated centrioles, suggesting that CP110 has also been depleted effectively (Fig. S1, C and D). However, we cannot completely rule out the possibility that the residual amount of CPAP and CP110 present might be sufficient for centrobin recruitment to the centrioles.
Figure 2.
Centrobin recruitment to the centriole biogenesis site is not dependent on CPAP and CP110. (A, B, C [first and second panels], and D) Centrobin recruitment to the centriole biogenesis site is not inhibited by CPAP and CP110 knockdown. HeLa cells transfected with control, CPAP, or CP110 siRNAs were treated with HU and stained using anti-centrobin, α-tubulin, CPAP, or CP110 antibodies. The percentage of centrobin-positive centrioles is shown in B and D. (C [first and third panels] and E) Knockdown of centrobin inhibits recruitment of CP110. HeLa cells transfected with control or centrobin siRNA and HU were stained using α-tubulin, centrobin, and CP110 antibodies. The percentage of CP110-positive centrioles is shown in E. Histograms are plotted as mean ± SEM (n = 3). The asterisk denotes that the difference is significant, and P < 0.001. Bars, 1 µm.
We further examined CP110 recruitment to procentrioles in centrobin RNAi cells. We found that only 30% of the centrobin RNAi–transfected cells exhibited four CP110 staining dots, whereas 90% of the control cells exhibited four CP110 staining dots (Fig. 2, C [first and third panels] and E), suggesting that CP110 recruitment requires prior recruitment of centrobin. A similar experiment was attempted to examine the recruitment of CPAP in the absence of centrobin. However, because CPAP is localized to the proximal end of the parental centrioles (Kleylein-Sohn et al., 2007; Kohlmaier et al., 2009; Schmidt et al., 2009), it was impossible to unambiguously distinguish the newly recruited CPAP protein from the CPAP existing on the parental centrioles by immunofluorescence. We indeed observed small CPAP staining dots as reported in the hSAS-6–depleted cells (Kohlmaier et al., 2009), but it could just be a consequence of inhibited centriole elongation. Hence, we were unable to determine whether the recruitment of CPAP is dependent on the presence of centrobin. Because both CPAP and CP110 are members of the early initiation proteins along with CEP135 and γ-tubulin (Kleylein-Sohn et al., 2007), our studies indicate that centrobin is also one of the early initiation proteins because it was recruited to the centriole biogenesis site before CPAP/CP110 and is required for the CP110 recruitment.
Centrobin is required for the elongation of centrioles in normal and CPAP-overexpressing cells
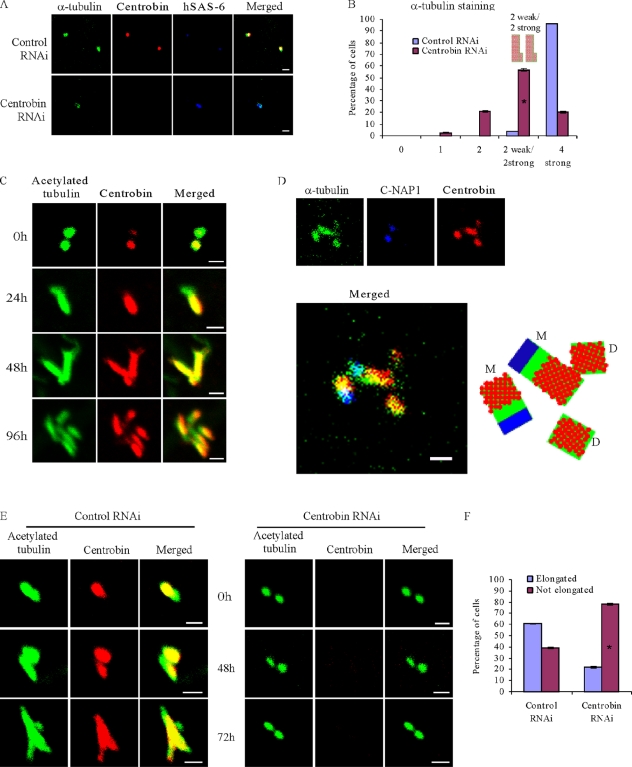
To determine whether centrobin is required only for assembly of the initiation structure or also for further elongation of the centriole, we examined the size of newly assembled daughter centrioles by staining one of the core centriole components, α-tubulin in HU-treated centrobin-depleted HeLa cells. Although nearly all of the control cells had four centrioles, 20% of centrobin-depleted cells exhibited only two centrioles, indicating that centrobin depletion inhibited centriole duplication. Detailed analysis of the centrobin-depleted cells still having four α-tubulin–stained centrioles revealed that in 58% of these cells, two of the four centrioles stained much weaker for α-tubulin, indicating that daughter centriole elongation is likely stunted in these centrobin-depleted cells (Fig. 3, A and B). This finding suggests that centrobin likely also functions during centriole elongation.
Figure 3.
Centrobin is required for the elongation of centrioles. (A and B) Centrobin is required for centriole elongation during normal centriole duplication. HeLa cells transfected with control or centrobin siRNA were treated with HU and stained with anti-centrobin, hSAS-6, and α-tubulin antibodies. The percentage of varied intensity of α-tubulin–positive centrioles was scored and plotted in B. (C and D) Centrobin was observed on the elongated portion of the mother centriole in addition to the elongating daughter centrioles. CPAP-induced PLSs were generated by inducing U2OS-CPAP cells with 1 µg/ml doxycycline for the indicated time points. Anti-centrobin and acetylated tubulin antibodies were used to stain centrioles/PLSs (C). In D, the cells were costained with anti–C-NAP1, centrobin, and α-tubulin antibodies. The illustration indicates the presence of centrobin on the elongated portion of the mother centriole as well. M and D refer to the mother and daughter centriole, respectively. (E and F) Knockdown of centrobin inhibits the elongation of PLSs. U2OS-CPAP cells were transfected with control or centrobin siRNAs for 72 h followed by doxycycline treatment for the indicated time points (E). The percentage of elongated versus nonelongated centrioles was quantified at the 72-h time point (F). Histograms are plotted as mean ± SEM (n = 3). Asterisks denote that the difference is significant, and P < 0.001. Bars, 1 µm.
Recently, it was reported that centrioles can elongate beyond the predetermined length of 500 nm upon CPAP overexpression or CP110 depletion (Kohlmaier et al., 2009; Tang et al., 2009). This system enables us to study the role of centrobin in centriole elongation specifically. The U2OS cells inducibly expressing CPAP reported by Kohlmaier et al. (2009) were induced to express CPAP for 24, 48, and 96 h by addition of doxycycline and then were stained with anticentrobin and antiacetylated tubulin antibodies to visualize the elongated procentriole-like structures (PLSs). As reported previously (Tang et al., 2009), we observed that at 24 h after induction (Fig. 3 C, second panel), mainly the daughter centriole started to elongate, and centrobin was present along the elongating centriole. At later time points (Fig. 3 C, second and third panels), both mother and daughter centrioles were elongated. Centrobin staining was found not only on the daughter centrioles but also on the elongated portion of mother centrioles, whereas the original portion of mother centrioles was devoid of centrobin staining (Fig. 3, C [bottom row] and D). The identity of the elongated mother centriole was confirmed by costaining with anti–C-NAP1 antibody together with anticentrobin and α-tubulin antibodies (Fig. 3 D; Fry et al., 1998). The presence of centrobin on the elongated portion of the mother also indicates a role of centrobin during centriole elongation.
