The syndecan Sds2 and the innate immunity inhibitor Sarm1 function together and in distinct pathways to promote proper neuronal morphogenesis.
Abstract
Dendritic arborization is a critical neuronal differentiation process. Here, we demonstrate that syndecan-2 (Sdc2), a synaptic heparan sulfate proteoglycan that triggers dendritic filopodia and spine formation, regulates dendritic arborization in cultured hippocampal neurons. This process is controlled by sterile α and TIR motif–containing 1 protein (Sarm1), a negative regulator of Toll-like receptor 3 (TLR3) in innate immunity signaling. We show that Sarm1 interacts with and receives signal from Sdc2 and controls dendritic arborization through the MKK4–JNK pathway. In Sarm1 knockdown mice, dendritic arbors of neurons were less complex than those of wild-type littermates. In addition to acting downstream of Sdc2, Sarm1 is expressed earlier than Sdc2, which suggests that it has multiple roles in neuronal morphogenesis. Specifically, it is required for proper initiation and elongation of dendrites, axonal outgrowth, and neuronal polarization. These functions likely involve Sarm1-mediated regulation of microtubule stability, as Sarm1 influenced tubulin acetylation. This study thus reveals the molecular mechanism underlying the action of Sarm1 in neuronal morphogenesis.
Introduction
Defects in neural development are known to result in complex neurodevelopmental disorders including mental retardation, epilepsy, attention deficient hyperactivity disorder, autism, and schizophrenia (Ehninger et al., 2008; Walsh et al., 2008). In addition to genetic factors, environmental factors also contribute to these neurodevelopmental disorders. For instance, prenatal infection and maternal immune responses have been suggested to modulate neural development of embryos and can lead to autistic or schizophrenic behaviors (Hornig and Lipkin, 2001; Smith et al., 2007; Patterson, 2009). Therefore, it has been hypothesized that innate immune responses influence neuronal development. Indeed, recent studies indicate that the Toll-like receptors (TLRs), key receptors for activation of innate immunity, regulate neurogenesis and neuritogenesis. TLR2 and TLR4 activation regulate hippocampal neurogenesis (Rolls et al., 2007), whereas TLR3 plays a negative role in the proliferation of embryonic neural progenitor cells (Lathia et al., 2008). Both TLR3 and TLR8 negatively modulate neurite outgrowth (Ma et al., 2006; Cameron et al., 2007). Collectively, these studies strengthen the notion that innate immunity plays a regulatory role in neural development.
Sterile α and TIR motif–containing protein 1 (Sarm1) is a multidomain adaptor molecule containing two sterile α motifs and one Toll/interleukin-1 receptor homology domain. Sarm1 was originally identified as a negative regulator of the TRIF-dependent TLR3 and TLR4 pathways in innate immunity (Mink et al., 2001; Carty et al., 2006). In addition, Sarm1 is known to function in the nervous system. Toll and interleukin 1 receptor domain protein (Tir-1), an orthologue of Sarm1 in Caenorhabditis elegans, is highly concentrated at synapses (Chuang and Bargmann, 2005). Tir-1 receives synaptic signals through its interaction with CaMK, regulates the downstream ASK1–MKK–JNK pathway, and thus regulates olfactory receptor expression (Chuang and Bargmann, 2005). In mammals, Sarm1 also regulates neuronal survival by targeting JNK3 to the mitochondria (Kim et al., 2007). Although TLR3 has been shown to regulate neurogenesis and neuritogenesis, it remains unclear whether Sarm1 plays a role in neural development.
Syndecan-2 (Sdc2), a member of the transmembrane heparan sulfate proteoglycan syndecan family, is highly concentrated at synapses (Hsueh et al., 1998). During neuronal differentiation, Sdc2 is expressed concurrently with synaptogenesis (Ethell and Yamaguchi, 1999; Hsueh and Sheng, 1999). In cultured hippocampal neurons, very low expression of Sdc2 begins 1 wk after plating and continuously increases during neuronal maturation. Overexpression of Sdc2 in cultured hippocampal neurons at 1–2 d in vitro (DIV) promotes dendritic filopodia formation at 4–5 DIV. These dendritic filopodia then transform into dendritic spines at 8–9 DIV (Ethell and Yamaguchi, 1999; Lin et al., 2007), ∼1 wk earlier than in the intrinsic process. To promote filopodia formation, an interaction between Sdc2 and neurofibromin is required; neurofibromin then activates protein kinase A (PKA) and triggers phosphorylation of the Ena/VASP proteins, thus promoting bundle formation of actin and filopodia formation (Lin et al., 2007). The extreme C-terminal tail of Sdc2, which contains a calcium/calmodulin-dependent serine protein kinase (CASK)-binding motif, is required for the transformation from dendritic filopodia to spine (Lin et al., 2007). CASK, a membrane-associated guanylate kinase, contains several protein–protein interacting domains and functions as an adaptor (Hsueh, 2006, 2009). CASK links Sdc2 to the actin cytoskeleton through its PDZ domain and protein 4.1-binding motif, thereby stabilizing spine morphology (Chao et al., 2008).
In this paper, we show that, in addition to spinogenesis, Sdc2 also regulates dendritic arborization. By investigating the molecular mechanism underlying Sdc2-induced dendrite outgrowth, we found that Sarm1 is a critical downstream effector of Sdc2 in dendritic arborization. Our observations also suggest multiple roles for Sarm1 in initiation and elongation of dendrites, axonal outgrowth, and neuronal polarity. This study thus provides new evidence that Sarm1, a molecule involved in innate immunity, contributes to the regulation of neuronal morphogenesis.
Results
Sdc2 regulates dendritic arbor morphogenesis
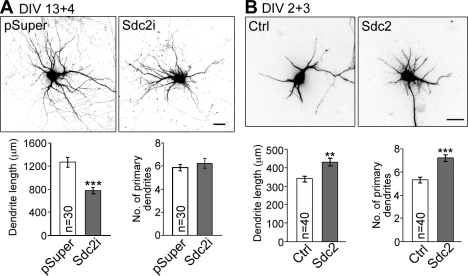
We noticed that dendritic arbor development seemed to be affected by Sdc2 while we were investigating the function of Sdc2 in neuronal morphology. To examine the function of endogenous Sdc2 in dendrite morphogenesis, we knocked down endogenous Sdc2 with a Sdc2-specific small hairpin RNA (shRNA) construct (Lin et al., 2007) for 4 d starting at 13 DIV. Although the primary dendrite number was not changed by knockdown of Sdc2, the total dendrite length was reduced (Fig. 1 A). Previous studies had demonstrated that overexpression of Sdc2 at 1–2 DIV, when endogenous Sdc2 was undetectable, accelerated the formation of dendritic filopodia and spines (Lin et al., 2007). To study the gain-of-function phenotype of Sdc2 on dendrite outgrowth, Sdc2 was again overexpressed in cultured hippocampal neurons at 1–2 DIV. Compared with vector control, Sdc2 overexpression increased the total length of dendrites as well as the number of primary dendrites (Fig. 1 B). Collectively, these observations suggest that Sdc2 not only regulates spine formation but also plays a role in dendritic arbor morphogenesis.
Figure 1.
Sdc2 regulates dendritic arborization. (A) Knockdown of Sdc-2 in mature neurons reduces dendritic arborization. At 13 DIV, cultured hippocampal neurons were transfected with either an Sdc2 shRNA (Sdc2i) or a vector control (pSuper.neo+GFP, abbreviated as pSuper). Dendritic arbors were analyzed at 17 DIV. Because pSuper co-expresses the GFP–neo fusion and the shRNA, GFP signals were used to outline cell morphology. (B) Sdc2 expression induces dendrite outgrowth in young neurons. Sdc2 and GFP were cotransfected into hippocampal neurons at 2 DIV, and cell morphology was analyzed at 5 DIV. Only GFP signal is shown here. The number of primary dendrites and the total dendritic length of transfected neurons were measured in three independent experiments. Equal numbers of transfected neurons were analyzed. Mean values ± SEM are shown (error bars). **, P < 0.005; ***, P < 0.0005. Bars, 30 µm.
Sarm1 interacts with Sdc2
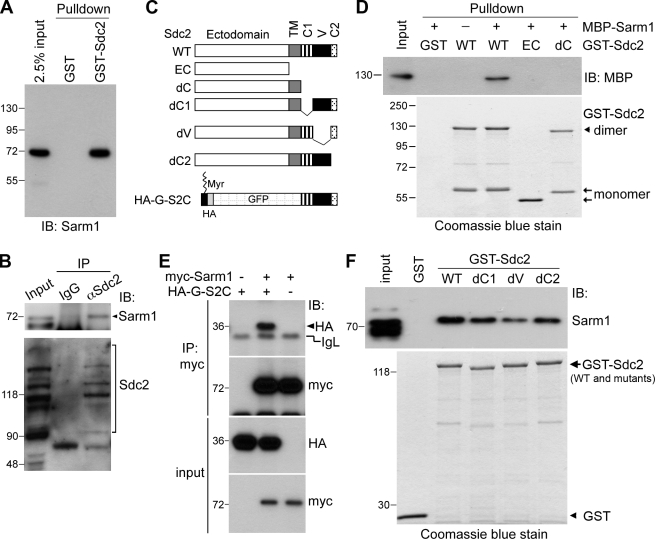
Although the Ena/VASP proteins act downstream of Sdc2 in dendritic filopodia formation (Lin et al., 2007), inhibition of Ena/VASP activity did not impair Sdc2-induced dendritic outgrowth (unpublished data). Therefore, we set out to identify the factors that function downstream of Sdc2 in dendritic arborization by searching for novel Sdc2-interacting proteins. In addition to known Sdc2-interacting proteins such as neurofibromin and CASK (Fig. S1 A), our GST–Sdc2 fusion protein also precipitated an ∼80-kD protein species from postnatal day 21 (P21) mouse brain extracts that was not present in the precipitate of GST alone or GST–Sdc2 fusion protein without incubation with brain extracts (Fig. S1 B). Analysis by mass spectrometry identified this protein species as Sarm1 (Fig. S1 C).
