The cilia-associated protein NPHP4 is a negative regulator of Hippo signaling that modulates cell proliferation in mammals.
Abstract
The conserved Hippo signaling pathway regulates organ size in Drosophila melanogaster and mammals and has an essential role in tumor suppression and the control of cell proliferation. Recent studies identified activators of Hippo signaling, but antagonists of the pathway have remained largely elusive. In this paper, we show that NPHP4, a known cilia-associated protein that is mutated in the severe degenerative renal disease nephronophthisis, acts as a potent negative regulator of mammalian Hippo signaling. NPHP4 directly interacted with the kinase Lats1 and inhibited Lats1-mediated phosphorylation of the Yes-associated protein (YAP) and TAZ (transcriptional coactivator with PDZ-binding domain), leading to derepression of these protooncogenic transcriptional regulators. Moreover, NPHP4 induced release from 14-3-3 binding and nuclear translocation of YAP and TAZ, promoting TEA domain (TEAD)/TAZ/YAP-dependent transcriptional activity. Consistent with these data, knockdown of NPHP4 negatively affected cellular proliferation and TEAD/TAZ activity, essentially phenocopying loss of TAZ function. These data identify NPHP4 as a negative regulator of the Hippo pathway and suggest that NPHP4 regulates cell proliferation through its effects on Hippo signaling.
Introduction
The Hippo signaling pathway was originally discovered in Drosophila melanogaster and has recently emerged as a potent regulator of cell proliferation and organ size (Badouel et al., 2009; Zhang et al., 2009b). Several components of the pathway act as tumor suppressors or as protooncogenes (Harvey and Tapon, 2007). Core components of the Hippo pathway include the upstream activator Merlin/Nf2 (Hamaratoglu et al., 2006), a gene that is mutated in tumors of nervous tissue (Trofatter et al., 1993; Ruttledge et al., 1994) and in renal cell carcinoma (Forbes et al., 2008; Morris and McClatchey, 2009; Dalgliesh et al., 2010), the Ser/Thr kinases MST1/2 (mammalian STE20 kinases 1 and 2) and Lats1/2 (large tumor suppressor 1 and 2), together with their coactivators WW45 and Mob. In the active state, Lats1/2 phosphorylates the transcriptional activators Yes-associated protein (YAP) and TAZ (transcriptional coactivator with PDZ-binding domain). This results in their cytoplasmic retention by binding to 14-3-3 (Kango-Singh and Singh, 2009), preventing TAZ- and YAP-dependent transcription, which is mediated predominantly by transcription factors of the TEA domain (TEAD) family (Wang et al., 2009). Although the upstream components of the Hippo signaling cascade Nf2, Fat4, MST1/2, Lats1/2, WW45, and Mob1 all function as tumor suppressors (Fernandez-L and Kenney, 2010), YAP and TAZ are highly expressed in several cancers and are considered to be oncogenes because overexpression of both TAZ and YAP results in enhanced proliferation and transformation of epithelial cells (Overholtzer et al., 2006; Zender et al., 2006; Chan et al., 2008; Lei et al., 2008). TAZ and YAP seem to have both overlapping and distinct functions in development. YAP knockout mice display embryonic lethality (Morin-Kensicki et al., 2006), whereas TAZ-null mice are viable but develop severe degenerative cystic kidney disease reminiscent of a severe human disorder called nephronophthisis (NPH; Hossain et al., 2007; Makita et al., 2008).
NPH, a genetically heterogeneous, autosomal recessive cystic kidney disease, is the most common genetic cause of end-stage renal disease in the first decades of life. Patients with NPH develop small-sized kidneys with multiple cysts at the renal corticomedullary boarder (Hildebrandt et al., 2009). Recessive mutations in 11 disease-causing genes (NPHP1–11) have been identified that encode for the so-called nephrocystins or NPH proteins (NPHPs). Just like the vast majority of proteins involved in cystic kidney disease, most of the NPHPs localize to the primary cilium (Fliegauf et al., 2007), a sensory organelle that is present on almost all cells of the human body and required to transmit extracellular signals to the interior of the cell (Berbari et al., 2009; Gerdes et al., 2009). Intriguingly, whereas loss of NPHPs results in kidney degeneration, several of these ciliary proteins, including members of the functional NPHP complex, have been found to be up-regulated in various tumors, such as breast cancer (Bowers and Boylan, 2004), pancreas carcinoma (Carter et al., 2010), and colorectal cancer (Sjöblom et al., 2006; Wood et al., 2007).