Furthermore, we depleted centrobin in the U2OS-CPAP cells using siRNA before inducing CPAP expression. As expected, 60% of the control cells exhibited elongated PLSs (Fig. 3, E [left panel, bottom row] and F). In comparison, only 20% of the centrobin-depleted cells had the elongated PLS in spite of the induction of CPAP expression for 72 h (Fig. 3 F). In summary, these findings demonstrate a critical role of centrobin in the centriole elongation process.
Centrobin interacts with tubulin in vivo
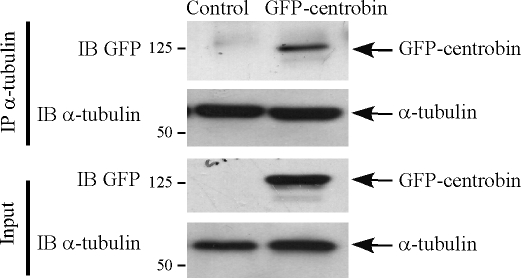
Because our finding proved a critical role of centrobin during the centriole elongation process, we next explored the mechanism by which centrobin functions during centriole elongation. The centriolar wall is composed of nine sets of microtubule triplets assembled from heterodimers of α- and β-tubulin, and centriole elongation is a process of adding tubulin dimers to the initiation structure underneath the CP110 cap (Kleylein-Sohn et al., 2007). Therefore, we examined whether centrobin interacts with tubulins. Centrobin interaction with tubulin was first examined by exogenously expressing GFP-centrobin in 293T cells and subjecting the lysates to immunoprecipitation using anti–α-tubulin antibody. Immunoblotting analysis revealed that a significant fraction of the overexpressed centrobin can be pulled down using anti–α-tubulin antibody, clearly indicating an interaction of centrobin and α-tubulin (Fig. 4). We further performed coimmunoprecipitation of endogenous centrobin and α-tubulin in lysates of 293T cells (Fig. S2). Indeed, we observed that anticentrobin but not the isotype control antibody could pull down α-tubulin (Fig. S2 A). The corollary stands true that anti–α-tubulin antibody can also immunoprecipitate endogenous centrobin (Fig. S2 B), indicating that centrobin interacts with α-tubulin in vivo. This interaction is likely important for the function of centrobin during centriole duplication.
Figure 4.
Centrobin interacts with α-tubulin in vivo. 293T cells transfected with control or GFP-centrobin vectors were lysed and subjected to immunoprecipitation (IP) using anti–α-tubulin antibody. Immunoblotting (IB) was performed with anti-GFP and anti–α-tubulin antibodies for detection of GFP-centrobin and endogenous α-tubulin, respectively. Molecular mass is indicated in kilodaltons.
Centrobin binds to tubulin in vitro, and its C terminus (aa 765–903) contains the tubulin-binding domain as well as centrosome localization ability
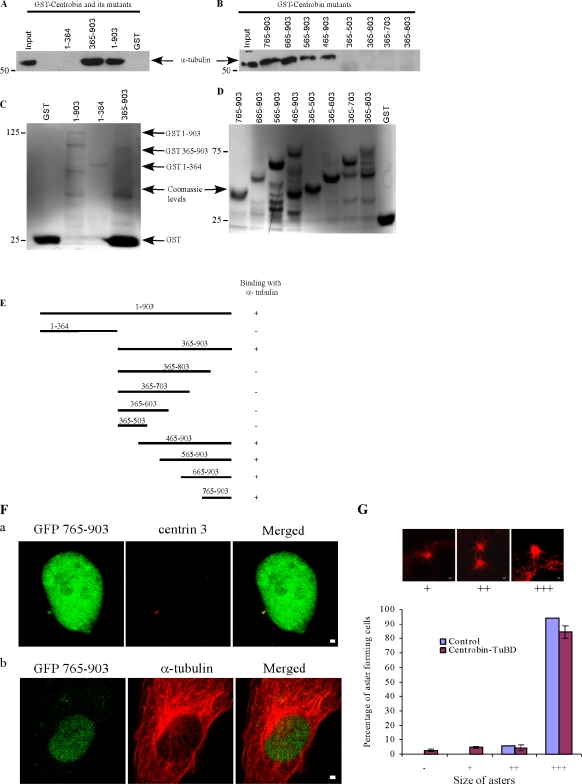
To identify the tubulin binding region of centrobin, an in vitro tubulin dimer binding assay was performed. A low concentration (1 µg/ml) of α/β-tubulin purified from HeLa cells was incubated with GST, GST-centrobin, or its mutants at 4°C. Under this condition, tubulin exists as dimers and does not polymerize into microtubules. As shown in Fig. 5 A, full-length centrobin bound to HeLa tubulins. In comparison, centrobin mutant 365–903 but not 1–364 bound to tubulins, suggesting that the tubulin-binding domain on centrobin maps to its C-terminal aa 365–903 (Fig. 5 A). A systematic mutagenesis of this region was then performed to precisely map the tubulin-binding domain on centrobin (Fig. 5, B, D, and E). We found that the binding site for tubulin on centrobin localizes to its C-terminal 139 residues (aa 765–903, referred to as centrobin-TuBD in the rest of the manuscript; Fig. 5, B and E).
Figure 5.
Centrosome localization ability and the tubulin-binding domain of centrobin map to its C terminus (aa 765–903). (A–D) Centrobin interacts with tubulin in vitro, and the tubulin-binding domain maps to aa 365–903. GST or GST-centrobin and its mutants were incubated with HeLa cell tubulin at 4°C. The bound proteins were fractionated and immunoblotted with anti–α-tubulin antibody. Input, an aliquot of 10% tubulin used in the binding reactions. The input of purified GST or GST-centrobin and its mutant proteins (C and D) were stained by Coomassie blue. Molecular mass is indicated in kilodaltons. (E) A schematic representation of centrobin and its mutants summarizing their binding with α-tubulin. (F) Centrobin 765–903 localizes to centrosomes but not the microtubules. HeLa cells transfected with GFP-centrobin 765–903 were stained with anti–centrin 3 and anti–α-tubulin antibodies to visualize the centrioles (panel a) and microtubules (panel b), respectively. (G) GFP–centrobin 765–903 or centrobin-TuBD does not inhibit the microtubule nucleation process. HeLa cells transfected with control or centrobin-TuBD vectors were subjected to aster formation assay and stained with anti–α-tubulin. Quantification of different sizes of asters is presented. Histograms are plotted as mean ± SEM (n = 3). Bars, 1 µm.
