Abstract
Nuclear bodies are dynamic structures that form at sites of specific activities associated with gene expression and genome maintenance. A paper in this issue (White et al. 2011. J. Cell Biol. doi: 10.1083/jcb.201012077) highlights key features of nuclear body biogenesis and suggests a unifying model in which formation of nuclear bodies is driven by nonrandom, biologically determined initial seeding events followed by stochastic self-assembly.
The spatial organization of the cell nucleus plays a fundamental role in genome function and maintenance (Misteli, 2007; Rajapakse and Groudine, 2011). Within the nuclear environment, which is characterized by a lack of membranes, chromosomes occupy specific nonrandom territories decorated by a variety of functionally distinct nuclear bodies (Zhao et al., 2009). Nuclear bodies are highly dynamic structures, whose components can rapidly exchange with the surrounding nucleoplasm. The molecular mechanisms of how nuclear bodies form and maintain their structural integrity are still poorly understood. More specifically, little is known about what triggers the formation of a nuclear body and how different assembly stages are orchestrated.
Two fundamentally different models of nuclear body formation have been considered (Dundr and Misteli, 2010). In one, referred to as the ordered assembly model, a nuclear body forms by a highly controlled series of sequential steps that follow an initial seeding event involving a very limited number of different molecules. Alternatively, a stochastic self-organization assembly model posits that a nuclear body is built by random, transient low affinity interactions of individual components without a strict hierarchical order of assembly. According to the self-organization model, any of the components of a nuclear body can initiate its formation. Strong evidence for this latter mode of assembly came from studies using an experimental assay to probe the assembly of the Cajal body (Kaiser et al., 2008), a nuclear body involved in the biogenesis and recycling of many small nuclear RNAs (Nizami et al., 2010). In these experiments, the authors tethered individual Cajal body components to an engineered random site in chromatin in living cells. Using this approach, they demonstrated that any Cajal body component can initiate the formation of the entire nuclear body, suggesting that, in line with self-organization, the order of component assembly is not important for Cajal body formation.
A critical step in nuclear body formation is the initial nucleation event that serves to immobilize critical components to provide a seeding platform to trigger recruitment and retention of building blocks. What might physiological seeding events be? It has been observed that several nuclear bodies form at sites of transcription (Matera et al., 2009), raising the intriguing possibility that RNAs may serve as physiological seeding agents. Two recent studies have shed light on the role of RNA in nuclear body formation, indicating that coding and noncoding RNAs indeed act as a structural element in their biogenesis. Shevtsov and Dundr (2011) and Mao et al. (2011) provided evidence, using different experimental approaches, that nuclear body–specific RNAs are sufficient to initiate the formation of nuclear bodies with which they are physiologically associated. Shevtsov and Dundr (2011) tethered coding histone H2b or β-globin pre-mRNAs or the noncoding NEAT1 or SatIII RNAs to an engineered locus in the genome in living cells; this led to de novo nucleation of the histone locus body (HLB) with the associated Cajal body, nuclear speckle, paraspeckle, and nuclear stress bodies, respectively, at these tethering sites. Mao et al. (2011) on the other hand visualized de novo formation of paraspeckles by inducing transcription of noncoding RNA Men ε/β (NEAT1 in humans), which provides the crucial structural framework for paraspeckle assembly (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). These results strongly suggest that RNA transcripts may act as a seeding scaffold to attract and retain RNA-associated components from a soluble nucleoplasmic pool, leading to the formation of specific nuclear bodies. RNA-mediated seeding of nuclear bodies is consistent with recent findings by Brody et al. (2011) indicating that splicing—and likely other pre-mRNA processing steps—can occur while transcripts are still attached to RNA polymerase II, even after they reach the termination stage. This suggests that when transcription is completed, but splicing has not been, the splicing machinery is retained at the site of transcription. This raises the interesting possibility that nuclear bodies may preferentially form around highly active gene loci with accumulated unprocessed transcripts that act as a seed for a nuclear body to speed up the pre-mRNA processing.
The study by White et al. (in this issue) now further highlights the involvement of a seeding step and random events in nuclear body formation. The authors performed an extensive analysis of the mechanism of assembly of the HLB, an evolutionarily conserved nuclear body associated with replication-dependent histone genes (Nizami et al., 2010). Using a genome-wide RNAi screen and proteomic analysis in Drosophila melanogaster, they identified histone transcription and pre-mRNA–processing factors Mxc (the Drosophila orthologue of mammalian NPAT) and FLASH as essential components for HLB formation. In addition, they found the transcription elongation factor Spt6 and the functionally uncharacterized Mute protein as new HLB components. They also demonstrated that Mxc is a substrate of cyclin E/Cdk2 kinase, which regulates G1/S transition, and its phosphorylation is responsible for Mute and Spt6 recruitment to HLB. Importantly, they find that Mxc and FLASH first colocalize into foci resembling the HLB during syncytial nuclear cycle 10 of Drosophila early embryos. However, these structures are not fully assembled HLBs, as they lack various components. Only in the next nuclear cycle, when zygotic histone gene expression begins, do fully functional HLBs containing Mxc/FLASH/Mute/U7 small nuclear RNP form. Thus, the authors conclude that foci containing Mxc and FLASH represent an early step in HLB assembly that serves as a seeding agent for the subsequent recruitment of other HLB components.