Results and discussion
NPHP4 is a novel antagonist of Hippo signaling that interacts with Lats1
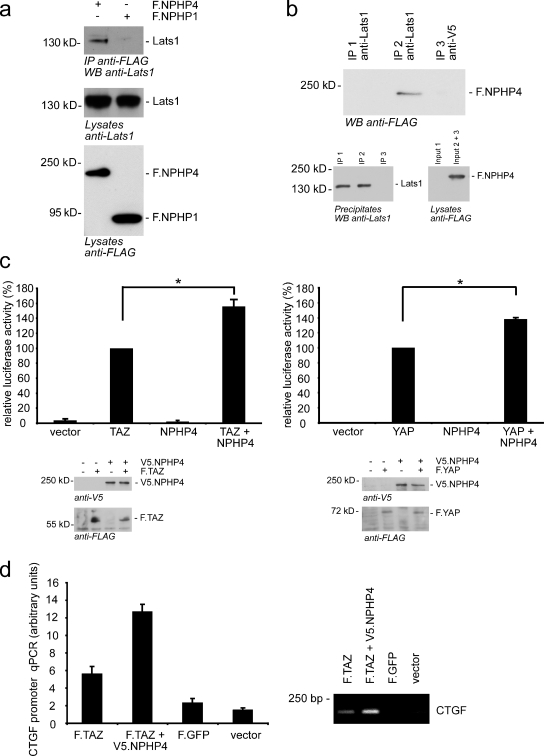
Because small kidney size is one of the most striking clinical features of NPH patients, we asked whether NPHPs might regulate Hippo signaling. To directly test whether NPHP1 or NPHP4 interacts with Lats1 and MST1, the two core components of the Hippo pathway, we immunoprecipitated FLAG-tagged NPHP1 and NPHP4 from HEK293T cells and analyzed the precipitates for coprecipitating Lats1 and MST1. Interestingly, endogenous Lats1 coprecipitated with NPHP4 but not with NPHP1 (Fig. 1 a), and this coprecipitation could be confirmed vice versa (Fig. 1 b). MST1 neither coprecipitated with NPHP4 nor with NPHP1 (Fig. S1 a). Next, we examined the functional consequences of the Lats1–NPHP4 interaction on Hippo signaling using an established GAL4-TEAD luciferase reporter system, the repression of which reflects activity of the Hippo pathway (Lei et al., 2008; Tian et al., 2010). As expected, exogenous TAZ or YAP dramatically increased GAL4-TEAD activity (Lei et al., 2008). Coexpression of NPHP4 with TAZ or YAP augmented TEAD4 transcriptional activity to 156 or 138%, respectively, in comparison with TAZ/YAP expression alone (Fig. 1 c). This effect of NPHP4 was dose dependent (Fig. S1 b). Interestingly, expression of NPHP4 without TAZ or YAP was not sufficient to induce TEAD activation, indicating that NPHP4 exerts its effect only if a certain threshold activation of TAZ/YAP is exceeded, suggesting that NPHP4 might act as an antagonistic enhancer of dysregulated Hippo signaling. Not only TAZ/YAP-dependent activation of the reporter plasmid but also binding of TAZ to the promoter region of connective tissue growth factor (CTGF), a previously described TAZ/TEAD target gene, was significantly increased as shown by chromatin immunoprecipitation emphasizing the biological significance of our results (Fig. 1 d).
Figure 1.
NPHP4 interacts with Lats1 and activates TAZ/YAP/TEAD transcription. (a) HEK293T cells were transiently transfected with the FLAG-tagged NPHPs NPHP1 (F.NPHP1) or NPHP4. After immunoprecipitation (IP) with FLAG (M2) beads, Western blot analysis revealed that endogenous Lats1 coprecipitated with NPHP4 but not with NPHP1. (b) Lysates from HEK293T cells transfected with either control vector (Input 1) or FLAG-tagged NPHP4 (Input 2 + 3) were subjected to immunoprecipitation using an anti-Lats1 (IP 1 and 2) or a control (anti-V5; IP 3) antibody. Western blot (WB) analysis showed that NPHP4 coprecipitated specifically with endogenous Lats1. (c) NPHP4 enhances the activity of the transcriptional coactivators TAZ and YAP. Plasmids encoding the indicated proteins or empty pcDNA6 vector were cotransfected in HEK293T cells together with the TEAD reporter plasmids as indicated in Materials and methods. Coexpression of NPHP4 increased the TAZ/YAP-dependent signaling to 156 and 138%, respectively (n = 3 for YAP and n = 5 for TAZ; *, P < 0.05). Expression of the indicated proteins was confirmed by Western blot analysis. (d) HEK293T cells were transfected with the indicated constructs. The chromatin immunoprecipitation assay revealed that NPHP4 enhances the binding of TAZ to the promoter region of CTGF, a known target gene of TAZ. Error bars represent SEM.
NPHP4 regulates the phosphorylation, localization, and activity of TAZ
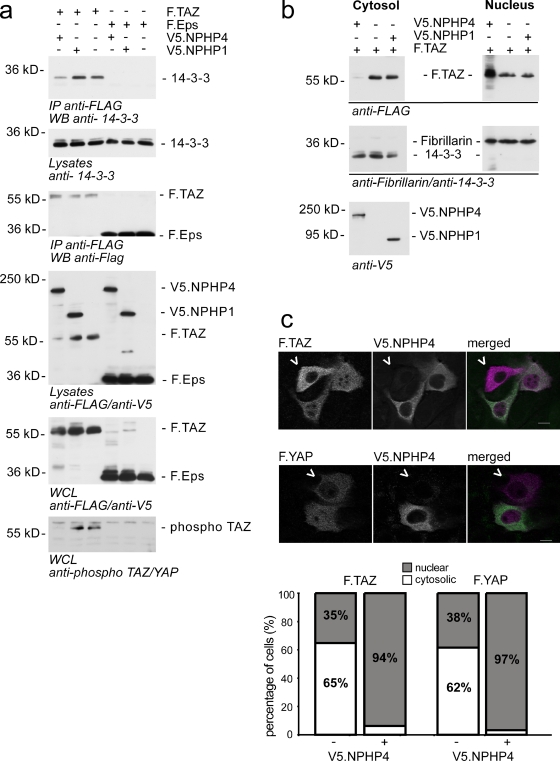
Lats1-mediated phosphorylation of TAZ on Ser-89 creates a phosphoepitope that is rapidly engaged by 14-3-3, leading to inactivation and nuclear exclusion of TAZ. To directly investigate whether NPHP4 abolished Lats1-mediated cytoplasmic retention of TAZ, we performed further coimmunoprecipitation experiments. These data revealed that the presence of NPHP4 considerably weakens the interaction of TAZ with endogenous 14-3-3 (Fig. 2 a), likely because of the reduced phosphorylation of TAZ (Fig. 2 a). Interestingly, total TAZ levels were markedly reduced in the cytoplasmic protein lysates not containing the nuclear fraction, whereas close to equal expression was found in whole cell lysates. Consistently, cell fractionation experiments showed a nuclear enrichment of TAZ in the presence of NPHP4 (Fig. 2 b). To confirm these findings by immunofluorescence, we used nontransformed immortalized breast epithelial cells (MCF-10A; Soule et al., 1990; Debnath et al., 2003). Exogenously expressed TAZ and YAP were localized predominantly in the cytoplasm, suggesting the presence of active Hippo signaling. Overexpression of NPHP4 shifted both TAZ and YAP to the nuclear compartment (Fig. 2 c), indicating NPHP4-dependent Hippo inactivation and TAZ/YAP derepression. Collectively, these data indicate that NPHP4, by interacting with and inhibiting the kinase Lats1, promotes nuclear targeting of TAZ and YAP, resulting in enhanced TEAD-dependent transcription.