Jeong et al. (2007) reported that the centrosome localization domain of centrobin is within aa 723–903. Our study indicates that the tubulin binding region of centrobin is restricted to aa 765–903 in its C terminus. In immunofluorescence experiments, we observed a diffuse cytoplasmic and strong nuclear localization for centrobin-TuBD. In addition, centrobin-TuBD also exhibited a clear centrosomal localization, as is evident by its colocalization with centrin staining (Fig. 5 F, panel a), suggesting that the tubulin-binding domain of centrobin is sufficient for its centrosomal localization. The nuclear localization of centrobin-TuBD is likely a result of the presence of an NLS in this fragment of centrobin. No microtubule localization of centrobin-TuBD was observed (Fig. 5 F, panel b). Furthermore, expression of centrobin-TuBD did not grossly affect the centrosome-mediated microtubule nucleation (Fig. 5 G).
Overexpression of centrobin-TuBD can disrupt the interaction of endogenous full-length centrobin with tubulin
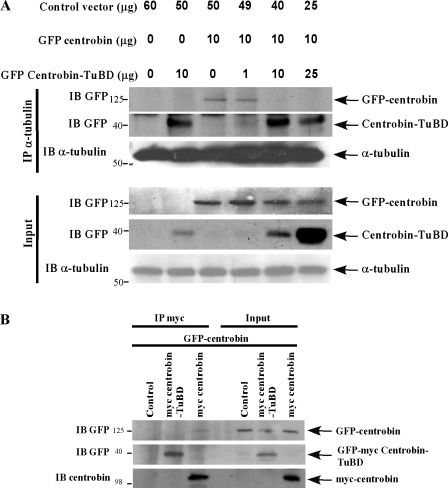
To test whether centrobin-TuBD expression can dominant negatively disrupt the endogenous centrobin–tubulin interaction, we cotransfected 293T cells with GFP-centrobin either alone or together with increasing amounts of centrobin-TuBD. Cells were then subjected to immunoprecipitation using anti–α-tubulin antibody followed by immunoblotting for centrobin. Though both full-length and centrobin-TuBD bound very well to α-tubulin when expressed by themselves (Fig. 6 A, second and third lanes), increased expression of centrobin-TuBD in the presence of a constant amount of full-length centrobin significantly hindered the binding of full-length centrobin to tubulin (Fig. 6 A, fifth and sixth lanes), indicating that the interaction of full-length centrobin with α-tubulin is disrupted in the presence of centrobin-TuBD.
Figure 6.
Centrobin-TuBD disrupts the interaction of full-length centrobin with tubulin. (A) Overexpression of centrobin-TuBD disrupts the interaction of endogenous full-length centrobin with a-tubulin. 293T cells transfected with vector, GFP-centrobin or GFP–centrobin-TuBD, or in combination as indicated were lysed and subjected to immunoprecipitation (IP) with anti–α-tubulin antibody. The bound proteins were fractionated and detected by immunoblotting (IB) using the indicated antibodies. (B) Centrobin-TuBD does not have the potential to dimerize/polymerize with full-length centrobin. 293T cells transfected with vector, myc-centrobin, or GFP-myc–centrobin-TuBD along with GFP-centrobin–expressing constructs were lysed and subjected to immunoprecipitation using anti-myc antibody. Immunoblotting was performed with anti-GFP and anti-myc antibodies. Molecular mass is indicated in kilodaltons.
To rule out the possibility that centrobin-TuBD may form a nonfunctional unit by dimerizing with the full-length centrobin, we coexpressed either myc-centrobin or GFP-myc–centrobin-TuBD along with GFP-centrobin in 293T cells to test the dimerization potential of centrobin and its mutants. Immunoprecipitation using the anti-myc antibody revealed that full-length myc-centrobin bound strongly to full-length GFP-centrobin (Fig. 6 B, first panel, third lane), but centrobin-TuBD did not (Fig. 6 B, first panel, first and second lanes), suggesting that though centrobin has the potential to dimerize/polymerize with itself, its C-terminal 139 residues are not responsible for this ability. Together, these findings prove that centrobin-TuBD is capable of disrupting the endogenous centrobin–tubulin interaction (Fig. 6) and can serve as a valuable tool to dissect the function of this interaction during the centriole duplication process.
Disruption of the interaction of centrobin and centrosomal tubulin by centrobin-TuBD inhibits centrobin recruitment to the centriole biogenesis site and centriole elongation
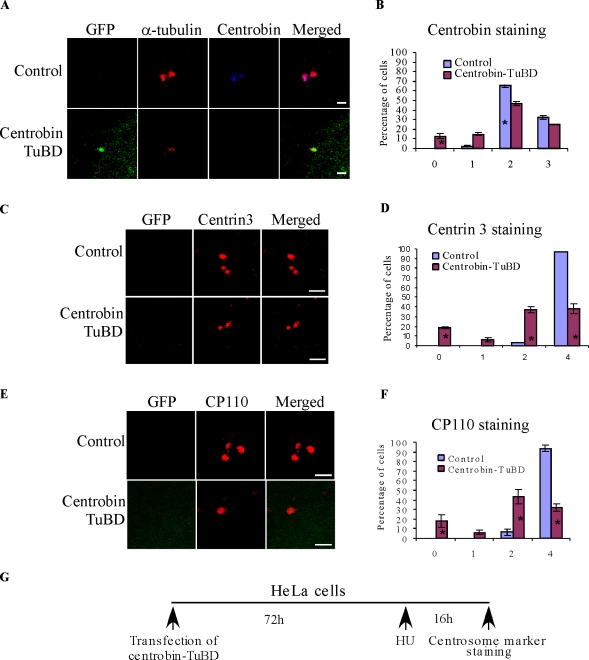
Because centrobin-TuBD can disrupt the centrobin–tubulin interaction, we examined whether centrobin-TuBD affected the recruitment of endogenous centrobin to the centrioles. For this purpose, HeLa cells were transfected with either GFP or GFP–centrobin-TuBD, treated with HU, and then immunostained using anticentrobin antibody (Fig. 7 G). Our anticentrobin antibody recognizes an epitope that is outside of the centrobin-TuBD region, and therefore, it can only stain the endogenous centrobin. As shown in Fig. 7 (A and B), although most of the control cells had either two or three centrobin-positive centrioles, ∼28% of centrobin-TuBD–expressing cells had less than two centrobin-positive centrioles (Fig. 7, A and B), suggesting that centrobin recruitment to the centrioles is affected in centrobin-TuBD–expressing cells. Observation of a significantly larger percentage of centrobin-TuBD–expressing cells with α-tubulin–positive centrioles but no centrobin (Fig. 7 A, second panel) suggests that the interaction of centrobin and centrosomal tubulins is responsible for recruitment of endogenous centrobin to the centriole and that centrobin-TuBD disrupts this interaction.
Figure 7.