To validate the interaction between Sdc2 and Sarm1, specific rabbit polyclonal and mouse monoclonal antibodies against Sarm1 were generated (Fig. S1, D and E). An immunoblot of the GST pull-down products from P21 mouse brain extracts indicated that Sarm1 does associate with GST–Sdc2 (Fig. 2 A). This interaction is specific because GST alone did not precipitate Sarm1 from the mouse brain (Fig. 2 A). The in vivo interaction between Sarm1 and Sdc2 was confirmed by coimmunoprecipitation of Sarm1 and Sdc2 from P21 mouse brain using Sdc2 antibody (Fig. 2 B). Control IgG precipitated neither Sarm1 nor Sdc2 (Fig. 2 B). Sdc2 appeared as a ladder of protein species on the SDS-PAGE gel because of the heterogeneity of heparan sulfate modification (Fig. 2 B, bottom). Some of the Sdc2 immunoreactivities in the input lane were not enriched in the precipitate of Sdc2 antibody. This result may be caused by a cross-reactivity of the Sdc2 antibody in immunoblotting. Alternatively, the accessibility of Sdc2 antibody to variant native Sdc2 proteins in brain extracts could differ because of the different heparan sulfate conjugation sites and various glycosylation levels. Some of the variants might therefore not be enriched by the Sdc2 antibody.
Figure 2.
Sarm1 interacts with the cytoplasmic domain of Sdc2. (A) GST–Sdc2 pull-down assay with extract prepared from P21 mouse brain and immunoblotted (IB) with Sarm1 antibody. GST alone was used as a control. (B) Co-immunoprecipitation of Sdc2 and Sarm1 from mouse brain. Sdc2 antibodies and nonimmune rabbit IgG were used in the immunoprecipitation. The precipitates were immunoblotted with Sarm1 and Sdc2 antibodies as indicated. The arrowhead indicates the position of Sarm1. Because of the heterogeneity of glycosylation, SDS-PAGE resulted in a ladder of Sdc2-immunoreactive bands. (C) Schematic of Sdc2 constructs. TM, transmembrane domain; C1, conserved region 1; V, variable region; C2, conserved region 2; Myr, myristoylation modification. (D) Sdc2 directly interacts with Sarm1. Sdc2-GST fusion proteins were used to pull down purified MBP-Sarm1 fusion protein in the presence of glutathione agarose. The presence of Sarm1 in the precipitate was examined by immunoblotting with MBP antibody. The relative amounts of GST fusions used in the pull-down assay are indicated by Coomassie blue staining. Sdc2 forms an SDS-resistant dimer through its transmembrane domain. The Sdc2 dimer and monomer are marked by the arrowhead and arrows, respectively. (E) The cytoplasmic domain of Sdc2 is sufficient for the interaction with Sarm1. HEK293T cells were cotransfected with HA-G-S2C, Myc-tagged Sarm1, and vector control, as indicated. Immunoprecipitation was performed using a Myc-tag antibody. The precipitates were then analyzed by immunoblotting with Myc and HA antibodies. The positions of HA-G-S2C and immunoglobulin light chain (IgL) are indicated. (F) Three regions (C1, V, and C2) are involved in the interaction with Sarm1. Various GST–Sdc2 fusion proteins were used to pull down Sarm1 from mouse extract. The results were immunoblotted with Sarm1 antibody. Coomassie blue stain indicated the relative amounts of GST fusions used in the pull-down assay. Molecular mass standards (kD) are indicated next to the gel blots.
The direct interaction between Sdc2 and Sarm1 was further interrogated with an in vitro binding assay using purified maltose-binding protein (MBP)-Sarm1 and various Sdc2 fusion proteins (Fig. 2 C). Glutathione agarose precipitated MBP-Sarm1 in the presence of wild-type GST–Sdc2 (Fig. 2 D). GST fusions containing the ectodomain of Sdc2 alone (EC) or containing an Sdc2 mutant lacking the cytoplasmic tail (dC) lost the ability to precipitate MBP-Sarm1 from the solution (Fig. 2 D), which indicates a direct interaction between Sarm1 and the cytoplasmic domain of Sdc2. To confirm this interaction, Myc-tagged Sarm1 was cotransfected into HEK293T cells with a GFP fusion protein containing the cytoplasmic domain of Sdc2 (HA-G-S2C). Coimmunoprecipitation revealed that the Myc tag antibody precipitated Sarm1 proteins as well as HA-G-S2C from cell extracts (Fig. 2 E). In the absence of Myc-tagged Sarm1, the Myc tag antibody did not precipitate HA-G-S2C (Fig. 2 E), which suggests the specificity of immunoprecipitation. Collectively, these data support the conclusion that the cytoplasmic domain of Sdc2 interacts with Sarm1.
To further define the region of Sdc2 that interacts with Sarm1, mutants specifically lacking the C1, V, or C2 regions were generated (Fig. 2 C). Among these mutants, the mutant missing the V region (dV) exhibited the weakest interaction with MBP-Sarm1 (Fig. 2 F). Although Sarm1 still interacted with the dC1 and dC2 mutants, these interactions were weaker than that of wild-type GST–Sdc2 (Fig. 2 F). These observations suggest that all three regions of Sdc2 contribute to Sarm1 binding, although the V region appears to be most critical for this interaction.
Distribution of Sarm1 in mouse brain and neurons
Previous studies have indicated that Sarm1 mRNA is mainly expressed in brain (Kim et al., 2007; Szretter et al., 2009), an observation we confirmed with Sarm1 protein in the brains of 2-mo-old mice (Fig. 3 A). Sarm1 protein was widely distributed in various brain regions, as revealed by immunoblotting (Fig. 3 B) and indirect immunofluorescence staining (Fig. 3 C). During development, Sarm1 protein was expressed at a higher level from embryonic day 18 (E18) to P14 in the mouse brain (Fig. 3 D).
Figure 3.
Sarm1 is widely expressed in rodent brain and neurons. (A) Immunoblot of Sarm1 in different mouse organs. GAPDH is used as an internal control. (B) Regional distribution of Sarm1 in mouse brain. Cx, cerebral cortex; Hi, hippocampus; St, striatum; Th, thalamus; Cb, cerebellum; BS, brain stem. α-Tubulin was used as an internal control. (C) Staining patterns of Sarm1 in mouse brain. The top right shows the merged image of the MAP2/Sarm1 double stain in the CA1 region of the hippocampus. The top left and bottom panels depict the Sarm1 patterns in brain regions including layer five of the somatosensory cortex (Cx), the posterior thalamic nuclear group (Th), and the caudate putamen of the striatum (St). 2-mo-old mice were used in A–C. (D) Developmental expression profile of Sarm1. The plotted relative Sarm1 protein expression levels were obtained by normalization to the corresponding α-tubulin protein amounts. The results are the means of three independent experiments. Error bars indicate SEM. (E) Distribution of Sarm1 protein in biochemical subcellular fractions of adult mouse brain. H, total homogenate; P1, nuclei and cell debris; S1, supernatant of P1; P2, crude synaptosomal fraction; S2, supernatant of P2; LP1, lysed synaptosomal membrane; LS1, supernatant of LP1; P3, light membrane fraction; S3, soluble cytosolic fraction. PSD-95 enriched in the P2 and LP1 fractions was used as a quality control of fraction preparation. Molecular mass standards (kD) are indicated next to the gel blots. (F) Distribution of PSD-95 (red) and Sarm1 (green) in cultured hippocampal neurons at 21 DIV. Representative high-magnification images are shown on the top right. Arrowheads indicate the Sarm1 puncta overlapping with PSD-95; arrows indicate the Sarm1 puncta adjacent to PSD-95 puncta. The percentage of overlapped Sarm1 and PSD-95 is shown on the bottom right. The original images and the overlays shifted for 1 and 1.66 µm were analyzed. Error bars indicate mean values ± SEM. **, P < 0.01; ***, P < 0.005. Bars: (C) 30 µm; (F, left) 20 µm (F, top right) 2 µm.
At the subcellular level, Sarm1 proteins were widely distributed in different regions, including the light membrane fraction, the lysed synaptosomal membrane fraction, and the crude synaptic cytoplasmic fractions (Fig. 3 E). Immunofluorescence staining also revealed a wide distribution of Sarm1 protein (Fig. 3, C and F) with a punctate pattern in soma and dendrites. Compared with PSD-95, a postsynaptic density marker, the Sarm1 puncta were smaller in size and widely distributed along dendrites (Fig. 3 F, left). However, some Sarm1 puncta colocalized with PSD-95 (Fig. 3 F, top right, arrowheads), whereas others were surrounded by or adjacent to PSD-95 (Fig. 3 F, top right, arrows). The correlation coefficient of PSD-95 and Sarm1 colocalization was 0.26. Although the colocalization coefficient was not high, shifting the image overlap by 1 or 1.66 µm significantly reduced the colocalization coefficient to 0.18 or 0.15, respectively (Fig. 3 F, bottom right; original vs. 1-µm shift, P = 0.0096; original vs. 1.66-µm shift, P = 0.0012). These analyses suggest that Sarm1 distributes along dendrites and that a fraction of Sarm1 is present at synapses.
