Although closely related, RhoA and RhoC have distinct molecular targets and functional roles in cell migration and invasion.
Abstract
Several studies suggest that RhoA and RhoC, despite their sequence similarity, have different roles in cell migration and invasion, but the molecular basis for this is not known. Using RNAi, we show that RhoA-depleted cells became elongated and extended multiple Rac1-driven narrow protrusions in 2D and 3D environments, leading to increased invasion. These phenotypes were caused by combined but distinct effects of the Rho-regulated kinases ROCK1 and ROCK2. Depletion of ROCK2 induced multiple delocalized protrusions and reduced migratory polarity, whereas ROCK1 depletion selectively led to cell elongation and defective tail retraction. In contrast, RhoC depletion increased cell spreading and induced Rac1 activation around the periphery in broad lamellipodia, thereby inhibiting directed migration and invasion. These effects of RhoC depletion are mediated by the formin FMNL3, which we identify as a new target of RhoC but not RhoA. We propose that RhoA contributes to migratory cell polarity through ROCK2-mediated suppression of Rac1 activity in lamellipodia, whereas RhoC promotes polarized migration through FMNL3 by restricting lamellipodial broadening.
Introduction
Rho family GTPases regulate cytoskeletal dynamics, thereby affecting multiple cellular functions including cell motility and polarity (Jaffe and Hall, 2005; Boureux et al., 2007). Their activation is tightly regulated in space and time. Most Rho GTPases cycle between an inactive GDP-bound and an active GTP-bound form, which interacts with its downstream targets (Vega and Ridley, 2008). Altered Rho GTPase activity or expression is implicated in cancer progression (Ellenbroek and Collard, 2007; Vega and Ridley, 2008). The Rho family comprises 20 members in humans grouped into eight subfamilies (Vega and Ridley, 2008). The Rho subfamily includes the isoforms RhoA, RhoB, and RhoC, which are 84% identical in sequence; most differences are concentrated near the C terminus (Wheeler and Ridley, 2004).
RhoA, RhoB, and RhoC can all induce stress fibers when overexpressed, and the Clostridium botulinum exoenzyme C3 transferase, which modifies all three isoforms, induces loss of stress fibers and inhibits cell migration (Aktories and Just, 2005). However, several lines of evidence indicate that the isoforms have different functions. For example, RhoA and RhoC localize to the plasma membrane or interact with RhoGDI in the cytoplasm, whereas RhoB localizes to endosomal membranes because of its unique C-terminal lipid modifications and regulates endosomal trafficking of membrane receptors (Adamson et al., 1992; Wheeler and Ridley, 2004; Heasman and Ridley, 2008). In addition, knockout mouse models indicate that RhoB has a potential tumor suppressor function, whereas RhoC is required specifically for metastasis (Liu et al., 2001; Hakem et al., 2005). Furthermore, RhoA often inhibits whereas RhoC enhances cancer cell invasion in vitro (Simpson et al., 2004; Bellovin et al., 2006), and RhoC is selectively up-regulated during epithelial-mesenchymal transition (Dietrich et al., 2009), which occurs in some cancers (Thiery, 2002).
Multiple targets have been identified for Rho proteins (Jaffe and Hall, 2005), but it is not clear whether the isoforms selectively act through different or common downstream targets to mediate specific responses. Where tested, the isoforms generally bind to the same targets in vitro; for example, both RhoA and RhoC can bind to the Rho-associated kinases ROCK1 and ROCK2 (Leung et al., 1996; Ishizaki et al., 1997). Recently, however, RhoC but not RhoA or RhoB has been suggested to bind specifically to the formin FMNL2 (Kitzing et al., 2010). Here we show that RhoA and RhoC have very different functions in regulating the localization of active Rac1, lamellipodial dynamics, cell morphology, migration, and invasion. We demonstrate that they act through different downstream targets—ROCKs and FMNL3, respectively—to mediate their distinct effects.
Results and discussion
RhoA and RhoC have different effects on cell morphology
To compare the effects of RhoA and RhoC on cell morphology, we used siRNA-mediated knockdown in PC3 prostate cancer cells and MDA-MB-231 breast cancer cells (Fig. S1, A–C). RhoA or RhoC knockdown did not affect the expression of the other isoform, but RhoA and to a lesser extent RhoC knockdown induced RhoB up-regulation, which is consistent with previous observations (Ho et al., 2008). RhoB levels were also increased by C3 transferase, which indicates that it is reduced RhoA and RhoC activity that induces RhoB up-regulation (Fig. S1 B). RhoB knockdown did not affect RhoA or RhoC levels. Down-regulation of each protein led to a decrease in the respective levels of active GTP-bound RhoA, RhoB, or RhoC (Fig. S1 A).
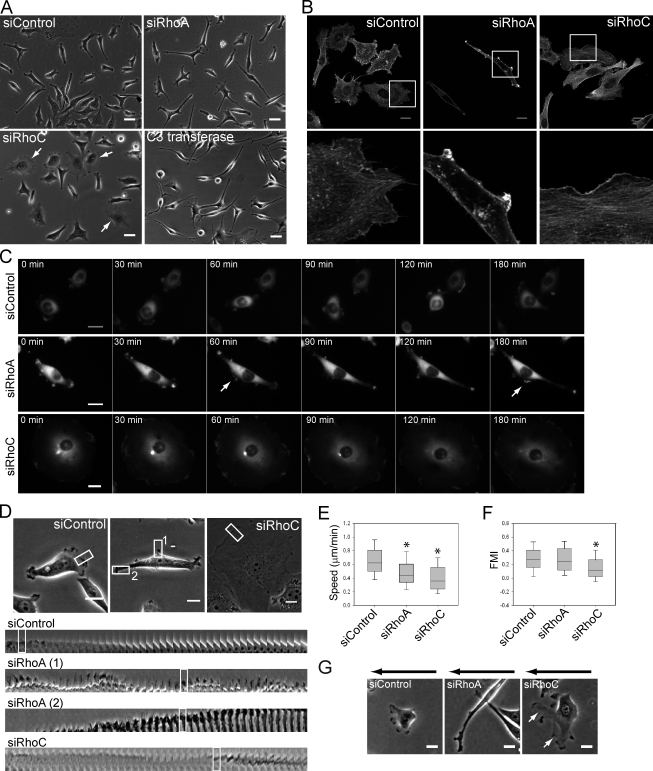
RhoA-depleted cells had an elongated cell shape compared with the siRNA control-treated cells, whereas RhoC depletion increased cell spread area (Fig. 1, A and B; Fig. S1 D; and Video 1). These phenotypes could be rescued by low-level expression of siRNA-resistant RhoA or RhoC (Fig. S1, E and F). C3 transferase–treated cells became highly elongated, and were thus most similar to RhoA-depleted cells (Fig. S1 G). These changes in morphology were confirmed with three different siRNAs (two are shown) and were similar in PC3 cells, MDA-MB-231 cells, and DU145 and LnCAP prostate cancer cell lines, which demonstrates that they are not cell type specific (Fig. S1 H and not depicted). Moreover, similar phenotypes were observed when cells were seeded on Matrigel-coated tissue culture plastic or uncoated plastic, and are thus not dependent on the extracellular matrix composition. The altered shapes of Rho isoform-depleted cells were not caused by a change in cell size or volume (not depicted), nor did Rho depletion lead to defects in cytokinesis, as indicated by the lack of multinucleate cells (Fig. 1 and Fig. S2 A).