Figure 2.
NPHP4 regulates the localization of TAZ. (a) HEK293T cells were transfected with the indicated constructs. Immunoprecipitation of TAZ or a control protein (Eps) with the FLAG antibody revealed that cotransfection of NPHP4, but not of NPHP1 or empty vector, decreased the interaction of TAZ with endogenous 14-3-3. Equal amounts of precipitated FLAG.TAZ were confirmed by staining the precipitates with anti-FLAG. Staining for TAZ in the cytoplasmic lysates used for immunoprecipitation (IP) showed reduced TAZ levels in the presence of NPHP4, whereas whole cell lysates (WCL), including the nuclear fraction, confirmed close to equal expression of TAZ. Staining for phosphorylation of the 14-3-3–binding motif of TAZ (phospho S89) revealed reduced phosphorylation of TAZ in the presence of NPHP4. (b) Cell fractionation experiments of HEK293T cells expressing the indicated proteins demonstrated that coexpression of NPHP4, but not NPHP1, increased the amount of TAZ in the nuclear compartment. Fibrillarin and 14-3-3 were used as nuclear and cytosolic marker proteins, respectively. (c) Breast epithelial cell MCF-10A was transfected with FLAG-tagged TAZ (F.TAZ) or FLAG-tagged YAP together with V5-tagged NPHP4 (V5.NPHP4) and stained with anti-FLAG and anti-V5 antibodies. In the absence of overexpressed NPHP4, F.TAZ/F.YAP showed a predominantly cytoplasmic localization (cells marked with arrowheads). In contrast, coexpression of V5.NPHP4 led to a highly significant increase in nuclear TAZ/YAP (bars, 10 µm; P < 0.01). WB, Western blot.
NPHP4 exerts its effect on Hippo signaling through relieving Lats1-mediated inhibition of TAZ
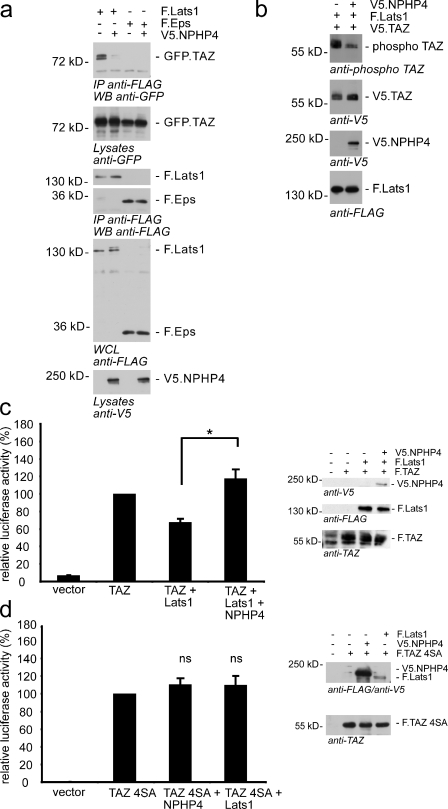
Lats1 physically interacts with TAZ and phosphorylates TAZ at Ser-89, which is part of the 14-3-3–binding motif (Kanai et al., 2000; Varelas et al., 2010). The identification of NPHP4 as a novel Lats1-interacting protein prompted us to evaluate the effect of NPHP4 on Lats1. We therefore coexpressed FLAG-tagged Lats1 with GFP-tagged TAZ in the presence or absence of V5-tagged NPHP4. Interestingly, NPHP4 inhibited the interaction of Lats1 with its target protein TAZ (Fig. 3 a), suggesting that the inhibitory role of NPHP4 on the Lats1 kinase activity involves inhibition of target protein binding. Accordingly, the Lats1-induced phosphorylation of TAZ was substantially reduced when NPHP4 was overexpressed (Fig. 3 b). Consistent with a role for NPHP4 in preventing substrate binding, autophosphorylation of Lats1 on Ser-909 was not affected by coexpression of NPHP4 (Fig. S2 a). These data implicate that NPHP4 does not affect the Hippo cascade upstream of Lats1. To examine whether NPHP4 could antagonize the repressive effect of Lats1 on TAZ/TEAD activity, we performed TAZ/TEAD reporter assays and included Lats1 into the assay. As expected, the presence of overexpressed Lats1 reduced TAZ activity (Fig. 3 c). The decline in TAZ/TEAD activity to 68% was reversed by coexpression of NPHP4, which not only derepressed the Lats1 effect but further increased TAZ/TEAD activity to 118% (Fig. 3 c). The fact that the interaction of NPHP4 with Lats1 abrogated the interaction of the latter protein with TAZ suggests that Lats1 and NPHP4 compete for TAZ binding under physiological conditions. This competitive inhibition might be the functional basis of the reduced Lats1-mediated TAZ phosphorylation that we have observed. To prove that Lats1 mediates the effect of NPHP4, we knocked down Lats1. Even a moderate decline in Lats1 expression resulted in a significant reduction of the NPHP4 effect on TAZ/TEAD activity (Fig. S2 b). To further confirm this finding, we generated a TAZ mutant (TAZ 4SA) lacking all Lats1 phosphorylation sites. Remarkably, the activity of this TAZ mutant in the reporter assay was not influenced by NPHP4 (Fig. 3 d). Collectively, these data indicate that NPHP4 acts as a potent negative regulator of Hippo signaling by modifying Lats1-dependent phosphorylation and, thereby, the localization of TAZ.
Figure 3.