Sarm1 is critical for maintenance of the dendritic arbor
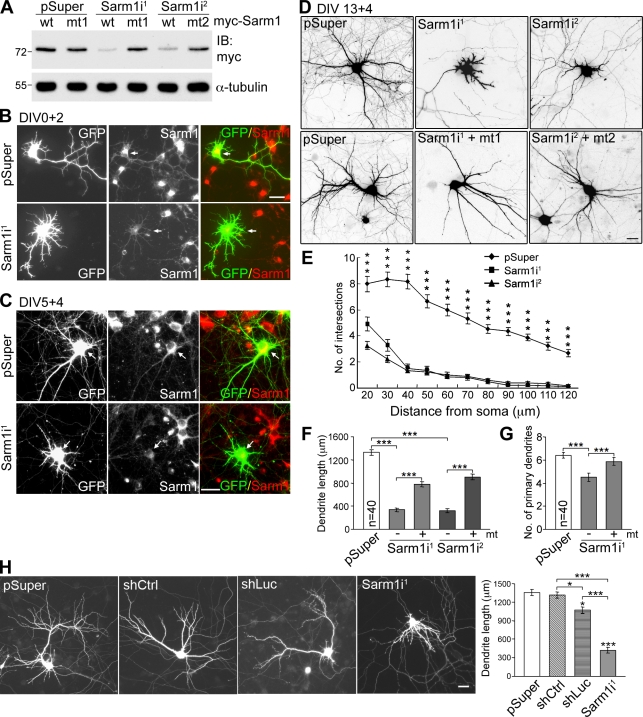
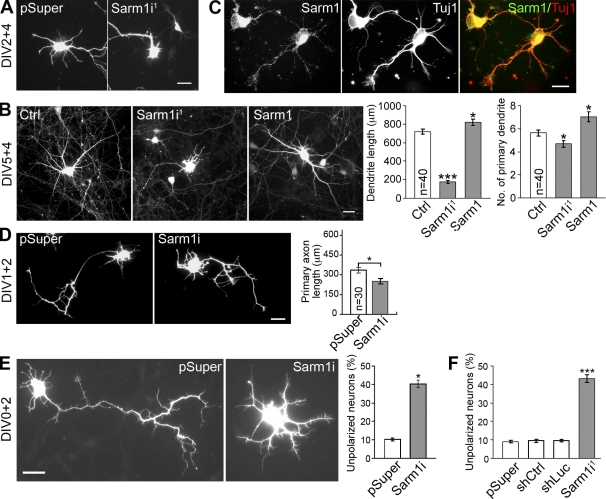
To investigate the biological function of Sarm1 in neurons, two Sarm1 shRNAs—Sarm1i1 and Sarm1i2—were used to knock down Sarm1 expression. The knockdown efficiency was first examined in COS-1 cells, where both Sarm1i1 and Sarm1i2 effectively reduced exogenous Sarm1 expression (Fig. 4 A). In cultured hippocampal neurons, immunofluorescence staining also revealed an obvious reduction in endogenous Sarm1 protein levels in Sarm1 shRNA-expressing cells. After expression for 2 (Fig. 4 B) or 4 d (Fig. 4 C), Sarm1i1 reduced the Sarm1 protein levels in cultured hippocampal neurons (not depicted for Sarm1i2). Compared with the pSuper vector control, Sarm1 knockdown neurons exhibited a dramatic reduction in the dendritic arbor (Fig. 4 D). We detected remarkably reduced dendrite length (Fig. 4 F) and dendrite number (Fig. 4 G) that led to a large reduction in the number of dendritic intersections, as revealed by Sholl analysis (Fig. 4 E). Expression of Sarm1i-resistant silent mutant constructs (Fig. 4 A) rescued the effect of the Sarm1 shRNAs to some extent (Fig. 4, D, F, and G). Possible causes for the partial rescues by these Sarm1 mutants are presented in the Discussion section.
Figure 4.
Sarm1 is critical for dendritic arborization in cultured hippocampal neurons. (A) Sarm1 knockdown in COS-1 cells via cotransfection of Sarm1i1, Sarm1i2, or pSuper control and Myc-tagged wild-type Sarm1 or specific silent mutants resistant to Sarm1i1 and Sarm1i2 (mt1 and mt2). Immunoblotting was performed with Myc tag and α-tubulin antibodies. Molecular mass standards (kD) are indicated next to the gel blots. (B and C) Knockdown of Sarm1 in cultured hippocampal neurons. Neurons were transfected with the indicated plasmids at 0 (B) or 5 DIV (C) and immunostained with Sarm1 and GFP antibodies at 2 (B) or 9 DIV (C). Arrows point to transfected neurons. (D) Sarm1 knockdown affects dendritic arbors. At 13 DIV, cultured hippocampal neurons were transfected using the plasmids indicated. Neuronal morphology was monitored by GFP signals at 17 DIV. (E) Sholl analysis of the effect of Sarm1 knockdown. ***, P < 0.001. (F) Total dendrite length. (G) The number of primary dendrites. ***, P < 0.0005. (H) The specificity of the effect of Sarm1 knockdown on dendrite outgrowth was confirmed by two additional controls. The experiment was performed as described in D, except that both shCtrl and shLuc were also included. Representative images and analysis of total dendrite length are shown. Equal numbers of transfected neurons, as indicated, were analyzed. Means ± SEM are shown (error bars). Experiments were performed in triplicate. Bars, 30 µm.
To further confirm the specificity of Sarm1i on dendritic arborization, we included two additional controls besides pSuper. One is a nonsilencing control (shCtrl) that expresses an shRNA sharing no significant sequence homology with mammalian genomes. The total dendritic length of neurons transfected with shCtrl was comparable with that of neurons transfected with vector alone (Fig. 4 H). The other is shLuc, which expresses an shRNA against the luciferase gene. Alvarez et al. (2006) found that the expression of shLuc activates the interferon pathway and reduces the number of dendritic branches and the number and size of dendritic spines in cultured brain slices (Alvarez et al., 2006). In cultured hippocampal neurons, shLuc also reduced the total dendrite length compared with either vector control or shCtrl (Fig. 4 H). However, Sarm1i1 had a much stronger effect than shLuc (Fig. 4 H), which supports a specific role of Sarm1 in dendritic arbor formation.
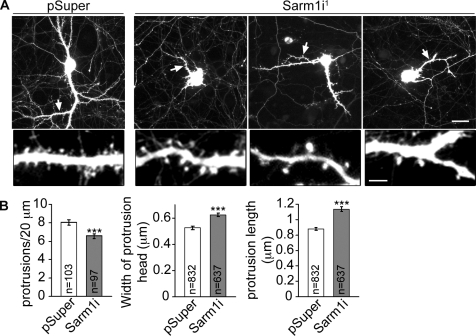
Sarm1 knockdown also influenced the number and morphology of dendritic spines in mature neurons. In Sarm1 knockdown neurons, mushroom-shaped or stubby dendritic spines were still present, but with lower density along dendrites (Fig. 5, A and B). We noticed that the remaining dendritic spines in Sarm1 knockdown neurons are longer and that their spine heads are larger compared with those in neurons transfected with vector control (Fig. 5, A and B). These data suggest that Sarm1 also influences the density and morphology of dendritic spines.
Figure 5.
Sarm1 knockdown influences spine morphology and density in mature neurons. (A) Dendritic spine morphology. At 13 DIV, cultured hippocampal neurons were transfected with Sarm1i1 or pSuper vector control. 4 d later, cells were fixed and stained with GFP antibodies. A region indicated by the arrows in the whole cell image is shown enlarged in on the bottom. Bars: (top) 30 µm; (bottom) 5 µm. (B) Quantification of the spine density, the width of the spine head, and the length of spines. Error bars indicate mean values ± SEM. ***, P < 0.0005.
Knockdown of Sarm1 reverses the ratio of dendrite growth rate to pruning rate
Time-lapse experiments were then performed to further investigate the effect of Sarm1 on dendritic arborization. Primary dendrites were relatively stable in control neurons (Fig. 6 A, top; and Video 1), and the total dendrite lengths of the control neurons were maintained almost constantly (Fig. 6 C). In contrast, the dendritic arbors of Sarm1 knockdown neurons gradually became smaller during the recording period (Fig. 6 A, bottom; and Video 2), and Sarm1 knockdown shortened the total dendritic lengths (Fig. 6 C). In Sarm1 knockdown neurons, some dendrites were constantly pruned back (Fig. 6 B, arrowheads); another population of dendrites tried to extend but were eventually pruned back (Fig. 6 B, arrows). The residual structures from retracting dendrites persisted as long and thin filopodium-like structures at the tips of the retracting dendrites (Video 3). Some dendrites extended back into these residual structures and then withdrew again. We also measured the dendrite growth and pruning rates of individual neurons. In control neurons, the dendrite growth rate was consistently higher than the pruning rate (Fig. 6 D), but the dendrite pruning rate was consistently higher than the growth rate in Sarm1 knockdown neurons (Fig. 6 D). Therefore, the ratio of growth rate to pruning rate was lower in Sarm1 knockdown neurons (Fig. 6 E). Collectively, these time-lapse analyses provide evidence for a critical role of Sarm1 in maintaining dendritic arbors by controlling dendritic growth and pruning rates.
Figure 6.
Time-lapse study of the effect of Sarm1 knockdown on dendrite morphology. Cultured hippocampal neurons were transfected with Sarm1i1 or pSuper at 13 DIV. The images were recorded 10 min/frame for 24 h starting from 15 DIV. (A) Representative images of a control neuron (pSuper; top) and a Sarm1 knockdown neuron (Sarm1i; bottom). Bar, 30 µm. (B) Enlarged local images of neurons transfected with Sarm1i1. Arrowheads indicate a continuously retracting dendrite, whereas the arrows denote a dendrite that outgrew first and then retracted. Bar, 5 µm. (C) The relative dendrite length of individual neurons during the recording period. (D) The growth and pruning rates of individual neurons; the rates of individual neurons are connected with a line. (E) The means of the ratio of growth rate to pruning rate (n = 5). Error bars indicate mean values ± SEM. **, P < 0.005.