Figure 1.
RhoA and RhoC have different effects of cell morphology and directional migration. PC3 cells were transfected with indicated siRNAs. (A) Cell morphology after siRNA transfection or C3 transferase treatment. Arrows indicate RhoC-depleted cells with increased spread area. (B) F-actin in cells on Matrigel. Boxed regions are shown at higher magnification below. (C) Images from movies of GFP–β-actin–expressing cells. Arrows indicate lateral protrusions. (D) Protrusion dynamics. Boxed regions on top are shown from movie frames (1 frame/min) in the bottom panels (boxed regions). (E) Migration speed. (F) Forward migration index (FMI) in a chemotaxis assay. *, P < 0.001. (G) Cell images during chemotaxis. White arrows indicate broader lamellipodia in RhoC-depleted cells. Black arrows: show gradient direction. Bars: (A) 40 µm; (B, C, D, and G) 20 µm.
The effect of RhoA depletion on cell morphology was not caused by RhoB up-regulation, as cells cotransfected with RhoA and RhoB siRNAs had a phenotype similar to that of RhoA depletion alone, whereas RhoB depletion alone led to a reduced cell spread area (Fig. S2 A). In addition, increased RhoB expression was also induced by RhoC suppression yet the phenotype of RhoA- and RhoC-depleted cells was very different.
RhoA and RhoC have different effects on distribution of lamellipodia and cell migration
To investigate how RhoA and RhoC depletion had different effects on cell shape, we analyzed cells by time-lapse microscopy. PC3 cells migrated by extending dynamic F-actin–rich protrusions, and frequently formed new protrusions and retracted old protrusions to change direction (Fig. 1, A and C; and Videos 1 and 2). In contrast, RhoA-depleted cells became elongated by extending long narrow protrusions in multiple directions, often from both ends of their long axis. Protrusions were also frequently observed along the elongated lateral sides of cells, and RhoA-depleted cells had more protrusions, which persisted for longer at the same position than those of control cells (Fig. 1 D, Fig. S3 A; Video 3). This was true both for lateral protrusions and protrusions at elongated tips. Multiple small protrusions were also observed in RhoA-depleted cells during spreading (Fig. S2 B and Video 4). RhoC-depleted cells had markedly different membrane dynamics. They often had broad lamellipodia that sometimes spread around most of the cell periphery. These showed dynamic regions of membrane ruffling but little net movement of the cell edge (Fig. 1, A and C; and Videos 1–3). These results indicate that RhoA is normally required to limit the number of protrusions, whereas RhoC restricts lamellipodial broadening.
Both RhoA and RhoC silencing reduced migration speed (Fig. 1 E) but through different mechanisms reflecting their effects on lamellipodia. RhoA-depleted cells extended narrow protrusions simultaneously in two or more opposing directions, resulting in reduced net movement, whereas RhoC-depleted cells with the spread phenotype were often unable to polarize (Video 1). However, RhoA-depleted cells underwent chemotaxis as efficiently as control cells with a similar persistence (Fig. 1 F and Video 5). They were still elongated but protruded more frequently toward the source of chemoattractant and preferentially elongated in the gradient direction (Fig. 1 G and Fig. S2 C). RhoC-depleted cells however migrated less efficiently toward the chemoattractant with reduced persistence (P < 0.005) and lower mean speed (P < 0.001; Fig. 1, E and F; Fig. S2 C; and Video 5). RhoC-depleted cells often had one or more abnormally extended and broad lamellipodia that split frequently (Fig. 1 G, Fig. S2 C, and Video 6), which is consistent with our observation that they have a defect in restricting lamellipodial broadening. This could be responsible for their reduced persistence.
RhoA depletion promotes invasion
RhoC is often required for cancer cell invasion, whereas results with RhoA vary depending on the system and cell type (Simpson et al., 2004; Bellovin et al., 2006). We investigated whether the effects of RhoA and RhoC on migratory polarity and lamellipodial distribution in 2D could contribute to cancer cell invasion in 3D. Cancer cells often have different morphologies in 3D environments compared with 2D (Sahai and Marshall, 2003; Sahai, 2005). The morphology of PC3 cells seeded on top of a thick 3D Matrigel layer was similar to that in 2D: RhoA-depleted cells were more elongated and RhoC-depleted cells were spread (Fig. S2 D).
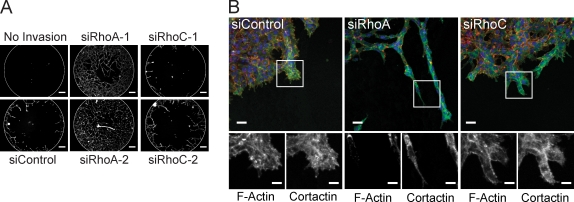
To observe the morphology of cells during directed invasion, we used MDA-MB-231 cells in a 3D invasion assay in Matrigel. RhoA-depleted cells extended very elongated protrusions and invaded the Matrigel more than control or RhoC-depleted cells (Fig. 2). Cortactin is a cytoskeletal protein that localizes in invading protrusions (Weaver, 2008). We observed enrichment of cortactin relative to F-actin at the invasive cell front particularly for RhoA knockdown cells, indicating the invasive nature of the protrusions (Fig. 2 B). Similar results were observed when a collagen I/Matrigel mix was used (Fig. S2 E). The elongated and protrusive phenotype of RhoA-depleted cells therefore correlates with an increase in invasive behavior.
Figure 2.
RhoA knockdown promotes invasive protrusions. (A) 3D invasion (ORIS assay) of siRNA-transfected MDA-MB-231 cells into Matrigel. “No invasion”: stopper removed at the end of assay. Bars, 100 µm. (B) Maximum intensity projection confocal images of F-actin, cortactin, and DAPI-stained cells at the invading front from A. Boxed areas are shown at higher magnification (bottom). Bars: (top) 40 µm; (bottom) 20 µm.