Disruption of centrobin–tubulin interaction inhibits centrobin recruitment to the centriole biogenesis site and centriole elongation. (A and B) Centrobin-TuBD overexpression blocks centrobin recruitment to the centrioles.HeLa cells transfected with control or GFP–centrobin-TuBD vectors and treated with HU were stained with anti-centrobin and anti-tubulin antibodies. The percentage of cells with indicated numbers of centrobin-positive centrioles is shown in B. (C–F) Centrobin-TuBD overexpression inhibits centriole duplication and elongation. Experiments were performed as depicted in the schematic (G), except that the cells were stained with anti–centrin 3 (C and D) and CP110 (E and F) antibodies. D and F represent the percentage of cells with the indicated number of centrin 3– and CP110-positive centrioles, respectively. Histograms are plotted as mean ± SEM (n = 3). Asterisks denote that the difference is significant, and P < 0.001. Bars, 1 µm.
Centrin staining has been found to be a good marker for centriole duplication (Middendorp et al., 2000; Salisbury et al., 2002; Strnad et al., 2007; Azimzadeh et al., 2009). Therefore, we stained the aforementioned centrobin-TuBD–expressing cells with anti–centrin 3 antibody. As shown in Fig. 7 (C and D), although 97% of the control cells had four centrioles per cell, only ∼40% of the centrobin-TuBD–transfected cells had four centrin-positive centrioles per cell. Similarly, when the cells were stained for CP110, only 32% of the centrobin-TuBD–transfected cells had four CP110-positive centrioles, whereas 94% of the control cells had four CP110-positive centrioles (Fig. 7, E and F). These findings indicated that centriole elongation was blocked in the centrobin-TuBD–expressing cells (Fig. 8 L, panel a).
Expression of centrobin-TuBD leads to centriole destabilization
Surprisingly, we also noticed that 15% of the centrobin-TuBD–expressing cells (Fig. 7, D and F) exhibited no centrin- or CP110-staining centrioles, which is different from what we observed in centrobin depletion experiments in which cells without centrin-staining centrioles were rarely observed (Zou et al., 2005). We think that three explanations are possible: first, as a result of two rounds of cell division without centriole duplication, centrobin-TuBD likely has a more potent inhibitory effect on centriole duplication but has a less potent effect on cell division than centrobin RNAi because it likely does not interfere with the other potential functions of centrobin. Second, centrobin-TuBD displaced the centrin and CP110 proteins from the existing centriole. Third, centrobin-TuBD can destabilize the existing centrioles.
To distinguish among these possibilities, HeLa cells were first synchronized in S phase by HU treatment for 8 h before transfection with centrobin-TuBD (Fig. 8 I). Anti–centrin 3 staining was performed 72 h after transfection. As shown in Fig. 8 (A and B), 20% of the centrobin-TuBD–transfected HeLa cells still exhibited zero centrioles. This finding was further confirmed in presynchronized U2OS cells (Fig. 8, C and D) in which the centrosome duplication process is uncoupled from the cell cycle but the centriole duplication proceeds unhampered (Stucke et al., 2002). As expected, ∼70% of the control cells had eight or more centrioles per cell. However, in the centrobin-TuBD–transfected U2OS cells, only 20% of the cells had eight or more centrioles per cell, indicating that centriole elongation is blocked in these cells as well. Again, we found that 20% of the centrobin-TuBD–transfected cells had zero centrioles (Fig. 8 D). Because the cells had been arrested in S phase for 8 h before and after transfection, no cell division was possible; therefore, the presence of cells with zero centrioles is not a result of two rounds of cell division without centriole duplication. Additionally, these data also confirm that the centrobin-TuBD–mediated effect was not a result of the effect of centrobin-TuBD on the cell cycle but rather a result of its direct effect on centrioles.
Both centrin and CP110 are proteins that localize to the distal portions of the centriole. To distinguish between displacement of these two proteins from the existing centrioles or the destabilization of the entire centriole, HeLa cells pretreated with HU and transfected with the centrobin-TuBD as described in this section were costained with anti–α-tubulin and CP110 antibodies. Z stack images were obtained to examine the presence of anti–α-tubulin– and anti-CP110–stained centrioles (Fig. 8, E and F). Our analysis revealed that in 24% of these cells, no visibly discernible centrioles were present (Fig. 8 F). It is noticeable that the cells lacking centrioles were not necessarily expressing the highest level of centrobin-TuBD (not depicted). Because α-tubulin forms the core structure of the centriole, this finding indicated that expression of centrobin-TuBD was able to destabilize the existing centrioles in addition to inhibiting the daughter centriole formation (Fig. 8 L, panel a).
To confirm the specificity of the observed effect of centrobin-TuBD, we tested whether coexpression of full-length centrobin can rescue the centrobin-TuBD–mediated centriole destabilization and inhibition of centrobin recruitment and centriole elongation. We found that upon high coexpression of full-length centrobin, the effect of centrobin-TuBD on centrobin recruitment, centriole elongation, and centriole stability was significantly reduced (Fig. 8, G and H). This partial but significant rescue by full-length centrobin suggests that the major effects of centrobin-TuBD on centrioles are specific, possibly through competition with endogenous centrobin for interaction with centrosomal tubulin.
To further confirm that some of the centrobin-TuBD–expressing cells indeed lost their centrioles, electron microscopic analysis of centrioles on thick 250-nm sections of HU-pretreated, GFP, or centrobin-TuBD–transfected HeLa cells was performed (Fig. 8 J). Although 12 morphologically normal centrioles were found in 300 control HeLa cells analyzed on randomly selected sections, only three centrioles were found in the 500 centrobin-TuBD–expressing cells analyzed. This finding indicated that a portion of the centrobin-TuBD–transfected cells indeed has no discernible centrioles, indicating that expression of centrobin-TuBD is able to destabilize the existing centrioles. The centrioles observed in centrobin-TuBD–expressing cells did not exhibit any obvious defects, likely because the destabilization process is a rapid process or the partially destabilized centrioles were not easily recognizable under EM. In summary, these findings indicate that disruption of centrobin–tubulin interaction leads to centriole destabilization, and centrobin–tubulin interaction is required to maintain the stability of the centriole (Fig. 8 L, panel a).
Centrobin-TuBD destabilizes elongated PLSs in CPAP-expressing cells
Next, we tested whether disruption of centrobin–tubulin interaction can destabilize the CPAP-induced PLS. First, the U2OS-CPAP cells were induced for 96 h to form the elongated PLS (Fig. 8 K, schematic), after which the cells were cotransfected with vector or GFP–centrobin-TuBD together with dsRed–centrin 1–expressing vectors to mark the transfected cells. At 72 h after transfection, cells were stained with antiacetylated tubulin antibody to quantify the percentage of cells with PLSs in the dsRed-centrin–positive cells. The elongated PLS in these cells was evaluated and shown in Fig. 3. Although 67% of the control cells had elongated PLSs, upon transfection with centrobin-TuBD, the number of cells with PLSs was reduced to 33% (Fig. 8 K). These findings indicate that disruption of centrobin–tubulin binding by the centrobin-TuBD can destabilize the preformed elongated PLS (Fig. 8 L, panel b), further corroborating the aforementioned finding that the centrobin–tubulin interaction is important for maintaining the stability of centrioles.