Sarm1 also controls dendrite initiation and elongation
We also examined whether Sarm1 regulates initiation and elongation of neuronal dendrite outgrowth. Sarm1i1 was transfected into cultured hippocampal neurons at 2 or 5 DIV, and neuronal morphology was monitored 3–4 d later. Similar to the data obtained from cultures ∼2 wk old (Fig. 4, D–G), knockdown of Sarm1 in younger neurons also resulted in shorter total dendrite length and fewer primary dendrites (Fig. 7, A and B; and Fig. 8). In contrast, overexpression of Sarm1 at 5 DIV led to an increase in the total dendrite length and dendrite number at 9 DIV (Fig. 7 B). These observations suggest that Sarm1 contributes to the initiation and elongation of dendritic growth.
Figure 7.
Sarm1 regulates development of dendrites and axons. Cultured hippocampal neurons were transfected with vector control, Sarm1i1, or Sarm1 at 2 (A), 5 (B), 1 (D), or 0 DIV (E), and harvested for immunostaining at 6 (A), 9 (B), 3 (D), or 2 (E) DIV. (A) Sarm1 regulates dendritic initiation. (B) Sarm1 contributes to dendrite elongation, as indicated by the total dendrite length and primary dendrite number. (C) Sarm1 is expressed in neurites. At DIV 2, cultured hippocampal neurons were immunostained with Sarm1 (viewed by Alexa Fluor 488) and Tuj1 (viewed by Alexa Fluor 555) antibodies. (D) Sarm1 knockdown shortens the length of the longest neurites. (E and F) Sarm1 regulates neuronal polarity. 4 h after plating, cultured hippocampal neurons were transfected with Sarm1i1 or vector control (E) or Sarm1i1, shCtrl, shLuc, or pSuper vector control (F), and the percentage of neurons that did not develop a dominant neurite (axon) was determined (see Materials and methods for details). Mean values ± SEM (error bars) are shown from three independent experiments in which equal numbers of transfected neurons were analyzed. Bars: (A, B, D, and E) 30 µm; (C) 20 µm. *, P < 0.05; ***, P < 0.0005.
Figure 8.
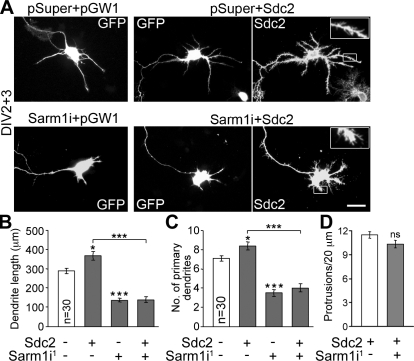
Sdc2 regulates dendrite outgrowth through Sarm1. (A) Cultured hippocampal neurons were transfected with the indicated plasmids at 2 DIV and then fixed for immunostaining with GFP and Sdc2 antibodies at 5 DIV. pSuper and pGW1 are the vectors for RNAi knockdown and gene expression, respectively. Insets (enlarged from the boxed regions) show the filopodia induced by Sdc2. Bar, 30 µm. (B) Total dendrite lengths. (C) Number of primary dendrites. (D) Density of dendritic filopodia. Because the vector control and Sarm1i1 alone did not obviously induce filopodia formation, only neurons transfected with Sdc2 alone or both Sdc2 and Sarm1i1 were subjected to quantitative analysis of filopodia. Thirty transfected neurons were collected randomly from each group. The experiments were repeated three times; data represent mean values ± SEM (error bar). *, P < 0.05; ***, P < 0.0005.
Sarm1 regulates axonal outgrowth and neuronal polarity
Significant Sarm1 signals were found along axons and at the growth cone in young neurons (Fig. 7 C). At 3 DIV, the lengths of the longest neurites, presumably the axons, of neurons transfected with Sarm1i1 at 1 DIV were measured and found to be shorter than the Sarm1 knockdown neurons (Fig. 7 D), which suggested that Sarm1 is also required for axon outgrowth.
Sarm1 knockdown also influenced the initiation of axon development. 4 h after plating, we transfected the Sarm1i1 construct and vector control into cultured hippocampal neurons and examined neuronal morphology at 2 DIV. Approximately 40% of Sarm1 knockdown neurons failed to establish neuronal polarity at 2 DIV; in contrast, only ∼10% of control neurons were still unpolarized (Fig. 7 E), which suggests that Sarm1 is also involved in the establishment of neuronal polarity.
To further confirm that Sarm1 influences neuronal polarity, we included shCtrl and shLuc in the experiment. The results showed that, unlike Sarm1i1, neither shLuc nor shCtrl resulted in an increase in the percentage of unpolarized neurons of cultured hippocampal neurons (Fig. 7 F), which supports the specificity of Sarm1i1 on impairment of neuronal polarization.
Sarm1 acts downstream of Sdc2
The preceding experiments provided evidence that Sarm1 plays multiple roles in dendritic and axonal outgrowth, that Sdc2 participates in dendritic arborization, and that Sdc2 interacts with Sarm1. We next addressed our original question of whether Sdc2 regulates dendritic arborization through Sarm1. Sarm1i1 was cotransfected with Sdc2 into cultured hippocampal neurons at 2 DIV (Fig. 8 A). At 5 DIV, compared with transfection of Sdc2 alone, knockdown of Sarm1 obviously reduced the total dendrite length and the number of primary dendrites induced by Sdc2 overexpression (Fig. 8, B and C), which suggested that Sarm1 mediates the effect of Sdc2 on dendritic arborization. The dendrite lengths of Sarm1 knockdown neurons were also shorter than those of control neurons (Fig. 8, B and C), which suggests that in addition to Sdc2, Sarm1 can also receive signal from other upstream factors to regulate dendritic arbor formation. The filopodia induced by Sdc2 overexpression appeared to be unaffected in Sarm1 knockdown neurons (Fig. 8, A and D). These observations support the hypothesis that Sarm1 acts downstream of Sdc2 to regulate dendritic arborization but not dendritic filopodia formation. Because Sdc2-induced filopodia are the precursors of dendritic spines (Lin et al., 2007), Sarm1 is likely not involved in the initiation of dendritic spine formation.
Sarm1 regulates microtubule stability
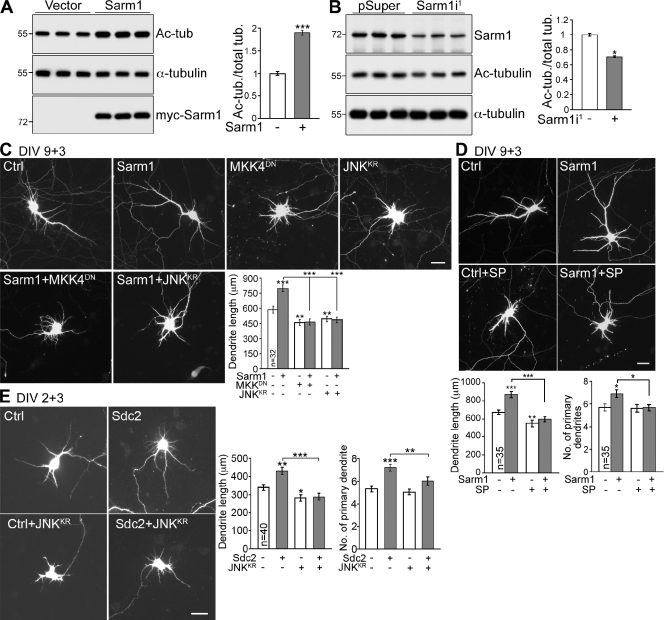
As microtubule dynamics has been implicated as critical for neurite outgrowth and differentiation (Witte et al., 2008; Poulain and Sobel, 2010; Stone et al., 2010), we wondered whether Sarm1 influences microtubule stability to control dendrite and axon outgrowth. In COS-1 cells, Sarm1 overexpression increased tubulin acetylation nearly twofold compared with vector control (Fig. 9 A). In mouse neuroblastoma Neuro-2A cells expressing endogenous Sarm1 proteins, tubulin acetylation was reduced when endogenous Sarm1 was knocked down (Fig. 9 B). Collectively, these analyses suggest that Sarm1 participates in microtubule stabilization in cells.
Figure 9.
The JNK pathway and microtubule stability are downstream of Sarm1 and Sdc2. (A) Sarm1 overexpression increases the acetylation levels of tubulin. COS-1 cells were transfected with vector control or Sarm1 and were immunoblotted with acetyl-tubulin and α-tubulin antibodies. The levels of tubulin acetylation were normalized to total α-tubulin. (B) Sarm1 knockdown reduces tubulin acetylation in mouse neuroblastoma Neuro-2A cells. 2 d after transfection, the acetylation levels of tubulin were determined by immunoblotting. (C) A JNK kinase dead mutant (JNKKR) and an MKK4 dominant-negative mutant (MKK4DN) suppress the effect of Sarm1 overexpression in dendritic arborization. (D) Sarm1-expressing neurons treated with JNK inhibitor SP600125 (SP) have shorter dendrites compared with Sarm1-expressing cells. In C and D, transfection was performed at 9 DIV and cells were harvested for immunostaining at 12 DIV. (E) Co-expression of JNKKR abolishes Sdc2–induced dendrite outgrowth. At 2 DIV, cultured hippocampal neurons were transfected with the indicated constructs, then fixed for immunostaining at 5 DIV. Three independent experiments were performed. Equal numbers of transfected neurons were analyzed for each experiment. The data represent mean values ± SEM (error bars). *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Bars, 30 µm.