NPHP4 derepresses the inhibition of TAZ by Lats1. (a) NPHP4 abrogates the interaction of Lats1 and TAZ. FLAG-tagged Lats1 (F.Lats1) was coexpressed with GFP-tagged TAZ (GFP.TAZ) in the presence or absence of V5-tagged NPHP4 (V5.NPHP4) in HEK293T cells. After immunoprecipitation (IP) with FLAG (M2) beads, Western blot (WB) analysis revealed that the interaction of Lats1 and TAZ was abolished in the presence of NPHP4. FLAG.Eps served as a control protein. Both FLAG.Lats1 and FLAG.Eps were stained in the precipitates to control the comparable efficiency of the immunoprecipitation. (b) Overexpression of NPHP4 reduces the phosphorylation of TAZ by Lats1. HEK293T cells were transfected with the indicated constructs. Cell lysates were analyzed by Western blotting using the pYAP–Ser-127/pTAZ–Ser-89 antibody. (c) NPHP4 derepresses the inhibitory effect of Lats1 on TAZ signaling. Plasmids encoding the indicated proteins or empty pcDNA6 vector were cotransfected into HEK293T cells together with the TEAD reporter plasmids as indicated in Material and methods. As expected, Lats1 reduced TEAD/TAZ activity. Additional coexpression of NPHP4 reversed this effect (n = 4; *, P < 0.05). Expression of the indicated proteins was confirmed by Western blotting. (d) The Lats1-resistant mutant of TAZ (TAZ 4SA) could not be activated by NPHP4, suggesting that NPHP4 regulates TAZ signaling through interference with Lats1-dependent phosphorylation of TAZ (n = 3). Error bars represent SEM. WCL, whole cell lysate.
NPHP4 and TAZ regulate the proliferative potential of tumor cells
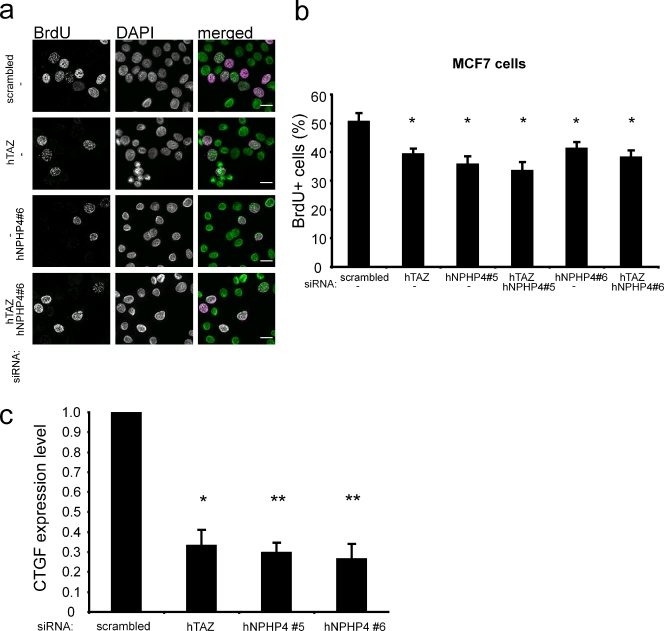
Down-regulation of Lats1 activity has been reported in various tumor entities (Hisaoka et al., 2002; Takahashi et al., 2005; Jiang et al., 2006), and Lats1-deficient mice spontaneously develop ovarian cancer at 3 mo and soft tissue sarcoma at 4–10 mo of age (St John et al., 1999). Therefore, we asked whether NPHP4 is indeed a driving force for proliferation in tumor cells. We therefore took advantage of the MCF-7 breast cancer cell line, in which small hairpin RNA–mediated knockdown of TAZ had been demonstrated to result in decreased cellular proliferation and delayed tumorigenesis in nude mice (Chan et al., 2008). We used RNAi to deplete TAZ and NPHP4 and examined the rate of DNA synthesis by analyzing incorporation of the thymidine analogue BrdU. Strikingly, just like knockdown of TAZ itself, loss of NPHP4 resulted in a decreased number of BrdU-positive cells, and the combined knockdown of both TAZ and NPHP4 had no further effect, suggesting that NPHP4 acts as an enhancer of TAZ signaling in this pathway (Fig. 4, a and b). In addition, we determined the TAZ/TEAD-dependent transcriptional activity and measured the expression of CTGF, a previously described TAZ/TEAD target gene (Zhang et al., 2009a). Knocking down NPHP4, just as depletion of TAZ, resulted in decreased expression of CTGF (Fig. 4 c). The efficiency of the NPHP4 and TAZ knockdown in these experiments was validated by quantitative PCR (qPCR; Fig. S3 a). The role of NPHP4 as a positive regulator of cell proliferation could be confirmed not only in a renal cell carcinoma cell line (A498) but also in differentiated kidney epithelial cells (HK2). These data strongly suggest that NPHP4 is a critical regulator, mediating the proliferation of both oncogenically transformed and normal epithelial cells (Fig. S3, b and c).
Figure 4.
TAZ and NPHP4 influence cell proliferation and TAZ/TEAD target gene expression in MCF-7 cells. (a and b) MCF-7 cells were transfected with the indicated siRNAs. After 72 h, the cells were serum starved for 5 h followed by BrdU labeling for 30 min in the presence of serum. BrdU-positive cells were detected using an anti-BrdU antibody, and nuclei were counterstained using DAPI. The reduction of TAZ or NPHP4 leads to a decrease in cell proliferation, and the combined knockdown had no additional effect (n = 3; *, P < 0.05 as compared with the negative control; bars, 20 µM) (c) Knockdown of TAZ or NPHP4 led to a decreased expression of the TAZ/TEAD downstream target CTGF. 72 h after transfection of the indicated siRNAs into MCF-7 cells, the CTGF expression levels were analyzed using qPCR (n = 3; *, P < 0.05; **, P < 0.01). The knockdown of NPHP4 and TAZ was validated using qPCR (Fig. S3 a). Error bars represent SEM. hTAZ, human TAZ; hNPHP4, human NPHP4.
Collectively, we found that NPHP4 is a potent activator of TAZ and YAP, as illustrated schematically in Fig. 5. Our data, for the first time, implicate an NPHP to have a proproliferative function and suggest that NPHP4 negatively regulates the tumor-suppressive Hippo pathway. Our findings further provide a possible explanation for the phenotype of small-sized kidneys and atrophic tubular epithelium in patients carrying loss-of-function mutations in NPHP4. In epithelial cells, loss of NPHP4 might lead to insufficient TAZ-dependent proliferative signaling under certain conditions. This idea is supported by the observation that two independently generated TAZ-deficient mouse models develop cystic kidneys and a phenotype that, at least partly, resembles human NPH (Hossain et al., 2007; Makita et al., 2008).