RhoA and RhoC have different effects on Rac1 activity localization
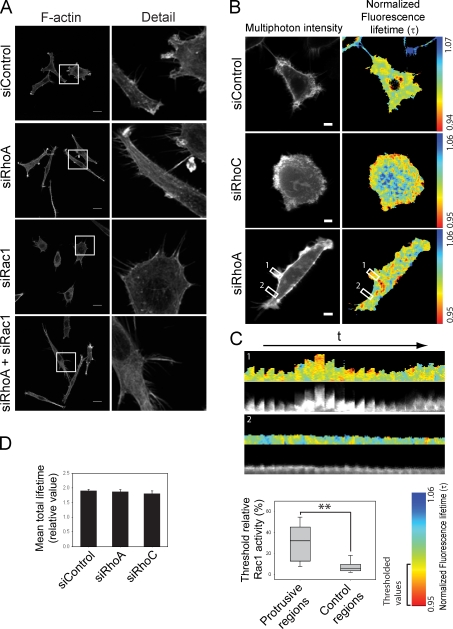
Despite the large changes in shape induced by RhoA or RhoC depletion, there was no difference in the expression or total activity of Rac1 (Fig. S2, G and H). Nevertheless, Rac1 was required for the dynamic protrusions in RhoA-depleted cells: cells depleted of both Rac1 and RhoA completely lacked actin-rich protrusions both at the ends and along the lateral edges (Fig. 3 A). Instead, cells had extensive filopodia along edges and at protrusion ends, in similar positions to where lamellipodia were observed after RhoA depletion alone. Rac1 depletion alone also led to an increase in filopodia (Fig. 3 A), perhaps because Cdc42-induced filopodia are more apparent after Rac inhibition (Nobes and Hall, 1995).
Figure 3.
RhoA inhibits Rac1-driven protrusive activity. (A) F-actin distribution in PC3 cells after transfection with siRNAs. Boxed regions are shown at higher magnification on the right. Bars, 20 µm. (B) FLIM images from PC3 cells cotransfected with the Rac1 biosensor and indicated siRNAs. Higher Rac1 activity is shown in “warmer” colors. Bars, 10 µm. (C, top) Kymographs of boxed regions in B: protruding membrane region (1) and control nonprotruding region (2); 1 frame/min (20 min), t = time. (C, bottom) Graph of threshold relative Rac1 activity values in protruding and control regions; **, P < 0.01. (D) Mean total Rac1 biosensor lifetime of cells transfected with the indicated siRNAs. Error bars indicate ± SEM.
We postulated that the differences in distribution and dynamics of lamellipodia in RhoA- versus RhoC-depleted cells could reflect changes in where Rac1 was active. We therefore investigated the localization of active Rac1 by live multiphoton fluorescence lifetime imaging microscopy (FLIM) using a Rac1 Raichu biosensor (Itoh et al., 2002; Makrogianneli et al., 2009). The biosensor localized mainly at the plasma membrane and did not induce any discernable morphological changes in PC3 cells (Fig. 3 B). In control cells, Rac1 activity was not strongly enriched at the periphery, but was observed sporadically in central regions of cells (Fig. 3 B and Videos 7 and 8), which might correspond to areas of dorsal protrusion and/or regions of substratum adhesion. In RhoA-depleted cells, Rac1 activity was significantly enriched at sites of membrane protrusive activity compared with nonprotruding areas, including the lateral protrusions (Fig. 3 C). In contrast, Rac1 activity in RhoC-depleted cells was more broadly distributed around the periphery (Fig. 3 B), which is consistent with the broad lamellipodia. Total Rac1 biosensor activity per cell was not altered by RhoA or RhoC knockdown (Fig. 3 D), corroborating the Rac1 activity pull-down results. Thus RhoA and RhoC regulate where Rac1 is active rather than how much is active. RhoA suppresses Rac1 activity to limit the number of protrusions, whereas RhoC restricts spreading of Rac1 activity along the membrane, thereby regulating lamellipodial broadening. The variation in cell morphology and behavior within the PC3 population (Fig. 1 and Video 1) could reflect cell-intrinsic differences on RhoA and RhoC activity.
ROCK1 and ROCK2 make different contributions to RhoA-regulated morphology and migration
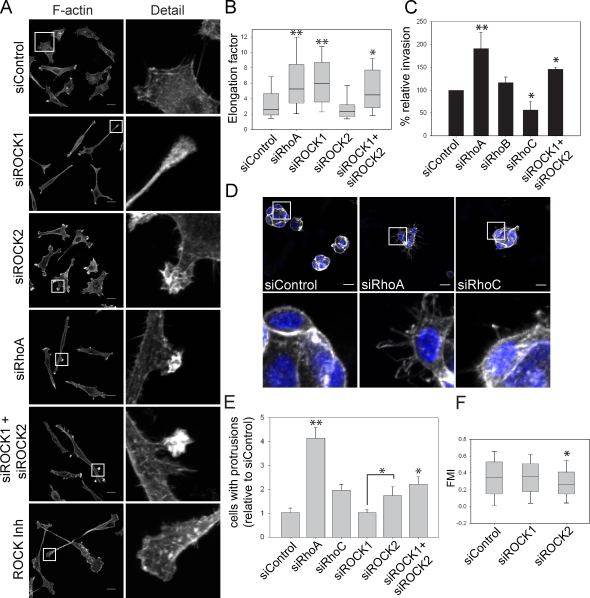
The Rho-associated protein kinases ROCK1 and ROCK2 are major mediators of Rho signaling and are best known as regulators of myosin light chain phosphorylation and actomyosin contractility (Riento and Ridley, 2003). Both RhoA and RhoC can bind to ROCKs but whether they are preferentially used by one Rho isoform is not known (Sahai and Marshall, 2002; Riento and Ridley, 2003). ROCK function is usually studied with chemical inhibitors such as Y-27632, which do not discriminate between ROCK1 and ROCK2 and target other kinases, at least in vitro (Bain et al., 2007). Different functions for ROCK1 and ROCK2 have been described using RNAi in fibroblasts (Yoneda et al., 2005, 2007). We used siRNAs to deplete ROCK1 or ROCK2 to determine if the phenotypes of RhoA or RhoC-depleted cells could be attributed to a specific ROCK isoform (Fig. S3 A). ROCK1 and ROCK2 were expressed similarly in PC3 cells (Fig. S3 A). ROCK1 but not ROCK2 suppression induced cell elongation, similar to RhoA knockdown but clearly distinct from RhoC knockdown (Fig. 4, A and B).
Figure 4.
ROCK1 and ROCK2 regulate different aspects of the RhoA morphological response. (A) F-actin distribution in PC3 cells after transfection with the indicated siRNAs or ROCK inhibitor Y-27632 treatment. Boxed regions are shown at higher magnification on the right. Bars, 20 µm. (B) Elongation of PC3 cells. (C) Transwell invasion assay. (D) PC3 cells embedded in Matrigel stained for F-actin (gray) and nuclei (DAPI; blue). Enlarged views of the boxed regions are shown below. Bars, 20 µm. (E) Quantification of cell groups (see D) with more than two F-actin–rich protrusions. (F) Forward migration index (FMI) of PC3 cells in a chemotaxis assay. **, P < 0.001; *, P < 0.01. Graphs show pooled results from experiments performed with at least two different siRNAs for each gene as indicated in Materials and methods. Error bars indicate mean ± SEM.