Sdc2 and Sarm1 regulate dendritic arborization through the MKK4–JNK pathway
In C. elegans, Sarm1 regulates the ASK1–MKK–JNK pathway to control expression of olfactory receptors (Chuang and Bargmann, 2005). In addition to phosphorylating transcription factors, JNK can phosphorylate microtubule-associated proteins and affect microtubule stability and dendrite morphology in mammals (Chang et al., 2003; Björkblom et al., 2005; Rosso et al., 2005; Tararuk et al., 2006; Podkowa et al., 2010). We therefore wondered whether Sarm1 regulates neuronal morphology through the JNK pathway. To examine this possibility, the JNK inhibitor SP600125, a JNK kinase dead mutant (JNKKR), and a microtubule-associated protein kinase kinase-4 (MKK4) dominant-negative mutant (MKK4DN) were included in our experiments. The total dendrite length induced by Sarm1 overexpression was shortened by all of these treatments (Fig. 9, C and D), which indicates that Sarm1 controls dendritic arborization through the MKK4–JNK pathway. Consistent with a role of Sarm1 in Sdc2–induced dendritogenesis, overexpression of the JNKKR mutant also abolished Sdc2–induced dendrite outgrowth (Fig. 9 E). Collectively, these observations demonstrate that Sdc2 and Sarm1 regulate dendrite morphogenesis via the JNK pathway.
Sarm1 regulates dendritic arborization in vivo
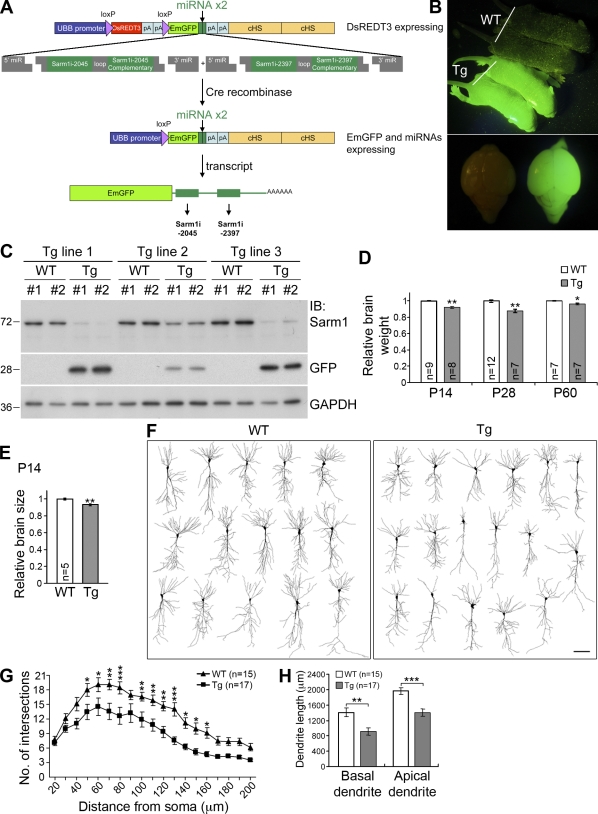
To confirm the function of Sarm1 in vivo, Sarm1 miRNA knockdown transgenic mice were generated (Fig. 10 A). In brief, two miRNA sequences directed against Sarm1 expression were inserted into the 3′ untranslated region of the emGFP gene, allowing use of the emGFP signals as indicators of expression of the artificial miRNAs (Fig. 10, B and C). Three independent transgenic lines were examined. Compared with the Sarm1 protein levels measured in wild-type littermates, Sarm1 levels were lower in all three transgenic lines, especially in transgenic lines 1 and 3 (Fig. 10 C). Accordingly, the GFP expression levels were higher in those two lines than in line 2 (Fig. 10 C). Although the gross morphological characteristics of the Sarm1 knockdown brains were comparable to those of wild-type littermates (Fig. S2 A), the total brain weights of Sarm1 knockdown mice were 5–10% lighter than those of wild-type littermates at the ages of P14, P28, and P60 (Fig. 10 D). Measurement of brain size by summing the total area of brain sections also indicated that Sarm1 knockdown brains were ∼10% smaller than those of wild-type littermates at P14 (Fig. 10 E). In some mice, Sarm1 knockdown resulted in smaller hippocampal volumes (Fig. S2 B). To investigate whether the dendritic arbors were affected in Sarm1 knockdown mice, we used Golgi-Cox staining to analyze neuronal morphology. Dendritic arbors of CA1 neurons were less complex in Sarm1 knockdown mice (Fig. 10 F), as reflected by a decrease in the number of dendritic intersections in the Sholl analysis (Fig. 10 G) and in the length of basal and apical dendrites (Fig. 10 H). Thus, these analyses provide clear evidence that Sarm1 regulates dendritic arborization in vivo.
Figure 10.
Reduction of Sarm1 in vivo impairs dendritic arborization. (A) Strategy used to generate Sarm1 knockdown mice; details are described in Materials and methods. (B) GFP expression in P5 Sarm1 knockdown transgenic (Tg) mice. (C) Reduction of Sarm1 protein levels in three independent Tg mouse lines. Immunoblotting was used to determine the protein levels of Sarm1 and GFP in the extracts containing forebrain and subcortical regions. For each line, two GFP-positive Tg and two wild-type (WT) littermates were examined. GAPDH was used as an internal control. Molecular mass standards (kD) are indicated next to the gel blots. (D) The relative brain weights of Sarm1 knockdown mice and WT littermates. P14, P28, and P60 mice were analyzed. *, P < 0.05; **, P < 0.005. (E) Total brain area of P14 mice. Sums of the total areas of a series of brain sections were determined to compare the brain size of Sarm1 knockdown mice and WT littermates. **, P < 0.005. (F) Cell morphology of CA1 neurons. 2-mo-old mice were used for Golgi-Cox staining. Camera lucida was then performed to examine neuronal morphology in the brain. Bar, 50 µm. (G) Sholl analysis of CA1 neurons. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (H) Total length of basal and apical dendrites of CA1 neurons. **, P < 0.005; ***, P < 0.0005. The data represent mean values ± SEM (error bars).
Discussion
The observations we describe here suggest multiple roles of Sarm1 in neuronal morphogenesis. Sarm1 contributes to the initiation, elongation, and maintenance of dendritic arbors, and influences axonal outgrowth and neuronal polarization. The MKK4–JNK pathway mediates the functions of Sarm1 in neuronal morphogenesis. Our investigation also demonstrates that Sdc2, a synaptic heparan sulfate proteoglycan, is one of the upstream effectors of Sarm1. Sdc2 may serve as a docking site for Sarm1 at synapses, after which Sarm1 may receive local signals and transduce the signals to the MKK–JNK pathway, thus maintaining the dendritic arbors of mature neurons.
In this study, we used two shRNA constructs to knock down Sarm1 expression in cultured hippocampal neurons. Both constructs efficiently reduced Sarm1 expression in cells. Although it is not clear why the rescue effect on dendritic arborization was only partial, splicing variants may have contributed to this phenomenon. A single Sarm1 gene encodes at least three different protein products (UniProtKB/Swiss-Prot; http://www.uniprot.org/uniprot/Q6PDS3). Although Sarm1i1 and Sarm1i2 target all of the splicing variants, in the rescue experiments, only isoform 1 was cotransfected with the Sarm1i constructs. Other splicing variants may also contribute to the function of Sarm1 in dendritic outgrowth, leading to the partial rescue by isoform 1. Another possibility is that dendritic arborization may be sensitive to Sarm1 protein levels; because it is very difficult to control exogenous Sarm1 protein levels, these levels may still be too low to fully rescue Sarm1 function in dendritic arborization.
Sarm1, the JNK pathway, and microtubule stability
Similar to its function in C. elegans (Chuang and Bargmann, 2005), Sarm1 also delivers signals to the MKK4–JNK pathway in mammalian neurons. Multiple JNK substrates have been identified, including the microtubule-associated proteins MAP1 and MAP2 and the transcription factors c-Jun and ATF2. Phosphorylation of MAP1 and MAP2 by JNK has been suggested to play a role in microtubule dynamics and dendrite outgrowth (Teng et al., 2001; Chang et al., 2003; Björkblom et al., 2005; Rosso et al., 2005; Tararuk et al., 2006; Podkowa et al., 2010). We also found that Sarm1 influences microtubule stability, and that it is possible that Sarm1 regulates local microtubule stability through the JNK pathway.
Sarm1 also affects axonal outgrowth and neuronal polarity. These effects may also be caused by JNK activity and microtubule stability. JNK and its interacting protein c-Jun N-terminal kinase-interacting protein-1 have been shown to control axon development (Dajas-Bailador et al., 2008; Barnat et al., 2010; Stone et al., 2010). Microtubule stabilization specifies neuronal polarity (Witte et al., 2008). In the axon, microtubule stability (indicated by tubulin acetylation levels) is higher than that in dendrites. Treatment with the microtubule stabilization reagent Taxol induces multiple axon formation (Witte et al., 2008). Because Sarm1 increases microtubule stability, it may therefore contribute to the establishment of neuronal polarity. In summary, JNK signaling and consequent microtubule stability may account for the multiple roles of Sarm1 in neuronal morphogenesis, although we cannot rule out the possibility that JNK may regulate gene expression through phosphorylation of transcription factors, thus having a long-term effect on neuronal morphology.
In mature neurons, Sarm1 knockdown reduces the spine density but enlarges the spine head and elongates the spines. However, in young neurons, Sarm1 does not obviously affect the density of Sdc2-induced filopodia, which suggests that Sarm1 is not involved in the initiation of spine formation. Thus far it is unclear how Sarm1 regulates spine density and morphology. Perhaps the JNK pathway is involved in this process. Alternatively, the changes in the dendritic spines may be indirectly caused by dendritic withdrawal. More investigations are required to address this issue.