Discussion
Previously, we identified centrobin as a centrosomal protein that is preferentially localized to the daughter centriole and is required for centriole duplication (Zou et al., 2005). In this study, we further elucidate the role of centrobin during centriole duplication. We found that centrobin recruitment to the procentrioles is dependent on the presence of hSAS-6 but not CPAP and CP110. The recruitment of CP110 but not hSAS-6 to the centriole biogenesis site requires the presence of centrobin, suggesting that centrobin is one of the important centriole initiation proteins. Furthermore, we found that centrobin is also required for centriole elongation and stability.
Centrioles are predominantly composed of α/β-tubulins. Centriole elongation, at least visually, is the process of assembling the nine microtubule triplets by adding α/β-tubulin dimers to an undefined initiating template structure. Here, we demonstrate that centrobin interacts directly with the core components of the centriole, α-tubulins via its C-terminal 139 residues (centrobin-TuBD). Centrobin-TuBD exhibited a clear centrosomal localization in addition to a diffused cytoplasmic and nuclear localization. It is noticeable that although centrobin-TuBD can bind strongly to tubulins, no detectable microtubular localization of centrobin-TuBD or adverse effect on microtubule nucleation was observed. Although surprising, this finding correlates with our previous observation that endogenous centrobin is not clearly detectable on microtubules (Zou et al., 2005). Jeong et al. (2007) had reported that centrobin is detectable in association with the roots or initiating points of microtubules in U2OS and MCF7 cells but not in HeLa cells. It is very likely that the tubulins at the initiating points of microtubules and tubulins at the centrosomes share similar conformation with the centrosomal tubulins, in which their centrobin-binding domain is exposed. When the tubulins are assembled into microtubules, the centrobin-binding domain is no longer accessible.
Importantly, overexpression of centrobin-TuBD can disrupt the endogenous full-length centrobin–tubulin interaction, providing us with a valuable tool to dissect the function of centrobin during centrobin duplication. Using centrobin-TuBD, we demonstrated that centrobin is recruited to centrioles and facilitates centriole elongation via its interaction with centrosomal tubulins. Moreover, we found that centrobin-TuBD overexpression destabilized the existing mother centrioles in addition to inhibiting the assembly of new centrioles (Fig. 8 L, panel a), indicating that centrobin is required for the stability of centrioles. Our previous findings (Zou et al., 2005) indicated that, in asynchronized cells, centrobin is mainly found on the daughter centrioles. Even in HU-treated cells, in which the degradation or displacement of centrobin from the daughter centriole is inhibited, there is at least one or two mother centrioles exhibiting no centrobin staining, which will suggest that centrobin should not be required for the stability of these mature centrioles. We propose two possible scenarios to explain these conflicting findings. First, centrobin is indeed required for the stability of both daughter and mature mother centrioles. On the mature mother centrioles, centrobin is still there to maintain their stability but becomes undetectable because additional mother centriole proteins block the access of centrobin antibody. Second, centrobin is only required to maintain the stability of the daughter centrioles. Once the daughter centrioles mature to become mother centrioles by recruiting additional mother centriole proteins and extensive modification of centriolar tubulins, centrobin is no longer required for their stability. Centrobin is then either degraded or displaced from the mother centrioles by the mother centriole proteins or the tubulin modifications. However, because of the small size of centrobin-TuBD and its presence at high concentration, it can still access the centrobin-binding domain on the tubulins and competes with the mother centriole proteins. Consequently, centrobin-TuBD will displace the mother centriole proteins and lead to destabilization of the mother centrioles. Although our current data cannot distinguish between these two scenarios, we speculate that the first scenario is more plausible because centrobin can indeed be present on the mother centriole as is evident in HU-treated cells. One seemingly conflicting piece of evidence against this hypothesis is that centrobin depletion inhibited the centriole duplication but did not destabilize the mother centrioles. The likely explanation is that centrobin assembled into the mother centrioles is stabilized and is impervious to depletion, as are most cellular structural proteins. Hence, centrobin depletion cannot destabilize the mother centriole, whereas centrobin-TuBD can displace the centrobin on the mother centrioles and lead to their destabilization. However, we do not have direct evidence for this, and further studies are required to prove this scenario. Furthermore, in our rescue experiments, the existing centrioles were still destabilized in 5% of cells and were not rescued by the wild-type centrobin expression; therefore, it is possible that both scenarios can coexist and account for the observed destabilization of existing centrioles.
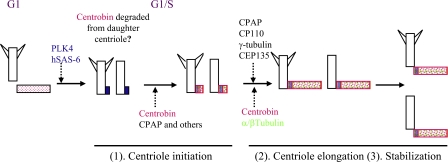
The key findings from this study suggest that centrobin functions at least at three stages of the centriole duplication pathway as depicted in Fig. 9. At the beginning of centriole biogenesis, hSAS-6 is first recruited to the proximal end of the mother centrioles in the G1/S phase. Centrobin is then recruited and likely participates in the undefined centriole initiation structure formation along with the other proposed centriole initiation proteins, including CPAP, CEP135, γ-tubulin, and CP110. During the elongation of procentrioles, centrobin may function as a scaffold protein via its interaction with tubulins to facilitate the addition of tubulin dimers to the centriole initiation complex. Centrobin also acts to stabilize the newly assembled daughter centrioles before their maturation. Whether centrobin is required to maintain the stability of the mature mother centriole remains to be determined. The function of centrobin during centriole elongation and its function to maintain the stability of the centriole are likely both mediated by its ability to bind to tubulins.
Figure 9.
Model of centrobin function during the centriole duplication process. The model summarizes our finding on the function of centrobin during centriole duplication. As the cell transitions from G1 to S phase, PLK4 activation leads to the recruitment of hSAS-6. Centrobin and other centriole initiation proteins are then recruited followed by centrobin, CPAP, CP110, γ-tubulin, and CEP135-mediated elongation of the centriole. Centrobin is also required to maintain the stability of the newly assembled centrioles before their maturation. Centrobin is likely degraded or displaced from the original daughter centriole once the centriole duplication starts.
CPAP also has the ability to bind to microtubules (Hung et al., 2004), and regulation of its cellular levels is required to maintain the centriole length (Tang et al., 2009). It has been suggested that as a result of its tubulin binding property, CPAP might act as a scaffold for tubulin addition during procentriole biogenesis. Because the property of tubulin binding is shared by CPAP and centrobin and both are required for centriole biogenesis (Cho et al., 2006; Kohlmaier et al., 2009), it would be interesting to study whether centrobin and CPAP cooperate to facilitate the assembly of the microtubule triplets that form the main structure of centrioles. So far, there is no convincing evidence indicating the existence of a lower eukaryotic centrobin homologue. If centrobin indeed cooperates with CPAP for centriole elongation, it will indicate that centrobin is functionally similar to C. elegans SAS-5. In summary, we conclude that centrobin–tubulin interaction is pivotal for centrobin recruitment to the centriole biogenesis site, centriole elongation, and stabilization of nascent centrioles until maturation.