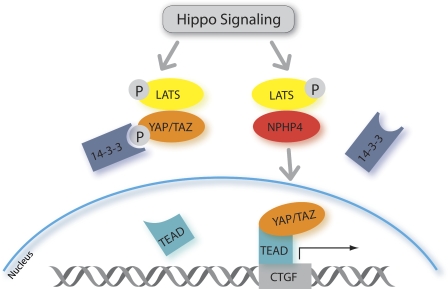
Figure 5.
NPHP4 antagonizes the Hippo pathway. Active Hippo signaling leads to phosphorylation (P) and activation of the large tumor suppressor (LATS) kinase, which in turn phosphorylates YAP at Ser-127 and TAZ at Ser-89. This creates phosphoepitopes that are rapidly engaged by 14-3-3, leading to cytoplasmic retention of YAP and TAZ and the inhibition of their transcriptional activation potential. NPHP4 abrogates the interaction of large tumor suppressor with its substrates YAP and TAZ, which abolishes 14-3-3 binding and allows for nuclear translocation of YAP and TAZ.
Although further studies are needed to clarify the in vivo importance of the NPHP4-mediated control of Hippo signaling, our findings may have two important consequences. First, normal organ development as well as repair after damage requires cellular proliferation to guarantee organ growth and tissue regeneration. Thus, lack of NPHP4 may lead to the overactivity of the Hippo signaling pathway, resulting in antiproliferative cellular signaling and, thus, leading to reduced growth or insufficient repair. This might explain some aspects of the phenotype of NPH. Our data show that the expression level of NPHP4 controls the activity of the Hippo mediator TAZ, suggesting that NPHP4 may act as a repressor of Hippo and enhancer of proliferative signaling, which may be critical for kidney development and repair.
Second, inappropriate activation of TAZ/TEAD through hyperregulated NPHP4 in mature tissues may contribute to tumorigenesis and explain why several NPHPs are overexpressed in various tumors. The ubiquitous expression of NPHP4, including in breast cancer cells, also implicates that this potential tumor-promoting function of NPHP4 is not restricted to the kidney but rather represents a general mechanism. Understanding the degree to which this plays a role in carcinogenesis in vivo will require further studies using cancer models in laboratory animals and gene expression analyses in human patient samples.
Materials and methods
Cell culture
HEK293T, A498, and MCF-7 cells were maintained in DME supplemented with 10% FBS. MCF-10A cells were cultured in DME/F12 supplemented with 5% horse serum, 0.5 µg/ml hydrocortisone, 100 ng/ml cholera toxin, 20 ng/ml EGF, and 10 µg/ml insulin (Debnath et al., 2003). HK2 cells were cultured in keratinocyte serum-free medium supplemented with 0.05 mg/ml pituitary extract and 5 ng/ml EGF.
Cell fractionation
HEK293T cells were transiently transfected with the indicated constructs using the calcium phosphate method. Cells were disrupted in a hypotonic buffer (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.05% NP-40, and protease inhibitors). After centrifugation at 3,000 g for 10 min, the cytoplasmic fraction was taken from the supernatant. The pellet was washed with PBS and, after centrifugation for 10 min at 10,000 g, treated with a hypertonic buffer (5 mM Hepes, pH 7.9, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 26% glycerol, 300 mM NaCl, and protease inhibitors) to yield the nuclear fraction. Both fractions were analyzed by Western blotting.
Immunofluorescence
MCF-10A cells were seeded onto coverslips and transfected with the indicated plasmids using transfection reagent (Genejuice; EMD). 24 h afterward, cells were rinsed with PBS several times and fixed with 4% PFA for 10 min. After blocking with 5% normal donkey serum and 0.1% Triton X-100 in Dulbecco’s PBS, cells were sequentially stained with the indicated antibodies (primary antibodies: rabbit anti-FLAG [pAb] and mouse anti-V5 [mAb]; secondary antibodies: Cy3-conjugated anti–rabbit IgG and Cy2-conjugated anti–mouse IgG obtained from Jackson ImmunoResearch Laboratories, Inc.). Afterward, the coverslips were mounted with Prolong gold with DAPI (Invitrogen) and subjected to immunofluorescence microscopy. Pictures were taken with a microscope (objectives: Plan Apochromat 20×/0.8 NA differential interference contrast, EC Plan Neofluar 40×/1.3 NA oil differential interference contrast, and C-Apochromat 63×/1.22 W; Axiovert 200; Carl Zeiss) equipped with a camera (AxioCam MRm; Carl Zeiss) and an imaging system (ApoTome; Carl Zeiss) using Axiovision 4.8 software for acquisition and subsequent image processing (Carl Zeiss). Figures were assembled using Macromedia Freehand 11 software (Adobe). About 200 cells were counted and quantified in regard to the localization of TAZ and YAP with or without NPHP4 coexpressed in the same cell. P-values were calculated using Fisher’s exact test.
Luciferase assays
The luciferase reporter plasmid (pGBD-Hyg-Luc) was transfected together with an activator plasmid (pGal4-TEAD), pGL4.74 (Promega) for normalization, and the indicated expression plasmids (TAZ, YAP, NPHP4, Lats1, and the control empty pcDNA6) into HEK293T cells in a 96-well format using Lipofectamine LTX (Invitrogen) as a transfection reagent. The total amount of DNA was always adjusted with empty pcDNA6. Renilla luciferase and firefly luciferase activities were measured by using a reporter assay system (Dual Luciferase; Promega) in a luminometer (Mithras LB 940; Berthold) 24 h after transfection. For the Lats1 siRNA experiment, the indicated plasmids and siRNAs were cotransfected into HEK293T cells. The measurement was performed 48 h after transfection. Transfections and measurements were performed in triplicates for each single experiment, and each experiment was repeated at least three times. Error bars shown in the figures represent SEM. P-values were calculated using an unpaired Student’s t test. Equal expression of the transfected proteins was confirmed by Western blot analysis.