However, in contrast to RhoA depletion, ROCK1-depleted cells had fewer small F-actin–rich protrusions, and the cell nucleus was predominantly localized in the first third of the cell body (in ∼70% of migrating cells, whereas in RhoA-depleted cells it was in the central third in 80% of cells). ROCK1-depleted cells also had a long tail, indicative of a tail retraction defect (63.5 ± 6% of cells). This phenotype was similar to that observed with the ROCK inhibitor Y-27632 (85 ± 4% of cells with elongated tails; Fig. 4, A and B; and Fig. S3 B). In contrast, ROCK2-depleted cells were not elongated and did not have a strong tail retraction defect (30.7 ± 6% of cells with elongated tails compared with 25 ± 6% of control cells) and instead frequently had multiple thin protrusions, each with a ruffling tip, distributed around the cell perimeter (Fig. 4, A and B; and Fig. S3 B). This was not observed in cells treated with Y-27632 or another ROCK inhibitor HA-1077 (Fig. 4 A and not depicted). This indicates that Y-27632 predominantly acts on the ROCK1 isoform in cells, as suggested previously (Darenfed et al., 2007). Low-level expression of mouse ROCK1 or ROCK2, resistant to human siRNAs, rescued these siRNA morphology phenotypes (Fig. S3 C; note that high-level ROCK1/ROCK2 expression induced cell contraction, as expected).
Interestingly, simultaneous knockdown of ROCK1 and ROCK2 induced a phenotype very similar to RhoA depletion (Fig. 4, A and B): cells were elongated with actin-rich protrusions at both ends and along lateral edges, and the nucleus was centrally localized. Each ROCK therefore has a distinct role downstream of RhoA, and the morphology of RhoA-depleted cells reflects a combination of ROCK1 and ROCK2 functions. Neither ROCK1 nor ROCK2 depletion phenotypes resembled that of RhoC suppression, thus it is unlikely that RhoC acts through ROCK1 or ROCK2 to regulate cell morphology in this context.
To determine whether the different effects of ROCK isoforms on 2D migration were similar in 3D invasion, PC3 cell invasion through Matrigel-coated transwells was analyzed. RhoA suppression increased invasion, whereas RhoC silencing reduced invasion (Fig. 4 C). As a control, RhoB depletion did not affect invasion. Only the simultaneous knockdown of ROCK1 and ROCK2 increased invasion, which is consistent with both being required for RhoA-mediated responses. To analyze the effects of ROCKs on morphology in 3D, PC3 cells were embedded in Matrigel. They mostly formed spherical groups of cells with cell–cell interactions (Fig. 4 D), in contrast to their single cell morphology in 2D. The morphology of cancer cell groups in 3D is indicative of their invasive potential and tumorigenicity (Kenny et al., 2007). Groups of RhoA-depleted cells had a more disorganized morphology with multiple dynamic extensions compared with control or RhoC-depleted cells (Fig. 4, D and E; and not depicted). Again, simultaneous ROCK1 and ROCK2 depletion induced a phenotype most similar to RhoA suppression (Fig. 4 E). RhoA-depleted cells also had an elongated morphology when embedded in a collagen I/Matrigel mix (Fig. S2 F).
The different roles of ROCK1 and ROCK2 downstream of RhoA could reflect differential localization in cells. We have not been able to detect endogenous ROCK1 or ROCK2 with the antibodies available. GFP-ROCK1 expressed at low levels showed a uniform distribution on the plasma membrane (Fig. S3 C). It is possible that active ROCK1 is preferentially localized to tails, but so far there is no method to localize active ROCK proteins. RFP-ROCK2 was enriched in protrusive areas in control or RhoA knockdown cells, which is consistent with a role in regulating protrusion.
The observation that ROCK2-depleted cells had unpolarized protrusive activity implies that ROCK2 normally acts to suppress Rac1 activation in protrusions. RhoA is active at the front of lamellipodial protrusions (Pertz et al., 2006; Machacek et al., 2009; Heasman et al., 2010), and it could therefore locally inhibit Rac in this region through ROCK2. When RhoA or ROCK2 levels are reduced, Rac1 can be active in more areas because of a lack of RhoA/ROCK2-mediated suppression, leading to multiple protrusions. In contrast, ROCK1 function appears to be restricted to tail retraction. Some RhoA-depleted cells transiently showed a tail retraction defect, but this phenotype was much stronger in ROCK1-depleted cells. RhoA is transiently activated in tails before retraction (Pertz et al., 2006; Heasman et al., 2010). The migratory phenotype of RhoA-depleted cells therefore reflects a combination of decreased ROCK1-mediated tail retraction and ROCK2 inhibition of Rac1-driven protrusions. The enhanced ability of RhoA/ROCK-depleted cells to invade through Matrigel correlates with their elongated morphology, which based on our results is likely to reflect increased Rac1 activation in the long invasive protrusions. This could be associated with production of matrix-degrading proteases required for invasion (Sahai, 2005).
ROCK2 regulates directional migration
We next investigated whether the distinct morphological phenotypes observed after ROCK1 or ROCK2 depletion correlated with different effects on cell migration behavior. Neither ROCK1 nor ROCK2 affected migration speed, but in chemotaxis assays ROCK2- but not ROCK1-depleted PC3 cells migrated with reduced persistence (forward migration index) compared with control cells, which indicates a specific involvement of ROCK2 in directional migration (Fig. 4 F and Fig. S3 D).
Although ROCK2 and RhoC knockdown both reduced directional migration (Fig. 1 F and Fig. 4 F), they act through different mechanisms: RhoC-depleted cells have broad lamellipodia whereas ROCK2-depleted cells have narrow protrusions (Fig. S3 E). Interestingly, RhoC suppression abolished the multiple small protrusions in ROCK2-depleted cells (Fig. S3 F), and instead cells were spread, often with broad lamellipodia, similar to RhoC depletion alone. Thus the function of RhoC in restricting lamellipodial broadening is required even in cells with multiple small protrusions.
RhoC acts through FMNL3 to regulate cell morphology and invasion
Given that the phenotype of ROCK-depleted cells did not resemble RhoC suppression, we tested other Rho partners to identify a potential target for RhoC. It was recently suggested that the formin family protein FMNL2 is a RhoC effector (Kitzing et al., 2010), whereas FMNL1 has been reported as a Cdc42 effector (Seth et al., 2006). We therefore investigated the effects of knocking down FMNL proteins on PC3 cell morphology (Fig. 5 A). Depletion of each protein induced a distinct phenotype. FMNL2 depletion did not resemble that of RhoC depletion but instead led to an increase in small ruffling protrusions, whereas FMNL1 and FMNL3 knockdown cells had an increased spread area, broad lamellipodia, and unpolarized morphology, resembling the effect of RhoC but not RhoA depletion. Expression of GFP-FMNL3 in FMNL3 knockdown cells rescued the increased spreading and broad lamellipodia (Fig. S3 H). When RhoC and each FMNL protein were depleted simultaneously, either a mixed or an additive phenotype was observed with FMNL2 and FMNL1, respectively, but no change was observed with FMNL3, which suggests that RhoC and FMNL3 act in the same pathway (Fig. 5 A). Down-regulation of FMNL3 also impaired invasion of PC3 cells through Matrigel, which indicates that it could contribute to RhoC signaling in promoting invasion (Fig. 5 B). Knockdown of FMNL1 or FMNL3 together with RhoA also induced large broad lamellipodia (Fig. S3 I); thus, the broad lamellipodium phenotype dominates over narrow protrusions.