Materials and methods
Cell lines and transfections
293T cells were cultured in DME supplemented with 10% FBS and transfected by the calcium phosphate method. HeLa and U2OS cells were grown in MEM α medium containing 10% FBS and were transfected by calcium phosphate precipitation. U2OS cells inducibly expressing CPAP (a gift from P. Gönczy, Swiss Institute for Experimental Cancer Research, Swiss Federal Institute of Technology, Lausanne, Switzerland) were grown in MEM α medium containing 10% FBS and were switched to the same medium containing 1 µg/ml doxycycline to induce the expression of CPAP. siRNAs were transfected using the Oligofectamine reagent (Invitrogen).
Antibodies and reagents
Anticentrobin polyclonal antibody has been described previously (Zou et al., 2005). Monoclonal anticentrobin antibody (clone 2E3) has been generated using the monoclonal antibody core facility at Northwestern University, purified, and characterized in our laboratory. Using centrobin knockdown cells, we conclusively demonstrated that this monoclonal anticentrobin antibody can recognize centrobin specifically in Western blotting and immunofluorescence (Fig. 2 C). Anti–α-tubulin antibodies (clone DM1A for Western blotting and immunofluorescence and clone B-5-1-2 for immunoprecipitation) and antiacetylated antibody (clone 6–11-B-1) were obtained from Sigma-Aldrich. Polyclonal anti-GFP antibody was obtained from Santa Cruz Biotechnology, Inc. Polyclonal anti–centrin 3 antibody was generated in the laboratory against full-length GST-tagged centrin 3 and affinity purified. Polyclonal antibodies for CPAP, CP110, and hSAS-6 were gifts from T.K. Tang (Institute of Biomedical Sciences, Taipei, Taiwan), B. Dynlacht (Department of Cell Biology, New York University, New York, NY), and P. Gönczy. T.K. Tang also provided us with polyclonal hSAS-6 antibody. Monoclonal antibody against hSAS-6 was also purchased from Santa Cruz Biotechnology, Inc. Polyclonal antibody against CP110 was purchased from Bethyl Laboratories, Inc. Monoclonal anti–C-NAP1 antibody was purchased from BD. Purified HeLa cell tubulin for in vitro binding studies was obtained from Cytoskeleton. RNAi-mediated depletion of proteins was performed using siRNA duplexes purchased from Thermo Fisher Scientific. The sequences for the siRNA against hSAS-6 were obtained from Leidel et al. (2005), and those for CPAP and CP110 were derived from Kleylein-Sohn et al. (2007), whereas the sequence for centrobin siRNA was obtained from Zou et al. (2005). Alexa Fluor 488/568/647–linked secondary antibodies as well as α-tubulin–Alexa Fluor 488 conjugate were purchased from Invitrogen.
Plasmid constructs
Cloning of centrobin (1–903) and centrobin-C (365–903) into GFP-N1 and pGEX vectors has been previously described (Zou et al., 2005). pET28a centrobin (1–903) was generated by PCR using restriction enzymes EcoRI and NotI. Mutants of centrobin (1–364, 365–803, 365–703, 365–603, 365–503, 465–903, 565–903, 665–903, and 765–903) were generated by PCR using full-length centrobin as the template and were cloned into the pGEX vector using the restriction enzymes XhoI and SacII. The construct for expressing the GST–centrin 3 protein that was used to generate the polyclonal centrin 3 antibody was cloned by PCR using the enzyme sites BamHI and EcoRI.
Immunofluorescence
For visualization of centrioles, cells grown on coverslips were left on ice for half an hour followed by treatment with an extraction buffer (20 mM Hepes, pH 7.4, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 0.5% Triton X-100) and fixed with ice-cold methanol at −20°C. The cells were then incubated with appropriate primary antibodies diluted in PBS containing 0.02% Tween 20 and then incubated with Alexa Fluor 488/568/647–linked secondary antibodies followed by DAPI staining for 10 min. Coverslips were mounted onto slides in mounting media containing antifade reagent (Invitrogen) and imaged at room temperature. For the centriole elongation assay, U2OS-CPAP cells were induced by doxycycline for different time points in a 24-well plate. To visualize the centrioles, cells were placed on ice for half an hour to depolymerize the microtubules. The same staining procedure was followed as mentioned previously in this section. α-Tubulin conjugated to Alexa Fluor 488 was used in the case of triple color staining. In the case of cells transfected with the centrobin-TuBD, cells were fixed with 4% PFA to avoid extraction of the mutated protein and then extracted with PBS buffer containing 0.5% Triton X-100 followed by blocking and staining with appropriate antibodies. In the aster-forming assay, cells were transfected with centrobin-TuBD for 72 h followed by nocodazole treatment for 2 h at 37°C. Subsequently, cells were washed extensively with PBS, and warm media were added to the cells for exactly 2 min, after which cells were fixed immediately with PFA and stained with α-tubulin. To quantify the PLS in U2OS-CPAP cells, dsRed–centrin 1 was cotransfected and used as a marker to identify the transfected cells. All images were acquired using a scanning confocal system (C1; Nikon) on a microscope (Eclipse 80i; Nikon) equipped with an APO PLAN 100×, 1.4 NA oil immersion objective (Nikon) except for images in Fig. 7 (C and E) and Fig. 8 (A and C), which were acquired using a confocal system (LSM 510 META; Carl Zeiss) equipped with an α Plan-FLUAR 100×, 1.45 NA oil immersion objective (Carl Zeiss). Images were acquired using EZ-C1 software (Nikon) and AIM 4.2 software (Carl Zeiss). Representative maximum projection images of Z stacks that were volume rendered using EZ-C1 freeviewer version 3.9 software are shown for Fig. 8 E.
Immunoprecipitation
For binding assay of overexpressed centrobin, cells were transfected with the indicated plasmids. At 48 h after transfection, the cells were washed in ice-cold PBS and then lysed using radioimmunoprecipitation assay lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Na-deoxycholate, 1% Triton X-100, and 100 mM PMSF) for 40 min on ice. The DNA was sheared using a 21-gauge needle, after which lysates were spun at 14 K for 20 min. Lysates were precleared twice with 50% slurry of protein G beads followed by incubation with primary antibodies overnight at 4°C and then with protein G beads for 40 min at 4°C to precipitate the immunocomplexes. After washing six times using lysis buffer, the bound proteins were fractionated on an SDS-PAGE gel, and Western blotting was performed to detect the bound proteins. To test the dimerization potential of centrobin, transfected 293T cells were lysed using radioimmunoprecipitation assay buffer containing 0.1% SDS. The rest of the protocol remained the same as described in this section, except that anti-myc antibody was used for the pull-down. For immunoprecipitations, 5 µg anti–α-tubulin and 5 µg anti-myc antibodies were used.