Coimmunoprecipitation
HEK293T cells were transiently transfected using the calcium phosphate method, and the total amount of DNA was always adjusted with empty pcDNA6. The following day, cells were harvested with ice-cold PBS. A small aliquot of this cell suspension was taken, and the cells of which were lysed directly in SDS-PAGE sample buffer (whole cell lysate). The harvested cells were lysed in a 1% Triton X-100 buffer (1% Triton X-100, 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 50 mM NaF, 15 mM Na4P2O7, 2 mM Na3VO4, and complete protease inhibitors [PIM; Roche]) for 15 min on ice. After centrifugation at 15,000 g for 15 min at 4°C and ultracentrifugation at 100,000 g for 30 min at 4°C, the supernatant was incubated at 4°C for 2 h with the anti-FLAG (M2) antibody covalently coupled to agarose beads (Sigma-Aldrich) or with 1 µg of the appropriate first antibody and 20 µl protein G–Sepharose beads (GE Healthcare). Before the addition of antibodies, a small aliquot of each supernatant was preserved and diluted with 2× SDS-PAGE sample buffer for later Western blot analysis (lysate). The beads were washed extensively with lysis buffer, and bound proteins were resolved by SDS-PAGE, blotted on to polyvinylidene fluoride membranes, and visualized with enhanced chemiluminescence after incubation of the blots with the respective antibodies.
Plasmids, reagents, and antibodies
TAZ, YAP, Salvador, and Lats1 cDNAs were provided by M. Yaffe (Massachusetts Institute of Technology, Boston, MA) and M. Sudol (Geisinger Clinic, Danville, PA). The TAZ 4SA mutant was generated using mutagenesis (QuickChange; Agilent Technologies) to mutate the serine residues in positions 66, 89, 117, and 306 to alanine. The GAL4-TEAD reporter system (pGBD-Hyg-Luc and pGal4-TEAD) was purchased from Biomyx. NPHP4 was cloned from a human kidney cDNA library. NPHP1 was cloned from a human kidney cDNA library into a modified pcDNA6 vector using standard PCR cloning techniques and has been described previously (Otto et al., 2003; Schermer et al., 2005). siRNAs used in knockdown experiments were directed against the following sequences: NPHP4 #5, 5′-AAGCAACGAGATGGTGCTACA-3′; NPHP4 #6, 5′-CAGATCTCGGGTCATCTCAAA-3′; TAZ, 5′-AGGTACTTCCTCAATCACA-3′ (previously described and validated by immunoblotting with endogenous TAZ in MCF-7 cells; Chan et al., 2008); scrambled/control, 5′-AAATGTACTGCGCGTGGAGAC-3′; Lats1 #1, 5′-GGAGTGTTACTCCTCCACC-3′; Lats1 #2, 5′-GGTTCTGAGAGTAAAATTA-3′, Lats1 #12, 5′-CAGCAGCGTCTACATCGTAAA-3′; and Lats1 #13, 5′-TTGGTGAGTGTTCCTAGGCTA-3′. siRNA strands were purchased from Biomers or QIAGEN (Lats1 #12 and Lats1 #13 siRNA). siRNAs against NPHP4 and TAZ have been tested by qPCR (Fig. S3 a), and the siRNA against TAZ has been previously described (Chan et al., 2008).
Antibodies were purchased from Sigma-Aldrich (anti-FLAG/M2, anti–β actin, anti-WWTR1/TAZ, and antitubulin), Cell Signaling Technology (anti-Lats1, antiphospho-Lats1 Ser-909, antiphospho-YAP Ser-127, and anti-MST1), AbD Serotec (anti-V5), Millipore (anti-V5), Abcam (antifibrillarin), and Santa Cruz Biotechnology, Inc. (anti-GFP and anti–14-3-3).
qPCR
MCF7 cells were transfected with the indicated siRNAs (final concentration of 20 nM) using a transfection reagent (Oligofectamine; Invitrogen). 72 h after transfection, cells were harvested in lysis reagent (QIAzol; QIAGEN), and RNA was isolated using the phenol-chloroform method. After DNase treatment (Invitrogen), the reverse transcription reaction was performed with a High-Capacity cDNA kit (Applied Biosystems). TaqMan assays (Applied Biosystems) were used to evaluate CTGF (Hs00170014_m1) levels. ActB (4326315E), HPRT1 (Hs02800695), and 18SrRNA (4319413E) served as endogenous controls. The efficiency of the NPHP4 siRNAs was confirmed using a TaqMan assay (Hs00296416_m1; Applied Biosystems), the TAZ siRNA was validated using SYBR green qPCR (TAZ forward primer, 5′-ACCCACCCACGATGACCCCA-3′, and TAZ reverse primer, 5′-GCACCCTAACCCCAGGCCAC-3′), and HPRT1 served as an endogenous control (HPRT1 forward primer, 5′-TGACACTGGCAAAACAATGCA-3′, and HPRT1 reverse primer, 5′-GGTCCTTTTCACCAGCAAGCT-3′). All qPCR experiments were performed on a real-time PCR system (7900HT; Applied Biosystems) and repeated at least three times. Error bars shown in the figures represent SEM. P-values were calculated using an unpaired Student’s t test.
BrdU assays
MCF-7, A498, and HK2 cells were transfected with the indicated siRNAs (final concentration of 20 nM) using Oligofectamine and processed 72 h after transfection. The MCF-7 cells were serum starved for 5 h followed by 30-min BrdU labeling in complete growth medium. After fixation in 100% methanol for 20 min, cells were stained with the BrdU Labeling and Detection Kit I (Roche). Nuclei were counterstained with DAPI. Pictures were randomly taken with a microscope (Axiovert 200) and the Axiovision software. At least 400 cells were counted for each point within each single experiment.