Signals upstream of Sarm1
There are several clues suggesting that Sarm1 also receives signals from other molecules in addition to Sdc2. First, Sarm1 expression in mouse brain or cultured hippocampal neurons occurs earlier than Sdc2 expression (Fig. 2; Hsueh et al., 1998; Ethell and Yamaguchi, 1999). In young neurons (at 2 and 5 DIV in this study), Sarm1 still regulates dendrite outgrowth, a regulation that appears to be independent of Sdc2 because Sdc2 is not yet expressed in young neurons. Second, Sarm1 knockdown substantially shortens the total dendrite length and reduces the number of dendrites below baseline. Finally, Sarm1 also controls axon development, which is apparently also independent of Sdc2. Of the four mammalian syndecans, Sdc2 and Sdc3 are the two major syndecans expressed in neurons (Hsueh and Sheng, 1999). However, Sdc2 is highly concentrated at synapses, whereas Sdc3 is present along the axon (Hsueh and Sheng, 1999). We therefore suspect that the axonal function of Sarm1 is likely to be independent of Sdc2. However, it is currently unclear whether Sdc3 is relevant to the axonal function of Sarm1. More investigations will be necessary to identify the other signal molecules upstream of Sarm1.
Sdc2-interacting proteins
Although the cytoplasmic domain of Sdc2 contains only 32 amino acid residues, at least seven intracellular proteins, including Sarm1, are known to interact with Sdc2: syntenin, CASK, synbindin, synectin, neurofibromin, and ezrin (Grootjans et al., 1997; Cohen et al., 1998; Hsueh et al., 1998; Ethell et al., 2000; Gao et al., 2000; Hsueh et al., 2001; Granés et al., 2003). Syntenin, CASK, synbindin, and synectin recognize the C2 region of Sdc2, whereas neurofibromin and ezrin require the C1 region for the interaction. Here we have shown that the C1, V, and C2 regions of Sdc2 participate in the interaction with Sarm1, although the V region seems to be most critical. Because the cytoplasmic domain of Sdc2 is very short, it is unlikely that a single Sdc2 molecule binds all these molecules simultaneously. However, because Sdc2 forms a dimeric or perhaps multimeric structure through its transmembrane domain (Asundi and Carey, 1995; Volta et al., 2010), it is very likely that the Sdc2 complex binds more than one interacting protein at the same time. Thus, it is possible that Sdc2 can couple on the extracellular matrix to multiple intracellular cytoskeletal pathways (through CASK or ezrin) and signaling pathways (including neurofibromin–PKA and Sarm1–JNK) simultaneously. Of course, it is also possible that Sdc2 interacts with these binding partners in a different protein complex.
Sarm1 mouse genetic models
Compared with the effect of Sarm1 knockdown in cultured hippocampal neurons, the effect of Sarm1 knockdown on dendritic arborization in transgenic mice is moderate. The brain weights and sizes of Sarm1 knockdown transgenic mice are only ∼5–10% less than those of wild-type littermates. This weaker effect in the brain may be caused by in vivo compensation. In addition to the Sarm1 knockdown transgenic mice reported here, two Sarm1 knockout mouse models have previously been developed. Kim et al. (2007) deleted exons 2–7 of mouse Sarm1 and found that Sarm1 is required for stress-induced apoptosis of hippocampal neurons. Szretter et al. (2009) removed exons 1 and 2 of mouse Sarm1 and demonstrated that Sarm1 is involved in restricting West Nile virus replication in the brainstem. However, because the global brain anatomy of Sarm1 knockdown mice is only slightly different from that of wild-type littermates, those studies may have missed the morphological distinction in Sarm1 mutant mice.
Sarm1 and TLR signaling
Sarm1 was originally identified as a negative regulator of the adaptor protein TRIF that acts downstream of TLR3 and TLR4 in the innate immune response (Carty et al., 2006). Because it has been shown that TLR3 is a negative regulator of axon growth in dorsal root ganglion neurons (Cameron et al., 2007), it will be interesting to investigate whether Sarm1 also acts downstream of TLR3 in axon outgrowth. We are also intrigued by the notion that TLR3 may be involved in the regulation of dendrite outgrowth. Because defects in neurodevelopment may lead to neuropsychiatric dysfunction, another issue worth exploring is the involvement of Sarm1 in neurodevelopmental disorders related to innate immunity.
Materials and methods
Animals
All animal experiments were performed with the approval of the Academia Sinica Institutional Animal Care and Utilization Committee and in strict accordance with its guidelines and those of the Council of Agriculture Guidebook for the Care and Use of Laboratory Animals. Animals were housed in the animal facility of the Institute of Molecular Biology, Academia Sinica, under controlled temperature and humidity.
Antibodies and chemicals
Sarm1 polyclonal and monoclonal antibodies were generated using MBP-Sarm1 (amino acids 1–410) recombinant proteins to immunize rabbits and mice, respectively. The Sdc2 antibody was generated using a GST–syndecan-2 (residues 33–211) recombinant protein as antigen and affinity purified using protein A–Sepharose (Lin et al., 2007). The following commercial antibodies were used: rabbit polyclonal Sarm1 (GeneTex Inc.), rabbit polyclonal GFP (Invitrogen), rabbit polyclonal glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology, Inc.), rabbit polyclonal MAP2 (Millipore), mouse monoclonal MBP (AbD Serotec), mouse monoclonal β-actin (Sigma-Aldrich), mouse monoclonal α-tubulin (Sigma-Aldrich), mouse monoclonal PSD-95 (Millipore), mouse monoclonal acetyl-tubulin (Sigma-Aldrich), mouse monoclonal MAP2 (Sigma-Aldrich), mouse monoclonal Myc (Cell Signaling Technology), HRP-conjugated secondary antibodies (GE Healthcare), and Alexa Fluor 488– and Alexa Fluor 555–conjugated secondary antibodies (Invitrogen). SP600125 was obtained from EMD.
Plasmid construction
Sdc2 shRNA, eukaryotic expression, and GST fusion constructs have been described and characterized previously (Hsueh et al., 2001; Lin et al., 2007; Chao et al., 2008). In brief, a nucleotide sequence 5′-GCTTCAGGATTATATCCTA-3′ was used for construction of Sdc2 shRNA. Full-length rat Sdc2 was subcloned into vector pGW1-CMV for expression in mammalian cells. GST–Sdc2 fusion protein contains the amino acid residues from 33 to 211 of rat syndecan-2. The mouse Sarm1 IMAGE clone was ordered from Invitrogen and further subcloned into the vector pGW1-CMV with a Myc tag (Wang et al., 2004) for expression in mammalian cells and into vector pMAL-c2X (New England BioLabs) for expression in bacteria. Two Sarm1 shRNA constructs were cloned into the pSuper.neo+GFP (pSuper) vector: Sarm1i1, 5′-GAACTCCAGACAGACCTAG-3′; and Sarm1i2, 5′-GAACATTGTGCCCATCATT-3′. Nonsilencing control (shCtrl), 5′-CCTAAGGTTAAGTCGCCCT-3′, and luciferase shRNA (shLuc), 5′-CGTACGCGGAATACTTCGA-3′, were also cloned into pSuper vector (Sarbassov et al., 2005; Alvarez et al., 2006). Myc-tagged Sarm1 mutants resistant to Sarm1i1 and Sarm1i2 were generated by site-directed mutagenesis with oligonucleotides carrying the following mutations: for Sarm1i1, 5′-GAACTCCAAACGGACCTAG-3′; for Sarm1i2, 5′-GAACATTGTCCCGATCATT-3′ (mutated residues are underlined). The JNKKR and MKK4DN constructs were gifts from M.-Z. Lai (Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan).
Cell culture, transfection, and drug treatment
HEK293T and COS-1 cells were grown in culture dishes in DME/10% FBS at 37°C and 5% CO2. For Neuro-2A, the culture medium was MEM/10% FBS/1 mM sodium pyruvate. Cells were transfected with the indicated plasmids with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Hippocampal neurons from E18–19 rat embryos were cultured and transfected via the calcium phosphate method as described previously (Lin et al., 2007). In addition to the indicated constructs, a GFP construct was cotransfected into neurons to outline cellular morphology. 20 µM SP600125 was added 24 h after transfection, and treatment was performed for 48 h.
Pull-down assay, mass spectrometry, immunoprecipitation, and in vitro binding assay
P21 mouse brains were lysed and homogenized in lysis buffer (PBS, pH 7.4, 1% Triton X-100, 1 mM PMSF, and 1 µg/ml each of aproptitin, leupeptin, and pepstatin). For the pull-down assay, 2 µg GST or GST–Sdc2 recombinant proteins were incubated with mouse brain lysate for 3 h. After washing, pull-down complexes were extracted with SDS sample buffer, boiled, and resolved by SDS-PAGE. Gels were then stained with Sypro-Ruby (Invitrogen). Bands of interest were excised from the gel, trypsinized, and analyzed by LC-MS/MS (Micromass Q-TOF Ultima API Mass Spectrometer; Waters). The Mascot search website (http://www.matrixscience.com/search_form_select.html) was used for protein identification. For immunoprecipitation, brain extracts were incubated with 2 µg of the indicated antibodies and protein A– and G–Sepharose beads (Sigma-Aldrich) overnight at 4°C. The precipitated complexes were washed and immunoblotted. For the in vitro binding assay, bead-conjugated GST–Sdc2 proteins (2 µg) were incubated with 1 µg purified MBP–Sarm1 in PBS containing 1% Triton X-100, 1 mM DTT, and 1 mM PMSF for 1 h at 4°C. Equal amounts of GST proteins were used as controls. After washing, samples were analyzed by immunoblotting with MBP antibody.