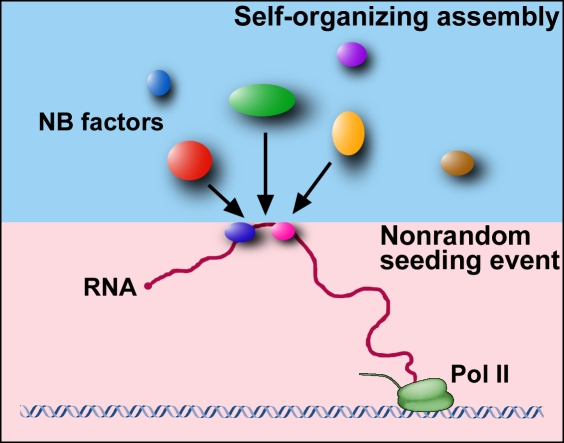
These observations highlight key aspects of nuclear body biogenesis and lead to a unifying model. The tethering experiment on the formation of Cajal bodies showed that a nuclear body can form by self-organization regardless of which components initiate the assembly. If nuclear body assembly required a strictly hierarchical order of components, tethering experiments could not work. Thus, this approach conclusively characterizes a fundamental property of nuclear structure formation, i.e., the contribution of self-organization to nuclear body assembly. However, the data by White et al. (2011) strongly emphasize the fact that not all events involved in nuclear body formation are random. In agreement with other experimental approaches, such as those performed by Mao et al. (2011), White et al. (2011) demonstrate that in a physiological situation, a seeding event plays a key role in initiating the assembly process. In the case of HLBs, the histone gene expression factors Mxc and FLASH recognize the histone gene cluster, and upon its activation, they act as a seed to initiate the random recruitment of other factors. For paraspeckles, the NEAT1 RNA appears to act as a seeding agent. Collectively, the data suggest that the most likely model for nuclear body formation is a hybrid model (Fig. 1), in which assembly of a nuclear body is triggered by an initial seeding event, which is nonrandom and driven by a biological process, such as transcription. Thus, seeding often involves RNA. Specific proteins may then bind to this seed, but these subsequent nuclear body assembly steps may occur in a largely random and self-organized fashion. This model should be useful to now uncover the full interaction network required to form a nuclear body.
Figure 1.
The two-step hybrid seeding model of nuclear body formation. In the first step, nonrandom, biologically determined RNAs (or proteins bound to the template) act as a seed. In the subsequent step, nuclear body (NB) formation is driven by random and stochastic self-organization. Pol II, RNA polymerase II.
References
- Brody Y., Neufeld N., Bieberstein N., Causse S.Z., Böhnlein E.M., Neugebauer K.M., Darzacq X., Shav-Tal Y. 2011. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 9:e1000573 10.1371/journal.pbio.1000573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., Carmichael G.G. 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 35:467–478 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., Lawrence J.B. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 33:717–726 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Misteli T. 2010. Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2:a000711 10.1101/cshperspect.a000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T.E., Intine R.V., Dundr M. 2008. De novo formation of a subnuclear body. Science. 322:1713–1717 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- Mao Y.S., Sunwoo H., Zhang B., Spector D.L. 2011. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13:95–101 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Izaguire-Sierra M., Praveen K., Rajendra T.K. 2009. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev. Cell. 17:639–647 10.1016/j.devcel.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2007. Beyond the sequence: cellular organization of genome function. Cell. 128:787–800 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- Nizami Z., Deryusheva S., Gall J.G. 2010. The Cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2:a000653 10.1101/cshperspect.a000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse I., Groudine M. 2011. On emerging nuclear order. J. Cell Biol. 192:711–721 10.1083/jcb.201010129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y.T., Ideue T., Sano M., Mituyama T., Hirose T. 2009. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA. 106:2525–2530 10.1073/pnas.0807899106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov S.P., Dundr M. 2011. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13:167–173 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L. 2009. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19:347–359 10.1101/gr.087775.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.E., Burch B.D., Yang X.-C., Gasdaska P.Y., Dominski Z., Marzluff W.F., Duronio R.J. 2011. Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 193:677–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Bodnar M.S., Spector D.L. 2009. Nuclear neighborhoods and gene expression. Curr. Opin. Genet. Dev. 19:172–179 10.1016/j.gde.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]