In vitro binding
GST-proteins were prepared by standard protocol using the BL21 strain of Escherichia coli and IPTG (Sigma-Aldrich) as the inducing agent. For the purification of GST-centrobin (1–903) and GST–α-tubulin, 2 M urea was added to the lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 2 mM MgCl2, 1% Triton X-100, 1% NP-40, and 100 mM PMSF). All the centrobin mutants were soluble and could be purified easily in the same buffer without urea. After induction, bacteria were pelleted and frozen overnight at −80°C, after which they were sonicated in lysis buffer. Glutathione beads (Pharmacia) were added to lysates to purify the proteins. Purified proteins were first quantified by Coomassie blue staining, after which equal amounts were used for in vitro binding with purified tubulins from HeLa cells (Cytoskeleton). In general, binding was performed for 2 h at 4°C to avoid polymerization of the tubulin dimers.
Centriole duplication assay
HeLa and U2OS cells were either treated with 16 mM HU for 16 h (72 h after transfection) or pretreated with HU for 8 h, after which they were transfected with the indicated plasmids for 72 h. The transfected cells were immunostained with the indicated antibodies to visualize the centrioles. Data presented are a mean of the number of centrioles per cell from three independent experiments with 300 cells counted in every experiment or as otherwise mentioned. SEM has been calculated for the graphs.
CPAP-induced PLS assay
U2OS-CPAP inducible cells were transfected with control or centrobin siRNA for 72 h, after which 1 µg/ml doxycycline was used to induce the elongation of centrioles. Cells were harvested at different time points and processed for immunofluorescence. To assay the effect of centrobin-TuBD overexpression on the stability of PLSs, cells were first induced with doxycycline for 96 h and then cotransfected with control or GFP–centrobin-TuBD together with dsRed–centrin 1–expressing vectors for 72 h. After staining with acetylated tubulin, 300 dsRed–centrin 1–positive cells in both control and mutant cells were analyzed for the presence of PLSs. The percentage of cells with PLSs was enumerated from three independent experiments and presented.
EM
HeLa cells were pretreated with HU for 8 h and then transfected with control or centrobin-TuBD expression vectors for 72 h. The cells on coverslips were then fixed with 2% glutaraldehyde solution for 30 min followed by staining with osmium tetroxide and uranyl acetate and dehydration, after which the nuclei were then embedded in Epon 812 resin. 250-nm-thick sections were prepared on copper grids and evaluated using a transmission electron microscope (1230; JEOL).
Statistical analysis
SEM has been calculated and plotted for all the graphs. Asterisks on all histograms indicate that the differences are significant. The p-value was calculated between the control and test groups using the Student’s paired t test and was found to be P < 0.001.
Depletion efficiency of CPAP and CP110 RNAi
To assess the depletion efficiency of CPAP and CP110 RNAi, HeLa cells were grown on coverslips and transfected with control or CPAP or CP110 RNAi for 48 h followed by treatment with HU for 16 h. For visualization of centrioles, cells grown on coverslips were left on ice for half an hour followed by treatment with an extraction buffer (20 mM Hepes, pH 7.4, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 0.5% Triton X-100) and were fixed with ice-cold methanol at −20°C. The cells were then incubated with appropriate primary antibodies diluted in PBS containing 0.02% Tween 20 and then incubated with Alexa Fluor 488/568/647–linked secondary antibodies followed by DAPI staining for 10 min. Coverslips were mounted onto slides in mounting media containing antifade reagent.
In vivo interaction of endogenous centrobin and tubulin
For endogenous binding studies, all the procedures were the same as described in the Immunoprecipitation section of Materials and methods, except that the cells were harvested when they were 80% confluent. For detection of the immunoprecipitated tubulins, mouse True blot HRP-conjugated anti–mouse IgG (eBioscience) was used as the secondary antibody to avoid detection of the heavy chain. To analyze the efficiency of pull-down, 5% of the input was loaded on the same gel and probed with the indicated antibodies.
Online supplemental material
The online supplemental material describes the detailed experimental procedure for evaluating the depletion efficiency of CPAP and CP110 RNAi (Fig. S1) as well as for studying the in vivo interaction of centrobin and tubulin (Fig. S2). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006135/DC1.
Acknowledgments
We would like to thank Dr. Yujie Bai, Dr. Lijun Cheng, Dr. Haixia Gong, and Dr. Deepak Rai for their technical contribution to this paper and Dr. Jayeeta Dhar for her efforts toward the purification of the monoclonal centrobin antibody (clone 2E3). We also thank Dr. Izolda Popova for her technical help in generating the anticentrobin monoclonal antibody and Dr. Vimla Band, Dr. Hamid Band, and Dr. Goberdhan Dimri for helpful suggestions and discussion. We would also like to thank Dr. Tang K. Tang, Dr. Brian Dynlacht, and Dr. Pierre Gönczy for kindly and promptly providing us with many important reagents used here and without which this study would have been impossible.
This work was supported by grants from the National Institutes of Health (1 R01 CA095221-01A1 and 1 R01 CA96986-01A1), Department of Defense Breast Cancer Research (DAMD17-00-1-0342), and American Cancer Society (RSG-03-048-01) to Q. Gao.