Figure 5.
RhoC acts through FMNL3 to regulate cell shape and invasion. (A) F-actin in PC3 cells after transfection with indicated siRNAs. Bar, 20 µm. (B) Transwell invasion assay with PC3 cells; invasion is relative to siControl. (C) GST-RhoA/C-V14 pull-down with lysates from 293T cells expressing GFP-FMNL3. MW, molecular weight markers. (D) F-actin and GFP localization in PC3 cells transfected with the indicated siRNAs and either GFP-FMNL3 or GFP alone. White asterisks indicate GFP-FMNL3 cells. The graph on the right shows quantification of GFP-positive cells with broad lamellipodia; *, P < 0.05. Error bars indicate mean ± SEM.
To test if FMNL3 was indeed a new RhoC-specific target, we tested its ability to bind to recombinant Rho proteins. FMNL3 bound to constitutively active GST-RhoC but not RhoA (Fig. 5 C). This was true for both N- and C-terminal GFP-tagged FMNL3 (Fig. S3 J). GFP-FMNL3 localized in actin-rich protrusions, and its expression inhibited the broad lamellipodium phenotype induced by RhoC knockdown (Fig. 5 D). Together, these results indicate that FMNL3 acts downstream of RhoC to restrict lamellipodial broadening. In contrast, FMNL2-GFP did not bind GST-RhoC or RhoA in vitro (Fig. S3 J). This, together with the knockdown phenotype, indicates that FMNL2 is not likely to act downstream of RhoC under these conditions.
Several formins are known to stimulate the nucleation and elongation of actin filaments, thereby contributing to cell motility, cytokinesis, and tissue morphogenesis (Chesarone et al., 2010). Not much is known about the function of FMNL3, but one study identifies it as being down-regulated in cells with reduced tumorigenic capacity (Martín-Rufián et al., 2006). It was also identified as overexpressed in a highly motile subpopulation of melanoma cells in vivo (Pinner et al., 2009). This is consistent with a role for FMNL3 downstream of RhoC in promoting cancer progression.
Our results show that the closely related Rho GTPases RhoA and RhoC have different effects on cell morphology, migration, and invasion by acting through different downstream targets. We identify for the first time a new RhoC-specific target, the formin FMNL3, involved in cancer cell invasion. RhoA depletion leads to elongated cells that extend narrow Rac1-driven protrusions in multiple directions in both 2D and 3D environments, stimulating invasion, and only migrate persistently when exposed to a gradient of chemoattractants. In contrast, RhoC-depleted cells spread with broad lamellipodia, and have reduced invasion and migrational persistence even during chemotaxis. These results could explain their opposing roles in tumor cell invasion. RhoA via ROCKs would inhibit Rac1-dependent protrusion required for invasion, whereas RhoC acting via FMNL3 would promote this process.
Materials and methods
Cell lines and reagents
PC3 prostate cancer cells were grown in RPMI containing 25 mM Hepes and 2 mM glutamine supplemented with 10% FCS, 100 µg/ml streptomycin, and 100 U/ml penicillin. MDA-MB-231 breast cancer cell line and 293T cells were grown in DME media supplemented with 25 mM Hepes and 2 mM glutamine, 10% FCS, 100 µg/ml streptomycin, and 100 U/ml penicillin. The following antibodies were used: RhoA (clone 26C4, sc-179), RhoB (119, sc-180) and RhoC (C-16, sc-12116), GFP (FL, sc-8334), and GST (B14, sc-138; all from Santa Cruz Biotechnology, Inc.). Also used were ROCK1 K-18 (sc-6056; Santa Cruz Biotechnology), mouse monoclonal ROCK1 (No. 611136; BD), ROCK2 C-20 (sc-1851; Santa Cruz Biotechnology, Inc.), mouse monoclonal ROCK2 (No. 610623; BD), Rac1 23A clone (No. 05-389; Millipore), mouse monoclonal anti-cortactin clone 4F11 (No. 05-180; Millipore), α-tubulin DM1A clone (T6199; Sigma-Aldrich), and rabbit polyclonal extracellular signal–regulated kinase (ERK), K-23 (sc-94; Santa Cruz Biotechnology). Secondary HRP-labeled antibodies were from GE Healthcare (anti–mouse and anti–rabbit) or Santa Cruz Biotechnology, Inc. (anti–goat). Cell-permeable C3 transferase (Cytoskeleton) was used at 1.1 µg/ml for 6 h. The ROCK inhibitor Y-27632 (EMD) was used at 5 µM for 12 h. Protease inhibitor cocktail (complete) was from Roche and Phosphatase inhibitor cocktail II and IV were from EMD. The EGFP–β-actin construct has been described previously (Ballestrem et al., 1998).
Transfections and Western blotting
All siRNAs were from Dharmacon (Thermo Fisher Scientific): siRhoA-1 (5′-AUGGAAAGCAGGUAGAGUU-3′), siRhoA-2 (5′-GAACUAUGUGGCAGAUAUC-3′), siRhoA-3 (5′-CGACAGCCCUGAUAGUUUA-3′), siRhoB-1 (5′-CAUCCAAGCCUACGACUAC-3′), siRnoB-2 (5′-GCAUCCAAGCCUACGACUA-3′), siRhoB-3 (5′-GCAUCCAAGCCUACGACUA-3′), siRhoC-1 (5′-AUAAGAAGGACCUGAGGCA-3′), siRhoC-2 (5′-GGAUCAGUGCCUUUGGCUA-3′), siRhoC-3 (5′-GAAAGAAGCUGGUGAUCGU-3′), on target plus siCONTROL non-targeting siRNA (D-001810-01, D-001810-02), siRac1-1 (5′-AGACGGAGCUGUAGGUAAA-3′), siRac1-2 (5′-CGGCACUGUCCCAACA-3′), siROCK1-1 (5′-GAAGAAACAUUCCCUAUUC-3′), siROCK1-2 (5′-GCCAAUGACUUACUUAGGA-3′), siROCK2-1 (5′-GCAAAUCUGUUAAUACUCG-3′), siROCK2-2 (5′-CAAACUUGGUAAAGAAUUG-3′), siFMNL3-1 (5′-GCGAGGAGGUCACGAAAUC-3′), siFMNL3-2 (5′-UGUCAGCCAUUCGAAUUAA-3′), and FMNL1, FMNL2, and FMNL3 pools (Dharmacon Smart pools). All siRNAs were initially tested for knockdown of the protein. FMNL1-3 siRNA knockdown was tested by RT-PCR as described previously (Kitzing et al., 2010). Cells were reverse-transfected with 50–75 nM siRNA using Lipofectamine 2000 (Invitrogen) in complete medium without antibiotics. Medium was changed to complete medium 6 h after transfection. Where indicated, cells were starved in 1% or 0.1% FCS at least 12 h before the relevant assay.