Immunohistochemistry, immunocytochemistry, and time-lapse recording
B6 mice were anesthetized and perfused with 4% PFA in PBS. Mouse brains were cut into 50-µm-thick sections with a microslicer (Dosaka). After permeabilization, brain sections were preincubated in 3% horse serum and 3% BSA in PBS for 1 h and then incubated with Sarm1 mouse monoclonal and MAP2 rabbit polyclonal antibodies overnight at 4°C. After washing, sections were incubated with anti-mouse and rabbit secondary antibodies conjugated with Alexa Fluor 488 and 555, respectively. For immunocytochemistry, cultured neurons were fixed in 4% PFA/4% sucrose in PBS for 10 min at room temperature followed by permeabilization with cold methanol at −20°C for 10 min. For staining with PSD-95 antibody, neurons were permeabilized with 0.2% Triton X-100 in PBS for 10 min. Fixed neurons were preincubated in 5% BSA in PBS, then incubated with primary antibodies for 2 h at room temperature or overnight at 4°C. After washing, neurons were further incubated with Alexa Fluor 488– and/or Alexa Fluor 555–conjugated secondary antibodies. DNA was counterstained with DAPI. Vectashield Mounting medium (H-1000; Vector Laboratories) was then used to mount the samples for imaging. Immunofluorescent images were visualized with either a fluorescence microscope (DM RE; Leica) equipped with a 20×/NA 0.70 (HC Plan-Apochromat; Leica) or a 40×/NA 1.25 (HCX Apochromat; Leica) objective lens or a confocal microscope (LSM 510 META or LSM 700; Carl Zeiss) equipped with a 63×/NA 1.4 oil (Plan-Apochromat; Carl Zeiss) objective lens. For regular fluorescence microscopy, images were acquired using a cooled charge-coupled device camera (RTE/CCD-1300-Y/HS; Roper Scientific) with MetaMorph software (Molecular Devices). For confocal microscopy, images were captured with LSM or Zen acquisition and analysis software (Carl Zeiss). All fixed cells or brain slices were imaged at 20–22°C. For time-lapse recording, the GFP signals were acquired by using an LSM 510 META-NLO confocal microscope equipped with a 20×/NA 0.8 objective lens (Plan-Apochromat; Carl Zeiss) and LSM software at 37°C with a 5% CO2 supplement. For publication, the images were processed with Photoshop (Adobe) with minimal adjustment of brightness or contrast applied to the whole images.
Golgi-Cox impregnation
Adult wild-type or Sarm1 knockdown transgenic mice (2 mo old) were anesthetized and perfused with saline. Brains were removed and immersed in Golgi-Cox solution (solution A + B mixture, FD Rapid GolgiStain kit; FD NeuroTechnologies). 5 d after incubation, brains were immersed in solution C for 2 d and subsequently sliced into 150-µm-thick sections using a vibratome. Sections were mounted on gelatinized slides, developed, fixed, and dehydrated according to the manufacturer’s instructions. Neuronal images were captured by the AxioCam HR system (Carl Zeiss) and the camera lucida drawings were performed with Photoshop (Adobe).
Neuronal morphometry
For cultured hippocampal neurons, cotransfected or coexpressed GFP was used to outline neuronal morphology. Three parameters were used to determine dendritic morphology: (1) the number of primary dendrites, where the primary dendrites are the processes directly emerging from the soma with a length >20 µm; (2) the total dendrite length, including primary dendrites and all dendritic branches; and (3) the number of intersections in the Sholl analysis (Sholl, 1953). For CA1 neurons revealed by Golgi-Cox staining and camera lucida, the lengths of the basal and apical dendrites were measured separately. To analyze axonal morphology, the lengths of the longest neurites were measured. Because knockdown of Sarm1 in young neurons impaired neuronal polarization, only neurites that are twofold longer than the rest of other neurites were included for the analysis of axon length. Neural polarization was defined as the establishment of neurons with a dominant neurite (presumably axon) that is at least twofold longer than the rest of individual neurites. Dendritic spine morphology was analyzed as described previously (Lin et al., 2007; Chao et al., 2008). All measurements were performed using the software ImageJ.
Sarm1 knockdown transgenic mice
To construct the Sarm1 miRNA knockdown transgene, the oligonucleotide cassettes Sarm1i-2045 (5′-TAAGCTTGTCCTCGAATTTGC-3′) and Sarm1i-2397 (5′-TAAGGCAGACCCATTGGCGTA-3′) were inserted into the vector cDNA6.2-GW/emGFP-miR (Invitrogen). The knockdown efficiency of the plasmid was examined in COS-1 cells coexpressing Sarm1. The transgene cassette (Fig. 10 A) under the control of the human ubiquitin promoter contains DsREDT3 flanked by two loxP sequences followed by an emGFP cassette (Wang et al., 2010). The DsREDT3 was kindly provided by B. Glick (Department of Molecular Genetics and Cell Biology, the University of Chicago, Chicago, IL). Two artificial miRNAs against mouse Sarm1 were inserted into the 3′ end of the emGFP transcript. The transgene cassette also contains insulator, which was a gift from G. Felsenfeld (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD). The entire transgene cassette was then dissected from the vector backbone and microinjected into the pronuclei of fertilized C57BL/6J oocytes. Before Cre-mediated recombination, DsREDT3 was expressed and used to identify four red fluorescent founders of the transgenic mice. After crossing with E2A-Cre mice, the DsREDT3 cassette excised itself, resulting in the expression of emGFP–miRNA fusion transcripts (Fig. 10 A). E2A-cre was then washed out by crossing transgenic mice with wild-type C57BL/6J mice. The emGFP signals were used as indicators of artificial miRNA expression. Genomic PCR and Southern blotting additionally confirmed the genotype. For brain size measurement, 50-µm-thick brain sections were collected and stained with hematoxylin and eosin. The brain area was measured in one of every three brain sections from brain region Bregma −0.94 to −2.54 mm using the software ImageJ.
Statistical analysis
Statistical analyses were performed with the unpaired Student’s t test with the exception of the Sholl analyses, which were performed by two-way analysis of variance. Data are presented as mean ± SEM, and n indicates the number of neurons analyzed in each experiment.
Online supplemental material
Fig. S1 contains the analysis of mass spectrometry and basic characterization of Sarm1 antibodies. Fig. S2 contains neuroanatomic analyses of Sarm1 knockdown mice. Video 1 is the time-lapse recording of a neuron transfected with pSuper vector control shown in Fig. 6 A. Video 2 is the time-lapse recording of a neuron transfected with Sarm1i1 that appears in Fig. 6 A. Videos 1 and 2 were recorded for 24 h starting at 15 DIV. Video 3 contains 6 h of high-magnification time-lapse imaging that highlights the withdrawal of dendrites and dendritic spines in a Sarm1i1-expressing neuron. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008050/DC1.
Acknowledgments
We thank Dr. Ming-Zong Lai for the JNKKR and MKK4DN constructs, Dr. Gary Felsenfeld for the insulator used in the transgenic cassette, Dr. Ben Glick for DsREDT3, and Drs. Ming-Zong Lai and Cheng-Ting Chien for valuable discussion. We also thank Dr. Heiko Kuhn for manuscript editing and the Proteomic Core Facility at the Institute of Biological Chemistry and the Transgenic Core Facility at the Institute of Molecular Biology, Academia Sinica, for technical assistance.
This work was supported by grants from Academia Sinica and the National Science Council (NSC 98-2321-B-001-002 and NSC 98-2311-B-001-012-MY3 to Y.-P. Hsueh). The authors declare that they have no conflicting financial interests.