The A498 and HK2 cells were labeled with BrdU in complete growth medium for 3 or 12 h, respectively. Afterward, the cells were fixed with 70% ethanol overnight after treatment with 2 N HCl/0.5% Triton X-100 and 0.1 M sodium tetraborate. Cells were stained with a mouse anti-BrdU antibody (Sigma-Aldrich) and Dylight 488–conjugated anti–mouse IgG (Jackson ImmunoResearch Laboratories, Inc.). DNA was counterstained with propidium iodide (Sigma-Aldrich), and cells were analyzed by FACS. The amount of BrdU + cells for each condition was calculated in comparison to the scrambled siRNA–treated control cells. Error bars shown in the figure represent SEM. P-values were calculated using an unpaired Student’s t test.
Chromatin immunoprecipitation
HEK293T cells were transfected with the plasmids indicated in the figures using the calcium phosphate method. 2 × 107 cells were cross-linked with 1% formaldehyde (Merck) for 10 min at RT. Cross-linking was quenched by cold PBS. Cells were scraped into 50 ml PBS containing protease inhibitors (PMSF and apotinin) and pelleted for 5 min at 300 g at 4°C. Cells were lysed, and the lysate was sonicated to a DNA fragment with a length of ∼1 kbp. Lysates were cleared by CL4B200 beads (Sigma-Aldrich). Then, 2 µg anti-FLAG antibody and anti–β actin as a negative control was added for overnight immunoprecipitation. 60 µl of preblocked GammaBind G Sepharose beads (GE Healthcare) was added for 1 h at 4°C with rotation. After washing, beads were eluted in 500 µl of buffer. The eluate was incubated for 65°C overnight to reverse cross-linking followed by proteinase K (Roche) digestion and DNA recovery with a PCR purification kit (QIAquick; QIAGEN). DNA was eluted in 50 µl H2O, and 4 µl of the eluate was used per well as the template for real-time PCR using SYBR green–based master mix (Applied Biosystems). Primers for the human CTGF promoter region close to the TAZ response element had the following sequences: 5′-GAGACTGCATCCTGAGTCACAC-3′ (forward primer) and 5′-GGCTCTTGAAACTCTCCAAAGA-3′ (reverse primer).
Online supplemental material
Fig. S1 shows that NPHP1 and NPHP4 do not interact with MST1 and demonstrates the dose-dependent effect of NPHP4 on TAZ signaling. Fig. S2 shows that NPHP4 does not affect autophosphorylation of Lats1 and that the effect of NPHP4 on TAZ activity depends on the presence of Lats1. Fig. S3 shows the validation of the siRNA against NPHP4 and TAZ by qPCR and the reduced cell proliferation after knockdown of TAZ or NPHP4 in renal cell carcinoma cells (A498) and in human kidney epithelial cells (HK2). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201009069/DC1.
Acknowledgments
We thank Stefanie Keller, Bettina Maar, Jasmin Manz, Ruth Herzog, Manuela Hochberger, and Katrin Walter for excellent technical assistance and members of the laboratories for helpful discussions. We thank Mike Yaffe for providing TAZ/YAP cDNAs and antibodies and Marius Sudol for the cDNA of Lats1. The MCF-7 cells were a kind gift from Jochen Fries (University Hospital of Cologne, Cologne, Germany).
This study was supported by the Deutsche Forschungsgemeinschaft (grants SCHE1562 to B. Schermer, BE2212 to T. Benzing, RE2246 to H.C. Reinhardt, and SFB832 to H.C. Reinhardt, T. Benzing, and B. Schermer), by the Deutsche Nierenstiftung (grant to R.U. Müller and H.C. Reinhardt), by the David H. Koch Foundation (grant to H.C. Reinhardt), and by Köln Fortune (grant to S. Habbig).
Footnotes
Abbreviations used in this paper:
- CTGF
- connective tissue growth factor
- NPH
- nephronophthisis
- NPHP
- NPH protein
- qPCR
- quantitative PCR
- TEAD
- TEA domain
- YAP
- Yes-associated protein
References
- Badouel C., Garg A., McNeill H. 2009. Herding Hippos: regulating growth in flies and man. Curr. Opin. Cell Biol. 21:837–843 10.1016/j.ceb.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Berbari N.F., O’Connor A.K., Haycraft C.J., Yoder B.K. 2009. The primary cilium as a complex signaling center. Curr. Biol. 19:R526–R535 10.1016/j.cub.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers A.J., Boylan J.F. 2004. Nek8, a NIMA family kinase member, is overexpressed in primary human breast tumors. Gene. 328:135–142 10.1016/j.gene.2003.12.002 [DOI] [PubMed] [Google Scholar]
- Carter H., Samayoa J., Hruban R.H., Karchin R. 2010. Prioritization of driver mutations in pancreatic cancer using cancer-specific high-throughput annotation of somatic mutations (CHASM). Cancer Biol. Ther. 10:582–587 10.4161/cbt.10.6.12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W., Lim C.J., Guo K., Ng C.P., Lee I., Hunziker W., Zeng Q., Hong W. 2008. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 68:2592–2598 10.1158/0008-5472.CAN-07-2696 [DOI] [PubMed] [Google Scholar]
- Dalgliesh G.L., Furge K., Greenman C., Chen L., Bignell G., Butler A., Davies H., Edkins S., Hardy C., Latimer C., et al. 2010. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 463:360–363 10.1038/nature08672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., Brugge J.S. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30:256–268 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- Fernandez-L A., Kenney A.M. 2010. The Hippo in the room: A new look at a key pathway in cell growth and transformation. Cell Cycle. 9:2292–2299 10.4161/cc.9.12.11919 [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Forbes S.A., Bhamra G., Bamford S., Dawson E., Kok C., Clements J., Menzies A., Teague J.W., Futreal P.A., Stratton M.R. 2008. The catalogue of somatic mutations in cancer (COSMIC). Curr. Protoc. Hum. Genet. Chapter 10:Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J.M., Davis E.E., Katsanis N. 2009. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 137:32–45 10.1016/j.cell.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8:27–36 10.1038/ncb1339 [DOI] [PubMed] [Google Scholar]
- Harvey K., Tapon N. 2007. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat. Rev. Cancer. 7:182–191 10.1038/nrc2070 [DOI] [PubMed] [Google Scholar]