Cell lysates for Western blot analysis were prepared 72 h after siRNA transfection using lysis buffer (50 mM Tris-HCl, pH 8, 0.5 mM EDTA, 150 mM NaCl, and 1% Triton X-100 plus protease and phosphatase inhibitors cocktails). Lysates were resolved on 4–12% polyacrylamide gels (Invitrogen), transferred to PVDF membranes, and blotted with the indicated antibodies.
The EGFP–β-actin construct was a gift from B. Wehrle-Haller (University of Geneva, Geneva, Switzerland). A stable EGFP–β-actin–expressing PC3 cell line was created by the transfection of PC3 cells using Lipofectamine 2000. Cells were selected in medium containing G418 until GFP–β-actin–expressing selected clones were visible. Clones were expanded and their morphology and migration was compared with control PC3 cells before being used.
Human FMNL3 cDNA cloned into pcDNA-DEST53 or pcDNA-DEST47 (Invitrogen) to create GFP-FMNL3 or FMNL3-GFP, respectively, were provided by S. Wiemann (DKFZ, Heidelberg, Germany) and M. Teresa Herrera-Abreu (King’s College London, London, England, UK). FMNL2-GFP in pcDNA-DEST-47 was a gift from K. Rottner (University of Bonn, Bonn, Germany). RhoA and RhoC cDNA with silent mutations that make them resistant to an siRNA for RhoA (siRhoA-1) or RhoC (siRhoC-1), respectively, were created using a QuikChange mutagenesis kit (Agilent Technologies) in expression vectors encoding GFP-RhoA and myc-RhoC. Mouse GFP-ROCK1 cDNA in pcDNA-DEST53 was a gift from C. Marshall and S. Kümper (Institute of Cancer Research, London, England, UK). Mouse ROCK2 cDNA in the pcDNA mRFP1 vector was described by Niault et al. (2009). Plasmids were transfected into PC3 cells using JETPRIME reagent (PolyPlus transfections) 48 h after siRNA transfection. Expression was assessed 15–20 h after DNA transfection.
Time-lapse microscopy
For bright field time-lapse microscopy, a fully motorized, multifield Nikon TE2000 microscope mounting an ORCA camera (Hamamatsu) and controlled by the MetaMorph software was used. A 10×/0.3 NA Plan Fluor ELWD objective lens was used. For random migration studies, cells were seeded on a multiwell plate 48–52 h after transfection, and 8 h later, images were acquired at 37°C every 5 min for 14 h. More than 50 cells per siRNA condition from three different experiments were tracked 58–60 h after transfection for 12–14 h. Still images are from 72 h after transfection. Unless indicated, cells were imaged in medium containing 1% FCS. To visualize cell spreading, images were taken at high speed (one image per second) immediately after plating. For time-lapse microscopy of EGFP–β-actin–expressing cells, cell images were acquired every 2 min for 3 h under similar conditions.
For chemotaxis assays, collagen IV precoated microslides (Ibidi) were used according to the manufacturer’s instructions using 10% FCS and 20 ng/ml hepatocyte growth factor (HGF) as chemoattractants. Cell images were acquired every 5 min for 14 h. More than 200 cells per condition from at least four different experiments using at least two different siRNAs for each gene were quantified and results were pooled in graphs. Chemotaxis and persistence were analyzed using the ImageJ Manual tracking and Chemotaxis tool plugins. Further two-sample comparison of migration velocity and persistence analysis was performed using the Mathematica “Chemotaxis_tests_M6 notebook” developed by G.A. Dunn (King’s College London; parameters used for motility analysis: δt=8, TR = 4; Zicha et al., 1997). These data were summarized in a rose plot that is a circular histogram showing the frequency of cell tracks with directions lying within each 20° interval. It displays the mean direction as a red arrow and the 95% confidence interval as a green sector when there is significant directional clustering. The Rayleigh test for unimodal clustering of directions (Mardia and Jopp, 2000) and the Moore rank test (Moore, 1980) were then applied to the data and a P ≤ 0.01 was chosen as the criterion for rejecting the null hypothesis of random directionality.
Immunofluorescence, confocal microscopy, and cell shape analysis
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS, and blocked with 1% BSA in PBS. Alexa fluor phalloidin (wavelengths 480 nm or 543 nm; Invitrogen) was used for F-actin visualization, and DAPI was used for nuclear staining. Confocal images were acquired with an inverted confocal microscope (LSM 510; Carl Zeiss) using 40× (1.2 NA) or 63× (1.4 NA) objective lenses. Images are from cells fixed 72 h after transfection unless indicated. Images show condition with a single siRNA but the same results were obtained with different siRNAs. Morphology analysis was performed using MetaMorph or Cell profiler software (Carpenter et al., 2006) from F-actin–stained fluorescence images. In brief, this involved segmentation of the images to obtain cell shape and area measurements from the corresponding thresholded objects. More than 100 cells from at least three different experiments were analyzed. The elongation factor is defined as the ratio between the major axis and the minor axis of the cell (1 = rounded, >1 = more elongated). The shape factor is calculated as 4A/p2, where A = area and p = perimeter (1 = circle, <1 = more elongated). Cells with broad lamellipodia (Fig. 5 D) were defined as cells with a flattened membrane extension with a narrow lamellipodium around all or part of the cell perimeter, as described for RhoC knockdown cells.
FLIM
Rac1 activity was localized with the Rac1 Raichu biosensor modified by M. Keppler (King’s College London) to express GFP and mRFP as the donor and acceptor fluorophores, respectively (Itoh et al., 2002; Makrogianneli et al., 2009). PC3 cells were transfected with the indicated siRNAs and seeded on a SmartSlide microincubator chamber (WaferGen Biosystems) after 24–36 h. After a further 24 h, cells were transfected with the expression plasmid encoding the Rac1 Raichu biosensor and incubated for 18 h. All FLIM measurements were performed at 37°C on a modified multiphoton microscopy system as described previously (Peter et al., 2005) with a Ti:sapphire Chameleon laser (Coherent, Inc.) tuned to 890 nm. A 510 ± 10 nm band-pass filter (Chroma Technology Corp) was used in the detector channel. Laser power was adjusted to give mean photon counting rates of the order of 104–105 photons/s (0.0001–0.001 photons/excitation event) and with peak rates approaching 106 photons/s, below the maximum counting rate afforded by the time-correlated single photon counting electronics to avoid pulse pile-up.