Footnotes
Abbreviations used in this paper:
- CASK
- calcium/calmodulin-dependent serine protein kinase
- DIV
- days in vitro
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- MBP
- maltose-binding protein
- MKK4
- microtubule-associated protein kinase kinase-4
- Sarm1
- sterile α and TIR motif–containing 1
- Sdc2
- syndecan-2
- shRNA
- small hairpin RNA
- TLR
- Toll-like receptor
References
- Alvarez V.A., Ridenour D.A., Sabatini B.L. 2006. Retraction of synapses and dendritic spines induced by off-target effects of RNA interference. J. Neurosci. 26:7820–7825 10.1523/JNEUROSCI.1957-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asundi V.K., Carey D.J. 1995. Self-association of N-syndecan (syndecan-3) core protein is mediated by a novel structural motif in the transmembrane domain and ectodomain flanking region. J. Biol. Chem. 270:26404–26410 10.1074/jbc.270.44.26404 [DOI] [PubMed] [Google Scholar]
- Barnat M., Enslen H., Propst F., Davis R.J., Soares S., Nothias F. 2010. Distinct roles of c-Jun N-terminal kinase isoforms in neurite initiation and elongation during axonal regeneration. J. Neurosci. 30:7804–7816 10.1523/JNEUROSCI.0372-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkblom B., Ostman N., Hongisto V., Komarovski V., Filén J.J., Nyman T.A., Kallunki T., Courtney M.J., Coffey E.T. 2005. Constitutively active cytoplasmic c-Jun N-terminal kinase 1 is a dominant regulator of dendritic architecture: role of microtubule-associated protein 2 as an effector. J. Neurosci. 25:6350–6361 10.1523/JNEUROSCI.1517-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J.S., Alexopoulou L., Sloane J.A., DiBernardo A.B., Ma Y., Kosaras B., Flavell R., Strittmatter S.M., Volpe J., Sidman R., Vartanian T. 2007. Toll-like receptor 3 is a potent negative regulator of axonal growth in mammals. J. Neurosci. 27:13033–13041 10.1523/JNEUROSCI.4290-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty M., Goodbody R., Schröder M., Stack J., Moynagh P.N., Bowie A.G. 2006. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat. Immunol. 7:1074–1081 10.1038/ni1382 [DOI] [PubMed] [Google Scholar]
- Chang L., Jones Y., Ellisman M.H., Goldstein L.S., Karin M. 2003. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell. 4:521–533 10.1016/S1534-5807(03)00094-7 [DOI] [PubMed] [Google Scholar]
- Chao H.W., Hong C.J., Huang T.N., Lin Y.L., Hsueh Y.P. 2008. SUMOylation of the MAGUK protein CASK regulates dendritic spinogenesis. J. Cell Biol. 182:141–155 10.1083/jcb.200712094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.F., Bargmann C.I. 2005. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 19:270–281 10.1101/gad.1276505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.R., Woods D.F., Marfatia S.M., Walther Z., Chishti A.H., Anderson J.M. 1998. Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J. Cell Biol. 142:129–138 10.1083/jcb.142.1.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas-Bailador F., Jones E.V., Whitmarsh A.J. 2008. The JIP1 scaffold protein regulates axonal development in cortical neurons. Curr. Biol. 18:221–226 10.1016/j.cub.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D., Li W., Fox K., Stryker M.P., Silva A.J. 2008. Reversing neurodevelopmental disorders in adults. Neuron. 60:950–960 10.1016/j.neuron.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell I.M., Yamaguchi Y. 1999. Cell surface heparan sulfate proteoglycan syndecan-2 induces the maturation of dendritic spines in rat hippocampal neurons. J. Cell Biol. 144:575–586 10.1083/jcb.144.3.575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell I.M., Hagihara K., Miura Y., Irie F., Yamaguchi Y. 2000. Synbindin, A novel syndecan-2-binding protein in neuronal dendritic spines. J. Cell Biol. 151:53–68 10.1083/jcb.151.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Li M., Chen W., Simons M. 2000. Synectin, syndecan-4 cytoplasmic domain binding PDZ protein, inhibits cell migration. J. Cell. Physiol. 184:373–379 [DOI] [PubMed] [Google Scholar]
- Granés F., Berndt C., Roy C., Mangeat P., Reina M., Vilaró S. 2003. Identification of a novel Ezrin-binding site in syndecan-2 cytoplasmic domain. FEBS Lett. 547:212–216 10.1016/S0014-5793(03)00712-9 [DOI] [PubMed] [Google Scholar]
- Grootjans J.J., Zimmermann P., Reekmans G., Smets A., Degeest G., Dürr J., David G. 1997. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc. Natl. Acad. Sci. USA. 94:13683–13688 10.1073/pnas.94.25.13683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig M., Lipkin W.I. 2001. Infectious and immune factors in the pathogenesis of neurodevelopmental disorders: epidemiology, hypotheses, and animal models. Ment. Retard. Dev. Disabil. Res. Rev. 7:200–210 10.1002/mrdd.1028 [DOI] [PubMed] [Google Scholar]
- Hsueh Y.P. 2006. The role of the MAGUK protein CASK in neural development and synaptic function. Curr. Med. Chem. 13:1915–1927 10.2174/092986706777585040 [DOI] [PubMed] [Google Scholar]
- Hsueh Y.P. 2009. Calcium/calmodulin-dependent serine protein kinase and mental retardation. Ann. Neurol. 66:438–443 10.1002/ana.21755 [DOI] [PubMed] [Google Scholar]
- Hsueh Y.P., Sheng M. 1999. Regulated expression and subcellular localization of syndecan heparan sulfate proteoglycans and the syndecan-binding protein CASK/LIN-2 during rat brain development. J. Neurosci. 19:7415–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y.P., Yang F.C., Kharazia V., Naisbitt S., Cohen A.R., Weinberg R.J., Sheng M. 1998. Direct interaction of CASK/LIN-2 and syndecan heparan sulfate proteoglycan and their overlapping distribution in neuronal synapses. J. Cell Biol. 142:139–151 10.1083/jcb.142.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y.P., Roberts A.M., Volta M., Sheng M., Roberts R.G. 2001. Bipartite interaction between neurofibromatosis type I protein (neurofibromin) and syndecan transmembrane heparan sulfate proteoglycans. J. Neurosci. 21:3764–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Zhou P., Qian L., Chuang J.Z., Lee J., Li C., Iadecola C., Nathan C., Ding A. 2007. MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J. Exp. Med. 204:2063–2074 10.1084/jem.20070868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia J.D., Okun E., Tang S.C., Griffioen K., Cheng A., Mughal M.R., Laryea G., Selvaraj P.K., ffrench-Constant C., Magnus T., et al. 2008. Toll-like receptor 3 is a negative regulator of embryonic neural progenitor cell proliferation. J. Neurosci. 28:13978–13984 10.1523/JNEUROSCI.2140-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-L., Lei Y.-T., Hong C.-J., Hsueh Y.P. 2007. Syndecan-2 induces filopodia and dendritic spine formation via the neurofibromin-PKA-Ena/VASP pathway. J. Cell Biol. 177:829–841 10.1083/jcb.200608121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Li J., Chiu I., Wang Y., Sloane J.A., Lü J., Kosaras B., Sidman R.L., Volpe J.J., Vartanian T. 2006. Toll-like receptor 8 functions as a negative regulator of neurite outgrowth and inducer of neuronal apoptosis. J. Cell Biol. 175:209–215 10.1083/jcb.200606016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink M., Fogelgren B., Olszewski K., Maroy P., Csiszar K. 2001. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics. 74:234–244 10.1006/geno.2001.6548 [DOI] [PubMed] [Google Scholar]
- Patterson P.H. 2009. Immune involvement in schizophrenia and autism: etiology, pathology and animal models. Behav. Brain Res. 204:313–321 10.1016/j.bbr.2008.12.016 [DOI] [PubMed] [Google Scholar]
- Podkowa M., Zhao X., Chow C.W., Coffey E.T., Davis R.J., Attisano L. 2010. Microtubule stabilization by bone morphogenetic protein receptor-mediated scaffolding of c-Jun N-terminal kinase promotes dendrite formation. Mol. Cell. Biol. 30:2241–2250 10.1128/MCB.01166-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain F.E., Sobel A. 2010. The microtubule network and neuronal morphogenesis: Dynamic and coordinated orchestration through multiple players. Mol. Cell. Neurosci. 43:15–32 10.1016/j.mcn.2009.07.012 [DOI] [PubMed] [Google Scholar]
- Rolls A., Shechter R., London A., Ziv Y., Ronen A., Levy R., Schwartz M. 2007. Toll-like receptors modulate adult hippocampal neurogenesis. Nat. Cell Biol. 9:1081–1088 10.1038/ncb1629 [DOI] [PubMed] [Google Scholar]
- Rosso S.B., Sussman D., Wynshaw-Boris A., Salinas P.C. 2005. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat. Neurosci. 8:34–42 10.1038/nn1374 [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 307:1098–1101 10.1126/science.1106148 [DOI] [PubMed] [Google Scholar]
- Sholl D.A. 1953. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 87:387–406 [PMC free article] [PubMed] [Google Scholar]
- Smith S.E., Li J., Garbett K., Mirnics K., Patterson P.H. 2007. Maternal immune activation alters fetal brain development through interleukin-6. J. Neurosci. 27:10695–10702 10.1523/JNEUROSCI.2178-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M.C., Nguyen M.M., Tao J., Allender D.L., Rolls M.M. 2010. Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol. Biol. Cell. 21:767–777 10.1091/mbc.E09-11-0967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter K.J., Samuel M.A., Gilfillan S., Fuchs A., Colonna M., Diamond M.S. 2009. The immune adaptor molecule SARM modulates tumor necrosis factor alpha production and microglia activation in the brainstem and restricts West Nile Virus pathogenesis. J. Virol. 83:9329–9338 10.1128/JVI.00836-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tararuk T., Ostman N., Li W., Björkblom B., Padzik A., Zdrojewska J., Hongisto V., Herdegen T., Konopka W., Courtney M.J., Coffey E.T. 2006. JNK1 phosphorylation of SCG10 determines microtubule dynamics and axodendritic length. J. Cell Biol. 173:265–277 10.1083/jcb.200511055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J., Takei Y., Harada A., Nakata T., Chen J., Hirokawa N. 2001. Synergistic effects of MAP2 and MAP1B knockout in neuronal migration, dendritic outgrowth, and microtubule organization. J. Cell Biol. 155:65–76 10.1083/jcb.200106025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volta M., Calza S., Roberts A.M., Roberts R.G. 2010. Characterisation of the interaction between syndecan-2, neurofibromin and CASK: dependence of interaction on syndecan dimerization. Biochem. Biophys. Res. Commun. 391:1216–1221 10.1016/j.bbrc.2009.12.043 [DOI] [PubMed] [Google Scholar]
- Walsh C.A., Morrow E.M., Rubenstein J.L. 2008. Autism and brain development. Cell. 135:396–400 10.1016/j.cell.2008.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.S., Hong C.J., Yen T.Y., Huang H.Y., Ou Y., Huang T.N., Jung W.G., Kuo T.Y., Sheng M., Wang T.F., Hsueh Y.P. 2004. Transcriptional modification by a CASK-interacting nucleosome assembly protein. Neuron. 42:113–128 10.1016/S0896-6273(04)00139-4 [DOI] [PubMed] [Google Scholar]
- Wang E., Hsieh-Li H.M., Chiou Y.Y., Chien Y.L., Ho H.H., Chin H.J., Wang C.K., Liang S.C., Jiang S.T. 2010. Progressive renal distortion by multiple cysts in transgenic mice expressing artificial microRNAs against Pkd1. J. Pathol. 222:238–248 10.1002/path.2765 [DOI] [PubMed] [Google Scholar]
- Witte H., Neukirchen D., Bradke F. 2008. Microtubule stabilization specifies initial neuronal polarization. J. Cell Biol. 180:619–632 10.1083/jcb.200707042 [DOI] [PMC free article] [PubMed] [Google Scholar]