- Hildebrandt F., Attanasio M., Otto E. 2009. Nephronophthisis: disease mechanisms of a ciliopathy. J. Am. Soc. Nephrol. 20:23–35 10.1681/ASN.2008050456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisaoka M., Tanaka A., Hashimoto H. 2002. Molecular alterations of h-warts/LATS1 tumor suppressor in human soft tissue sarcoma. Lab. Invest. 82:1427–1435 [DOI] [PubMed] [Google Scholar]
- Hossain Z., Ali S.M., Ko H.L., Xu J., Ng C.P., Guo K., Qi Z., Ponniah S., Hong W., Hunziker W. 2007. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc. Natl. Acad. Sci. USA. 104:1631–1636 10.1073/pnas.0605266104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Li X., Hu J., Zhou W., Jiang Y., Li G., Lu D. 2006. Promoter hypermethylation-mediated down-regulation of LATS1 and LATS2 in human astrocytoma. Neurosci. Res. 56:450–458 10.1016/j.neures.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Kanai F., Marignani P.A., Sarbassova D., Yagi R., Hall R.A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L.C., Yaffe M.B. 2000. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19:6778–6791 10.1093/emboj/19.24.6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M., Singh A. 2009. Regulation of organ size: insights from the Drosophila Hippo signaling pathway. Dev. Dyn. 238:1627–1637 10.1002/dvdy.21996 [DOI] [PubMed] [Google Scholar]
- Lei Q.Y., Zhang H., Zhao B., Zha Z.Y., Bai F., Pei X.H., Zhao S., Xiong Y., Guan K.L. 2008. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 28:2426–2436 10.1128/MCB.01874-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita R., Uchijima Y., Nishiyama K., Amano T., Chen Q., Takeuchi T., Mitani A., Nagase T., Yatomi Y., Aburatani H., et al. 2008. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am. J. Physiol. Renal Physiol. 294:F542–F553 10.1152/ajprenal.00201.2007 [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki E.M., Boone B.N., Howell M., Stonebraker J.R., Teed J., Alb J.G., Magnuson T.R., O’Neal W., Milgram S.L. 2006. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol. Cell. Biol. 26:77–87 10.1128/MCB.26.1.77-87.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris Z.S., McClatchey A.I. 2009. Aberrant epithelial morphology and persistent epidermal growth factor receptor signaling in a mouse model of renal carcinoma. Proc. Natl. Acad. Sci. USA. 106:9767–9772 10.1073/pnas.0902031106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E.A., Schermer B., Obara T., O’Toole J.F., Hiller K.S., Mueller A.M., Ruf R.G., Hoefele J., Beekmann F., Landau D., et al. 2003. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat. Genet. 34:413–420 10.1038/ng1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M., Zhang J., Smolen G.A., Muir B., Li W., Sgroi D.C., Deng C.X., Brugge J.S., Haber D.A. 2006. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. USA. 103:12405–12410 10.1073/pnas.0605579103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttledge M.H., Sarrazin J., Rangaratnam S., Phelan C.M., Twist E., Merel P., Delattre O., Thomas G., Nordenskjöld M., Collins V.P., et al. 1994. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat. Genet. 6:180–184 10.1038/ng0294-180 [DOI] [PubMed] [Google Scholar]
- Schermer B., Höpker K., Omran H., Ghenoiu C., Fliegauf M., Fekete A., Horvath J., Köttgen M., Hackl M., Zschiedrich S., et al. 2005. Phosphorylation by casein kinase 2 induces PACS-1 binding of nephrocystin and targeting to cilia. EMBO J. 24:4415–4424 10.1038/sj.emboj.7600885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science. 314:268–274 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- Soule H.D., Maloney T.M., Wolman S.R., Peterson W.D., Jr, Brenz R., McGrath C.M., Russo J., Pauley R.J., Jones R.F., Brooks S.C. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075–6086 [PubMed] [Google Scholar]
- St John M.A., Tao W., Fei X., Fukumoto R., Carcangiu M.L., Brownstein D.G., Parlow A.F., McGrath J., Xu T. 1999. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 21:182–186 10.1038/5965 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Miyoshi Y., Takahata C., Irahara N., Taguchi T., Tamaki Y., Noguchi S. 2005. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin. Cancer Res. 11:1380–1385 10.1158/1078-0432.CCR-04-1773 [DOI] [PubMed] [Google Scholar]
- Tian W., Yu J., Tomchick D.R., Pan D., Luo X. 2010. Structural and functional analysis of the YAP-binding domain of human TEAD2. Proc. Natl. Acad. Sci. USA. 107:7293–7298 10.1073/pnas.1000293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofatter J.A., MacCollin M.M., Rutter J.L., Murrell J.R., Duyao M.P., Parry D.M., Eldridge R., Kley N., Menon A.G., Pulaski K., et al. 1993. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 72:791–800 10.1016/0092-8674(93)90406-G [DOI] [PubMed] [Google Scholar]
- Varelas X., Miller B.W., Sopko R., Song S., Gregorieff A., Fellouse F.A., Sakuma R., Pawson T., Hunziker W., McNeill H., et al. 2010. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell. 18:579–591 10.1016/j.devcel.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Wang K., Degerny C., Xu M., Yang X.J. 2009. YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem. Cell Biol. 87:77–91 10.1139/O08-114 [DOI] [PubMed] [Google Scholar]
- Wood L.D., Parsons D.W., Jones S., Lin J., Sjöblom T., Leary R.J., Shen D., Boca S.M., Barber T., Ptak J., et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science. 318:1108–1113 10.1126/science.1145720 [DOI] [PubMed] [Google Scholar]
- Zender L., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., et al. 2006. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 125:1253–1267 10.1016/j.cell.2006.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu C.Y., Zha Z.Y., Zhao B., Yao J., Zhao S., Xiong Y., Lei Q.Y., Guan K.L. 2009a. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem. 284:13355–13362 10.1074/jbc.M900843200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yue T., Jiang J. 2009b. Hippo signaling pathway and organ size control. Fly (Austin). 3:68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]