Each FLIM image was acquired over 44 s at low excitation power to achieve sufficient photon statistics for fitting while avoiding either pulse pile-up and keeping photobleaching to a minimum. The imaging interval per cell was 60 s and the total imaging period was at least 20 min. Analysis for time-resolved data was performed essentially as described previously (Peter et al., 2005). The multiphoton intensity images were thresholded to remove the background autofluorescence and areas of the cell that did not have sufficient photon counts for fitting. To better visualize the intracellular fluorescence lifetime changes, cell images were normalized by the mean fluorescence lifetime of the cell of interest and pseudocolored with a rainbow look-up table, in which warmer color represents areas with higher than average Rac1 activity (lower fluorescence lifetimes) and colder color areas with lower Rac1 activity. Total mean lifetime over each whole cell in each condition was calculated from still images.
For analysis of Rac1 activity in protruding areas, kymographs of 30 × 10 pixel regions on protruding membrane areas or control nonprotruding areas in RhoA knockdown cells were plotted from the normalized lifetime images. Relative lifetimes were threshold as indicated and the relative percentage of pixels within the threshold during the movie was calculated for 17 regions per condition in eight different cells from different experiments. Protruding areas were identified from the membrane dynamics in the GFP intensity images.
Rho GTPase activity assays and GST pull-down
RhoA, RhoB, and RhoC activity assays were performed using Rho activation assay beads (GST-Rhotekin-RBD on glutathione beads) according to the manufacturer’s instructions (Cytoskeleton). Rac1 activity assays were performed using Rac1/Cdc42 assay reagent (GST-PAK1-PBD on glutathione beads) according to the manufacturer’s instructions (Millipore).
GST-RhoA/C pull-down assays were performed with 5 µg of bacterially expressed GST-RhoA/C V14 protein or GST protein bound to glutathione-Sepharose beads (GE Healthcare). They were incubated with 293T cell lysates overexpressing the indicated GFP-tagged FMNL3 or FMNL2 protein. Incubation was performed for 2 h at 4°C in lysis buffer (1% NP-40, 20 mM Tris-HCl, pH 8, and 130 mM NaCl). After washing the beads, proteins bound were resolved by electrophoresis and detected with the indicated antibodies.
Invasion assay and 3D culture
The transwell-based BD Biocoat invasion system (8 µm pore diameter; BD) was used to evaluate invasion through a Matrigel layer. The same BD inserts without the Matrigel coating were used as migration controls. To measure cell invasion, 105 cells in medium containing 0.1% FCS were added to the transwells, and 2 × 104 cells were used for migration. Medium containing 1% FCS was used as an attractant in the lower chamber. Cells on the bottom of the coated transwell were counted after 24 h for invasion and 12 h for migration. The final time point was always 72 h after siRNA transfection.
To study morphology in 3D matrices, cells were reverse transfected with siRNA in suspension in complete medium and mixed after 6 h with a final concentration of 3 mg/ml of Matrigel or a mix of collagen I and Matrigel (2:1 mg/ml; BD). 72 h after transfection, cells were fixed with 4% paraformaldehyde and stained for F-actin and DNA as described above. Z-stack confocal images were acquired on a confocal microscope (LSM 510 Meta; Carl Zeiss) with a 20× objective lens. Images shown are maximum intensity projections of a ∼100-µm z stack of images acquired every 5 µm.
The Oris 3D Invasion system (Platypus technologies) was used as described by the manufacturer. In brief, cells were seeded on a 96-well plate previously coated with a Matrigel or a collagen I/Matrigel mix (2:1) layer and with silicon stoppers covering the central area of the wells to create a cell-free gap. The stoppers were removed after 12 h and cells were immediately overlaid with a thick layer of the same matrix. Cells were fixed and stained after 48 h and cells invading toward the gap in the center of the well were imaged.
Statistical analysis
For statistical significance analysis, one-way analysis of variance with a Dunn’s multiple comparison after test or unpaired t test were used. All significances indicated are compared with siCsontrol condition unless otherwise stated. All box and whisker plots boxes show median and 25th and 75th percentiles; whiskers show the 95th percentile.
Online supplemental material
Fig. S1 and Fig. S2 show Rho isoform-specific knockdowns, morphology phenotypes, and Rac1 activity. Fig. S3 shows the effects of depleting Rho GTPases and downstream targets on cell morphology and migration, and FMNL3 overexpression experiments. Videos 1–3 show cell shape and migration after Rho isoform knockdown. Video 4 shows spreading of a RhoA-depleted cell. Videos 5 and 6 show chemotaxis after different knockdowns. Videos 7 and 8 show FLIM acquired images. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201011038/DC1.
Acknowledgments
We thank Bernard Wehrle-Haller, Sandra Kümper, Chris Marshall, Melanie Keppler, Klemens Rottner, Stefan Wiemann, and Maria Teresa Herrera-Abreu for cDNAs and plasmids.
This work was funded by the Breast Cancer Campaign, Cancer Research UK, and Ludwig Institute for Cancer Research. F. Vega was supported in part by a Marie Curie fellowship (European Commission Project No. 24441).
Footnotes
Abbreviations used in this paper:
- FLIM
- fluorescence lifetime imaging microscopy
- HGF
- hepatocyte growth factor
References
- Adamson P., Paterson H.F., Hall A. 1992. Intracellular localization of the P21rho proteins. J. Cell Biol. 119:617–627 10.1083/jcb.119.3.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktories K., Just I. 2005. Clostridial Rho-inhibiting protein toxins. Curr. Top. Microbiol. Immunol. 291:113–145 10.1007/3-540-27511-8_7 [DOI] [PubMed] [Google Scholar]
- Bain J., Plater L., Elliott M., Shpiro N., Hastie C.J., McLauchlan H., Klevernic I., Arthur J.S., Alessi D.R., Cohen P. 2007. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408:297–315 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestrem C., Wehrle-Haller B., Imhof B.A. 1998. Actin dynamics in living mammalian cells. J. Cell Sci. 111:1649–1658 [DOI] [PubMed] [Google Scholar]
- Bellovin D.I., Simpson K.J., Danilov T., Maynard E., Rimm D.L., Oettgen P., Mercurio A.M. 2006. Reciprocal regulation of RhoA and RhoC characterizes the EMT and identifies RhoC as a prognostic marker of colon carcinoma. Oncogene. 25:6959–6967 10.1038/sj.onc.1209682 [DOI] [PubMed] [Google Scholar]
- Boureux A., Vignal E., Faure S., Fort P. 2007. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol. Biol. Evol. 24:203–216 10.1093/molbev/msl145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M.A., DuPage A.G., Goode B.L. 2010. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11:62–74 10.1038/nrm2816 [DOI] [PubMed] [Google Scholar]
- Darenfed H., Dayanandan B., Zhang T., Hsieh S.H., Fournier A.E., Mandato C.A. 2007. Molecular characterization of the effects of Y-27632. Cell Motil. Cytoskeleton. 64:97–109 10.1002/cm.20168 [DOI] [PubMed] [Google Scholar]