Footnotes
Abbreviations used in this paper:
- CPAP
- centrosomal P4.1–associated protein
- HU
- hydroxyurea
- PLS
- procentriole-like structure
References
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 426:570–574 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- Anderson C.T., Stearns T. 2009. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr. Biol. 19:1498–1502 10.1016/j.cub.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Bornens M. 2007. Structure and duplication of the centrosome. J. Cell Sci. 120:2139–2142 10.1242/jcs.005231 [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Hergert P., Delouvée A., Euteneuer U., Formstecher E., Khodjakov A., Bornens M. 2009. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 185:101–114 10.1083/jcb.200808082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., Glover D.M. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Bornens M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14:25–34 10.1016/S0955-0674(01)00290-3 [DOI] [PubMed] [Google Scholar]
- Bornens M., Azimzadeh J. 2007. Origin and evolution of the centrosome. Adv. Exp. Med. Biol. 607:119–129 10.1007/978-0-387-74021-8_10 [DOI] [PubMed] [Google Scholar]
- Cho J.H., Chang C.J., Chen C.Y., Tang T.K. 2006. Depletion of CPAP by RNAi disrupts centrosome integrity and induces multipolar spindles. Biochem. Biophys. Res. Commun. 339:742–747 10.1016/j.bbrc.2005.11.074 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Maddox P.S., Desai A., Oegema K. 2008. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin-mediated addition of centriolar microtubules. J. Cell Biol. 180:771–785 10.1083/jcb.200709102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M., Canard C., Gönczy P. 2006. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16:1844–1849 10.1016/j.cub.2006.07.059 [DOI] [PubMed] [Google Scholar]
- Doxsey S. 2001. Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol. 2:688–698 10.1038/35089575 [DOI] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., Mikule K. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15:303–311 10.1016/j.tcb.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Fry A.M., Mayor T., Meraldi P., Stierhof Y.D., Tanaka K., Nigg E.A. 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141:1563–1574 10.1083/jcb.141.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Jurczyk A., Sillibourne J., Halilovic E., Mogensen M., Groisman I., Blomberg M., Doxsey S. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161:535–545 10.1083/jcb.200301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.D., Wilkinson C.J., Nigg E.A. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Sluder G. 2001. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 15:1167–1181 10.1101/gad.894001 [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Li C., Thompson E.A., Maller J.L., Sluder G. 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 283:851–854 10.1126/science.283.5403.851 [DOI] [PubMed] [Google Scholar]
- Hung L.Y., Tang C.J., Tang T.K. 2000. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the γ-tubulin complex. Mol. Cell. Biol. 20:7813–7825 10.1128/MCB.20.20.7813-7825.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L.Y., Chen H.L., Chang C.W., Li B.R., Tang T.K. 2004. Identification of a novel microtubule-destabilizing motif in CPAP that binds to tubulin heterodimers and inhibits microtubule assembly. Mol. Biol. Cell. 15:2697–2706 10.1091/mbc.E04-02-0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., Tsukita S. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517–524 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- Jeffery J.M., Urquhart A.J., Subramaniam V.N., Parton R.G., Khanna K.K. 2010. Centrobin regulates the assembly of functional mitotic spindles. Oncogene. 29:2649–2658 10.1038/onc.2010.37 [DOI] [PubMed] [Google Scholar]
- Jeong Y., Lee J., Kim K., Yoo J.C., Rhee K. 2007. Characterization of NIP2/centrobin, a novel substrate of Nek2, and its potential role in microtubule stabilization. J. Cell Sci. 120:2106–2116 10.1242/jcs.03458 [DOI] [PubMed] [Google Scholar]
- Kemp C.A., Kopish K.R., Zipperlen P., Ahringer J., O’Connell K.F. 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell. 6:511–523 10.1016/S1534-5807(04)00066-8 [DOI] [PubMed] [Google Scholar]
- Kirkham M., Müller-Reichert T., Oegema K., Grill S., Hyman A.A. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 112:575–587 10.1016/S0092-8674(03)00117-X [DOI] [PubMed] [Google Scholar]
- Kitagawa D., Busso C., Flückiger I., Gönczy P. 2009. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev. Cell. 17:900–907 10.1016/j.devcel.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., Nigg E.A. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Kohlmaier G., Loncarek J., Meng X., McEwen B.F., Mogensen M.M., Spektor A., Dynlacht B.D., Khodjakov A., Gönczy P. 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19:1012–1018 10.1016/j.cub.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R., Borisy G.G. 1981. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 91:814–821 10.1083/jcb.91.3.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey K.R., Jackson P.K., Stearns T. 1999. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA. 96:2817–2822 10.1073/pnas.96.6.2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B.M., Gull K. 1995. A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 130:919–927 10.1083/jcb.130.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Gönczy P. 2003. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell. 4:431–439 10.1016/S1534-5807(03)00062-5 [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- Loncarek J., Khodjakov A. 2009. Ab ovo or de novo? Mechanisms of centriole duplication. Mol. Cells. 27:135–142 10.1007/s10059-009-0017-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Stearns T. 2007. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8:161–167 10.1038/nrm2100 [DOI] [PubMed] [Google Scholar]
- Meraldi P., Lukas J., Fry A.M., Bartek J., Nigg E.A. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1:88–93 10.1038/10054 [DOI] [PubMed] [Google Scholar]
- Middendorp S., Küntziger T., Abraham Y., Holmes S., Bordes N., Paintrand M., Paoletti A., Bornens M. 2000. A role for centrin 3 in centrosome reproduction. J. Cell Biol. 148:405–416 10.1083/jcb.148.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. 2007. Centrosome duplication: of rules and licenses. Trends Cell Biol. 17:215–221 10.1016/j.tcb.2007.03.003 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., Raff J.W. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- O’Connell K.F., Caron C., Kopish K.R., Hurd D.D., Kemphues K.J., Li Y., White J.G. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 105:547–558 10.1016/S0092-8674(01)00338-5 [DOI] [PubMed] [Google Scholar]
- Ou Y.Y., Mack G.J., Zhang M., Rattner J.B. 2002. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 115:1825–1835 [DOI] [PubMed] [Google Scholar]
- Pelletier L., Ozlü N., Hannak E., Cowan C., Habermann B., Ruer M., Müller-Reichert T., Hyman A.A. 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14:863–873 10.1016/j.cub.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Pelletier L., O’Toole E., Schwager A., Hyman A.A., Müller-Reichert T. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Pihan G.A., Purohit A., Wallace J., Knecht H., Woda B., Quesenberry P., Doxsey S.J. 1998. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 58:3974–3985 [PubMed] [Google Scholar]
- Pihan G.A., Wallace J., Zhou Y., Doxsey S.J. 2003. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 63:1398–1404 [PubMed] [Google Scholar]
- Robbins E., Gonatas N.K. 1964. The ultrastructure of a mammalian cell during the mitotic cycle. J. Cell Biol. 21:429–463 10.1083/jcb.21.3.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E., Jentzsch G., Micali A. 1968. The centriole cycle in synchronized HeLa cells. J. Cell Biol. 36:329–339 10.1083/jcb.36.2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., Glover D.M. 2007. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17:1465–1472 10.1016/j.cub.2007.07.034 [DOI] [PubMed] [Google Scholar]
- Salisbury J.L., Suino K.M., Busby R., Springett M. 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12:1287–1292 10.1016/S0960-9822(02)01019-9 [DOI] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.D., Nigg E.A. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Strnad P., Gönczy P. 2008. Mechanisms of procentriole formation. Trends Cell Biol. 18:389–396 10.1016/j.tcb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gönczy P. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke V.M., Silljé H.H., Arnaud L., Nigg E.A. 2002. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 21:1723–1732 10.1093/emboj/21.7.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.J., Fu R.H., Wu K.S., Hsu W.B., Tang T.K. 2009. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11:825–831 10.1038/ncb1889 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Stearns T. 2006a. Controlling centrosome number: licenses and blocks. Curr. Opin. Cell Biol. 18:74–78 10.1016/j.ceb.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Stearns T. 2006b. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 442:947–951 10.1038/nature04985 [DOI] [PubMed] [Google Scholar]
- Vorobjev I.A., Chentsov Yu.S. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93:938–949 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Li J., Bai Y., Gunning W.T., Wazer D.E., Band V., Gao Q. 2005. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J. Cell Biol. 171:437–445 10.1083/jcb.200506185 [DOI] [PMC free article] [PubMed] [Google Scholar]