- Dietrich K.A., Schwarz R., Liska M., Grass S., Menke A., Meister M., Kierschke G., Längle C., Genze F., Giehl K. 2009. Specific induction of migration and invasion of pancreatic carcinoma cells by RhoC, which differs from RhoA in its localisation and activity. Biol. Chem. 390:1063–1077 10.1515/BC.2009.110 [DOI] [PubMed] [Google Scholar]
- Ellenbroek S.I., Collard J.G. 2007. Rho GTPases: functions and association with cancer. Clin. Exp. Metastasis. 24:657–672 10.1007/s10585-007-9119-1 [DOI] [PubMed] [Google Scholar]
- Hakem A., Sanchez-Sweatman O., You-Ten A., Duncan G., Wakeham A., Khokha R., Mak T.W. 2005. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 19:1974–1979 10.1101/gad.1310805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman S.J., Ridley A.J. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- Heasman S.J., Carlin L.M., Cox S., Ng T., Ridley A.J. 2010. Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J. Cell Biol. 190:553–563 10.1083/jcb.201002067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.T., Merajver S.D., Lapière C.M., Nusgens B.V., Deroanne C.F. 2008. RhoA-GDP regulates RhoB protein stability. Potential involvement of RhoGDIalpha. J. Biol. Chem. 283:21588–21598 10.1074/jbc.M710033200 [DOI] [PubMed] [Google Scholar]
- Ishizaki T., Naito M., Fujisawa K., Maekawa M., Watanabe N., Saito Y., Narumiya S. 1997. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett. 404:118–124 10.1016/S0014-5793(97)00107-5 [DOI] [PubMed] [Google Scholar]
- Itoh R.E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., Matsuda M. 2002. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22:6582–6591 10.1128/MCB.22.18.6582-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Kenny P.A., Lee G.Y., Myers C.A., Neve R.M., Semeiks J.R., Spellman P.T., Lorenz K., Lee E.H., Barcellos-Hoff M.H., Petersen O.W., et al. 2007. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 1:84–96 10.1016/j.molonc.2007.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzing T.M., Wang Y., Pertz O., Copeland J.W., Grosse R. 2010. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene. 29:2441–2448 10.1038/onc.2009.515 [DOI] [PubMed] [Google Scholar]
- Leung T., Chen X.Q., Manser E., Lim L. 1996. The p160 RhoA-binding kinase ROK alpha is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol. Cell. Biol. 16:5313–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.X., Rane N., Liu J.P., Prendergast G.C. 2001. RhoB is dispensable for mouse development, but it modifies susceptibility to tumor formation as well as cell adhesion and growth factor signaling in transformed cells. Mol. Cell. Biol. 21:6906–6912 10.1128/MCB.21.20.6906-6912.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G.L., Hahn K.M., Danuser G. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature. 461:99–103 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrogianneli K., Carlin L.M., Keppler M.D., Matthews D.R., Ofo E., Coolen A., Ameer-Beg S.M., Barber P.R., Vojnovic B., Ng T. 2009. Integrating receptor signal inputs that influence small Rho GTPase activation dynamics at the immunological synapse. Mol. Cell. Biol. 29:2997–3006 10.1128/MCB.01008-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardia K.V., Jopp P.E. 2000. Directional Statistics. Wiley, Chichester, England: 429 pp [Google Scholar]
- Martín-Rufián M., Segura J.A., Lobo C., Matés J.M., Márquez J., Alonso F.J. 2006. Identification of genes downregulated in tumor cells expressing antisense glutaminase mRNA by differential display. Cancer Biol. Ther. 5:54–58 10.4161/cbt.5.1.2238 [DOI] [PubMed] [Google Scholar]
- Moore B.R. 1980. A Modification of the Rayleigh Test for Vector Data. Biometrika. 67:175–180 10.1093/biomet/67.1.175 [DOI] [Google Scholar]
- Niault T., Sobczak I., Meissl K., Weitsman G., Piazzolla D., Maurer G., Kern F., Ehrenreiter K., Hamerl M., Moarefi I., et al. 2009. From autoinhibition to inhibition in trans: the Raf-1 regulatory domain inhibits Rok-alpha kinase activity. J. Cell Biol. 187:335–342 10.1083/jcb.200906178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes C.D., Hall A. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 81:53–62 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R.L., Hahn K.M. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 440:1069–1072 10.1038/nature04665 [DOI] [PubMed] [Google Scholar]
- Peter M., Ameer-Beg S.M., Hughes M.K., Keppler M.D., Prag S., Marsh M., Vojnovic B., Ng T. 2005. Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophys. J. 88:1224–1237 10.1529/biophysj.104.050153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinner S., Jordan P., Sharrock K., Bazley L., Collinson L., Marais R., Bonvin E., Goding C., Sahai E. 2009. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer Res. 69:7969–7977 10.1158/0008-5472.CAN-09-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riento K., Ridley A.J. 2003. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 4:446–456 10.1038/nrm1128 [DOI] [PubMed] [Google Scholar]
- Sahai E. 2005. Mechanisms of cancer cell invasion. Curr. Opin. Genet. Dev. 15:87–96 10.1016/j.gde.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Sahai E., Marshall C.J. 2002. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat. Cell Biol. 4:408–415 10.1038/ncb796 [DOI] [PubMed] [Google Scholar]
- Sahai E., Marshall C.J. 2003. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5:711–719 10.1038/ncb1019 [DOI] [PubMed] [Google Scholar]
- Seth A., Otomo C., Rosen M.K. 2006. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLα and mDia1. J. Cell Biol. 174:701–713 10.1083/jcb.200605006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson K.J., Dugan A.S., Mercurio A.M. 2004. Functional analysis of the contribution of RhoA and RhoC GTPases to invasive breast carcinoma. Cancer Res. 64:8694–8701 10.1158/0008-5472.CAN-04-2247 [DOI] [PubMed] [Google Scholar]
- Thiery J.P. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2:442–454 10.1038/nrc822 [DOI] [PubMed] [Google Scholar]
- Vega F.M., Ridley A.J. 2008. Rho GTPases in cancer cell biology. FEBS Lett. 582:2093–2101 10.1016/j.febslet.2008.04.039 [DOI] [PubMed] [Google Scholar]
- Weaver A.M. 2008. Cortactin in tumor invasiveness. Cancer Lett. 265:157–166 10.1016/j.canlet.2008.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler A.P., Ridley A.J. 2004. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp. Cell Res. 301:43–49 10.1016/j.yexcr.2004.08.012 [DOI] [PubMed] [Google Scholar]
- Yoneda A., Multhaupt H.A., Couchman J.R. 2005. The Rho kinases I and II regulate different aspects of myosin II activity. J. Cell Biol. 170:443–453 10.1083/jcb.200412043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A., Ushakov D., Multhaupt H.A., Couchman J.R. 2007. Fibronectin matrix assembly requires distinct contributions from Rho kinases I and -II. Mol. Biol. Cell. 18:66–75 10.1091/mbc.E06-08-0684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicha D., Dunn G., Jones G. 1997. Analyzing chemotaxis using the Dunn direct-viewing chamber. Methods Mol. Biol. 75:449–457 [DOI] [PubMed] [Google Scholar]